Abstract
Membrane proteins are of particular biological and pharmaceutical importance, and computational modeling and structure prediction approaches play an important role in studies of membrane proteins. Developing an accurate model quality assessment program is of significance to the structure prediction of membrane proteins. Few such programs are proposed that can be applied to a broad range of membrane protein classes and perform with high accuracy. We developed a new model scoring function IQ, based on the analysis of four types of inter-residue interactions within the transmembrane domains of helical membrane proteins. This function was tested using three high-quality model sets: all 206 models of GPCR Dock 2008, all 284 models of GPCR Dock 2010, and all 92 helical membrane protein models of the HOMEP set. For all three sets, the scoring function can select the native structures among all of the models with the success rates of 93%, 85%, and 100% respectively. For comparison, these three model sets were also adopted for a recently published model assessment program for membrane protein structures, ProQM, which gave the success rates of 85%, 79%, and 92% separately. These results suggested that IQ outperforms ProQM when only the transmembrane regions of the models are considered. This scoring function should be useful for the computational modeling of membrane proteins.
Keywords: membrane proteins, structure quality, inter-residue interactions, frequency score, average number of interactions
Introduction
Computational modeling and prediction of 3D structures of membrane proteins plays a valuable role in the biological and pharmacological studies of this important class of proteins (1, 2, 3, 4). Membrane proteins represent ~30% of the human genome and perform diverse cellular functions such as cross-cell signal transduction, membrane transport, and photosynthesis (5, 6, 7). Membrane proteins are also the largest class of drug targets with the G-protein coupled receptor (GPCR) superfamily alone accounting for approximately 27% of drugs currently available on the market (8). Despite the significant progress in experimental techniques (9), structure determination of membrane proteins significantly lags behind that of soluble proteins (10), representing less than 1% of the structures in the Protein Data Bank (PDB). It is thus very desirable to develop computational tools for the structure prediction of membrane proteins.
A prime challenge of membrane protein structure prediction is to develop accurate scoring functions to assess the quality of a set of model structures, the so-called MQAP (model quality assessment program) (11). Because of the paucity of available membrane protein structures, scoring functions developed from small-molecule data or soluble protein structures are traditionally adopted for this purpose (12). Given the clear differences between membrane and soluble proteins, in terms of amino acid propensity, packing density, and side-chain rotamer frequencies (13, 14, 15, 16), it is unsurprising that the application of these scoring functions in quality assessment of membrane proteins is far from perfect (12).
The steady accumulation of X-ray structures of membrane proteins in recent years facilitates the effort of developing more accurate MQAPs by enabling the direct analysis of experimental structures of membrane proteins (11, 17, 18, 19). Early attempts by Fleishman and Ben-Tal and by Park et al. were limited by the number of experimental structures available. As a result, the application of such scoring functions is generally limited to proteins with the relatively small numbers of TM helices (17, 18). In the works from our group, a scoring function derived from the overall analysis of the total number of specific inter-residue interactions of 19 high-resolution, non-redundant TM protein structures was reported (19). Most recently, Ray et al. developed a learning-based MQAP called ProQM, which takes both general and membrane protein specific features into consideration (11). The MQAPs based on both the total number of inter-residue interactions (19, 20, 21) and the machine learning approach (11) are applicable to helical membrane proteins of various class and are thus of promise as general MQAPs for helical membrane proteins.
Direct studies of inter-residue interactions in helical membrane proteins have revealed interesting findings in the packing of helical membrane proteins (22). Here, we seek to uncover insight into the pattern of specific inter-residue interactions in helical membrane proteins and to explore its application as a high quality scoring function. To achieve our goals, we first compiled a high-resolution, non-redundant X-ray structure dataset of 18 helical membrane proteins. Four types of specific inter-residue interactions in the transmembrane (TM) domain of these structures were identified and classified. The histogram distribution of these interactions is proposed as an additional component to the original scoring function, which was based on the average number of interactions per residue (19). The application of the resulting scoring function, called IQ (Interaction-based Quality assessment), is validated using three test model sets: the GPCR Dock 2008 (23), GPCR Dock 2010 (24), and the HOMEP benchmark (25) datasets, and the results were compared with those of the learning-based program ProQM.
Materials and Methods
High-resolution structure dataset of helical membrane proteins
A total of 18 high-resolution, non-redundant TM domains was compiled for this study (Table I). This dataset was built from the previously compiled set of 19 high-resolution TM domains (19). Newly reported X-ray structures of helical membrane proteins were identified from an online membrane protein resource (http://blanco.biomol.uci.edu/mpstruc/listAll/list, April 11th, 2011), and those with the resolution of 2.5Å or better were included in the dataset. Next, sequence alignment was carried out for each non-identical chain in the dataset using ClustalW2 (http://www.ebi.ac.uk/Tools/msa/clustalw2/) (26). If any chain has > 35% sequence identity with another, only the one with the better resolution was retained. The remaining members in the dataset were then searched against the CATH database to ensure that each member in this dataset represented a unique membrane protein superfamily based on its classification in the CATH database (27). If two members belong to the same CATH superfamily, only the one with the better resolution was retained. This TM protein dataset was further winnowed using the following criteria: (i) The TM domain contains at least four TM helices, with each helix having at least 18 amino acid residues; and (ii) The domain has more than approximately 50% of its surface in the TM region being in contact with the lipid. One protein (PDB ID: 2UUH) was removed from the dataset based on these criteria. In addition, for the protein (PDB ID: 2WJN), the individual helices in either of its two TM chains arrange in a crescent form when looking along the axis that is perpendicular to the membrane. There are no interactions formed between any two helices that are not neighbors in the crescent form. Thus, this protein was also removed from the dataset.
Table I.
