Abstract
Diaphyseal medullary stenosis with malignant fibrous histiocytoma (DMS-MFH) is an autosomal-dominant syndrome characterized by bone dysplasia, myopathy, and bone cancer. We previously mapped the DMS-MFH tumor-suppressing-gene locus to chromosomal region 9p21–22 but failed to identify mutations in known genes in this region. We now demonstrate that DMS-MFH results from mutations in the most proximal of three previously uncharacterized terminal exons of the gene encoding methylthioadenosine phosphorylase, MTAP. Intriguingly, two of these MTAP exons arose from early and independent retroviral-integration events in primate genomes at least 40 million years ago, and since then, their genomic integration has gained a functional role. MTAP is a ubiquitously expressed homotrimeric-subunit enzyme critical to polyamine metabolism and adenine and methionine salvage pathways and was believed to be encoded as a single transcript from the eight previously described exons. Six distinct retroviral-sequence-containing MTAP isoforms, each of which can physically interact with archetype MTAP, have been identified. The disease-causing mutations occur within one of these retroviral-derived exons and result in exon skipping and dysregulated alternative splicing of all MTAP isoforms. Our results identify a gene involved in the development of bone sarcoma, provide evidence of the primate-specific evolution of certain parts of an existing gene, and demonstrate that mutations in parts of this gene can result in human disease despite its relatively recent origin.
Introduction
Diaphyseal medullary stenosis with malignant fibrous histiocytoma (DMS-MFH [MIM 112250]) is a rare, autosomal-dominant bone dysplasia and cancer syndrome of unknown etiology.1–3 The disorder has a unique bone-dysplasia phenotype characterized by cortical growth abnormalities, including diffuse diaphyseal medullary stenosis with overlying endosteal cortical thickening, metaphyseal striations, and scattered infarctions within the bone marrow. Affected individuals endure pathologic fractures that subsequently heal poorly, progressive wasting, bowing of the lower extremities, painful debilitation, and the development of presenile cataracts. We recently expanded the known clinical features of the syndrome by characterizing two new unrelated families affected by a progressive form of muscular disease consistent with facioscapulohumeral muscular dystrophy (FSHD [MIM 158900]) (see below). Among DMS-MFH-affected individuals, approximately 35% develop a form of bone sarcoma consistent with the diagnosis of malignant fibrous histiocytoma (MFH).1–4
Using a positional-cloning approach, we originally localized the disease-associated allele locus to chromosomal region 9p21–22 and established a 3.5 cM critical locus between markers D9S1778 and D9S171.4 Given the cancer component of the syndrome, the 9p21–22 region is of particular interest in that it is one of the most frequently deleted and/or translocated chromosomal regions in human cancer.5 A diverse group of human cancers demonstrate loss of this region and include gliomas,6,7 melanomas,8 non-small-cell lung cancers,9 acute leukemias,10,11 and, of direct significance to this study, osteosarcomas.12,13 In an attempt to further narrow the region as well as establish a link between hereditary and sporadic tumor forms, we performed loss of heterozygosity (LOH) analysis of sporadic MFH samples. This analysis supported a shared genetic etiology between hereditary and sporadic MFH cases and mapped the smallest region of overlap to the 2.9 Mb region between markers D9S736 and D9S171.14
A number of DMS-MFH candidate genes were originally screened by DNA sequencing and were excluded because they lacked mutations. These genes included the cyclin-dependent kinase inhibitor 2A (CDKN2A [p16] [MIM 600160]) and its alternatively spliced product p14-ARF, CDKN2B (p15), members of the interferon (IFN) superfamily, and the methylthioadenosine phosphorylase gene (MTAP [MIM 156540]).4 MTAP has been thought to consist of eight exons and seven introns15 and encode a ubiquitously expressed enzyme that plays a crucial role in the salvage pathway for adenine and methionine in all tissues.16 In the salvage pathways, methylthioadenosine (MTA), a by-product of the polyamine pathway, is recovered through its phosphorolysis into adenine and methylthioribose-1-phosphate by MTAP.17 Through a series of reactions, methylthioribose-1-phosphate is then converted into methionine.17,18 It is suggested that loss of MTAP activity plays a role in human cancer because its loss has been reported in a number of cancers, including osteosarcoma,12,13 leukemia,19 non-small-cell lung cancer,20 malignant melanoma,21 biliary-tract cancer,22 breast cancer,23 pancreatic cancer,24 and gastrointestinal stromal tumors.25 Reintroduction of MTAP expression into the MCF7 breast adenocarcinoma cell line, which lacks endogenous MTAP gene expression and enzymatic activity, inhibits the cells' ability to grow both in vitro and in vivo;23 the fact that MTAP inhibits cell growth is consistent with its presumed role as a tumor suppressor.
We have now identified and characterized the genetic defect underlying DMS-MFH. All affected members of five unrelated DMS-MFH-affected families possess synonymous mutations in the most proximal of three terminal MTAP exons identified and characterized in these studies. Interestingly, DNA-sequence analysis revealed that at least two of the exons are remnants of retroviral insertions into the primate genome. Both disease-causing mutations in exon 9, the most proximal of these exons, result in exon skipping and subsequent loss of this exon in alternatively spliced, biologically active isoforms. Biochemical studies provide evidence that isoforms containing exon 7 have MTAP activity. Altogether, these findings identify a gene associated with both hereditary bone dysplasia and osteosarcoma and also highlight the importance of evolutionarily co-opted gene parts for both health and disease.
Material and Methods
Linkage and Haplotype Mapping
After participants gave informed consent for these studies, which were approved by the Human Research Protection Program at the Mount Sinai School of Medicine, blood samples were obtained from affected and unaffected family members. Genomic DNA was extracted with the Puregene kit according to the manufacturer's (Minneapolis, MN) protocol, and individuals were genotyped with a panel of markers spanning the length of chromosome 9; included were markers from the Single Chromosome Scan Human Screening Set (Research Genetics, Carlsbad, CA) and a number of polymorphic markers that were custom generated. Family 1 from the original study was not included in this reanalysis. Allelic determination was performed with the ABI 3130xl Genetic Analyzer and GeneMapper 4.0 software (Applied Biosystems, Foster City, CA). We handled the generated data by using Mega 2,26 and linkage analysis was computed by SIMWALK2 v.2.8327 with the following parameters: all families are genetically homogenous, inheritance pattern is autosomal dominant and has 80% penetrance, there are no phenocopies, and the disease allele frequency is 0.0001. Marker positions were obtained from the Marshfield database and the UCSC Genome Browser March 2006 assembly. We further refined areas of positive location scores by using custom-generated microsatellite markers within the defined region. In brief, we designed the microsatellite markers by identifying simple tandem repeats (STRs) in DNA sequences of clone fragments from the Human Genome Assembly (UCSC Genome Browser) by using the Tandem Repeats Finder Program.28 Fluorescently labeled primers were then designed to amplify these repeat regions, and allele separation and analyses were performed as previously described29 (Table S1, available online).
DNA Sequence Analysis
We used PCR to amplify all MTAP exons by using Amplitaq-Gold (Applied Biosystems, Foster City, CA), and we purified them by using the QIAquick Spin PCR Purification Kit (QIAGEN, Valencia, CA); the exons were amplified and purified according to the manufacturers' protocol, and they were directly sequenced on an ABI Prism 3700 automated DNA Analyzer (Applied Biosystems, Foster City, CA). Data were analyzed with the program Sequencher v3.0 (Gene Codes Corporation, Ann Arbor, MI). Sets of intronic primers that we used to amplify the coding region and intron and exon boundaries are listed in Table S1. The PCR cycling conditions were the following: 94°C (10 min) for 1 cycle, 94°C (30 s), 55°C (30 s), and 72°C (60 s) for 35 cycles each, and a final extension of 72°C (10 min).
Computational DNA Analysis
We performed computational analysis to detect aberrant splice sites by using the NetGene2 Server30,31 and the splicing-enhancer motif-prediction program ESEfinder Release 2.0,32 for which we used the default parameters for the following SR proteins: SF2/ASF, SC35, SRp40, and SRp55.
RNA Isolation, Semiquantitative Reverse Transcription PCR, and Quantitative Real Time PCR
We extracted cultured-cell-line and tumor RNA by using the RNeasy Mini kits, and we treated it with DNase according to the manufacturer's (QIAGEN, Valencia, CA) protocol. For semiquantitative reverse transcription PCR (RT-PCR), we reverse transcribed a total of 1 μg of RNA per reaction by using first-strand cDNA synthesis with random primers (Promega, Madison, WI). To evaluate the transcription level of the MTAP splice variants, we performed RT-PCR by using combinations of isoform-specific primers listed in Table S1. We electrophoretically separated and visualized RT-PCR products on a 1.5% agarose gel by using ethidium bromide. We excised the bands and cloned and inserted them into the pCR4-TOPO vector (Invitrogen, Carlsbad, CA), and we sequenced 20 independent clones from each of the isolated bands to establish their identity. For quantitative RT-PCR (qRT-PCR), we reverse transcribed a total of 1 μg of RNA per reaction by using iSCRIPT cDNA Synthesis according to the manufacturer's (Bio-Rad, Hercules, CA) protocol. We performed qRT-PCR by using iQ SYBR Green Supermix (Bio-Rad, Hercules, CA), according to the manufacturer's protocol, on an ABI PRISM 7900HT Sequence Detection System (Applied Biosystems, Foster City, CA) and by using the primers listed in Table S1. All values were normalized to either GAPDH or HPRT levels. All experiments were done in triplicate, and all cell-culture experiments were independently validated at least three times.
