Abstract
Hematologic maligancies exhibit a growth advantage by up-regulation of components within the molecular apparatus involved in cell-cycle progression. The SCF (Skip-Cullin1-F-box protein) E3 ligase family provides homeostatic feedback control of cell division by mediating ubiquitination and degradation of cell-cycle proteins. By screening several previously undescribed E3 ligase components, we describe the behavior of a relatively new SCF subunit, termed FBXL2, that ubiquitinates and destabilizes cyclin D2 protein leading to G0 phase arrest and apoptosis in leukemic and B-lymphoblastoid cell lines. FBXL2 expression was strongly suppressed, and yet cyclin D2 protein levels were robustly expressed in acute myelogenous leukemia (AML) and acute lymphoblastic leukemia (ALL) patient samples. Depletion of endogenous FBXL2 stabilized cyclin D2 levels, whereas ectopically expressed FBXL2 decreased cyclin D2 lifespan. FBXL2 did not bind a phosphodegron within its substrate, which is typical of other F-box proteins, but uniquely targeted a calmodulin-binding signature within cyclin D2 to facilitate its polyubiquitination. Calmodulin competes with the F-box protein for access to this motif where it bound and protected cyclin D2 from FBXL2. Calmodulin reversed FBXL2-induced G0 phase arrest and attenuated FBXL2-induced apoptosis of lymphoblastoid cells. These results suggest an antiproliferative effect of SCFFBXL2 in lymphoproliferative malignancies.
Introduction
Studies suggest that D-type cyclins are dysregulated in hematologic malignancies making them potentially suitable molecular targets for chemotherapeutic intervention.1,2 D-type cyclins play integral roles in G1/S cell-cycle progression where they act through 4 cyclin-dependent kinases (Cdks): Cdk 2, Cdk 4, Cdk 5, and Cdk 6. Active cyclin D/Cdk 4 and cyclin D/Cdk 6 partially phosphorylate retinoblastoma tumor suppressor protein (Rb) thereby reducing its binding to E2F, and thus promoting cell progression to the S-phase by allowing E2F-mediated activation of cyclin E gene transcription.3 There are 3 major cyclin D family members: cyclin D1, cyclin D2, and cyclin D3 of which cyclin D1 has been well studied.4 As an important regulator for G1 to S phase progression, cyclin D1 plays an important role in multiorgan tumorogenesis and its expression is elevated in mantle cell lymphoma, multiple myeloma, and some leukemias.5–10 Accordingly, cyclin D1 degradation through its site-specific ubiquitination and disposal by the proteasome results in G1 phase arrest.11 Specifically, the SCF (Skp1-Cullin1-F-box) E3 ligase family members FBXO4, FBXW8, and FBXO31 directly interact with cyclin D1 and mediate its ubiquitination.12–14 Another D-type cyclin, cyclin D2 is selectively expressed at very high levels in Epstein-Barr virus positive lymphoma cell lines.15,16 Cyclin D2 transcripts are also highly expressed in chronic lymphocytic leukemia, whereas cyclin D1 and cyclin D3 mRNA are almost not detectable.17 In nonhematologic malignancies, overexpression of cyclin D2 is linked to increased invasiveness of human squamous carcinoma.18 However, compared with cyclin D1, relatively limited data are available regarding the function and molecular regulation of cyclin D2.
Ubiquitination of proteins brands them for degradation either by the proteasome or via the lysosome that regulates diverse processes.19 The conjugation of ubiquitin to a target protein is orchestrated by a series of enzymatic reactions involving an E1 ubiquitin-activating enzyme, ubiquitin transfer from an E1-activating enzyme to an E2-conjugating enzyme, and last, generation of an isopeptide bond between the substrate's ϵ-amino lysine and the carboxyl-terminus of ubiquitin catalyzed by a E3-ubiquitin ligase.20 Of the many E3 ligases, the SCF superfamily is among the best studied. The SCF complex has a catalytic core complex consisting of Skp1, Cullin1, and the E2 ubiquitin-conjugating (Ubc) enzyme.21 The SCF complex also contains an adaptor receptor subunit, termed F-box protein, that targets hundreds of substrates through phosphospecific domain interactions.22 F-box proteins have 2 domains: an NH2-terminal F-box motif and a carboxyl-terminal leucine-rich repeat (LRR) motif or WD repeat motif. The SCF complex uses the F-box motif to bind Skp1, whereas the leucine-rich/WD repeat motif is used for substrate recognition.23 Of the nearly 70 F-box proteins described, only ∼ 6 have defined roles in cellular processes.24 The recent discovery that disruption of the gene encoding the SCF member, SCFFBW7, results in T-cell acute lymphoblastic leukemia suggests that this class of ubiquitin E3 complexes may play an indispensible role in the pathogenesis of some hematologic malignancies.25 We have recently identified an orphan protein that belongs to the SCF family, termed FBXL2, which may exhibit tumor suppressor activity. The gene encoding FBXL2, was strongly repressed in human lung adenocarcinoma.26 After its initial description,23 FBXL2 was shown to interact with the hepatitis C virus nonstructural protein 5A (NS5A); we showed that FBXL2 is involved in the ubiquitination of a lipogenic enzyme, cytidylyltransferase.27,28 Aside from these studies, there have been no studies investigating the biologic role of FBXL2.
Calmodulin (CaM; 16.7 kD) is a highly conserved calcium-sensing protein that antagonizes some proteinases and modulates stability of regulatory proteins.29 CaM binds its targets in a calcium bound (holoCaM) or calcium-1 free (apopCaM) form and thus its interactions with partners may be calcium-dependent or calcium-independent.29 Many CaM binding proteins harbor recognition motifs characterized by a basic amphipathic helix, moderate to high helical hydrophobic moment, and a net positive charge. Other motifs described include an IQ motif (I/LQXXXRGXXXR), a 1-8-14, and 1-5-10 CaM-binding motif.29 As a major intracellular calcium receptor, CaM exerts multiple modes of regulatory control within the cell cycle.30 For example, CaM appears essential for cyclin-dependent kinase 4 (Cdk4) activities and nuclear accumulation of cyclin D1-Cdk4 during the G1 phase.31 Interestingly, CaM also directly interacts with cyclin E and mediates calcium sensitive G1/S phase transition.32,33 We showed that CaM binds and stabilizes cytidylyltransferase required for phospholipid synthesis during cellular membrane formation suggesting that its combinatorial effects regulate processes intrinsic to cellular growth and repair.34
In this study, by first using a SV40-tumorogenic cell line,35 we show that cyclin D2 is targeted for its ubiquitination by SCFFBXL2 leading to its degradation within the proteasome. Unlike other F-box proteins that target phosphodegrons, SCFFBXL2 recognizes a canonical CaM-binding motif within cyclin D2 to mediate its ubiquitination and degradation. CaM also binds within this molecular signature and opposes SCFFBXL2 ubiquitination and disposal of cyclin D2. SCFFBXL2 activity against cyclin D2 was sufficient to induce G0 arrest and apoptosis of Epstein-Barr virus (EBV)–transformed human B lymphocytes and 4 leukemic cell lines indicative of the ability of this orphan F-box protein to exert antiproliferative activity. Importantly, we detected very low FBXL2 immunoreactive levels coupled with high cyclin D2 content in leukemic cells isolated from subjects with acute myelogenous leukemia (AML) and acute lymphoblastic leukemia (ALL), which suggests antiproliferative activity for this F-box protein.
Methods
Materials
The sources of the transformed murine lung epithelial (MLE) cell line, CaM, Erk, and GST antibodies were previously described.34,36 HCC1739 BL cells were from ATCC. Purified SCFFBXL2 was purchased from Abnova. Purified bovine calmodulin, ubiquitin, E1, E2, MG132, leupeptin, and cycloheximide were purchased from Calbiochem. Rabbit polyclonal ubiquitin, cyclin and Cdk sampler kits, were purchased from Cell Signaling. Mouse monoclonal cyclin D2 antibodies were from Abcam. The F-box protein cDNAs were purchased from OpenBiosystems. GenomONE HVJ-E transfection kit was from Cosmo bio USA. HilyMax was from Dojindo. Adenoviral CaM constructs were generated as described.34 Goat polyclonal FBXL2 antibody and siRNAs were from Santa Cruz Biotechnology. Rabbit polyclonal FBXL2 antibody was custom made from Rockland Immunochemicals. All DNA sequencing was performed by the University of Pittsburgh DNA Core Facility.