List of PDB entries for membrane proteins included in the high-resolution structure dataset.
| No. | Protein Name | PDB ID |
CATH Domain |
Resolution (Å) |
CATH Code | Number of TM Helices |
|---|---|---|---|---|---|---|
| 1 | Formate dehydrogenase-N: E. coli | 1KQF | C | 1.6 | 1.20.950.20.1 | 4 |
| 2 | K+ channel | 1ORS | C | 1.9 | 1.20.120.350.1 | 4 |
| 3 | Cytochrome C oxidase: R. sphaeroides | 1M56 | C01/ 02 |
2.3 | 1.10.287.70.8/ 1.20.120.80.1 |
5 |
| 4 | Quinol-fumarate reductase: W. succinogenes |
2BS2 | C | 2.2 | 1.20.950.10.1 | 5 |
| 5 | Aqy1 yeast aquaporin: P. pastoris | 2W2E | A | 1.15 | 1.20.1080.10.1 | 6 |
| 6 | GlpG rhomboid protease: E. coli | 2XOV | A | 1.65 | Not Available | 6 |
| 7 | Bacteriorhodopsin | 1C3W | A | 1.55 | 1.20.1070.10.1 | 7 |
| 8 | Polysulfide Reductase: T. thermophilus | 2VPZ | C | 2.4 | 1.20.1630.10.1 | 8 |
| 9 | Cytochrome b-cl: S. cerevisiae | 3CX5 | C | 1.9 | 1.20.810.10.1 | 8 |
| 10 | C1C Cl channel: E. coli | 1OTS | A | 2.51 | 1.10.3080.10.1 | 10 |
| 11 | Ca2+ ATPase | 1WPG | A02 | 2.3 | 1.20.1110.10.1 | 10 |
| 12 | Putative metal-chelate-type ABC Transporter: H. influenzae |
2NQ2 | A | 2.4 | 1.10.3470.10.1 | 10 |
| 13 | SLAC1 TehA homolog: H. influenzae | 3M71 | A | 1.2 | Not Available | 10 |
| 14 | Rh transport protein: N. europaea | 3B9W | A | 1.3 | 1.10.3430.10.1 | 11 |
| 15 | Cytochrome C oxidase: R. sphaeroides | 1M56 | A | 2.3 | 1.20.210.10.1 | 12 |
| 16 | LeuT Leucine transporter: A. aeolicus | 2QEI | A | 1.85 | Not Available | 12 |
| 17 | Carnitine transporter: P. mirabilis | 2WSW | A | 2.3 | Not Available | 12 |
| 18 | ApcT Transporter: M. jannaschii | 3GIA | A | 2.35 | Not Available | 12 |
For each protein chain retained in this dataset, its TM helical boundaries were identified based on the PDBTM database (http://pdbtm.enzim.hu/) (28). The TM helix assignments in the PDBTM database are determined using only structural information. All loops in the soluble regions, cofactors, ligands, and H2O molecules were manually removed to keep only the alpha helices in the TM regions, since they are the primary focus of this study.
Testing datasets
In order to test the usefulness of the proposed scoring function, it was applied to three sets of models of helical membrane proteins: the GPCR Dock 2008 (23), GPCR Dock 2010 (24), and HOMEP (25) datasets. The two GPCR model sets represent submission to the first two rounds of the community-wide assessment of GPCR structure modeling and ligand docking. Participating in these blind competitions, independent labs and groups from around the world had produced GPCR models before the experimental structures of the model targets were released. GPCR Dock 2008 contains 206 models of the human adenosine A2A receptor (PDB ID: 3EML), and GPCR Dock 2010 contains 284 models of the human CXCR4 chemokine receptor (PDB ID: 3ODU, 3OE0, 3OE6, 3OE8, and 3OE9) and the human D3 dopamine receptor (PDB ID: 3PBL).
The HOMEP benchmark dataset is a carefully compiled set of homologous models of 94 membrane protein query-template pairs of known structures and covers a wide range of sequence identities from <10% to 80% (25). Among the 188 models obtained, 92 are helical membrane proteins, representing 46 query-template pairs. For each pair, a homology model based on their sequence-to-sequence alignment and a model based on their structural alignment were constructed for the query protein using Modeller 6v2 (29). These 92 models of helical membrane proteins were studied here. For comparison, their corresponding X-ray structures were downloaded from the PDB (30) and analyzed in parallel.
For each model from the three test sets, its TM helical boundaries were identified based on the definition for its corresponding X-ray structure in the PDBTM database (28). The loops of the soluble regions and ligands were again manually removed to keep only alpha helices that lie within the TM regions.
Development of scoring function IQ
The scoring function IQ was derived based on the analysis of four specific types of inter-residue interactions in the high-resolution structure dataset. These include hydrophobic interactions, hydrogen bonds, ionic bonds, and disulfide bonds. The definition of hydrogen bonds used here was given as described by Stickle et al. (31). It includes both distance and geometry criteria. For the distance criterion, the hydrogen bond radii for electronegative heavy atoms including N, O, and S atoms were defined first. If the distance between a potential donor-acceptor pair in the structure was less than or equal to the sum of the hydrogen bond radii of its respective atoms, this pair was subjected to the further geometric evaluation. For the geometric criterion, two angle criteria were applied depending on the hybridization state of the heavy atoms (31). For sp2 atoms, there are two additional planarity tests on top of the angle criterion; hydrophobic interactions and ionic bonds were based only on proximity, and the default cutoff was 4.5 Å; and disulfide bonds were defined to exist between explicitly bonded sulfur pairs or non-bonded sulfur pairs within the distance cutoff of 2.5 Å.
For a pair of amino acid residues, their interactions could satisfy the criteria of both hydrogen bond and ionic bond interactions. In that case, both types of interactions were counted. These four interactions were determined using the Protein Contacts function in MOE (Molecular Computing Group Inc., version 2010.10) using the default parameters as was previously shown to be effective (32). Employing the default setting, histidine residues are automatically protonated, and interactions between residues that are less than four positions along the sequence were excluded in the calculation.