RACE: Rapid Amplification of cDNA 3′ Ends
Total RNA was isolated from five primary human fibroblast cell lines. We used a total of 1 μg of RNA per cell line, the PowerScript Reverse Transcriptase, and the BD SMART IIA primer from the BD SMART RACE cDNA amplification kit (according to the manufacturer's [Franklin Lakes, NJ] protocol) to prepare corresponding first-strand cDNAs. In brief, each of the 3′ RACE-ready cDNAs was used in PCR-amplification reactions with the SMART RACE kit universal primers and sense gene-specific primers (GSP). First-round PCR and second-round PCR were performed with the specific primers listed in Table S1, Outer Primer (SMART RACE kit), and Inner Primer (SMART RACE kit).
Cell Culture and Transfections
Patient-derived osteosarcoma, fibroblast, and lymphoblast cell lines and other commercially available cell lines (obtained from ATCC) were maintained in Dulbecco's modified Eagle's medium (DMEM) supplemented with 10% fetal bovine serum, 100 U/ml penicillin, and 100 μg/ml streptomycin and were grown at 37°C in 5% CO2. For the expression studies, we transfected the cells 24 hr after plating by using lipofectamine 2000 according to the manufacturer's (Invitrogen, Carlsbad, CA) recommended protocol.
Expression Constructs
GST and V5 Expression Vectors
To generate the vectors expressing each of the MTAP splice variants (archetype MTAP and MTAP_v1, _v2, _v3, _v4, _v5, and _v6), we used RT-PCR to amplify the cDNA of each splice variant from normal fibroblasts by using the relevant primers listed in Table S1. In brief, the exon 1 forward primer was used for the amplification of all variants, and the reverse primers were designed to delete the stop codon so that a fusion protein would be generated with the V5 epitope. The amplified products were cloned and inserted into the pcDNA3.1/V5-His TOPO TA expression vector (Invitrogen, Carlsbad, CA). The resulting clones were completely sequenced in both orientations prior to their use.
Partial Minigene Expression Vectors
We used genomic DNAs obtained from a normal fibroblast cell line and two patient-derived fibroblast cell lines to amplify the following fragments: an 8 kb fragment containing exons 6–8 (flanked by introduced SalI and NheI restriction sites; subcloned into the pcDNA3.1/V5-His TOPO TA vector), 2.1 kb fragments containing exon 9 and carrying the wild-type (WT) DNA or either the c.813-2A>G or c.885A>G mutation (flanked by introduced SpeI and XhoI restriction sites; cloned into the pCR4-TOPO vector), and a 3 kb fragment containing exons 10 and 11 (flanked by introduced XhoI and SacII restriction sites; cloned into the pCR4-TOPO vector). Each 2.1 kb fragment was digested with SpeI and XhoI, and we cloned the fragments and inserted them into a SpeI/XhoI-digested pcDNA3.1 (exons 6–8) vector to generate the following three pcDNA3.1 (exons 6–8 and 9) constructs: WT, c.813-2A>G, and c.885A>G. Finally, the 3 kb fragment was digested with XhoI and SacII and cloned and inserted into each one of the three pcDNA3.1 (exons 6–8 and 9) vectors digested with XhoI and SacII. The following three pcDNA3.1 (exons 6–8 and 9–11) minigene expression vectors were created: WT, c.813-2A>G, and c.885A>G. The primers that we used are listed in Table S1. We performed PCR amplifications by using the EXPAND Long Template PCR system according to the manufacturer's (Roche, Indianapolis, IN) protocol. We sequenced all exons and approximately 500 base pairs of the intron-exon flanking boundaries of the generated plasmids.
Coimmunoprecipitation Assay
Relevant combinations of GST- and V5-tagged constructs for archetype MTAP and MTAP_v1, _v2, _v3, _v4, _v5, and _v6 were cotransfected into PC3M cells. All of the following procedures were performed at 4°C. Cell extracts for immunoblotting were harvested in NP40 lysis buffer (Santa Cruz Biotechnology, standard protocol), and insoluble material was removed by centrifugation (10 min at 13,000 rpm). One tenth of the cell extracts were reserved for subsequent immunoblot analysis. Cell extracts were incubated with V5 antibody (1 μg/ml) or GST antibody (1 μg/ml) for 4 hr and Protein A-Sepharose (Invitrogen, Carlsbad, CA). Beads were washed three times with 1% lysis buffer, and coimmunoprecipitates were released by being boiled in 100 μl 2× SDS Reducing Sample Buffer (Invitrogen, Carlsbad, CA) for 5 min. Coimmunoprecipitates were analyzed by immunoblot analysis (see below).
Immunoblot and Densitometric Analysis
Cell extracts for immunoblotting were harvested in radioimmunoprecipitation assay (RIPA) buffer (Santa Cruz Biotechnology, standard protocol). Equal amounts of protein (50 μg) as determined by the BioRad DC Protein quantification assay were loaded and separated by polyacrylamide gel electrophoresis and transferred to nitrocellulose membranes. We performed immunoblotting by using a goat polyclonal antibody to actin (SC-1615), a monoclonal (0.2 mg/ml) antibody to GST (BD PharMingen), and a monoclonal antibody to the V5 tag (Santa Cruz Biotechnology). We analyzed enhanced chemiluminescent images of immunoblots by using a scanning densitometer and quantifying the bands (BIOQUANT NOVA imaging system). All values were normalized to actin and expressed as fold changes relative to the control.
MTA Quantification
Blood-serum and cell-extract lysates were mixed with 63 pmol [5′-2H3] MTA, neutralized with KOH, and centrifuged. MTA fractions were purified by HPLC (SymmetryShield RP18 column), concentrated, dissolved in 10% methanol with 0.1% TFA, and subjected to LCQ ESI-MS analysis. MTA was quantitated with an internal mass standard of [5′-2H3] (301 amu) relative to the peak area for authentic MTA (298 amu). Samples were analyzed in triplicate.
MTAP Enzymatic Activity Assay
Cells were trypsinized and washed twice with PBS, and the cell pellets were frozen at −80°C. On the day of the analysis, the pellets were thawed on ice and resuspended in MTAP lysis buffer (20 mM potassium phosphate, pH 7.4, 1 mM dithiothreitol, and Roche complete, EDTA-free protease cocktail in a dilution equivalent to 1 tablet for 125 ml of lysate buffer), sonicated three times for 15 s each and cooled on ice between sonications, and centrifuged at 13,000 × g for 15 min at 4°C. Protein concentrations were measured with the Bio-Rad DC Protein Assay. The MTAP-activity assay was as described33 and had the following modifications: Cell lysates or an enzyme blank consisting of an equal volume of lysis buffer was preincubated at 37°C for 5 min in 86 μl of a solution containing 116 mM potassium phosphate, 58 mM KCl, and 0.23 mM dithiothreitol in quartz microcuvettes. The enzyme reactions were started with the addition of 4 μl xanthine oxidase (0.15 units) and 10 μl of 5 mM MTA. Data was collected for 1,300 s, and the rates were calculated by a linear fit to the data between 800 and 1,100 s. The blank rate was subtracted from all the enzyme rates. MTAP-assay rates were linear; the lysate protein concentration ranged from 10 to 500 ng protein per assay. One unit of MTAP activity is the amount of enzyme that catalyzed the formation of 1 μmole of adenine per minute under the conditions of the assay.
Results
Positional Cloning of the DMS-MFH-Associated Gene
Previously, we performed genome-wide linkage analysis and haplotype reconstruction on three unrelated families to establish the critical region of the DMS-MFH-associated gene as a 3.5 cM locus in chromosomal region 9p21–22. The centromeric boundary was defined by D9S1778, and the telomeric end was defined by D9S171.4 Since the original mapping, we identified two additional families (families 4 and 5) (Figure 1). Interestingly, both of the new families had evidence of a progressive myopathic disease (Martignetti et al., unpublished observations34) not previously noted in the three original families. The disease-associated gene for one of these later multigenerational families (this family was originally described by Henry et al.35 as having a history of bone disease, pathologic fractures, fibrosarcoma, cataracts, and myopathy (MIM 609940]) had been independently mapped to a 15 Mb region at 9p21–22.34 This region broadly overlapped the DMS-MFH critical region. To determine whether these two additional families affected by myopathic disease were allelic and supported the previously identified DMS-MFH critical region, we performed multipoint parametric linkage analysis by using markers spanning chromosome 9.
Figure 1.