Human samples
THP1, MOLM-13, K562, and U937 cells were obtained from ATCC, and human AML and ALL samples were obtained from the University of Pittsburgh Health Sciences Tissue Bank. THP1 cells were derived from the peripheral blood of a male subject with acute monocytic leukemia.37 U937 cells were isolated from a male subject with histiocytic lymphoma.38 K562 cells were derived from a female subject with chronic myelogenous leukemia (CML) in blast crisis.39 MOLM-13 cells were established from the peripheral blood of a patient during relapse with acute monocytic leukemia.40 The study was approved both by the University of Pittsburgh Tissue Utilization Committee and University of Pittsburgh Institutional Review Board. Subjects were diagnosed with leukemia and clinical data presented (supplemental Table 1, available on the Blood Web site; see the Supplemental Materials link at the top of the online article).
Cell culture
MLE cells were cultured in DMEM-F12 (Gibco) supplemented with 2%-10% fetal bovine serum (FBS; DMEM-2 or 10). Cells in some studies were synchronized using serum starvation (DMEM-F12) for 48 hours or treatment with nocodazole or aphidicolin. In other studies, cells were treated with leupeptin or MG132 at 1:2000 dilutions. For half-life studies, cells were treated with cycloheximide (40 μg/mL) at different time points. HCC1739 B lymphocyte, THP1, MOLM-13, K562, and U937 cells were cultured in RPMI/1640 (Gibco) supplemented with 10% FBS with 2mM Glutamine, 25mM HEPES (N-2-hydroxyethylpiperazine-N′-2-ethanesulfonic acid), 1mM sodium pyruvate, 1% nonessential amino acid. Cells lysates were prepared by brief sonication in 150mM NaCl, 50mM Tris, 1.0mM EDTA, 2mM dithiothreitol, 0.025% sodium azide, and 1mM phenylmethylsulfonyl fluoride (buffer A) at 4°C.
Expression of recombinant protein and RNAi
All plasmids were delivered into cells using nucleofection, Lipofectmine 2000, HilyMax, or a GenomONE HVJ-E transfection kit.41,42 Cellular expression of green fluorescent tagged plasmids using nucleofection was achieved at > 90%. For overexpression of CaM, 4 × 106 cells were plated in 100 mm dishes for 24 hours, and then infected with Adv-CaM or an empty vector (Adv-Con) at MOI = 40 for 12 hours followed by FBXL2 plasmid expression. For siRNA studies, 1 × 106 cells were transfected using lipofectamine 2000 with 10 μg of RNA and harvested after an additional 48 hours.
Coimmunoprecipitation and binding assays
Total protein (250 μg) from cell lysates was precleared with 20 μL of protein A/G beads for 1 hour at 4°C. Primary antibody(5 μg) was added for 18 hours of incubation at 4°C. Protein A/G beads (40μL) were added for an additional 6 hours of incubation. Beads were slowly centrifuged and washed 5 times using 50mM HEPES, 150mM NaCl, 0.5mM EGTA, 50mM NaF, 10mM Na3VO4, 1mM phenylmethylsulfonyl fluoride, 20μM leupeptin, and 1% (vol/vol) Triton X-100 (RIPA) buffer, as described.43 The beads were heated at 100°C for 5 minutes with 80 μL of protein sample buffer before sodium dodecyl sulfate (SDS)-polyacrylamide gel electrophoresis (PAGE) and immunoblotting. For CaM-binding assays, CaM sepharose beads were incubated with V5-FBXL2 or cyclin D2 transfected cell lysates (50 μg), with or without Ca2+ at 4°C for 2 hours. Eluted products were processed for SDS-PAGE and immunoblotting as described.34
Immunostaining
Cells (2 × 105) were plated at 70% confluence on 35 mm MatTek glass-bottom culture dishes. Immunofluorescent cell imaging was performed on a Nikon A1 confocal microscope. Cells were washed with PBS and fixed with 4% paraformaldehyde for 20 minutes, then exposed to 15% BSA, 1:500 primary antibodies, and 1:1000 Alexa 488– or Alexa 647– labeled goat anti–mouse or rabbit secondary antibody sequentially for immunostaining.
In vitro ubiquitin conjugation assay
The ubiquitination of V5-cyclin D2 was performed in a volume of 25 μL containing 50mM Tris pH 7.6, 5mM MgCl2, 0.6mM DTT, 2mM ATP, 1.5 ng/μL E1, 10 ng/μL Ubc5, 10 ng/μL Ubc7, 1μg/μL ubiquitin (Calbiochem), 1μM ubiquitin aldehyde, 4-16 μL of purified Cullin1, Skp1, Rbx1, and FBXL2. Reaction products were processed for V5 immunoblotting.
Quantitative RT-PCR, cloning, and mutagenesis
Total RNA was isolated and reverse transcription was performed followed by quantitative real-time PCR with SYBR Green qPCR mixture as described.44 All mutant constructs were generated using PCR-based approaches using appropriate primers or site-directed mutagenesis as described previously.28
Cell-cycle and apoptosis analysis
Transfected cells were incubated with BrdU (5-bromo-2′-deoxyuridine; 20μM) for 40 minutes, fixed, and stained following the manufacturer's protocols (BD Biosciences). Fluorescence-activated cell sorter (FACS) samples were analyzed with the Accuri C6 system. DNA content was analyzed using FCS3 Version 3.0 express software (De Novo Software). Cells were stained with annexin V before analysis by FACS. Viable cells were counted and expressed as a percentage of total cells.
Statistical analysis
Statistical comparisons were performed with the Prism Version 4.03 (GraphPad Software) using an ANOVA 1 or an unpaired t test with P < .05 indicative of significance.
Results
Ectopic expression of FBXL2 induces cyclin D2 degradation
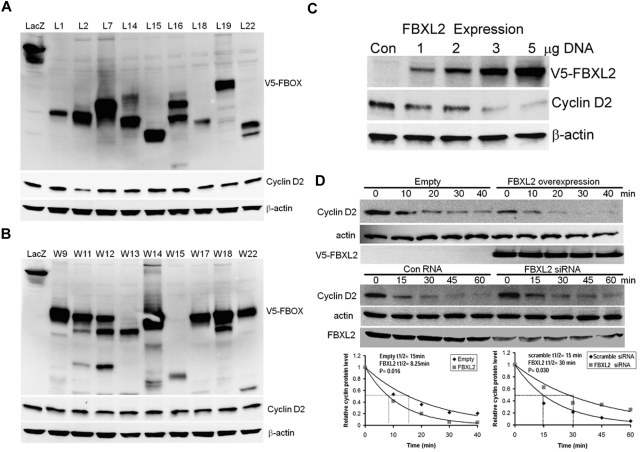
We first tested more than thirty F-box proteins that might be involved in cyclin D2 degradation. Several randomly selected F-box protein (FBXL and FBXW family) members were ectopically expressed in a transformed MLE cell line (Figure 1A-B). Cells were collected and cell lysates were analyzed for expression of F-box proteins and endogenous cyclin D2 levels. Although all F-box proteins were effectively expressed, only expression of FBXL2 was sufficient to decrease immunoreactive cyclin D2 levels. To further evaluate this, cells were transfected with increased amounts of FBXL2 plasmid revealing that the F-box protein triggered cyclin D2 degradation in a dose-dependent manner (Figure 1C). We next examined cyclin D2 half-life after FBXL2 overexpression and knockdown. Ectopic FBXL2 expression significantly decreased cyclin D2 t1/2, whereas knockdown of FBXL2 markedly increased cyclin D2 lifespan in cells (Figure 1D). Consistent with a posttranslational effect, FBXL2 overexpression did not significantly alter CCND2 steady-state mRNA levels (data not shown). To further assess the specificity of these effects, a random F-box protein (FBXL12) from the FBXL family was selected and ectopically expressed in cells demonstrating lack of ability of this protein to disrupt cyclin D2 stability (supplemental Figure 1). Moreover, loss- and gain-of-function studies of FBXL2 did not alter cyclin D1 t1/2 (supplemental Figure 2). These studies show that cyclin D2 is a potential molecular target for SCFFBXL2 for its ubiquitination and degradation.