Two types of analysis were subsequently performed from the resulting inter-residue interaction lists. The first is to calculate the average number of interactions within each structure. A linear correlation between the average number of inter-residue interactions and the quality of helical membrane protein model/structures has been reported previously (20). To apply this analysis to the models in the test datasets, the best model should be the one that has the highest value of the average number of inter-residue interactions. The calculation is shown below:
| (1) |
where c̅ defines the average number of inter-residue interactions, Ninteractions is the total number of interactions in the model, and Nresidues is the total number of residues in the model.
For the second type of analysis, the distributions of the four types of interactions were studied. First, interactions within each type were further classified based on the grouping of the two residues involved in each interaction (33). The 20 typical amino acids can be classified by their physico-chemical properties into six groups: 1) Arginine and Lysine; 2) Aspartic Acid and Glutamic Acid; 3) Histidine, Phenylalanine, Tryptophan, and Tyrosine; 4) Asparagine, Glutamine, Serine, and Threonine; 5) Alanine, Isoleucine, Leucine, Methionine, Valine, and Cysteine; and 6) Glycine and Proline (33). With such groups defined, the classifications of each type of inter-residue interactions were further sub-divided into 21 possible categories, determined by the two groups to which the two interacting residues belong. In total, there should be 84 categories of possible inter-residue interactions. The number of interactions in each group was normalized for each structure, and the frequency distribution was generated. This frequency distribution was averaged for all structures in the high-resolution structure dataset and regarded as the standard.
To apply this type of the analysis to the models in the test datasets, the best model should have the least difference in frequency distribution from the average distribution derived from the high-resolution structure dataset. The calculation for this analysis is shown below:
| (2) |
where, fs defines the frequency score, fm,i is the frequency of a specific interaction type i in the model, fc,i is the average frequency of a specific interaction type i in the high-resolution set, and N is the number of interaction types. fs ranges from 0 for identical distributions to 2 for mutually exclusive distributions so a lower score is a better score.
Results from these two types of analyses were then combined to generate a single score IQ. In the IQ score, the c̅ score was transformed using a log-normal probability density so that it falls in the range of 0–0.5. The fs score is already in the range of 0–2. So it was merely divided by four. The ultimate IQ score varies from zero to one with decreasing model quality. The calculation for the IQ score is shown below:
| (3) |
where d is set to the high-resolution structure dataset’s maximum c̅ value and s is set to the range of the c̅ value derived from the same dataset.
Comparison with scoring function ProQM
To compare the performance of IQ with other MQAPs specifically developed for membrane protein structures, the recently published learning-based model assessment program, ProQM, was chosen because of its high-quality performance (11). Tailored models from all three testing datasets described above and their corresponding X-ray structures were submitted to the ProQM server (http://wallner.theophys.kth.se/index.php). The Global Quality Score for each model derived from the server was compared with that of the corresponding X-ray structure. A model with a worse score was considered having worse quality than the X-ray structure.
Results
The work presented here is to gain novel insight into inter-residue interactions in membrane protein structures and to develop an accurate scoring function for membrane protein structure assessment.
Structures in the high-resolution protein dataset display a similar pattern of inter-residue interactions
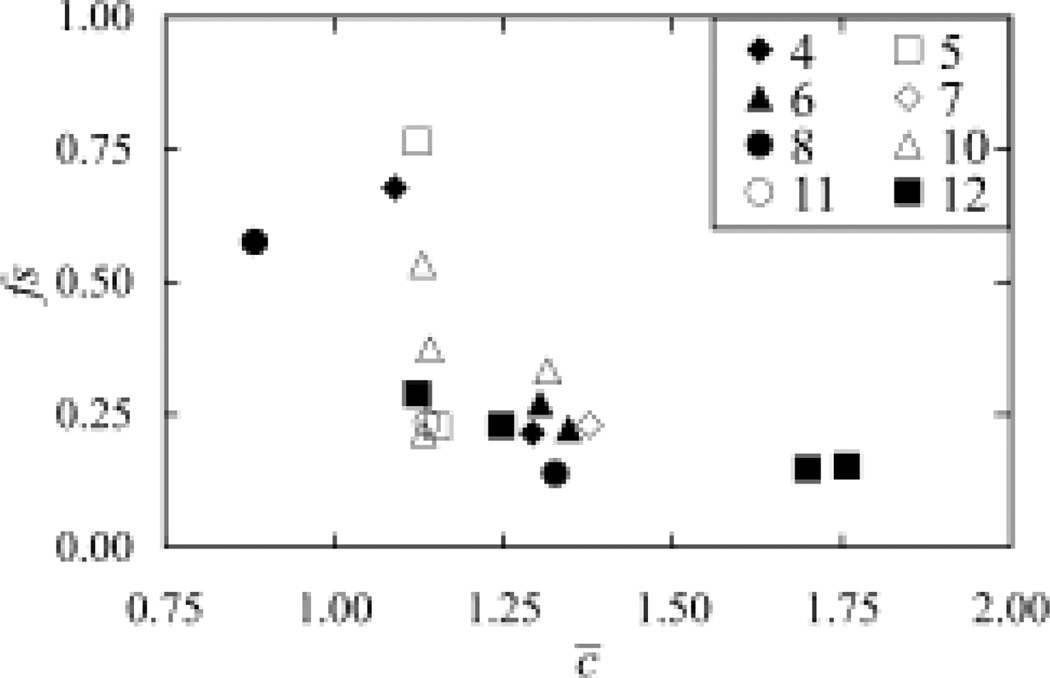
The high-resolution structure dataset of membrane proteins includes a total of 18 high-resolution, non-redundant TM domains. Each structure domain within the dataset belongs to a unique superfamily classified in the CATH database (27). The number of TM helices contained in this dataset varies quite significantly, ranging from four to twelve. In contrast, the average number of inter-residue interactions and the distribution of different types of inter-residue interactions among these structures are quite similar (Figure 1). The mean value of the average number of inter-residue interactions, c̅, across the current set is 1.29 with a standard deviation of 0.24. The mean frequency score, fs, across the dataset is 0.28 with a standard deviation of 0.16.
Fig. 1.