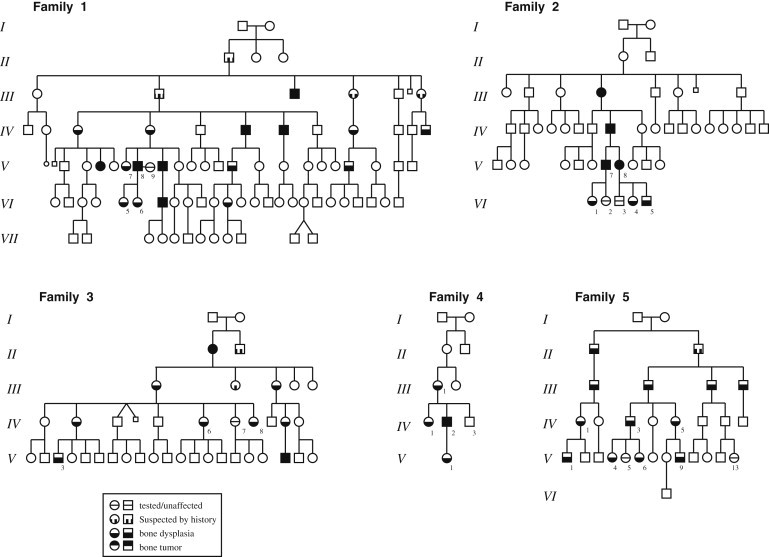
Pedigrees of the Five DMS-MFH-Affected Families
Families 1, 2, and 3 were originally described as the “American,” “Australian,” and “New York” families, respectively.1–3 Family 4 is a previously undescribed DMS-MFH-affected family from New York. Family 5 has been described as having autosomal-dominant bone fragility and limb-girdle myopathy.34 Family members classified as “suspected by history” were unavailable for radiological diagnosis but had a history of multiple (>2) pathological fractures and/or had children who were clinically diagnosed with DMS-MFH on the basis of plain-film X-rays and family history.
We analyzed all five families by using a combination of previously described and laboratory-developed microsatellite markers. A maximal combined location score of 4.27 was obtained for marker D9SB3 (Figure 2A). We then performed haplotype analysis of additional markers within the shared region and reconstruction of haplotypes to further narrow the region. The minimal disease locus was established by recombination events between markers AL882 and D9S1749 in unaffected individual F3 IV-6 and between markers D9S916 and D9S976 in affected individual F3 IV-7. These events narrowed the critical region to 1.2 Mb (Figure 2B).
Figure 2.
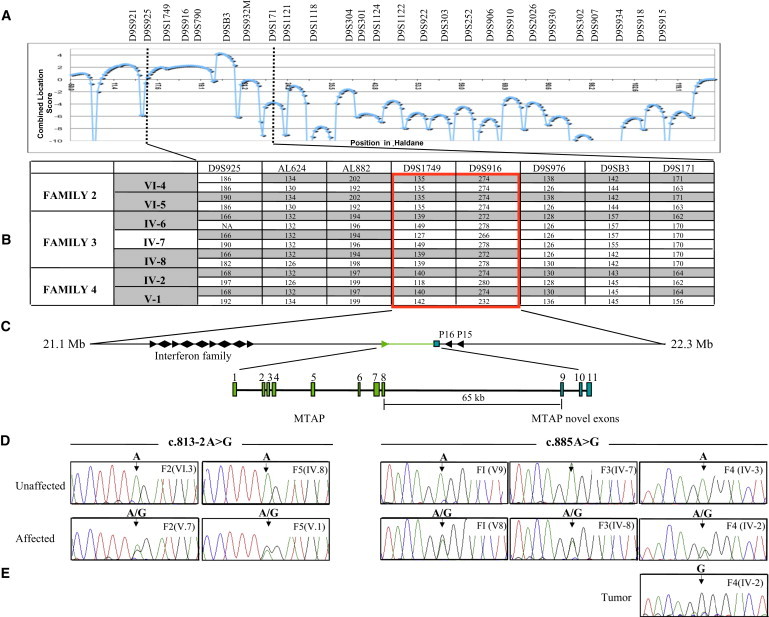
Identification of the Critical Region and the DMS-MFH-Associated Gene
(A) The results of a combined multipoint parametric linkage analysis for families 2, 3, and 4. The maximal location score is 4.27 at marker D9SB3.
(B) Haplotype analysis of informative individuals from families 2, 3, and 4 narrowed the DMS-MFH critical region (boxed).
(C) Physical map of the DMS-MFH critical region, which spans 1.3 Mb between the flanking markers AL882 and D9S976. Arrows indicate transcriptional direction of candidate genes. The exon-intron structure of MTAP highlights the eight exons of MTAP (green) and the three terminal exons (blue). Maps are not drawn to scale.
(D) Representative DNA-sequence chromatograms from selected affected and unaffected individuals from each family.
(E) Tumor DNA sequence revealing homozygosity of the diseased allele and loss of the unaffected allele.
DNA-sequence analysis of known candidate genes within the original critical region had previously failed to identify causative mutations.4 In particular, this analysis included the known eight exons and corresponding intron-exon boundaries of MTAP.4 Having exhausted all known genes, we next sought and analyzed predicted genes and putative open reading frames (ORF) from within the region. In silico analysis of the region with the use of the UCSC Genome Browser identified an ORF (GenBank accession number AF216650) located 65 Kb downstream of the known MTAP termination site within a truncated expressed sequence tag (EST). DNA-sequence analysis of the putative 192 bp ORF, which we termed MTAP exon 9, and its intron-exon boundaries revealed the presence of one of two heterozygous A>G substitutions for all affected members of the first four DMS-MFH-affected families (Figure 2D). Using the EST clone (GenBank accession number AK309365) as a reference, we determined that one mutation was a synonymous change at position c.885A>G (p.(=)), effectively R100R, and was present in affected families 1, 3, and 4. The second mutation, c.813-2A>G, was an intronic change present in affected family 2. The sequence changes segregated appropriately with the disease phenotype within all respective family members in each family. We then analyzed affected and unaffected individuals from family 5; all affected individuals possessed the c.813-2A>G mutation. To test the possibility that these changes represented polymorphisms and not pathogenic mutations, we screened a control population. Neither mutation was identified in 1,000 chromosomes from 500 unaffected control individuals. Similarly, the mutations were not present in dbSNP build 131.
The DMS-MFH Mutation Is Homozygously Present in a Patient-Derived Osteosarcoma
MFH is a rare, highly aggressive bone tumor of uncertain histogenesis but whose histologic appearance, treatment, and response are similar to those of osteosarcoma.36,37 Indeed, the diagnosis of MFH has been controversial, and in cases where osteoid is present, the diagnosis of osteosarcoma is favored.38 Approximately one-third of affected individuals within our families developed bone sarcomas arising between the second and fifth decades of life. The diagnoses were either MFH3 or bone fibrosarcoma.1 As shown in Figure 3, given the presence of osteoid, the histopathological analysis of a tumor from a DMS-MFH-affected individual (III-3 from family 4; c.885A>G) is consistent with the diagnosis of osteosarcoma. Thus, inherited MTAP alternative-splicing mutations can result in histology-proven osteosarcoma.
Figure 3.
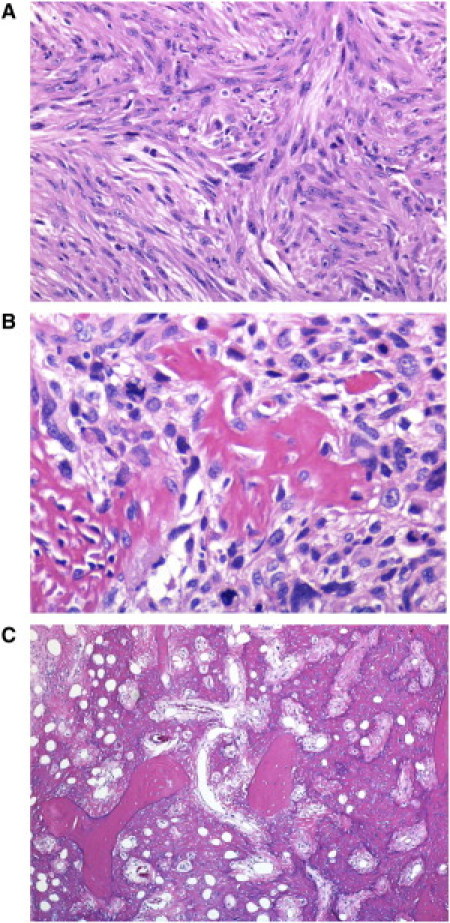
Histopathological Analysis of an Osteosarcoma from Patient F4 IV-2
(A) Histologic analysis revealed that 95% of the studied tumor specimen displayed the typical pattern of malignant spindle (fibroblastic) cells of bone MFH.
(B) Malignant cells forming neoplastic bone.
(C) Focal sheets of neoplastic bone within the tumor are shown to be entrapping pre-existing bone trabeculae, and the overtly malignant cells are shown to produce bone. All sections were stained with hematoxylin and eosin.
Moreover, and in agreement with Knudson's two-hit hypothesis for a tumor-suppressing gene,39 direct sequencing of this patient's osteosarcoma genomic DNA demonstrated homozygosity for the c.885A>G mutation (Figure 2E). LOH analysis with microsatellite markers spanning the originally defined 2.9 Mb DMS-MFH critical region revealed complete loss of the WT allele from the unaffected chromosome (data not shown).
DMS-MFH Mutations Result in Exon Skipping and Altered Expression of MTAP Isoforms
Given the identification of these disease-specific genomic DNA mutations, we sought to understand the relationship between the previously uncharacterized ninth exon and the MTAP RNA transcripts. Therefore, we performed 3′ RACE on the total RNA isolated from control and patient-derived fibroblast and lymphoblast cell lines and also from a patient-derived tumor cell line that we established. For 3′ RACE, we used intron-spanning primers anchored in exons 5 and 6 of the WT gene and sequenced the resulting cDNA products. Using this approach, we identified, as expected, the archetype MTAP transcript and, in accord with our hypothesis, additional isoforms containing exon 9. In total, six additional isoforms were identified; none contained the WT terminal exon 8, and all affected the C terminus of the protein product in different ways (Figure S1). Four contained either a short (9S; 103 nt) or long (9L; 192 nt) form of exon 9. Additionally, four of the isoforms contained a unique sequence that was mappable to two additional downstream exons, 10 and 11 (Figure 2C and Figure S1). On the basis of the electrophoretic mobility of the MTAP transcripts generated, we named the six alternative splice variants MTAP_v1 (exons 1–7 and 9S–11), _v2 (exons 1–7 and 9L), _v3 (exons 1–7, 10, and 11), _v4 (exons 1–6 and 9S–11), _v5 (exons 1–6 and 9L), and _v6 (exons 1–6, 10, and 11). Splice variants 1–3 contained the WT exon 7 sequence; variants 4–6 did not.