Figure 1.
Ectopic expression of FBXL2 induces the degradation of cyclin D2. (A-B) MLE cells were transfected with control plasmid lacZ or other V5 tagged F-box proteins (L and W families). Cells were collected and cell lysates were analyzed for V5, cyclin D2 and β-actin immunoblotting. (C) Cells were transfected with increasing amounts of V5-tagged FBXL2 plasmid. Cells were collected and cell lysates were analyzed for V5, cyclin D2, and β-actin immunoblotting. (D) Cyclin D2 protein half-life determination after FBXL2 overexpression (top panel), or FBXL2 knockdown using siRNA (middle panel). Bottom panel shows levels of cyclin D2 on immunoblots that were quantified densitometrically and are shown graphically. The data from each panel represent at least n = 2 separate experiments.
FBXL2 targets cyclin D2 for ubiquitination during mitosis
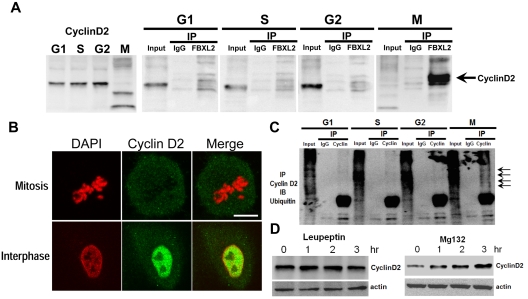
To test whether FBXL2 interacts with cyclin D2, we performed coimmunoprecipitation (coip) experiments. MLE cells were first synchronized separately to G1, S, and the G2 phase; mitotic cells were also collected through shake-off. Cells were immediately lysed and subjected to FBXL2 ip. Cyclin D2 immunoreactive levels oscillated during the cell cycle; low levels of endogenous cyclin D2 occur during mitosis (Figure 2A left panel). In addition, cyclin D2 levels in the FBXL2 immunoprecipitates were only detected in mitotic cells (Figure 2A). Thus, endogenous FBXL2 protein targets cyclin D2 during mitosis. When a panel of cells was immunostained for cyclin D2 and counterstained with DAPI to visualize the nucleus, the fluorescent intensity representing endogenous cyclin D2 levels was lower in identified cells undergoing mitosis compared with cells in interphase (Figure 2B). In other experiments, synchronized cells were subjected to cyclin D2 coip; the immunoprecipitates were then resolved in SDS-PAGE and probed with ubiquitin. Polyubiquitinated products (high molecular weight bands) of cyclin D2 were detected during mitosis (Figure 2C arrows). Hence, endogenous FBXL2 targets cyclin D2 for polyubiquitination not during interphase but with mitosis. We next determined the subcellular pathway for FBXL2-mediated degradation of cyclins. Addition of the proteasomal inhibitor, MG132, to cells led to accumulation of cyclin D2 protein, whereas this was not seen with the lysosomal inhibitor, leupeptin (Figure 2D). These observations indicate that cyclin D2 is bound, polyubiquitinated, and targeted for elimination in the proteasome in a cell cycle–specific manner by FBXL2.
Figure 2.
FBXL2 targets cyclin D2 for ubiquitination during mitosis. (A) MLE cells were synchronized to each cell phase followed by coimmunoprecipitation of endogenous FBXL2 and then cyclin D2 immunoblotting. (B) Cells were immunostained for cyclin D2 and counterstained with DAPI to visualize nucleus. (C) In vivo ubiquitination assays. Polyubiquitinated cyclin D2 was detected during cell-cycle progression by immunoprecipitation of endogenous cyclins followed by immunoblotting for ubiquitin. The arrows show polyubiquitinated cyclin D2. (D) Cyclin D2 levels in cells treated with leupeptin or MG132. The data from each panel represent at n = 2 separate experiments.
Cyclin D2 is polyubiquitinated within its C-terminus
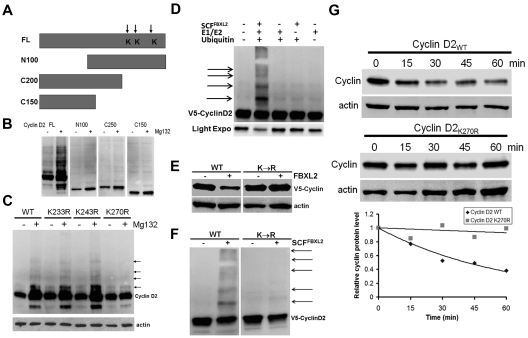
To determine the ubiquitination acceptor site within cyclin D2, a deletional mapping approach was used where truncated cyclin mutants were constructed and expressed in cells before MG132 treatment (Figure 3A-B). MG132 treatment triggered appearance of robust polyubiquitinated wild-type (WT) cyclin D2; in contrast, the proteasomal inhibitor failed to increase accumulation of mutants that lacked either NH2-terminal (N100) or C-terminal (C200, C150) fragments, the latter that contain 3 potential lysine residues. Of note, the NH2-terminal truncation variant lacks a FBXL2 dock site that explains its inability to generate slower migrating products after MG132 treatment. Further, cyclin D2K270R mutants accrued minimally after MG132 exposure in cells suggesting that Lys270 is a putative ubiquitination site for cyclin D2 (Figure 3C arrows). Importantly, inclusion of purified SCFFBXL2 with the full complement of E1 and E2 enzymes plus ubiquitin was sufficient to generate polyubiquitinated cyclin D2 species in vitro (Figure 3D). In addition, coexpression of FBXL2 with cyclins resulted in the degradation of WT cyclin D2, but not the Lys270R mutant (Figure 3E). In vitro ubiquitination assays were used again in which WT or mutant cyclin D2 were reacted with the purified ubiquitin SCFFBXL2 complex, and here the Lys270R mutant was not ubiquitinated (Figure 3F). Finally, the Lys270R mutant exhibited significantly extended t1/2 compared with the WT cyclin D2 (Figure 3G).
Figure 3.
Cyclin D2 is polyubiquitinated at carboxyl-terminal acceptor sites. (A) Diagram of cyclin D2 deletion mutants constructed. Arrows indicate putative ubiquitin receptor sites. (B) V5-immunoblotting for levels of full-length (FL) or truncated cyclin D2 constructs after cellular expression and culture of cells in the absence (−) or presence (+) of MG132. (C) Cells were transfected with cyclin D2 point mutants before culture in the absence (−) or presence (+) of MG132. Immunoblotting was then performed for detection of accumulated (polyubiquitinated) cyclin D2 proteins in cells. The arrows indicate lack of polyubiquitinated signals after expression of the cyclin D2K270R construct. (D) In vitro ubiquitination assays. Purified SCF complex components were incubated with V5-cyclin D2 and the full complement of ubiquitination reaction components (second lane from left) showing polyubiquitinated cyclin D2. (E) Cyclin D2 protein levels in cells after cotransfection with either WT cyclin D2 or Lys270R cyclin D2 with or without ectopic FBXL2 expression. (F) In vitro ubiquitination assays. Purified SCF complex were incubated with WT V5-cyclin D2, or a Lys270R V5-cyclin D2 mutant and the full complement of ubiquitination reaction components. (G) Cyclin D2 protein half-life determination using cyclohexamide after expression of WT V5-cyclin D2, or Lys270R (V5-cyclin D2) mutant (data are from n = 2 experiments).