Analysis of the inter-residue interactions in the high-resolution structure dataset, showing the distribution of the frequency scores vs. distribution of the average number of interactions. The PDB IDs for structures 1–18 are listed in Table I. Structures with different numbers of TM helices were represented differently.
There are only few outliers with the Polysulfide Reductase (PDB ID: 2VPZ) having the lowest average number of interactions of 0.88, and the ApcT Transporter (PDB ID: 3GIA) and the LeuT Leucine transporter (PDB ID: 2QEI) having the highest average number of interactions of 1.70 and 1.76 respectively. Careful inspection of their X-ray structures suggests these abnormal values were resulted mainly from the packing of their individual TM helices. For the Polysulfide Reductase, its eight TM helices formed two packing groups instead of one, with four helices in each group. These two groups were arranged in parallel with few interactions forming between them. While for the two transporters, several kinks were observed inside their TM helices. These kinks broke the whole helix and resulted in the re-orientations of partial helices, thus allowing the formation of more interactions with other helices.
In addition, proteins having the different numbers of TM helices can have similar c̅ and fs values. Overall, these results demonstrate that the distribution of various types of favorable interactions across different helical membrane proteins is relatively homogeneous.
GPCR Dock 2008 test set
In order to test the ability of the new scoring function IQ to discriminate models, it was applied to data sets of models of known structures. The first set comprises the 206 models of the A2A adenosine receptor submitted to the Critical Assessment of GPCR Structure Modeling and Docking 2008 (23). These models were generated by different research groups using state-of-art modeling techniques with extensive refinement. As a result, a handful of models have >70% residues in correct position. This set thus poses a challenge for the high-quality MQAP.
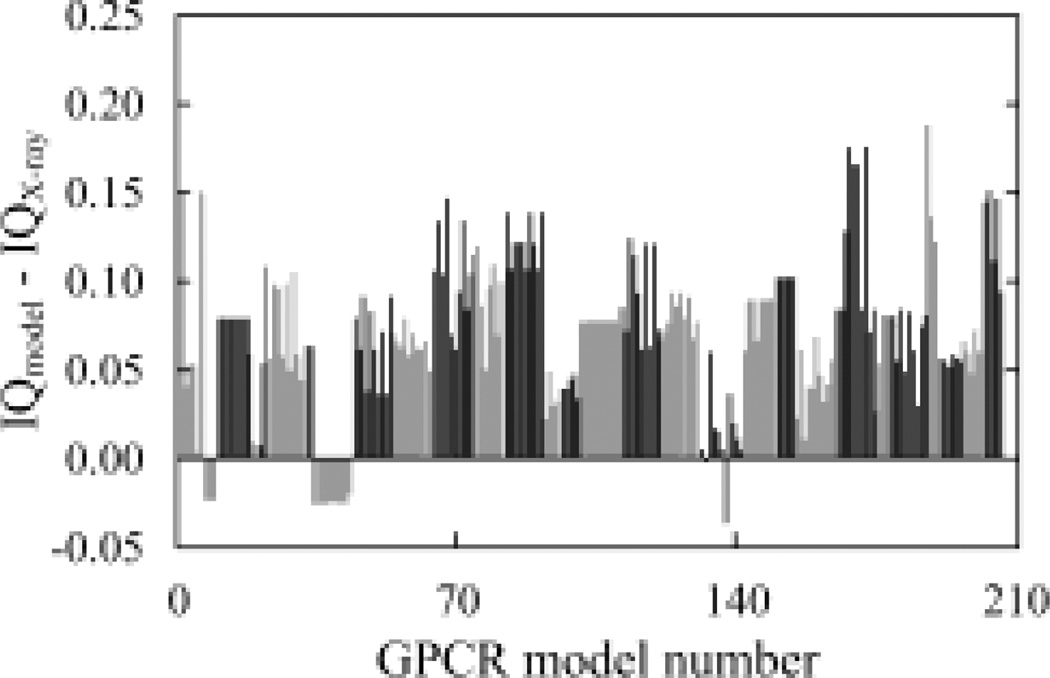
Applying IQ to the models in this dataset showed that 191 models (92.7%) of the 206 models scored worse than the X-ray structure (PDB ID: 3EML) (Figure 2). Mostly the ten models from the group with accession ID 1600 scored better. Consistently, one of the ten models from this group has been ranked in the top 10 by Z-score (23). Further, most of the 206 models scored worse than the X-ray structure in the frequency score test (Supplementary Figure 1), suggesting that the distribution of the inter-residue interactions in these models is less consistent with the structures in the high-resolution dataset than their corresponding X-ray structure (PDB ID: 3EML).
Fig. 2.
Analysis of the inter-residue interactions in the GPCR Dock 2008 set. Models are sorted from low to high by their Accession ID from reference (23). Models with the same Accession ID are grouped together and have the same gray scale, which is toggled between black and gray as the Accession ID changes.
GPCR Dock 2010 test set
The second test set comprises the 284 models of the CXCR4 chemokine receptor (PDB ID: 3ODU, 3OE0, 3OE6, 3OE8, and 3OE9) and the D3 dopamine receptor (PDB ID: 3PBL) submitted to the second round of the community-wide assessment of GPCR Structure Modeling and Docking (24). Of these X-ray structures, only one (PDB ID: 3ODU) had its structure determined to a resolution of 2.5 Å or better. Similar to the GPCR Dock 2008 set, models in this set also reflect state-of-art modeling for GPCRs and pose a challenge for high-quality MQAP.