The two DMS-MFH mutations, one intronic and the other exonic, did not predict amino acid changes. We hypothesized that one possible pathogenic mechanism could be through an effect on alternative splicing. We analyzed the DNA sequence for the presence of known donor and acceptor splice motifs and intronic and exonic cis elements that could direct splice-site identification. The intronic c.813-2A>G mutation was predicted to result in the loss of a canonical splice acceptor site.31 The c.885A>G transition abolished a predicted exonic splicing enhancer (ESE) sequence.32
To validate these in silico findings, we generated three MTAP minigene constructs, WT, c.813-2A>G, and c.885A>G, containing ∼11.5 Kb of genomic sequence and differing at only the single nucleotide position being interrogated (Figure 4A). The minigene constructs were transiently transfected into MCF7 cells, and the relative expression levels of each isoform were determined by qRT-PCR. The identity of each transcript was established by RT-PCR, subcloning, and the sequencing of at least 20 independent clones from gel-isolated bands.
Figure 4.
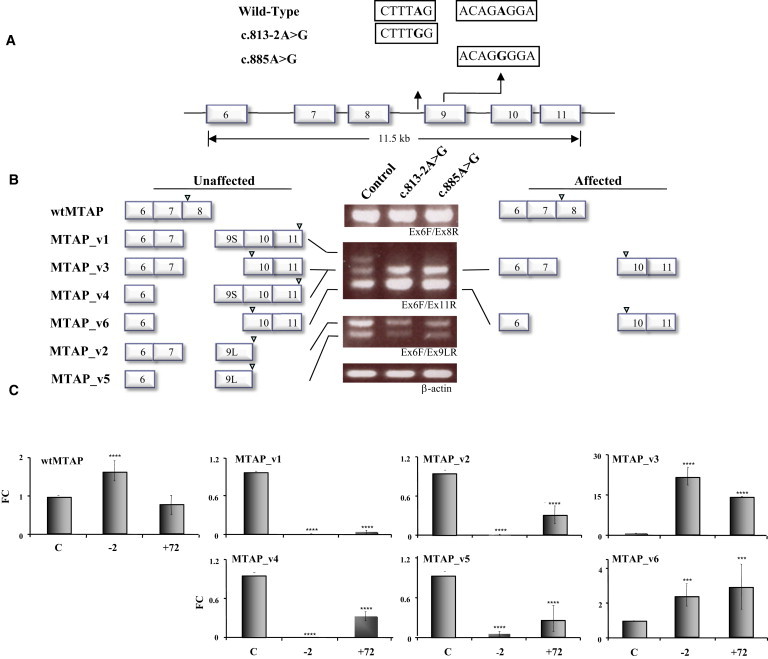
Patient-Derived MTAP Mutations Result in Exon Skipping
(A) Sequence differences of the three minigene constructs, WT, c.813-2A>G, and c.885A>G, are highlighted.
(B) Schematic representation of the major sequence-verified isoforms flanking the electrophoretic profile of each minigene construct. Arrowheads above exons depict the positions of translational stop codons. The WT construct expresses all alternative splice variants that are detectable with this combination of primers (exon 6 forward [Ex6F] and exon 11 reverse [Ex11R]) (left), whereas patient-derived mutant constructs, c.813-2A>G and c.885A>G, expressed relatively very low levels of exon-9S- and -9L-containing variants, v1/v4 and v2/v5, respectively. By comparison, the expression of isoforms v3 and v6 was markedly elevated in both mutant constructs.
(C) qRT-PCR analysis of archetype MTAP and isoform expression in cells that express each of the minigene constructs. Expression analysis of the three minigene constructs demonstrated that the c.813-2A>G mutant construct resulted in significantly increased archetype MTAP expression levels, whereas both the c.813-2A>G and c.885A>G mutant constructs were associated with an absence of and/or significantly decreased levels of MTAP_v1, _v2, _v4, and _v5. Both mutant constructs resulted in significantly increased expression levels of MTAP_v3 and _v6. The following abbreviation is used: wtMTAP, archetype MTAP. The error bars represent the averages of three independent experiments.
The three different MTAP constructs revealed clear differences in the expression pattern of splice variants, whereas expression of the WT form was unchanged (Figure 4B). Although the WT gene construct directed expression of all seven MTAP transcripts, the single-nucleotide changes at c.813-2A>G and c.885A>G resulted in markedly decreased expression of the four exon-9-containing transcripts, namely MTAP_v1, _v2, _v4, and _v5. In contrast, there was a significant increase in the two isoforms lacking exon 9, namely MTAP_v3 and _v6. Differences were noted in the transcription effects of the two mutations. Quantification of the resultant isoforms by qRT-PCR demonstrated that the c.813-2A>G mutation ablated expression of all isoforms containing exon 9 and significantly increased the expression of archetype MTAP (Figure 4C). In addition, the first three amino acids were lacking from both 9S isoforms. The c.885A>G mutation decreased the expression levels of all exon 9 isoforms by approximately 70% but had no effect on archetype-MTAP expression. Both patient-derived mutations significantly increased the expression of MTAP_v3 and _v6 to the same degree (Figure 4C).
The mutation-dependent isoform expression pattern that we identified by using the engineered minigene expression constructs was also present in patient-derived tissues. As shown in Figure 5, patient-derived fibroblast and lymphoblast cell lines, constitutively heterozygous for the mutations, demonstrated MTAP isoform expression patterns similar to each other but markedly different from the expression patterns of control cells (Figures 5A and 5B). Quantitative RT-PCR analysis revealed that in patient-derived cells, expression levels of MTAP_v1 and expression levels of MTAP_v3 and _v6 were approximately 50% lower and nearly five to ten times higher, respectively, than those of the controls (Figures 5A and 5B). In both these cell-line types, we were unable to consistently detect quantifiable levels of MTAP_v2, _v4, and _v5 (results not shown).
Figure 5.
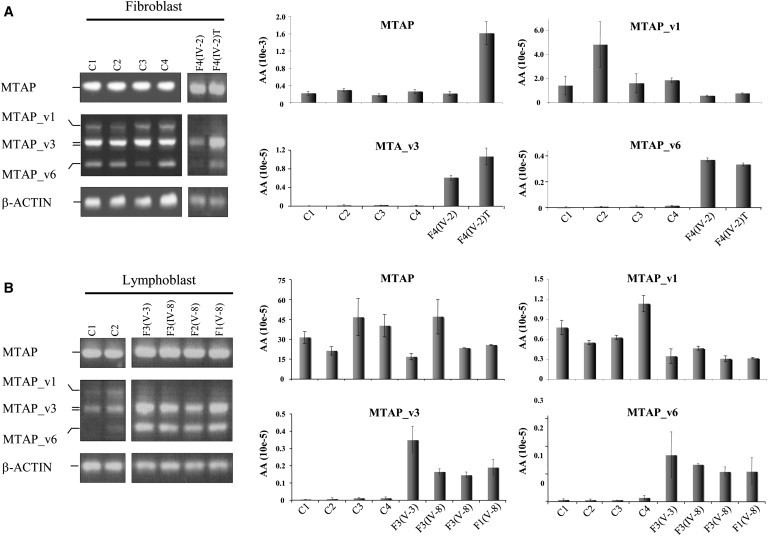
Dysregulated MTAP Splicing Patterns in Patient-Derived Cell Lines
MTAP expression patterns in patient-derived and control fibroblast (A) and lymphoblast (B) cell lines. Semiquantitative RT-PCR analysis is on the left, and qRT-PCR analysis of archetype MTAP is on the right.
(A) C1, C2, C3, and C4 are controls. Fibroblast and tumor cell lines are from affected individual F4 III-3.
(B) C1 and C2 are controls. Lymphoblast cell lines are from individuals F3 IV-5, F3 IV-6, F3 IV-7, F4 III-5, and F4 III-6. The following abbreviation is used: wtMTAP, archetype MTAP.
MTAP Splice Variants Can Physically Interact with Archetype MTAP and Are Biologically Active
We began exploring the function of the different MTAP splice variants by first determining their respective protein stabilities. The transient transfection of expression vectors (which contained each isoform fused with a 3′ V5 and/or a 5′ GST terminal tag) into MCF7 cells demonstrated that all six MTAP splice variants were translated. Although RNA levels were essentially equivalent (data not shown), there was, however, a large variability in protein expression levels (Figure S2). Archetype-MTAP and MTAP_v1, _v2, and _v3 proteins (i.e., those containing exon 7) were expressed at comparable levels, whereas MTAP_v4, _v5, and _v6 were expressed at markedly lower levels.