FBXL2 and CaM both vie for cyclin D2 docking
Calmodulin binds and protects some regulatory proteins and is needed for cell-cycle progression.30 Cyclin D2 harbors 2 potential CaM binding IQ motifs within its NH2-terminus suggesting that these motifs may be required for CaM interaction (supplemental Figure 3). WT or NH2-terminal deleted cyclin D2 were transfected in cells, cell lysates were then applied to CaM-sepharose beads to test protein interaction. The pull-down experiments show that only WT cyclin interacts with CaM optimally with inclusion of EDTA, and this interaction was significantly disrupted by even low micromolar calcium concentrations (supplemental Figure 4A). A cyclin D2 variant devoid of the amino-terminus failed to interact with CaM, supporting the premise that an IQ motif within this region is required for molecular interaction between the cyclin D2 and CaM (supplemental Figure 4A). Interestingly, FBXL2 also failed to interact with the NH2-terminal deletion cyclin D2 (supplemental Figure 4B). We next identified which of the 2 potential IQ motifs within cyclin D2 are required for CaM binding. Glu98 of cyclin D2 was essential for CaM binding (supplemental Figure 4C top panel). Importantly, FBXL2 also uses this molecular site within this motif to target cyclin D2 (supplemental Figure 4C bottom panel). For confirmation, in vitro ubiquitination assays demonstrated that the point mutant of cyclin D2 (Q98A) was not ubiquitinated (supplemental Figure 4D). Coexpression of FBXL2 with cyclins resulted in the degradation of WT cyclin but not the cyclin D2Q98A point mutant (supplemental Figure 4E). Importantly, this variant exhibited a significantly longer t1/2 compared with WT cyclin D2 (supplemental Figure 4F). FBXL family proteins contain leucine-rich repeats (LRR) for substrate targeting and residues 80-423 contain 12 LRRs that display extensive internal homology (supplemental Figure 5). In mapping studies, cell lysates expressing his-tagged FBXL2 truncation mutants were copurified with GST-cyclin D2 using his-pull downs. The data show that deletion of the last 5 LLRs (C250) or the last 2 LLRs (C350) markedly disrupted FBXL2-cyclin D2 interaction (supplemental Figure 5). Thus, cyclin D2 binds FBXL2 within its last 2 LLR domains (350-423).
FBXL2 degradation of cyclin D2 is antagonized by CaM
CaM overexpression significantly increased cyclin D2 half-life, whereas CaM knockdown decreased the stability of this protein (supplemental Figure 6A). Our recent studies indicate that CaM also directly interacts with FBXL2, as several bulky and hydrophobic residues within the F-box amino-terminus appear important for CaM interaction.28 Thus, we next tested the functionality of mutant FBXL2 proteins. Cells transfected with either WT FBXL2 or FBXL2F79A (that does not bind CaM) effectively decreased cyclin D2 levels (supplemental Figure 6B left panel). When cells were preinfected with a replication-deficient adenovirus expressing CaM (Adv-CaM) and then transfected with these FBXL2 plasmids, CaM gene transfer was not able to totally rescue cyclin D2 protein levels (supplemental Figure 6B right panel), when cells were transfected with FBXL2F79A. These results suggest that in addition to competition for IQ motif occupancy within cyclins, CaM's ability to directly bind and sequester FBXL2 to protect cyclin D2 might also be functionally important.
We also investigated the FBXL2, CaM, and cyclin interactions during cell-cycle progression. Lysate from synchronized cells were subjected to FBXL2, cyclin D2 ip, the immunoprecipitates were then resolved in SDS-PAGE and probed with CaM. There was significant loss of FBXL2/CaM binding during mitosis and CaM associates with cyclin D2 at the highest levels during the S-phase with limited, if any, binding during mitosis (supplemental Figure 6C). Because both CaM and FBXL2 interact with cyclins through the same IQ motif, we next tested the concept that there exists intermolecular competition between FBXL2 and CaM for cyclin D2 binding. This was analyzed using pull-down experiments in the presence or absence of calcium in which FBXL2 was immobilized on beads and used as bait for cyclin D2 and CaM (supplemental Figure 6D). Three negative controls were included: (1) FBXL2-agarose alone was assayed to control for cyclin and CaM contamination; (2) Cyclin D2 and CaM with calcium were run over empty talon beads, eluted, and proteins were resolved by SDS-PAGE followed by cyclin and CaM immunoblotting to ensure that associations were FBXL2 specific; and (3) V5 immunoblotting was used as a loading control, to ensure that pull-down experiments had equivalent amounts of FBXL2. The results indicate that not only does FBXL2 directly interact with cyclin D2 and CaM, but that excess CaM disrupts FBXL2 interaction with cyclin D2 (supplemental Figure 6D). Interestingly, excess calcium promotes cyclin D2 binding to FBXL2. Using a synthetic cyclin D2 peptide (LQLLGAVCMF) and FBXL2 peptide (79FLRKLSLRGCI89) encoding the putative CaM-binding motif, we observed tight binding between CaM and cyclin D2 (Kd = 0.31μM,), and FBXL2 (Kd = 0.8μM) using isothermal calorimetry (supplemental Figure 6E).
FBXL2/CaM regulate cell cycle in leukemic cells
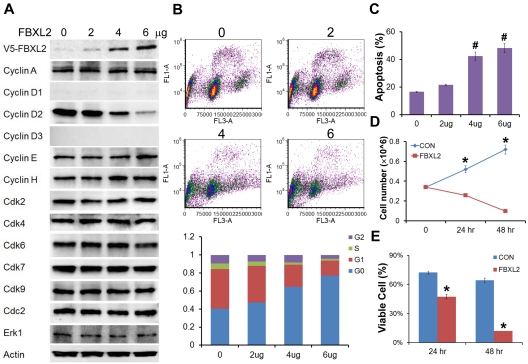
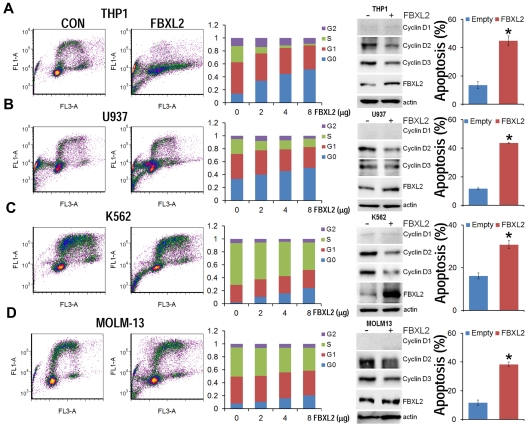
To evaluate the biologic relevance of these interactions, we examined models of hematologic malignancies. First, HCC1739 BL cells transfected with increasing amounts of FBXL2 plasmid resulted in decreased levels of the G1/S phase regulator, cyclin D2 (Figure 4A). In contrast, levels of other G1 cyclins and the G2/M regulator cyclin A were unaffected, and ectopic FBXL2 expression did not alter levels of other Cdks or negative control proteins. Of note, EBV-transformed B lymphocytes have low levels of cyclin D1 and D3, thus we were not able to detect these proteins by immunoblotting. Next, cells transfected with FBXL2 were labeled with BrdU and analyzed by 2-color flow cytometry (Figure 4B). Interestingly, we observed significant increases in the G0 phase in a dose-dependent manner coupled with an increase in apoptosis (Figure 4C). A time course study was also conducted where HCC1739 BL cells transfected with FBXL2 were analyzed using trypan blue staining, viable cells were quantified showing that FBXL2 markedly inhibited cell growth and viability in culture (Figure 4D-E). We next extended our studies to assess FBXL2 activity in several leukemic cells lines (Figure 5A-D). Cells were transfected with increasing amounts of FBXL2 plasmids, after 48 hours, cells were labeled with BrdU and analyzed by 2-color flow cytometry. Of note, there were somewhat variable baseline patterns within the cell cycle between the leukemic lines; eg more rapidly proliferative cells such as K562 and MOLM-13 contained large portions of cells within the S-phase cells (50% to ∼ 70%), whereas slower growing THP1 and U937 lines contained a larger fraction of cells in the G0-G1 phase (60%-70%). Nevertheless, as with HCC1739 BL cells, ectopic expression of FBXL2 consistently increased the G0 phase and decreased S-phase at variable degrees in all cells lines tested. Further, all 4 cells tested displayed extremely low levels of cyclin D1 mass (Figure 5 immunoblots on right). However, we did detect increased FBXL2 protein levels, consistently decreased concentrations of cyclin D2, and variably decreased cyclin D3 levels after F-box plasmid transfection. Cells transfected with FBXL2 were also labeled with annexin V, and analyzed by 2-color flow cytometry. Interestingly, we observed significant increases in apoptosis in all 4 cell lines studied (Figure 5 far right panels).