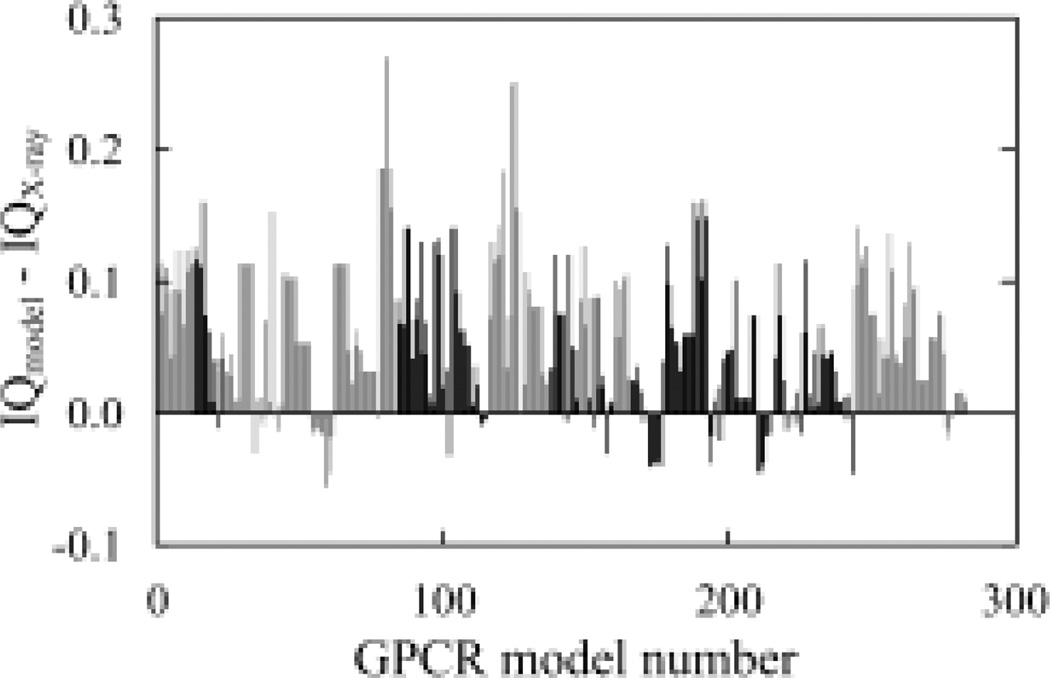
The IQ function has also performed quite well when applied to the models in this dataset, showing that 242 models (85.2%) of the 284 models scored worse than their X-ray structures (Figure 3). Only 42 models scored better. Furthermore, if only the models of the high-resolution X-ray structure target (PDB ID: 3ODU) were analyzed, only one of the 23 models scored better than the X-ray structure itself. In addition, different from GPCR Dock 2008 set, for the 284 models in this test set, most of them (83%) scored worse in the test of the average number of interactions (Supplementary Figure 2).
Fig. 3.
Analysis of the inter-residue interactions in the GPCR Dock 2010 set. Models of the same target are grouped together and have the same gray scale, which is toggled between black and gray as the Accession ID changes. The PDB ID and chain number of the X-ray structure of each GPCR target are: 1, 3ODU, chain A; 2, 3ODU, chain B; 3, 3OE0, chain A; 4, 3OE6, chain A; 5, 3OE8, chain A; 6, 3OE8, chain B; 7, 3OE8, Chain C; 8, 3OE9, chain A; 9, 3OE9, chain B; 10, 3PBL, chain A; 11, 3PBL, chain B.
HOMEP test set
To test the broad application of the proposed scoring function, it was applied to the analysis of 92 homology models of helical membrane proteins in the HOMEP benchmark dataset (25). HOMEP is a compiled dataset of homology models of various helical membrane proteins with a wide range of sequence identities. The 92 models represent 46 query-template pairs, and their quality varies quite significantly (20).
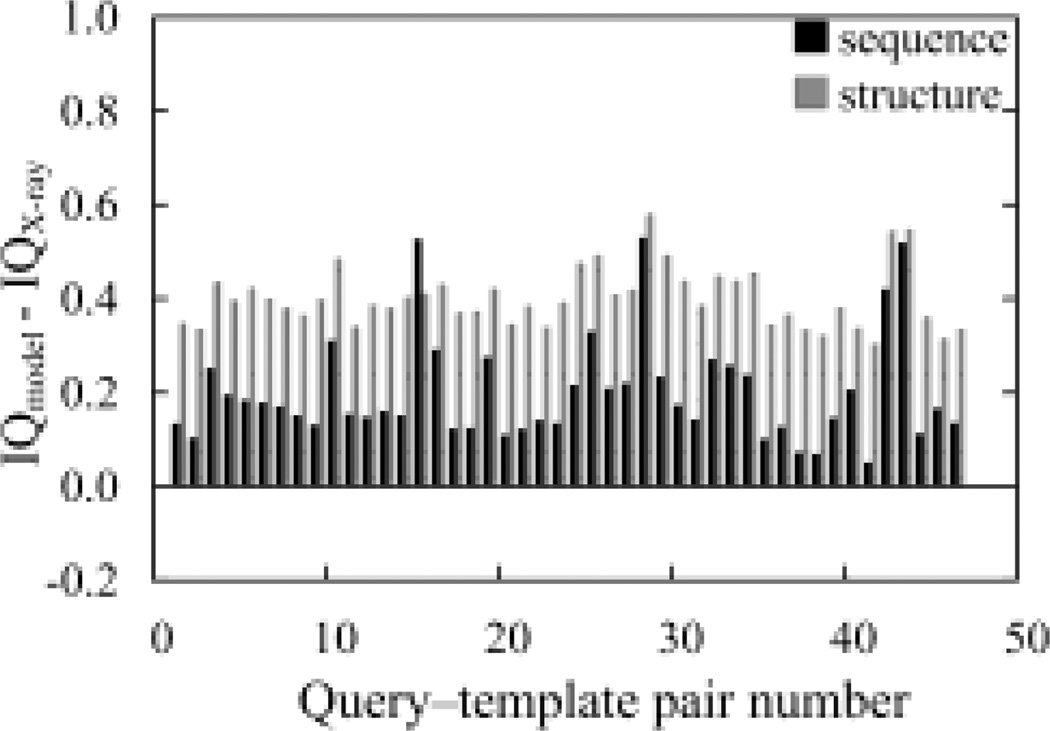
Upon applying both tests of the IQ, 92 models (100%) scored worse than their corresponding X-ray structures (Figure 4). This indicated that all native structures will be selected from among all of the models of it. For this test set, both individual tests have performed well (Supplementary Figure 3). For the frequency score test, 73 of the 92 models scored worse than their X-ray structures. For the test of the average number of interactions, 90 of the 92 models scored worse than their X-ray structures.