These findings suggest a possible difference in protein stability that might be regulated through the proteosome pathway. To test this hypothesis, we treated the transfected cells with the proteosome inhibitor MG132. The v4, v5, and v6 isoforms accumulated, whereas archetype MTAP and variants v1, v2, and v3 were relatively unaffected (Figure S2A). The half-life of each splice variant was estimated by cyclohexamide treatment. The half-life of archetype MTAP and MTAP_v1, _v2, and _v3 was ≥12 hr. It was significantly less than 6 hr for MTAP_v4, _v5, and _v6 (Figure S2B).
Given these findings and the knowledge that MTAP exists as a trimeric40–42 protein complex, we tested the ability of the MTAP splice variants to physically interact with archetype MTAP. We performed coimmuprecipitations on cell extracts from cotransfected PC3M cells by using combinations of V5- and GST-tagged fusion proteins. All MTAP splice variants were able to physically interact with archetype MTAP (Figure S3).
We next sought to determine whether MTAP splice variants have MTAP activity. We directly measured the ability of each isoform to convert MTA to adenine by using a biochemical assay after we stably transfected them into two MTAP-null cell lines, MCF7 and MNNG-HOS (a human-osteosarcoma-derived cell line). Cellular lysates from mock-transfected cells demonstrated negligible MTAP activity. Against this null-activity background, only MTAP isoforms v1, v2, and v3 demonstrated MTAP activity (Figure S4). Given that variants v4, v5, and v6 had appreciably shorter half-lives, we also performed these studies in the presence of MG132. Despite the presence of these isoforms, as determined by immunoblot analysis, we were unable to detect MTAP activity within the limits of our assay system (data not shown).
MTA Serum Levels Are Increased in DMS-MFH
MTA is not normally present in human serum. Cells lacking MTAP activity are unable to metabolize MTA,43 and functional inhibition44 or dysregulation of MTAP activity would therefore be expected to result in intracellular MTA accumulation and secretion. When tumor cells are MTAP deficient, excess MTA would be expected to be cleared by surrounding MTAP-normal stromal cells. If MTAP expression or activity is globally affected in all tissues, for example, in an individual with an inherited germline MTAP deficiency, then serum levels of MTA should be increased. To establish whether decreased expression of exon-9-containing isoforms affects MTAP activity, we measured MTA serum levels in DMS-MFH-affected family members and controls. Serum samples from two affected adults (F4 III-1 and F4 IV-1), one unaffected family member (F4 IV-3), and two unrelated controls were analyzed in a blinded fashion. All three serum samples from unaffected individuals had no detectable MTA levels. In marked contrast, both affected individuals had accumulations of MTA detectable in their serum (Table 1).
Table 1.
MTA Serum Levels
| Serum Donor | MTA Level (pmol/100 ul) |
|---|---|
| F4 III-1 | 11.5 |
| F4 IV-2 | 4.3 |
| F4 IV-3 | not detected |
| Control 1 | not detected |
| Control 2 | not detected |
Molecular Modeling of MTAP Isoforms
The high-resolution structure of human archetype MTAP has been previously determined by X-ray crystallography with several ligands. Most relevant is the trimeric enzyme complex including 5′-deoxy-5′-methylthioadenosine sulfate (PDB ID 1CG6) and the MTAP apoprotein (PDB ID 1CB0).36 These structures of human 5′-deoxy-5′-methylthioadenosine phosphorylase at 1.7 Å resolution provide insights into substrate binding (Figure 6A) and catalysis and serve as a template for modeling potential interactions among the subunits. On the basis of this model, the amino acid sequences providing the substrate binding site are encoded primarily by exons 6 and 7. Only the L279 residue is provided by exon 8 (Figure S5). The structure reveals that each of the three identical subunits is comprised of an alpha-beta domain containing an eight-stranded and a five-stranded mixed beta sheet with six dispersed alpha helices similar to the family of purine nucleoside phosphorylases (Figure 6B). On the basis of the results of our coimmunoprecipitation experiments, the MTAP trimer could exist as a heterologous assembly of different subunits comprised of archetype and splice variants. In the MTAP splice variants, the 75 amino acids of v1 are inserted after K271, and those of v4 are inserted after A230. In the MTAP monomer, v1–v3 and v4–v6 oppose each other, but most importantly, at the interfaces of different subunits of the trimer, all of the splice variants are relatively close to each other spatially, regardless of in which exon amino acids are inserted within the symmetrically equivalent subunits. As seen in Figure 6C, exon 6 is in juxtaposition with exon 7 of the adjacent subunit. The trimeric subunit interface of MTAP does appear to be affected by the alternate splicing events or, possibly, the MTA active site (Figure 6C). Secondary-structure and disulfide-bond prediction analyses predict the generation of a disulfide bond between cysteine residues in exons 9 and 10; these prediction analyses require future biochemical analysis for validation.
Figure 6.
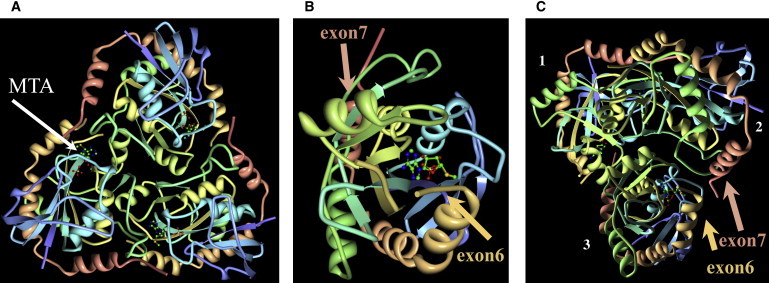
Molecular Modeling of MTAP
Modeling is based on the coordinates of the trimeric enzyme complex including 5′-deoxy-5′-methylthioadenosine sulfate (PDB ID 1CG6) and the MTAP apoprotein (PDB ID 1CB0).42 Rendering was done with the program O v.13 (courtesy of Dr. Alwyn Jones, University of Uppsala, Uppsala, Sweden), and visualization was done with the PyMOL molecular graphics system v.1.3 (Delano Scientific).
(A) Overview of the top of the MTAP trimer. The stick representation (arrow) indicates the MTA substrate.
(B) View of an MTAP monomer. The insertion positions of exon 7 (salmon) and exon 6 (tan) are at opposite ends.
(C) View of the bottom of the MTAP trimer. Subunits and the insertion positions of exon 7 (salmon) and exon 6 (tan; discontinuous electron density) are indicated. Displayed is the close proximity of one of these junctions: exon 7 in subunit 2 with exon 8 in subunit 3. Because of the three-fold symmetry, there is the possibility of three splice-variant insertion points in the trimer.
Ancestral Retroposition Events Gave Rise to Specific MTAP Exons in Humans and Possibly Other Anthropoid Primates
Sequence analysis of the three terminal exons revealed that exons 9 and 10 shared high homology with different primate-specific retroviral sequences, which are known to have integrated multiple times into different chromosomes throughout the genome. In general, human endogenous retrovirus (HERV) sequences are the result of ancestral infection events that can become incorporated into the genome and transmitted vertically through speciation events.45 As shown in Figure 7A, an analysis (with the use of programs RepeatMasker and Retrosearch) of exons 9 and 10 suggested that these exons arose from integrated retroviruses from distinct subfamilies. Exon 9 arose from part of a MER50I element, and exon 10 arose from part of a THE1A element, one of several families of primate-specific long terminal repeat (LTR) retrotransposons.46 Both the c.813-2A>G and c.885A>G mutations in exon 9 represent G>A nucleotide transitions from the consensus MER50I sequence.
Figure 7.
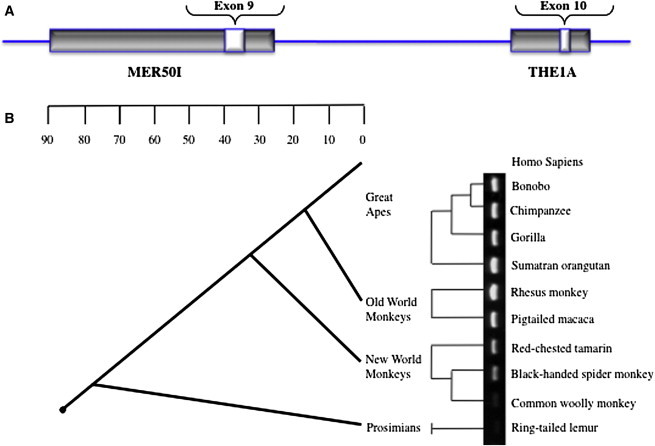
Evolution of MTAP Exon 9
(A) Schematic representation demonstrating that exons 9 and 10 arose from distinct families of retroviral integration events.
(B) The PCR results of exon 9 in a primate genomic DNA panel are superimposed adjacent to a phylogenetic tree. The results demonstrate that exon 9 was integrated into the primate genome at some point in evolution between the divergence of the ring-tailed lemur and the common woolly monkey approximately 40 million years ago.
To more precisely establish the evolutionary age of exon 9, which contains the DMS-MFH mutations, we amplified and sequenced this exon and its intron-exon boundaries from a panel of primate genomic DNA. PCR amplicons were obtained and sequenced for confirmation in great apes and Old and New World monkeys. No product was amplified from the ring-tailed lemur, a member of an ancestral extant primate lineage (Figure 7). From this analysis, we could determine that the MER50I remnant, which now encodes MTAP exon 9, was integrated over 40 million years ago into the lineage leading to anthropoid primates.