Figure 4.
FBXL2 inhibits cell-cycle progression in transformed B-lymphoblastoid cells. (A) Immunoblotting showing levels of cyclins, CDKs, and negative control proteins, actin, and Erk after control (0) plasmid or ectopic FBXL2 expression. (B-C) FACS analysis in cells transfected with either empty plasmid or with increasing amount of FBXL2 plasmid. (D-E) Proliferation and viability studies of HCC1739 BL cells after FBXL2 overexpression. HCC1739 BL cells were transfected with FBXL2 for either 24 hours or 48 hours, cells were stained with trypan blue, and viable cells were analyzed by cell counting and graphed. n = 3 experiments, in control (C) #P < .01 versus empty plasmid, in panel D *P < .01 versus 0 hours, and in panel E *P < .01 versus control (CON).
Figure 5.
FBXL2 induces G0 arrest in leukemic cell lines. (A-D) THP1, U937, K562, and MOLM-13 cells were cultured in RPMI medium followed by overexpression of FBXL2 using a GenomONE HVJ-E transfection kit. Cells were labeled with BrdU and analyzed by 2-color flow cytometry (left panels). Quantification of each cell phase is presented in the bar graphs (middle panels). Cells were also collected after transfection, cyclin Ds and FBXL2 protein levels were analyzed by immunoblotting (right immunoblots). Cells were also labeled with annexin V and analyzed by 2-color flow cytometry (far right panels). Apoptotic cells were quantified. *P < .05 versus empty plasmid.
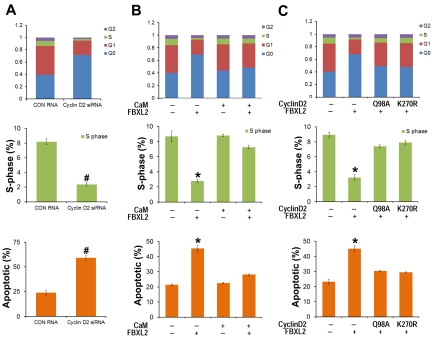
As a complementary approach, HCC1739 BL cells were also transfected with siRNA targeting cyclin D2, cells were labeled with BrdU and analyzed by 2-color flow cytometry. Compared with control RNA, cyclin D2 knockdown caused G0 arrest, thus recapitulating effects of FBXL2 overexpression in cells (Figure 6A). To test whether CaM antagonizes FBXL2 activity in this system, HCC1739 BL cells were preinfected with a replication-deficient adenovirus expressing CaM (Adv-CaM) and then transfected with FBXL2 plasmid. FBXL2 effectively caused G0 arrest, decreased S-phase, and induced apoptosis (Figure 6B); however CaM gene transfer was able to totally rescue these phenotypes. To further confirm the specificity of cyclin D2 degradation by FBXL2, HCC1739 BL cells were cotransfected with FBXL2 and with or without 2 cyclin D2 mutants Q98A and K270R (Figure 6C). Cyclin D2 mutants Q98A and K270R are resistant in FBXL2 ubiquitination and degradation, and FBXL2 effectively promotes G0 arrest, decreased S-phase, and triggered apoptosis; however coexpression of either Q98A or K270R mutants was able to antagonize the F-box protein's activity in this pathway (Figure 6C).
Figure 6.
FBXL2 and CaM differentially regulate the cell cycle in transformed B lymphoblastoid cells. (A) HCC1739 BL cells were transfected with either control RNA or cyclin D2 siRNA. Forty-eight hours later, cells were labeled with BrdU and analyzed through FACS or stained with annexin V and analyzed using a cell counter (n = 3 experiments, #P < 0.01 versus control (CON) RNA. (B) Cells were preinfected with a replication-deficient Adv-CaM or an empty virus and then transfected with FBXL2 plasmid. Forty-eight hours later, cells were either labeled with BrdU and analyzed through FACS or stained with annexin V and analyzed using a cell counter (n = 3 experiments, *P < .01 versus empty). (C) Cells were cotransfected with FBXL2 with or without 2 cyclin D2Q98A and cyclin D2K270R mutants. Forty-eight hours later, cells were either labeled with BrdU and analyzed through FACS or stained with annexin V and analyzed as previously described (n = 3 experiments, *P < .01 versus empty).
FBXL2 induces apoptosis in AML and ALL cells
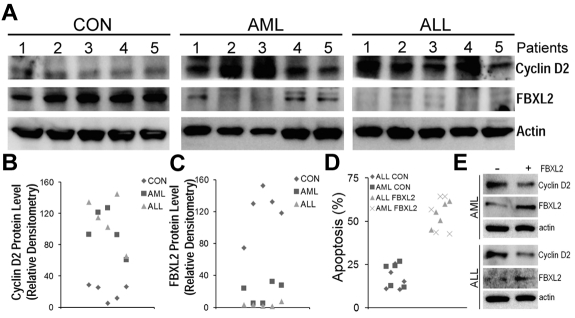
To further test our hypothesis that effects of FBXL2 were more widespread, we cultured peripheral blood mononuclear cells (PBMCs) isolated from control, AML and ALL subjects for 18 hours, cells were then collected, lysed and assayed for FBXL2 and cyclin D2 by immunoblotting (Figure 7A-C). Interestingly, we observed a remarkable decrease in immunoreactive FBXL2 levels from AML samples, and in particular, almost no FBXL2 protein was detected in ALL samples. In contrast, cyclin D2 was highly expressed in both AML and ALL samples compared with controls. There appears to be a inverse correlation between FBXL2 and cyclin D2 protein levels as in AML samples, as expression of FBXL2 was almost completely lost in preparations from subjects 2 and 3, where cyclin D2 expression was the highest among all samples tested. Subjects 1, 4, and 5 expressed minimal FBXL2 levels, and accordingly cyclin D2 expression was less pronounced compared with subjects 2 and 3. Finally, to confirm antiproliferation activity of FBXL2, we transfected FBXL2 into PBMCs. Twenty-four hours after transfection, cells were stained with annexin V and processed by flow cytometry. Apoptotic cells were quantified and graphed in Figure 7D. We observed a significant increase in apoptosis in AML or ALL cells transfected with FBXL2, compared with vehicle transfected control cells.
Figure 7.
FBXL2 and cyclin D2 are differentially expressed in AML and ALL subjects. (A-C) PBMCs from 5 controls, AML, and ALL subjects were cultured in RMPI medium for 18 hours. Cells were then collected, lysed, and assayed for FBXL2 and cyclin D2 by immunoblotting (A). FBXL2 and cyclin D2 protein levels were quantified by densiometry using ImageJ Version 1.45 software, and the distribution of cyclin D2 (B) and FBLX2 protein (C) in each individual subject was graphed (n = 5, P < .005 vs control). (D) Leukemic cells were transfected with FBXL2 using a HVJ-E transfection kit for 24 hours. Cells were stained with annexin V and processed by flow cytometry. Apoptotic cells were quantified and graphed. (n = 5, P < .01 vs control). (E) FBXL2 and cyclin D2 protein levels were also analyzed from representative of AML and ALL transfected PBMCs.
These results were also supported by immunoblot analysis showing increased FBXL2 and decreased cyclin D2 protein levels in transfected samples (Figure 7E). Hence, these data suggest that FBXL2 markedly inhibits AML or ALL cell proliferation in vitro.