Fig. 4.
Analysis of the inter-residue interactions in the HOMEP set. Gray scale indicates the type of alignment used in generating a model. The PDB IDs of each query-template pair are: 1, 1E12-1H68; 2, 1E12-1M0L; 3, 1E12-1U19; 4, 1EYS-1OGY; 5, 1EYS-1PRC; 6, 1FX8-1J4N; 7, 1FX8-1RC2; 8, 1H68-1E12; 9, 1H68-1M0L; 10, 1H68-1U19; 11, 1J4N-1FX8; 12, 1J4N-1RC2; 13, 1KB9-1BCC; 14, 1KB9-1NTM; 15, 1KQF-1L0V; 16, 1KQF-1QLA; 17, 1M0L-1E12; 18, 1M0L-1H68; 19, 1M0L-1U19; 20, 1M56-1AR1; 21, 1M56-1V54; 22, 1NTM-1BCC; 23, 1NTM-1KB9; 24, 1OGV-1EYS; 25, 1OGV-1PRC; 26, 1PRC-1EYS; 27, 1PRC-1OGV; 28, 1QLA-1KQF; 29, 1QLA-1L0V; 30, 1RC2-1FX8; 31, 1RC2-1J4N; 32, 1U19-1E12; 33, 1U19-1H68; 34, 1U19-1M0L; 35, 1V54-1AR1; 36, 1V54-1M56; 37, 1AR1-1M56; 38, 1AR1-1V54; 39, 1BCC-1KB9; 40, 1BCC-1NTM; 41, 1KPL-1OTS; 42, 1L0V-1KQF; 43, 1L0V-1QLA; 44, 1OTS-1KPL; 45, 1PV6-1PW4; 46, 1PW4-1PV6.
Performance comparison with ProQM
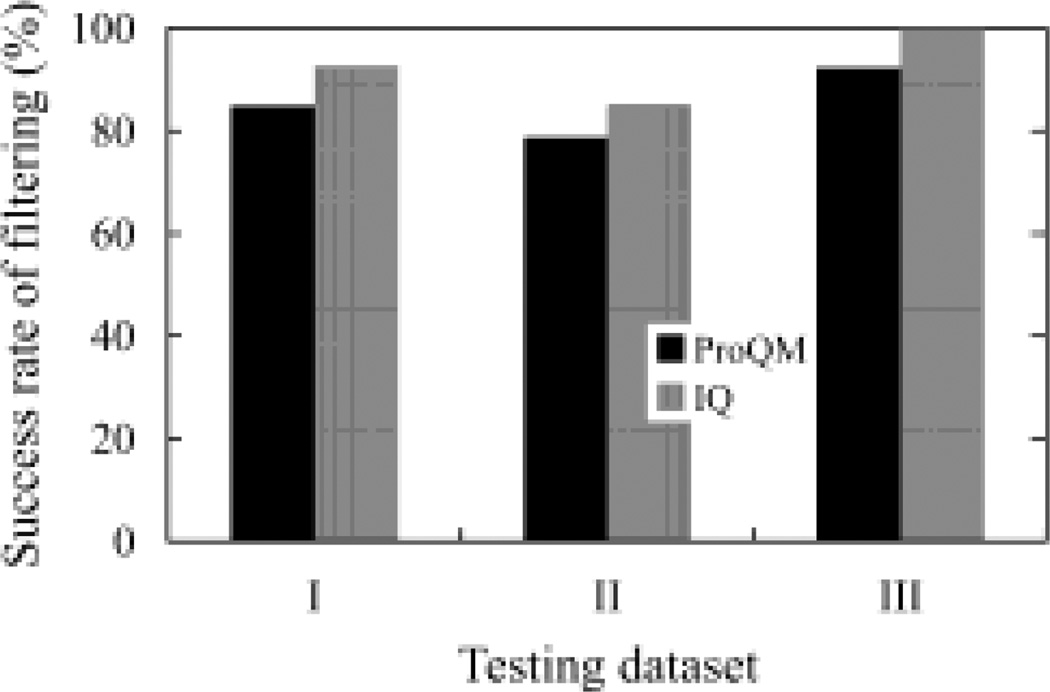
Recently, Ray et al. developed a learning-based MQAP, ProQM, that takes both general and membrane protein specific features into consideration (11). It was shown that this program has clearly outperforms all methods developed for generic proteins (11). Applying ProQM to the three test sets analyzed above indicates that the program can select the native structures among all of the models with the success rates of 85%, 79% and 92% respectively (Figure 5). These results suggested that our program, IQ, performed better than the ProQM program regarding the transmembrane domains of membrane proteins.
Fig. 5.
Comparison of the proposed scoring function IQ with the learning-based model assessment program ProQM (11). I. GPCR Dock 2008 dataset; II. GPCR Dock 2010 dataset; and III. HOMEP dataset.