Discussion
Hereditary cancer syndromes represent a powerful and tractable biologic system for identifying cancer-causing mutations.47 Although the syndromes themselves are rare, their study can provide insight into the basis of the sporadic forms of the cancers. DMS-MFH represents the only known hereditary form of MFH, which has been thought to exist along a spectrum of bone sarcomas with osteosarcoma. Osteosarcoma itself has been linked to several hereditary disorders, including Li-Fraumeni syndrome (MIM 151623), Rothmund-Thomson syndrome (MIM 268400), Bloom syndrome (MIM 210900), Werner syndrome (MIM 277700), Paget disease (MIM 602080), and retinoblastoma, albeit the incidence of cancer development associated with these syndromes is lower when compared to the >30% associated with DMS-MFH. Recently, one of our affected individuals developed histologically proven osteosarcoma (Figure 3), thus further supporting a genetic link between these tumor types.
The 9p21 region containing the MTAP locus is one of the most frequently deleted and/or translocated chromosomal regions in human cancer.5 The facts that MTAP is more complex than previously recognized and that its terminal coding exon lies within 25 kb of the p15/p16 locus has immediate significance to LOH mapping and copy number variation (CNV) studies in human cancer. Deletions including the p15/p16 locus will more than likely also include the 3′ region of MTAP and therefore might affect MTAP biochemical activity. Thus, the interpretation of many of these studies with regard to the genes being affected should be reevaluated. Ultimately, the identification of the MTAP splice variants and the fact that their genetic loss results in DMS-MFH provide a possible in vivo demonstration that MTAP can act as a tumor suppressor. Given our findings that the DMS-MFH mutations also result in overexpression of two splice variants, MTAP_v3 and _v6, the possibility that at least two of the MTAP isoforms could represent oncogenic variants must also be considered at this time.
Of possible related significance, the 9p21 chromosomal region containing the full-length MTAP has been linked to coronary artery disease (CAD [MIM 611139]) and myocardial infarction in three independent genome-wide association studies (GWASs).48–50 Because of a paucity of known transcripts in the linkage-disequilibrium block, the region has been viewed as a “gene desert.” MTAP might represent an intriguing heart-disease candidate gene for several reasons. First, in DMS-MFH-affected family 1, two male family members died of heart disease in their early forties without other known risk factors. A third family member has been recently diagnosed with early CAD (J.A.M., unpublished data). As such, CAD might represent a previously unrecognized aspect of the disease phenotype in this syndrome. Second, two of our families have been diagnosed with myopathic disease and have features overlapping the symptoms of facioscapulohumeral muscular dystrophy and limb-girdle muscular dystrophy. Each of these chronic myopathic disorders is associated with an increased risk of heart disease.51,52 Third, defects in polyamine metabolism have been associated with defects in angiogenesis53 and altered myocyte function,54,55 whereas a nearly pathognomonic feature of DMS-MFH bone dysplasia is the presence of scattered infarctions throughout the medullary cavity.4 Finally, the CAD risk alleles identified in the original GWASs have been shown to localize to a STAT1-dependent enhancer element that can interact with MTAP and affect its transcription.56 Future studies will be required for exploring the possible role(s) of MTAP in CAD.
An intriguing evolutionary aspect of these studies is the origin of the three terminal MTAP exons. Mammalian chromosomes are interspersed with remnants of ancient retroviral integration events. It is estimated that 8% of the human genome consists of retroelements containing LTRs that flank regions corresponding to the gag, pol, and env genes.57 Although they usually infect somatic cells, retroviruses can also infect germ cells and establish permanent residence and future vertical transmission through a species in a Mendelian fashion.46 Indeed, the majority of HERVs are present in apes and Old World monkeys, suggesting that their original integrations took place more than 25 million years ago,58 and evidence now exists that some HERV proteins have been co-opted into functional roles.59 Two of the clearest examples are provided by the expression of two fusogenic proteins by trophoblasts. Syncytin 1 had been derived from a HERV-W envelope glycoprotein, is expressed in all trophoblastic cells, and mediates trophoblast cell fusion into the multinucleated syncytiotrophoblast layer. Syncytin 2, derived from a HERV-FRD envelope glycoprotein, is expressed in human placenta.60–62 Our studies reveal that two of the three terminal exons of MTAP are derived from independent retroviral integration events during primate evolution. Although the integration of exon-9-associated material most likely occurred after prosimian and New World monkey divergence more than 40 million years ago (Figure 7), the timing of its being functionally co-opted is presently unknown.
The retroviral origins of the terminal exons of MTAP suggest a number of interesting a priori conclusions. First, given the evolutionary species restriction of these HERVs, the existence of MTAP variants and possible biochemical regulation resulting from their expression must be unique to primates. Second, the exon 9 mutations that result in DMS-MFH highlight not only the existence of MTAP isoforms but also that their heterozygous loss (v1, v2, v4, and v5) and/or overexpression (v3 and v6) results in disease. This functionally demonstrates the importance of the acquired domains in MTAP function and their fixation into normal human physiology. This phenomenon, the recruitment and fixation over evolutionary time of a nucleic-acid sequence as a functional gene, or part thereof, is termed exaptation, and a number of examples are known.63–65 However, we are unaware of any other example wherein the loss of a co-opted gene and/or protein domain results in a disease phenotype.
Finally, our results continue to demonstrate the dynamic nature of the genome and the dual reality of disease associations and beneficial implications for both alternative splicing and retroelements. Future studies will now be required for defining the exact biochemical function(s) and regulation of these isoforms, their association with bone dysplasia, cancer initiation, and CAD, and their interaction with archetype MTAP.
Acknowledgments
The authors gratefully acknowledge the families that participated in this study. We also thank D. Springfield and J. Brosius for their thoughtful comments about the patients and manuscript, respectively, and X. Xu, J. Solis, J. Moorjani, C. Meret, S. Tam, and N. Kham for technical assistance.
Supplemental Data
Web Resources
The URLs for data presented herein are as follows:
ESEfinder Release 3.0, http://rulai.cshl.edu/cgi-bin/tools/ESE3/esefinder.cgi
Marshfield database, http://research.marshfieldclinic.org/genetics/GeneticResearch/compMaps.asp
NetGene2 Server, http://www.cbs.dtu.dk/services/NetGene2/
Online Mendelian Inheritance in Man (OMIM), http://www.omim.org
RepeatMasker, http://www.repeatmasker.org/
Retrosearch, http://www.daimi.au.dk/∼biopv/herv/
UCSC Genome Browser, March 2006 assembly, http://genome.ucsc.edu/
Accession Numbers
The EMBL-Bank accession numbers for the translated protein MTAP variants reported in this paper are: HE654772 (MTAP_v1), HE654773 (MTAP_v2), HE654774 (MTAP_v3), HE654775 (MTAP_v4), HE654776 (MTAP_v5), and HE654777 (MTAP_v6).