Discussion
Cyclin D2 is overexpressed in hematopoietic malignancies and pathobiologically it appears to play a critical role in the proliferation and transformation of hematopoietic cells.45–47 However, there is a paucity of data on the molecular regulation of cyclin D2 expression. Cyclin D2 elimination via the proteasome is regulated by site-specific phosphorylation catalyzed by p38 kinase, but the identity of the ubiquitin E3 ligase that mediates this process is unknown.48 Here, we show that both ectopically expressed and endogenous FBXL2 regulate viablity and proliferation of transformed HCC1739 BL cells by mediating ubiquitination and degradation of cyclin D2. We also observe opposing activities of SCFFBXL2 and a universal calcium sensor, CaM, on cyclin D2 stability. This antiproliferative activity of FBXL2 was also confirmed in several leukemia cell lines and in PBMCs from human AML and ALL subjects. In human leukemic samples, there appears to be an inverse correlation between F-box protein levels and immunoreactive cyclin D2 content. FBXL2 acting as the receptor component of a classic SCF ubiquitin E3 ligase, uniquely recognizes a CaM binding signature; this is an unusual characteristic of FBXL2 as other F-Box proteins recognize phosphodegrons within target substrates.49–51 In this regard, Thr286 within cyclin D1 is recognized by the E3 ligase subunits FBXO4, FBXW8, and FBXO31 to facilitate their activities.12–14 Although this Thr site is highly conserved among the D cyclins and may also be important in cyclin D2 protein stability, FBXL2 did not use this molecular signal for targeting but rather docks within a consenus IQ signature to facilitate ubiquitination. In support of this, ectopic expression of cyclin D2 point mutants within the IQ motif were resistant to ubiquitination and were sufficient to extend cyclin D2 half-life (supplemental Figure 4F). As a whole, these results provide the first evidence that this F-box protein appears to be a major regulator of cyclin D2 lifespan and thus might serve as a key feedback inhibitory signal for leukemic cell proliferative activity. How FBXL2 expression in hematologic malignancies is dysregulated requires further investigation.
Our work identifies functionally distinct domains that regulate intermolecular competition between the effectors, FBXL2 and CaM, and their putative target, cyclin D2. We first show that cyclin D2 contains a canonical IQ motif typical of calcium independent CaM binding proteins (supplemental Figure 4A). Unexpectedly, both FBXL2 and CaM target this signature within cyclin D2 and bind in the absence of calcium (supplemental Figure 4A). Each of these proteins also interact in vitro (supplemental Figure 6D). Hence, cyclin D2 levels in cells would probably depend on the relative binding affinities not only between FBXL2 and CaM, but also the interaction of these binding partners for their common substrate. Interestingly, despite overexpression of an Adv5-CaM in cells, this was inefficient at restoring cyclin D2 levels when coexpressing an FBXL2 mutant that lacks the ability to interact with CaM. These results suggest that CaM's ability to act as a decoy to directly antagonize and sequester the F-box protein represents a mechanistically relevant interaction in addition to FBXL2 and CaM intermolecular competition for occupancy within the cyclin IQ motif. However, our isothermal calorimetry studies, demonstrating very low binding constants between CaM and cyclin D2 (Kd = 0.27μM; supplemental Figure 6E) versus relatively higher values between CaM and FBXL2 (Kd = 0.81μM), suggest that CaM competition with FBXL2 might be a more functionally relevant mechanism in vivo.
To investigate cyclin D2 protein regulation, we initially used transformed MLE cells because of their very high replicative capacity, their ability to be used to readily assess expression of checkpoint elements within the mitotic program, and their relative ease of transfectability of exogenous plasmids. These cells were used to interrogate SCFFBXL2 functionality, relevant molecular signatures within cyclin D2 for protein interaction, and binding associations between interacting partners. Because MLE cells also express cyclin D1 and cyclin D3 that might confound data interpretation, we extended studies to HCC1739 BL cells where cyclin D2 was robustly expressed and yet little, if any, cyclin D1 and cyclin D3 were detected.15,16 This phenotype mimics chronic lymphocytic leukemia, where cyclin D2 is highly expressed but cyclin D1 and cyclin D3 mRNA are almost undetectable.17 Importantly, using HCC1739 BL cells and several leukemic cell lines, we were able to recapitulate observations initially identified in MLE cells showing that FBXL2 ubiquitination and proteasomal degradation of cyclin D2 results in G0 arrest and impaired proliferative capacity, an effect antagonized by CaM.
In the process of preparing this paper, we recently showed that the chemotherapuetic agent vinorelbine up-regulates FBXL2 protein levels in vitro.52 In addition, FBXL2 targets cyclin D3 for ubiquitination and degradation (Figure 5). However, cyclin D3 appears to exert a key functional role in mitosis, and its role during G/S phase transition may be less predominant.53 Still, the observation here that FBXL2 triggers G0 arrest was unexpected, as usually cyclin D2 regulates G1/S transition.54 One explanation for this is that because cyclin D2 translocates the cyclin-dependent kinase inhibitor, p27, out of the nucleus thereby promoting its degradation, cyclin D2 might also be necessary for G0/G1 phase progression.55 Indeed, our data suggest that both cyclin D2 knockdown or ectopic expression of FBXL2 in HCC1739 BL cells significantly decreases the S-phase and causes G0 arrest (Figures 4B and 6A). Although FBXL2 targets cyclin D2 during mitosis in MLE cells (Figure 2A), it is unclear during which phase of the cell cycle FBXL2 degrades cyclin D2 in transformed B lymphocytes. Specifically, HCC1739 BL cells are a suspension cell line posing significant technical challenges in synchronization and isolation of mitotic cells to elucidate FBXL2 activity within the cell cycle. Regardless, when FBXL2 is ectopically expressed in these cells or in several leukemic cell lines, cyclin D2 is degraded, and its depletion results in G0 arrest and a decrease in the S-phase (Figure 5). CaM gene transfer was able to effectively rescue the effect of FBXL2 on G0 arrest and apoptosis (supplemental Figure 6B), and coexpression of either the cyclin D2Q98A or cyclin D2K270R mutants were able to antagonize FBXL2 function in HCC1739 BL cells (Figure 6C). These results coupled with our data from human AML and ALL samples suggest that cyclin D2 protein stability are diametrically regulated by the SCFFBXL2 complex and perhaps CaM activity. Because cyclin D2 is the dominant D-type cyclin in EBV associated B-cell lymphoma,15,16 our data suggest that tight interplay between these effectors will regulate B lymphocyte proliferative activity through control of cyclin D2 abundance. Hence, although speculative, SCFFBXL2 may exhibit antiproliferation activity in some B-cell lymphomas and leukemias.
Supplementary Material
Acknowledgments
The authors thank A. F. Stewart for critical review of the manuscript and helpful suggestions.
This material is based on work supported, in part, by the US Department of Veterans Affairs, Veterans Health Administration, Office of Research and Development, Biomedical Laboratory Research and Development. This work was supported by a Merit Review Award from the US Department of Veterans Affairs and National Institutes of Health R01 grants HL096376, HL097376, and HL098174 (to R.K.M.). The contents presented do not represent the views of the Department of Veterans Affairs or the United States Government.
Footnotes
The online version of this article contains a data supplement.
The publication costs of this article were defrayed in part by page charge payment. Therefore, and solely to indicate this fact, this article is hereby marked “advertisement” in accordance with 18 USC section 1734.
Authorship
Contribution: B.B.C., J.R.G., T.A.C., and C.Z. performed experiments directly related to the current studies; B.B.C. directed the study and wrote the paper; H.L.M., M.F., J.F.M., and M.B. provided editorial guidance and expertise with use of leukemic cell lines and human AML and ALL samples; and R.K.M. reviewed the data, helped direct the study, and assisted with editorial revisions.
Conflict-of-interest disclosure: The authors declare no competing financial interests.
Correspondence: Rama K. Mallampalli, The University of Pittsburgh, Pulmonary, Allergy, & Critical Care Medicine, UPMC Montefiore, NW 628 Dept of Medicine, Pittsburgh, PA 15213; e-mail: mallampallirk@upmc.edu.