An example illustrating the difference in the pattern of inter-residue interactions between a model and its X-ray structure
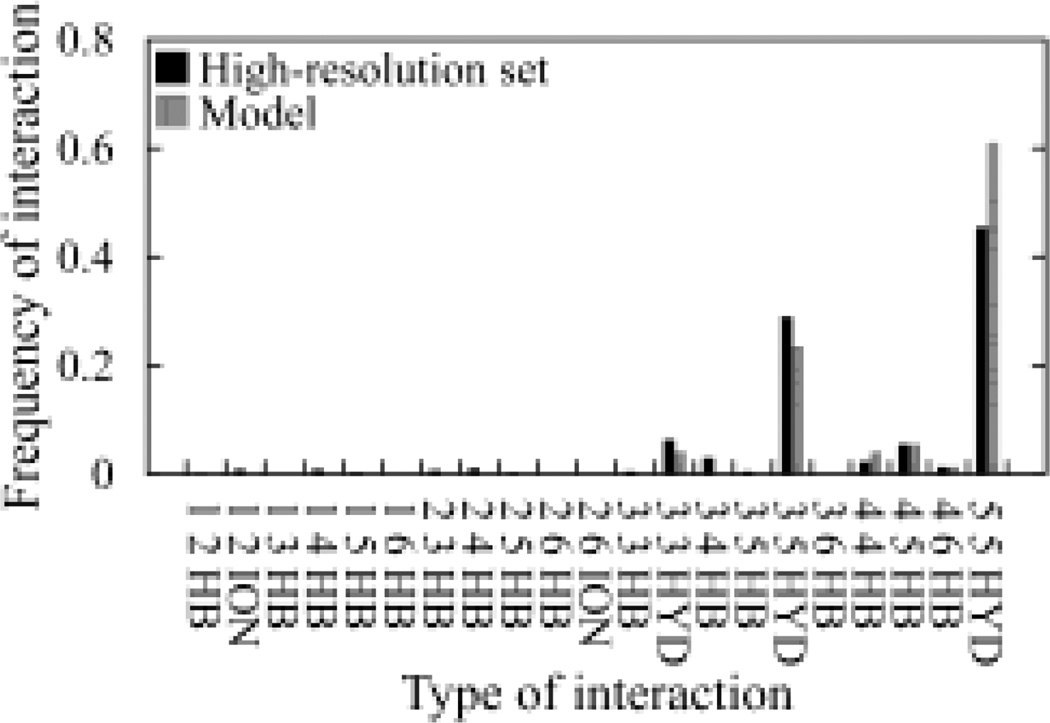
The above analyses clearly demonstrated that IQ can be applied for the discrimination of models of helical membrane proteins. To better understand the detailed difference in the frequency distribution of inter-residue interactions between a model and the high-solution dataset, we randomly selected a model from the GPCR Dock 2008 test set that scored worse than its X-ray structure and compared the frequency distribution. It is quite clear that the model has a greater number of the type 5-5 hydrophobic and a lower number of the 3–5 hydrophobic interactions (Figure 6). This observation could be helpful in that it suggests that regions that form these types of interactions may contain potential modeling errors and should be examined carefully.
Fig. 6.
Comparison of the frequency distribution of various types of inter-residue interactions between a GPCR model (Accession ID: 2200) and the high-resolution dataset. In total, there should be 84 categories of possible inter-residue interactions. Only those categories of interactions greater than zero are presented.
Discussion
Developing accurate scoring functions for discriminating the native structure from a body of constructed models represents an eminent challenge in computational structure prediction of membrane proteins (11). Despite the importance of computational modeling in the biological and pharmaceutical studies of membrane proteins, few efforts have been reported in this aspect (11, 17, 18, 19). Inter-residue interactions play a key role in the folding of both soluble and membrane proteins (34). Studying inter-residue interactions in membrane proteins may facilitate the development of such high-quality scoring functions.
In our previous works, we have demonstrated that a simple measure (c̅) that calculates the average number of four types of favorable inter-residue interactions could be used for a qualitative assessment of membrane protein models (19). Analyses of a high-resolution, non-redundant X-ray structure dataset of membrane proteins showed that all the structures have a c̅ value in the range of 1.05–1.48. Subsequently, we proposed that this range could be adopted as a quality measure of structure models of membrane proteins. Applying this to several different decoy datasets showed that it works well, but further refinement is necessary (19). In our following work, a linear correlation between this measure and the quality of helical membrane protein model/structures has been reported (20). This finding is understandable. When a protein folds, numerous favorable and unfavorable inter-residue interactions form within the structure. The greater the number of favorable interactions formed in the structure, the better the structure quality. Applying this finding to model discrimination, the best structure model should have the highest c̅ value and this value should also fall into the range derived from the high-resolution structure dataset. For this study, if a model has a lower c̅ value than its X-ray structure, it was regarded as having scored worse than the X-ray structure even though its c̅ value may fall into the range derived from the high-resolution dataset.
Ideally, the c̅ measurement alone should work sufficiently. However, the current definition of the four types of favorable interactions used here has not been perfected. In addition, a protein model can be constructed using a variety of modeling methods. As a result, an error in modeling (e.g. rotating a TM helix incorrectly) could result in a higher number of hydrophobic or other types of interactions.
In this work, we proposed another measure (fs) based on the analysis of the same subset of favorable interactions that could complement the c̅ measure. This measure examines the relative distribution of those interactions in the same high-resolution, non-redundant X-ray structure dataset of membrane proteins (Table I) and compares the difference in the relative distribution of various types of interactions in a structure relative to that derived from the high-resolution dataset. Membrane proteins showed clear preference in terms of amino acid propensity (13, 14, 15, 16). We hypothesized that a good quality structure model should display the same preference. For this second measure (fs), the lower this fs value, the more consistent in the distribution and the better the quality of the structure is.
These two measures, c̅ and fs, were combined to obtain a comprehensive scoring function (IQ), which was tested on three high-quality, independently constructed test sets, GPCR Dock 2008 (206 models), GPCR Dock 2010 (284 models), and HOMEP (92 models). The success rates of selecting the native structures from all related models for the three datasets are 93%, 85% and 100% respectively. As models in these test sets were generated for the community-wide modeling and structure prediction tests by various research groups all over the world, they represent the highest quality models using state-of-art modeling techniques (23). The results of the scoring function on these test sets are clearly very encouraging. More importantly, since models in these datasets cover a broad range of helical membrane proteins, these results demonstrate that the proposed MQAP has a broad range of application.