References
- 1.Arnold W.H. Hereditary bone dysplasia with sarcomatous degeneration. Study of a family. Ann. Intern. Med. 1973;78:902–906. doi: 10.7326/0003-4819-78-6-902. [DOI] [PubMed] [Google Scholar]
- 2.Hardcastle P., Nade S., Arnold W. Hereditary bone dysplasia with malignant change. Report of three families. J. Bone Joint Surg. Am. 1986;68:1079–1089. [PubMed] [Google Scholar]
- 3.Norton K.I., Wagreich J.M., Granowetter L., Martignetti J.A. Diaphyseal medullary stenosis (sclerosis) with bone malignancy (malignant fibrous histiocytoma): Hardcastle syndrome. Pediatr. Radiol. 1996;26:675–677. doi: 10.1007/BF01356833. [DOI] [PubMed] [Google Scholar]
- 4.Martignetti J.A., Desnick R.J., Aliprandis E., Norton K.I., Hardcastle P., Nade S., Gelb B.D. Diaphyseal medullary stenosis with malignant fibrous histiocytoma: A hereditary bone dysplasia/cancer syndrome maps to 9p21-22. Am. J. Hum. Genet. 1999;64:801–807. doi: 10.1086/302297. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Mitelman F. Wiley/Liss; New York: 1994. Catalog of Chromosome Aberrations in Cancer. [Google Scholar]
- 6.Miyakoshi J., Dobler K.D., Allalunis-Turner J., McKean J.D., Petruk K., Allen P.B., Aronyk K.N., Weir B., Huyser-Wierenga D., Fulton D. Absence of IFNA and IFNB genes from human malignant glioma cell lines and lack of correlation with cellular sensitivity to interferons. Cancer Res. 1990;50:278–283. [PubMed] [Google Scholar]
- 7.Olopade O.I., Jenkins R.B., Ransom D.T., Malik K., Pomykala H., Nobori T., Cowan J.M., Rowley J.D., Diaz M.O. Molecular analysis of deletions of the short arm of chromosome 9 in human gliomas. Cancer Res. 1992;52:2523–2529. [PubMed] [Google Scholar]
- 8.Fountain J.W., Karayiorgou M., Ernstoff M.S., Kirkwood J.M., Vlock D.R., Titus-Ernstoff L., Bouchard B., Vijayasaradhi S., Houghton A.N., Lahti J. Homozygous deletions within human chromosome band 9p21 in melanoma. Proc. Natl. Acad. Sci. USA. 1992;89:10557–10561. doi: 10.1073/pnas.89.21.10557. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Lukeis R., Irving L., Garson M., Hasthorpe S. Cytogenetics of non-small cell lung cancer: Analysis of consistent non-random abnormalities. Genes Chromosomes Cancer. 1990;2:116–124. doi: 10.1002/gcc.2870020207. [DOI] [PubMed] [Google Scholar]
- 10.Diaz M.O., Ziemin S., Le Beau M.M., Pitha P., Smith S.D., Chilcote R.R., Rowley J.D. Homozygous deletion of the alpha- and beta 1-interferon genes in human leukemia and derived cell lines. Proc. Natl. Acad. Sci. USA. 1988;85:5259–5263. doi: 10.1073/pnas.85.14.5259. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Diaz M.O., Rubin C.M., Harden A., Ziemin S., Larson R.A., Le Beau M.M., Rowley J.D. Deletions of interferon genes in acute lymphoblastic leukemia. N. Engl. J. Med. 1990;322:77–82. doi: 10.1056/NEJM199001113220202. [DOI] [PubMed] [Google Scholar]
- 12.García-Castellano J.M., Villanueva A., Healey J.H., Sowers R., Cordon-Cardo C., Huvos A., Bertino J.R., Meyers P., Gorlick R. Methylthioadenosine phosphorylase gene deletions are common in osteosarcoma. Clin. Cancer Res. 2002;8:782–787. [PubMed] [Google Scholar]
- 13.Miyazaki S., Nishioka J., Shiraishi T., Matsumine A., Uchida A., Nobori T. Methylthioadenosine phosphorylase deficiency in Japanese osteosarcoma patients. Int. J. Oncol. 2007;31:1069–1076. [PubMed] [Google Scholar]
- 14.Martignetti J.A., Gelb B.D., Pierce H., Picci P., Desnick R.J. Malignant fibrous histiocytoma: Inherited and sporadic forms have loss of heterozygosity at chromosome bands 9p21-22-evidence for a common genetic defect. Genes Chromosomes Cancer. 2000;27:191–195. [PubMed] [Google Scholar]
- 15.Nobori T., Takabayashi K., Tran P., Orvis L., Batova A., Yu A.L., Carson D.A. Genomic cloning of methylthioadenosine phosphorylase: A purine metabolic enzyme deficient in multiple different cancers. Proc. Natl. Acad. Sci. USA. 1996;93:6203–6208. doi: 10.1073/pnas.93.12.6203. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Kamatani N., Nelson-Rees W.A., Carson D.A. Selective killing of human malignant cell lines deficient in methylthioadenosine phosphorylase, a purine metabolic enzyme. Proc. Natl. Acad. Sci. USA. 1981;78:1219–1223. doi: 10.1073/pnas.78.2.1219. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Trackman P.C., Abeles R.H. The metabolism of 1-phospho-5-methylthioribose. Biochem. Biophys. Res. Commun. 1981;103:1238–1244. doi: 10.1016/0006-291x(81)90255-2. [DOI] [PubMed] [Google Scholar]
- 18.Trackman P.C., Abeles R.H. Methionine synthesis from 5′-S-Methylthioadenosine. Resolution of enzyme activities and identification of 1-phospho-5-S methylthioribulose. J. Biol. Chem. 1983;258:6717–6720. [PubMed] [Google Scholar]
- 19.Kamatani N., Yu A.L., Carson D.A. Deficiency of methylthioadenosine phosphorylase in human leukemic cells in vivo. Blood. 1982;60:1387–1391. [PubMed] [Google Scholar]
- 20.Schmid M., Malicki D., Nobori T., Rosenbach M.D., Campbell K., Carson D.A., Carrera C.J. Homozygous deletions of methylthioadenosine phosphorylase (MTAP) are more frequent than p16INK4A (CDKN2) homozygous deletions in primary non-small cell lung cancers (NSCLC) Oncogene. 1998;17:2669–2675. doi: 10.1038/sj.onc.1202205. [DOI] [PubMed] [Google Scholar]
- 21.Stevens A.P., Spangler B., Wallner S., Kreutz M., Dettmer K., Oefner P.J., Bosserhoff A.K. Direct and tumor microenvironment mediated influences of 5′-deoxy-5′-(methylthio)adenosine on tumor progression of malignant melanoma. J. Cell. Biochem. 2009;106:210–219. doi: 10.1002/jcb.21984. [DOI] [PubMed] [Google Scholar]
- 22.Karikari C.A., Mullendore M., Eshleman J.R., Argani P., Leoni L.M., Chattopadhyay S., Hidalgo M., Maitra A. Homozygous deletions of methylthioadenosine phosphorylase in human biliary tract cancers. Mol. Cancer Ther. 2005;4:1860–1866. doi: 10.1158/1535-7163.MCT-05-0103. [DOI] [PubMed] [Google Scholar]
- 23.Christopher S.A., Diegelman P., Porter C.W., Kruger W.D. Methylthioadenosine phosphorylase, a gene frequently codeleted with p16(cdkN2a/ARF), acts as a tumor suppressor in a breast cancer cell line. Cancer Res. 2002;62:6639–6644. [PubMed] [Google Scholar]
- 24.Subhi A.L., Tang B., Balsara B.R., Altomare D.A., Testa J.R., Cooper H.S., Hoffman J.P., Meropol N.J., Kruger W.D. Loss of methylthioadenosine phosphorylase and elevated ornithine decarboxylase is common in pancreatic cancer. Clin. Cancer Res. 2004;10:7290–7296. doi: 10.1158/1078-0432.CCR-04-0972. [DOI] [PubMed] [Google Scholar]
- 25.Huang H.Y., Li S.H., Yu S.C., Chou F.F., Tzeng C.C., Hu T.H., Uen Y.H., Tian Y.F., Wang Y.H., Fang F.M. Homozygous deletion of MTAP gene as a poor prognosticator in gastrointestinal stromal tumors. Clin. Cancer Res. 2009;15:6963–6972. doi: 10.1158/1078-0432.CCR-09-1511. [DOI] [PubMed] [Google Scholar]
- 26.Mukhopadhyay N., Almasy L., Schroeder M., Mulvihill W.P., Weeks D.E. Mega2: Data-handling for facilitating genetic linkage and association analyses. Bioinformatics. 2005;21:2556–2557. doi: 10.1093/bioinformatics/bti364. [DOI] [PubMed] [Google Scholar]
- 27.Sobel E., Lange K. Descent graphs in pedigree analysis: Applications to haplotyping, location scores, and marker-sharing statistics. Am. J. Hum. Genet. 1996;58:1323–1337. [PMC free article] [PubMed] [Google Scholar]
- 28.Benson G. Tandem repeats finder: A program to analyze DNA sequences. Nucleic Acids Res. 1999;27:573–580. doi: 10.1093/nar/27.2.573. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Dowling O., Difeo A., Ramirez M.C., Tukel T., Narla G., Bonafe L., Kayserili H., Yuksel-Apak M., Paller A.S., Norton K. Mutations in capillary morphogenesis gene-2 result in the allelic disorders juvenile hyaline fibromatosis and infantile systemic hyalinosis. Am. J. Hum. Genet. 2003;73:957–966. doi: 10.1086/378781. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Hebsgaard S.M., Korning P.G., Tolstrup N., Engelbrecht J., Rouzé P., Brunak S. Splice site prediction in Arabidopsis thaliana pre-mRNA by combining local and global sequence information. Nucleic Acids Res. 1996;24:3439–3452. doi: 10.1093/nar/24.17.3439. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Brunak S., Engelbrecht J., Knudsen S. Prediction of human mRNA donor and acceptor sites from the DNA sequence. J. Mol. Biol. 1991;220:49–65. doi: 10.1016/0022-2836(91)90380-o. [DOI] [PubMed] [Google Scholar]
- 32.Cartegni L., Wang J., Zhu Z., Zhang M.Q., Krainer A.R. ESEfinder: A web resource to identify exonic splicing enhancers. Nucleic Acids Res. 2003;31:3568–3571. doi: 10.1093/nar/gkg616. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Savarese T.M., Crabtree G.W., Parks R.E., Jr. 5′-Methylthioadenosine phosphorylase-L. Substrate activity of 5′-deoxyadenosine with the enzyme from Sarcoma 180 cells. Biochem. Pharmacol. 1981;30:189–199. doi: 10.1016/0006-2952(81)90077-0. [DOI] [PubMed] [Google Scholar]