References
- 1.Buchakjian MR, Kornbluth S. The engine driving the ship: metabolic steering of cell proliferation and death. Nat Rev Mol Cell Biol. 2010;11(10):715–727. doi: 10.1038/nrm2972. [DOI] [PubMed] [Google Scholar]
- 2.Siebert R, Willers CP, Opalka B. Role of the cyclin-dependent kinase 4 and 6 inhibitor gene family p15, p16, p18, and p19 in leukemia and lymphoma. Leuk Lymphoma. 1996;23(5-6):505–520. doi: 10.3109/10428199609054859. [DOI] [PubMed] [Google Scholar]
- 3.Ohtani K, DeGregori J, Nevins JR. Regulation of the cyclin E gene by transcription factor E2F1. Proc Natl Acad Sci U S A. 1995;92(26):12146–12150. doi: 10.1073/pnas.92.26.12146. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Lew DJ, Dulic V, Reed SI. Isolation of three novel human cyclins by rescue of G1 cyclin (Cln) function in yeast. Cell. 1991;66(6):1197–1206. doi: 10.1016/0092-8674(91)90042-w. [DOI] [PubMed] [Google Scholar]
- 5.Gautschi O, Ratschiller D, Gugger M, Betticher DC, Heighway J. Cyclin D1 in non-small cell lung cancer: a key driver of malignant transformation. Lung Cancer. 2007;55(1):1–14. doi: 10.1016/j.lungcan.2006.09.024. [DOI] [PubMed] [Google Scholar]
- 6.Zhang Q, Tian L, Mansouri A, Korapati AL, Johnson TJ, Claret FX. Inducible expression of a degradation-resistant form of p27Kip1 causes growth arrest and apoptosis in breast cancer cells. FEBS Lett. 2005;579(18):3932–3940. doi: 10.1016/j.febslet.2005.06.012. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Zi X, Agarwal R. Silibinin decreases prostate-specific antigen with cell growth inhibition via G1 arrest, leading to differentiation of prostate carcinoma cells: implications for prostate cancer intervention. Proc Natl Acad Sci U S A. 1999;96(13):7490–7495. doi: 10.1073/pnas.96.13.7490. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Gladkikh A, Potashnikova D, Korneva E, Khudoleeva O, Vorobjev I. Cyclin D1 expression in B-cell lymphomas. Exp Hematol. 2010;38(11):1047–1057. doi: 10.1016/j.exphem.2010.08.002. [DOI] [PubMed] [Google Scholar]
- 9.Ngo BT, Felthaus J, Hein M, et al. Monitoring bortezomib therapy in multiple myeloma: screening of cyclin D1, D2, and D3 via reliable real-time polymerase chain reaction and association with clinico-pathological features and outcome. Leuk Lymphoma. 2010;51(9):1632–1642. doi: 10.3109/10428194.2010.496014. [DOI] [PubMed] [Google Scholar]
- 10.Ishigaki T, Sasaki K, Watanabe K, et al. Amplification of IGH/CCND1 fusion gene in a primary plasma cell leukemia case. Cancer Genet Cytogenet. 2010;201(1):62–65. doi: 10.1016/j.cancergencyto.2010.05.006. [DOI] [PubMed] [Google Scholar]
- 11.Shan J, Zhao W, Gu W. Suppression of cancer cell growth by promoting cyclin D1 degradation. Mol Cell. 2009;36(3):469–476. doi: 10.1016/j.molcel.2009.10.018. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Lin DI, Barbash O, Kumar KG, et al. Phosphorylation-dependent ubiquitination of cyclin D1 by the SCF(FBX4-alphaB crystallin) complex. Mol Cell. 2006;24(3):355–366. doi: 10.1016/j.molcel.2006.09.007. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Okabe H, Lee SH, Phuchareon J, Albertson DG, McCormick F, Tetsu O. A critical role for FBXW8 and MAPK in cyclin D1 degradation and cancer cell proliferation. PLoS One. 2006;1:e128. doi: 10.1371/journal.pone.0000128. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Santra MK, Wajapeyee N, Green MR. F-box protein FBXO31 mediates cyclin D1 degradation to induce G1 arrest after DNA damage. Nature. 2009;459(7247):722–725. doi: 10.1038/nature08011. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Sinclair AJ, Palmero I, Holder A, Peters G, Farrell PJ. Expression of cyclin D2 in Epstein-Barr virus-positive Burkitt's lymphoma cell lines is related to methylation status of the gene. J Virol. 1995;69(2):1292–1295. doi: 10.1128/jvi.69.2.1292-1295.1995. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Palmero I, Holder A, Sinclair AJ, Dickson C, Peters G. Cyclins D1 and D2 are differentially expressed in human B-lymphoid cell lines. Oncogene. 1993;8(4):1049–1054. [PubMed] [Google Scholar]
- 17.Delmer A, Ajchenbaum-Cymbalista F, Tang R, et al. Overexpression of cyclin D2 in chronic B-cell malignancies. Blood. 1995;85(10):2870–2876. [PubMed] [Google Scholar]
- 18.Liu SC, Bassi DE, Zhang SY, Holoran D, Conti CJ, Klein-Szanto AJ. Overexpression of cyclin D2 is associated with increased in vivo invasiveness of human squamous carcinoma cells. Mol Carcinog. 2002;34(3):131–139. doi: 10.1002/mc.10057. [DOI] [PubMed] [Google Scholar]
- 19.Tanaka Y, Tanaka N, Saeki Y, et al. c-Cbl-dependent monoubiquitination and lysosomal degradation of gp130. Mol Cell Biol. 2008;28(15):4805–4818. doi: 10.1128/MCB.01784-07. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Hochstrasser M. Biochemistry. All in the ubiquitin family. Science. 2000;289(5479):563–564. doi: 10.1126/science.289.5479.563. [DOI] [PubMed] [Google Scholar]
- 21.Cardozo T, Pagano M. The SCF ubiquitin ligase: insights into a molecular machine. Nat Rev Mol Cell Biol. 2004;5(9):739–751. doi: 10.1038/nrm1471. [DOI] [PubMed] [Google Scholar]
- 22.Cenciarelli C, Chiaur DS, Guardavaccaro D, Parks W, Vidal M, Pagano M. Identification of a family of human F-box proteins. Curr Biol. 1999;9(20):1177–1179. doi: 10.1016/S0960-9822(00)80020-2. [DOI] [PubMed] [Google Scholar]
- 23.Ilyin GP, Rialland M, Glaise D, Guguen-Guillouzo C. Identification of a novel Skp2-like mammalian protein containing F-box and leucine-rich repeats. FEBS Lett. 1999;459(1):75–79. doi: 10.1016/s0014-5793(99)01211-9. [DOI] [PubMed] [Google Scholar]
- 24.Skaar JR, Pagan JK, Pagano M. SnapShot: F-box proteins I. Cell. 2009;137(6):1160. doi: 10.1016/j.cell.2009.05.039. [DOI] [PubMed] [Google Scholar]
- 25.Inuzuka H, Shaik S, Onoyama I, et al. SCF(FBW7) regulates cellular apoptosis by targeting MCL1 for ubiquitylation and destruction. Nature. 2011;471(7336):104–109. doi: 10.1038/nature09732. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Malard V, Berenguer F, Prat O, Ruat S, Steinmetz G, Quemeneur E. Global gene expression profiling in human lung cells exposed to cobalt. BMC Genomics. 2007;8(147):147. doi: 10.1186/1471-2164-8-147. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Wang C, Gale M, Jr., Keller BC, et al. Identification of FBL2 as a geranylgeranylated cellular protein required for hepatitis C virus RNA replication. Mol Cell. 2005;18(4):425–434. doi: 10.1016/j.molcel.2005.04.004. [DOI] [PubMed] [Google Scholar]
- 28.Chen BB, Coon TA, Glasser JR, Mallampalli RK. Calmodulin antagonizes a calcium-activated SCF ubiquitin E3 ligase subunit, FBXL2, to regulate surfactant homeostasis. Mol Cell Biol. 2011;31(9):1905–1920. doi: 10.1128/MCB.00723-10. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Rhoads AR, Friedberg F. Sequence motifs for calmodulin recognition. FASEB J. 1997;11(5):331–340. doi: 10.1096/fasebj.11.5.9141499. [DOI] [PubMed] [Google Scholar]
- 30.Kahl CR, Means AR. Regulation of cell cycle progression by calcium/calmodulin-dependent pathways. Endocr Rev. 2003;24(6):719–736. doi: 10.1210/er.2003-0008. [DOI] [PubMed] [Google Scholar]
- 31.Taules M, Rius E, Talaya D, Lopez-Girona A, Bachs O, Agell N. Calmodulin is essential for cyclin-dependent kinase 4 (Cdk4) activity and nuclear accumulation of cyclin D1-Cdk4 during G1. J Biol Chem. 1998;273(50):33279–33286. doi: 10.1074/jbc.273.50.33279. [DOI] [PubMed] [Google Scholar]
- 32.Choi J, Chiang A, Taulier N, Gros R, Pirani A, Husain M. A calmodulin-binding site on cyclin E mediates Ca2+-sensitive G1/s transitions in vascular smooth muscle cells. Circ Res. 2006;98(10):1273–1281. doi: 10.1161/01.RES.0000223059.19250.91. [DOI] [PubMed] [Google Scholar]
- 33.Choi J, Husain M. Calmodulin-mediated cell cycle regulation: new mechanisms for old observations. Cell Cycle. 2006;5(19):2183–2186. doi: 10.4161/cc.5.19.3265. [DOI] [PubMed] [Google Scholar]
- 34.Chen BB, Mallampalli RK. Calmodulin binds and stabilizes the regulatory enzyme, CTP: phosphocholine cytidylyltransferase. J Biol Chem. 2007;282(46):33494–33506. doi: 10.1074/jbc.M706472200. [DOI] [PubMed] [Google Scholar]
- 35.Wikenheiser KA, Clark JC, Linnoila RI, Stahlman MT, Whitsett JA. Simian virus 40 large T antigen directed by transcriptional elements of the human surfactant protein C gene produces pulmonary adenocarcinomas in transgenic mice. Cancer Research. 1992;52(19):5342–5352. [PubMed] [Google Scholar]
- 36.Ray NB, Durairaj L, Chen BB, et al. Dynamic regulation of cardiolipin by the lipid pump Atp8b1 determines the severity of lung injury in experimental pneumonia. Nat Med. 2010;16(10):1120–1127. doi: 10.1038/nm.2213. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Tsuchiya S, Yamabe M, Yamaguchi Y, Kobayashi Y, Konno T, Tada K. Establishment and characterization of a human acute monocytic leukemia cell line (THP-1). Int J Cancer. 1980;26(2):171–176. doi: 10.1002/ijc.2910260208. [DOI] [PubMed] [Google Scholar]
- 38.Lehmann MH. Recombinant human granulocyte-macrophage colony-stimulating factor triggers interleukin-10 expression in the monocytic cell line U937. Mol Immunol. 1998;35(8):479–485. doi: 10.1016/s0161-5890(98)00043-1. [DOI] [PubMed] [Google Scholar]
- 39.Lozzio CB, Lozzio BB. Human chronic myelogenous leukemia cell-line with positive Philadelphia chromosome. Blood. 1975;45(3):321–334. [PubMed] [Google Scholar]
- 40.Matsuo Y, MacLeod RA, Uphoff CC, et al. Two acute monocytic leukemia (AML-M5a) cell lines (MOLM-13 and MOLM-14) with interclonal phenotypic heterogeneity showing MLL-AF9 fusion resulting from an occult chromosome insertion, ins(11;9)(q23;p22p23). Leukemia. 1997;11(9):1469–1477. doi: 10.1038/sj.leu.2400768. [DOI] [PubMed] [Google Scholar]
- 41.Agassandian M, Chen BB, Schuster CC, Houtman JC, Mallampalli RK. 14-3–3zeta escorts CCTalpha for calcium-activated nuclear import in lung epithelia. FASEB J. 2010;24(4):1271–1283. doi: 10.1096/fj.09-136044. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Agassandian M, Chen BB, Schuster CC, Houtman JC, Mallampalli RK. {zeta} escorts CCTα for calcium-activated nuclear import in lung epithelia. Faseb J. 2010;24(4):1271–1283. doi: 10.1096/fj.09-136044. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Mallampalli RK, Ryan AJ, Salome RG, Jackowski S. Tumor necrosis factor-alpha inhibits expression of CTP:phosphocholine cytidylyltransferase. J Biol Chem. 2000;275(13):9699–9708. doi: 10.1074/jbc.275.13.9699. [DOI] [PubMed] [Google Scholar]
- 44.Butler PL, Mallampalli RK. Cross-talk between remodeling and de novo pathways maintains phospholipid balance through ubiquitination. J Biol Chem. 2010;285(9):6246–6258. doi: 10.1074/jbc.M109.017350. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Deininger MW, Vieira SA, Parada Y, et al. Direct relation between BCR-ABL tyrosine kinase activity and cyclin D2 expression in lymphoblasts. Cancer Res. 2001;61(21):8005–8013. [PubMed] [Google Scholar]
- 46.Parada Y, Banerji L, Glassford J, et al. BCR-ABL and interleukin 3 promote haematopoietic cell proliferation and survival through modulation of cyclin D2 and p27Kip1 expression. J Biol Chem. 2001;276(26):23572–23580. doi: 10.1074/jbc.M101885200. [DOI] [PubMed] [Google Scholar]
- 47.Jena N, Deng M, Sicinska E, Sicinski P, Daley GQ. Critical role for cyclin D2 in BCR/ABL-induced proliferation of hematopoietic cells. Cancer Res. 2002;62(2):535–541. [PubMed] [Google Scholar]
- 48.Kida A, Kakihana K, Kotani S, Kurosu T, Miura O. Glycogen synthase kinase-3beta and p38 phosphorylate cyclin D2 on Thr280 to trigger its ubiquitin/proteasome-dependent degradation in hematopoietic cells. Oncogene. 2007;26(46):6630–6640. doi: 10.1038/sj.onc.1210490. [DOI] [PubMed] [Google Scholar]
- 49.Liu C, Kato Y, Zhang Z, Do VM, Yankner BA, He X. beta-Trcp couples beta-catenin phosphorylation-degradation and regulates Xenopus axis formation. Proc Natl Acad Sci U S A. 1999;96(11):6273–6278. doi: 10.1073/pnas.96.11.6273. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Hansen DV, Loktev AV, Ban KH, Jackson PK. Plk1 regulates activation of the anaphase promoting complex by phosphorylating and triggering SCFbetaTrCP-dependent destruction of the APC Inhibitor Emi1. Mol Biol Cell. 2004;15(12):5623–5634. doi: 10.1091/mbc.E04-07-0598. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Watanabe N, Arai H, Iwasaki J, et al. Cyclin-dependent kinase (CDK) phosphorylation destabilizes somatic Wee1 via multiple pathways. Proc Natl Acad Sci U S A. 2005;102(33):11663–11668. doi: 10.1073/pnas.0500410102. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Chen BB, Glasser JR, Coon TA, Mallampalli RK. F-box protein FBXL2 exerts human lung tumor suppressor-like activity by ubiquitin-mediated degradation of cyclin D3 resulting in cell cycle arrest [published online ahead of print October 24, 2011]. Oncogene. doi: 10.1038/onc.2011.432. doi: 10.1038/onc.2011.432. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Chen BB, Glasser JR, Coon TA, Mallampalli RK. FBXL2 is a ubiquitin E3 ligase subunit that triggers mitotic arrest. Cell Cycle. 2011;10(20):3487–3494. doi: 10.4161/cc.10.20.17742. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Sicinska E, Aifantis I, Le Cam L, et al. Requirement for cyclin D3 in lymphocyte development and T cell leukemias. Cancer Cell. 2003;4(6):451–461. doi: 10.1016/s1535-6108(03)00301-5. [DOI] [PubMed] [Google Scholar]
- 55.Susaki E, Nakayama K, Nakayama KI. Cyclin D2 translocates p27 out of the nucleus and promotes its degradation at the G0-G1 transition. Mol Cell Biol. 2007;27(13):4626–4640. doi: 10.1128/MCB.00862-06. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.