- 34.Watts G.D., Mehta S.G., Zhao C., Ramdeen S., Hamilton S.J., Novack D.V., Mumm S., Whyte M.P., Mc Gillivray B., Kimonis V.E. Mapping autosomal dominant progressive limb-girdle myopathy with bone fragility to chromosome 9p21-p22: A novel locus for a musculoskeletal syndrome. Hum. Genet. 2005;118:508–514. doi: 10.1007/s00439-005-0075-z. [DOI] [PubMed] [Google Scholar]
- 35.Henry E.W., Auckland N.L., McINTOSH H.W., Starr D.E. Abnormality of the long bones and progressive muscular dystrophy in a family. Can. Med. Assoc. J. 1958;78:331–336. [PMC free article] [PubMed] [Google Scholar]
- 36.Picci P., Bacci G., Ferrari S., Mercuri M. Neoadjuvant chemotherapy in malignant fibrous histiocytoma of bone and in osteosarcoma located in the extremities: Analogies and differences between the two tumors. Ann. Oncol. 1997;8:1107–1115. doi: 10.1023/a:1008283516969. [DOI] [PubMed] [Google Scholar]
- 37.Jeon D.G., Song W.S., Kong C.B., Kim J.R., Lee S.Y. MFH of Bone and Osteosarcoma Show Similar Survival and Chemosensitivity. Clin. Orthop. Relat. Res. 2010;469:584–590. doi: 10.1007/s11999-010-1428-z. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Ghandur-Mnaymneh L., Zych G., Mnaymneh W. Primary malignant fibrous histiocytoma of bone: Report of six cases with ultrastructural study and analysis of the literature. Cancer. 1982;49:698–707. doi: 10.1002/1097-0142(19820215)49:4<698::aid-cncr2820490418>3.0.co;2-7. [DOI] [PubMed] [Google Scholar]
- 39.Knudson A.G. Two genetic hits (more or less) to cancer. Nat. Rev. Cancer. 2001;1:157–162. doi: 10.1038/35101031. [DOI] [PubMed] [Google Scholar]
- 40.Della Ragione F., Cartenì-Farina M., Gragnaniello V., Schettino M.I., Zappia V. Purification and characterization of 5′-deoxy-5′-methylthioadenosine phosphorylase from human placenta. J. Biol. Chem. 1986;261:12324–12329. [PubMed] [Google Scholar]
- 41.Della Ragione F., Oliva A., Gragnaniello V., Russo G.L., Palumbo R., Zappia V. Physicochemical and immunological studies on mammalian 5′-deoxy-5′-methylthioadenosine phosphorylase. J. Biol. Chem. 1990;265:6241–6246. [PubMed] [Google Scholar]
- 42.Appleby T.C., Erion M.D., Ealick S.E. The structure of human 5′-deoxy-5′-methylthioadenosine phosphorylase at 1.7 A resolution provides insights into substrate binding and catalysis. Structure. 1999;7:629–641. doi: 10.1016/s0969-2126(99)80084-7. [DOI] [PubMed] [Google Scholar]
- 43.Williams-Ashman H.G., Seidenfeld J., Galletti P. Trends in the biochemical pharmacology of 5′-deoxy-5′-methylthioadenosine. Biochem. Pharmacol. 1982;31:277–288. doi: 10.1016/0006-2952(82)90171-x. [DOI] [PubMed] [Google Scholar]
- 44.Schramm V.L. Enzymatic transition state theory and transition state analogue design. J. Biol. Chem. 2007;282:28297–28300. doi: 10.1074/jbc.R700018200. [DOI] [PubMed] [Google Scholar]
- 45.Bannert N., Kurth R. The evolutionary dynamics of human endogenous retroviral families. Annu. Rev. Genomics Hum. Genet. 2006;7:149–173. doi: 10.1146/annurev.genom.7.080505.115700. [DOI] [PubMed] [Google Scholar]
- 46.Smit A.F. Identification of a new, abundant superfamily of mammalian LTR-transposons. Nucleic Acids Res. 1993;21:1863–1872. doi: 10.1093/nar/21.8.1863. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Fearon E.R. Human cancer syndromes: Clues to the origin and nature of cancer. Science. 1997;278:1043–1050. doi: 10.1126/science.278.5340.1043. [DOI] [PubMed] [Google Scholar]
- 48.McPherson R., Pertsemlidis A., Kavaslar N., Stewart A., Roberts R., Cox D.R., Hinds D.A., Pennacchio L.A., Tybjaerg-Hansen A., Folsom A.R. A common allele on chromosome 9 associated with coronary heart disease. Science. 2007;316:1488–1491. doi: 10.1126/science.1142447. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Helgadottir A., Thorleifsson G., Manolescu A., Gretarsdottir S., Blondal T., Jonasdottir A., Jonasdottir A., Sigurdsson A., Baker A., Palsson A. A common variant on chromosome 9p21 affects the risk of myocardial infarction. Science. 2007;316:1491–1493. doi: 10.1126/science.1142842. [DOI] [PubMed] [Google Scholar]
- 50.Willer C.J., Sanna S., Jackson A.U., Scuteri A., Bonnycastle L.L., Clarke R., Heath S.C., Timpson N.J., Najjar S.S., Stringham H.M. Newly identified loci that influence lipid concentrations and risk of coronary artery disease. Nat. Genet. 2008;40:161–169. doi: 10.1038/ng.76. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.van der Kooi A.J., Ledderhof T.M., de Voogt W.G., Res C.J., Bouwsma G., Troost D., Busch H.F., Becker A.E., de Visser M. A newly recognized autosomal dominant limb girdle muscular dystrophy with cardiac involvement. Ann. Neurol. 1996;39:636–642. doi: 10.1002/ana.410390513. [DOI] [PubMed] [Google Scholar]
- 52.Kimonis V.E., Kovach M.J., Waggoner B., Leal S., Salam A., Rimer L., Davis K., Khardori R., Gelber D. Clinical and molecular studies in a unique family with autosomal dominant limb-girdle muscular dystrophy and Paget disease of bone. Genet. Med. 2000;2:232–241. doi: 10.1097/00125817-200007000-00006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Takigawa M., Nishida Y., Suzuki F., Kishi J., Yamashita K., Hayakawa T. Induction of angiogenesis in chick yolk-sac membrane by polyamines and its inhibition by tissue inhibitors of metalloproteinases (TIMP and TIMP-2) Biochem. Biophys. Res. Commun. 1990;171:1264–1271. doi: 10.1016/0006-291x(90)90822-5. [DOI] [PubMed] [Google Scholar]
- 54.Harris S.P., Patel J.R., Marton L.J., Moss R.L. Polyamines decrease Ca(2+) sensitivity of tension and increase rates of activation in skinned cardiac myocytes. Am. J. Physiol. Heart Circ. Physiol. 2000;279:H1383–H1391. doi: 10.1152/ajpheart.2000.279.3.H1383. [DOI] [PubMed] [Google Scholar]
- 55.Tantini B., Fiumana E., Cetrullo S., Pignatti C., Bonavita F., Shantz L.M., Giordano E., Muscari C., Flamigni F., Guarnieri C. Involvement of polyamines in apoptosis of cardiac myoblasts in a model of simulated ischemia. J. Mol. Cell. Cardiol. 2006;40:775–782. doi: 10.1016/j.yjmcc.2006.03.002. [DOI] [PubMed] [Google Scholar]
- 56.Harismendy O., Notani D., Song X., Rahim N.G., Tanasa B., Heintzman N., Ren B., Fu X.D., Topol E.J., Rosenfeld M.G., Frazer K.A. 9p21 DNA variants associated with coronary artery disease impair interferon-γ signalling response. Nature. 2011;470:264–268. doi: 10.1038/nature09753. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57.Lander E.S., Linton L.M., Birren B., Nusbaum C., Zody M.C., Baldwin J., Devon K., Dewar K., Doyle M., FitzHugh W., International Human Genome Sequencing Consortium Initial sequencing and analysis of the human genome. Nature. 2001;409:860–921. doi: 10.1038/35057062. [DOI] [PubMed] [Google Scholar]
- 58.Mayer J., Meese E. Human endogenous retroviruses in the primate lineage and their influence on host genomes. Cytogenet. Genome Res. 2005;110:448–456. doi: 10.1159/000084977. [DOI] [PubMed] [Google Scholar]
- 59.Volff J.N., Brosius J. Modern genomes with retro-look: Retrotransposed elements, retroposition and the origin of new genes. In: Volff J.-N., editor. Gene and Protein Evolution. Genome Dynamics. Karger; Basel: 2007. pp. 175–190. [DOI] [PubMed] [Google Scholar]
- 60.Blaise S., de Parseval N., Bénit L., Heidmann T. Genomewide screening for fusogenic human endogenous retrovirus envelopes identifies syncytin 2, a gene conserved on primate evolution. Proc. Natl. Acad. Sci. USA. 2003;100:13013–13018. doi: 10.1073/pnas.2132646100. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 61.Blond J.L., Lavillette D., Cheynet V., Bouton O., Oriol G., Chapel-Fernandes S., Mandrand B., Mallet F., Cosset F.L. An envelope glycoprotein of the human endogenous retrovirus HERV-W is expressed in the human placenta and fuses cells expressing the type D mammalian retrovirus receptor. J. Virol. 2000;74:3321–3329. doi: 10.1128/jvi.74.7.3321-3329.2000. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 62.Mi S., Lee X., Li X., Veldman G.M., Finnerty H., Racie L., LaVallie E., Tang X.Y., Edouard P., Howes S. Syncytin is a captive retroviral envelope protein involved in human placental morphogenesis. Nature. 2000;403:785–789. doi: 10.1038/35001608. [DOI] [PubMed] [Google Scholar]
- 63.Brosius J., Gould S.J. On “genomenclature”: A comprehensive (and respectful) taxonomy for pseudogenes and other “junk DNA”. Proc. Natl. Acad. Sci. USA. 1992;89:10706–10710. doi: 10.1073/pnas.89.22.10706. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 64.Krull M., Brosius J., Schmitz J. Alu-SINE exonization: En route to protein-coding function. Mol. Biol. Evol. 2005;22:1702–1711. doi: 10.1093/molbev/msi164. [DOI] [PubMed] [Google Scholar]
- 65.Baertsch R., Diekhans M., Kent W.J., Haussler D., Brosius J. Retrocopy contributions to the evolution of the human genome. BMC Genomics. 2008;9:466. doi: 10.1186/1471-2164-9-466. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.