Among the three test sets, the GPCR Dock 2010 set gave the least perfect success rate. Considering the relatively low resolution of the X-ray structures of the target proteins in this dataset, this result is understandable. X-ray structures of lower resolution often have more errors in the packing of their amino acid residues, which could result in the overall lower IQ score. As pointed out previously, if only the models of the high-resolution X-ray structure target (PDB ID: 3ODU) in the dataset were analyzed, only one of the 23 models scored better than the X-ray structure itself. Furthermore, careful examination of the individual results by applying the c̅ and fs measures respectively showed that although both worked less well on this set than on the other two test sets, they complement each other to some extent (Supplementary Figure 1–3). As a result, the overall success rate is still quite good.
The assessment of a MQAP method by its ability to rank a native structure higher than decoys has been questioned recently (35). It was suggested that models need to be well refined prior to the application of this approach. As mentioned above, models contained in these three test sets were constructed by various groups and have been subjected to various refinement procedures. Therefore, the refinement requirement is held true and further refinement is not necessary for the works performed here.
Recently, a MQAP method based on a machine-learning approach has been developed for helical membrane protein modeling (11). Comparison of our scoring function with this MQAP program showed that IQ clearly performed better in selecting native structures among all models (Figure 5). Furthermore, compared to the machine-learning based approach, for which the interpretation of the outcome is often not very straight forward, our knowledge-based approach has the advantage that clearly indicates why one structure is favored over others (Figure 6). The difference in the pattern of the underlying inter-residue interactions of models, on which our scoring function is based, is easily obtained. On the other hand, ProQM has the advantage of scoring individual residues and also can potentially pinpoint problem areas of a model, which could be quite useful.
In summary, through the analysis of specific types of inter-residue interactions in membrane proteins, we have developed a knowledge-based scoring function for model discrimination. The discrimination power of this scoring function was tested using three large-scale, independently generated, high-quality model sets. The success rates of selecting native structures among the models were 93%, 85% and 100%. The diversity of models of these test sets suggests this scoring function can be applied to models of a broad range of membrane proteins.
Acknowledgements
The authors thank Dr. Vagmita Pabuwal at our group at the University of the Sciences in Philadelphia for helpful discussion and technical support. We thank Drs. Irina Kufareva and Raymond Stevens for sharing with us the GPCR Dock 2010 dataset of membrane protein models, and Drs. Lucy R. Forrest and Barry Honig for sharing with us the HOMEP dataset of membrane protein models. We thank our anonymous reviewers for constructive comments. This work was supported by the National Institutes of Health grant R15-GM084404.
References
- 1.Visiers I, Ballesteros JA, Weinstein H. Three-Dimensional Representations of G Protein-Coupled Receptor Structures and Mechanisms. Methods Enzymol. 2002;343:329–371. doi: 10.1016/s0076-6879(02)43145-x. [DOI] [PubMed] [Google Scholar]
- 2.Oliveira L, Hulsen T, Lutje Hulsik D, Paiva AC, Vriend G. Heavier-than-Air Flying Machines are Impossible. FEBS Lett. 2004;564:269–273. doi: 10.1016/S0014-5793(04)00320-5. [DOI] [PubMed] [Google Scholar]
- 3.Fanelli F, De Benedetti PG. Computational Modeling Approaches to Structure-Function Analysis of G Protein-Coupled Receptors. Chem. Rev. 2005;105:3297–3351. doi: 10.1021/cr000095n. [DOI] [PubMed] [Google Scholar]
- 4.Kontoyianni M, DeWeese C, Penzotti JE, Lybrand TP. Three-Dimensional Models for Agonist and Antagonist Complexes with Beta 2 Adrenergic Receptor. J. Med. Chem. 1996;39:4406–4420. doi: 10.1021/jm960241a. [DOI] [PubMed] [Google Scholar]
- 5.Wallin E, von Heijne G. Genome-Wide Analysis of Integral Membrane Proteins from Eubacterial, Archaean, and Eukaryotic Organisms. Protein Sci. 1998;7:1029–1038. doi: 10.1002/pro.5560070420. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Liu Y, Engelman DM, Gerstein M. Genomic Analysis of Membrane Protein Families: Abundance and Conserved Motifs. Genome Biol. 2002;3 doi: 10.1186/gb-2002-3-10-research0054. research0054. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Fleming KG. Riding the Wave: Structural and Energetic Principles of Helical Membrane Proteins. Curr. Opin. Biotechnol. 2000;11:67–71. doi: 10.1016/s0958-1669(99)00056-7. [DOI] [PubMed] [Google Scholar]
- 8.Overington JP, Al-Lazikani B, Hopkins AL. How Many Drug Targets are there? Nat. Rev. Drug Discov. 2006;5:993–996. doi: 10.1038/nrd2199. [DOI] [PubMed] [Google Scholar]
- 9.Xu F, Wu H, Katritch V, Han GW, Jacobson KA, Gao ZG, Cherezov V, Stevens RC. Structure of an Agonist-Bound Human A2A Adenosine Receptor. Science. 2011;332:322–327. doi: 10.1126/science.1202793. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Karnik SS, Gogonea C, Patil S, Saad Y, Takezako T. Activation of G-Protein-Coupled Receptors: A Common Molecular Mechanism. Trends Endocrinol. Metab. 2003;14:431–437. doi: 10.1016/j.tem.2003.09.007. [DOI] [PubMed] [Google Scholar]
- 11.Ray A, Lindahl E, Wallner B. Model Quality Assessment for Membrane Proteins. Bioinformatics. 2010;26:3067–3074. doi: 10.1093/bioinformatics/btq581. [DOI] [PubMed] [Google Scholar]
- 12.Gao C, Stern HA. Scoring Function Accuracy for Membrane Protein Structure Prediction. Proteins. 2007;68:67–75. doi: 10.1002/prot.21421. [DOI] [PubMed] [Google Scholar]
- 13.Chamberlain AK, Bowie JU. Analysis of Side-Chain Rotamers in Transmembrane Proteins. Biophys. J. 2004;87:3460–3469. doi: 10.1529/biophysj.104.044024. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Adamian L, Liang J. Helix-Helix Packing and Interfacial Pairwise Interactions of Residues in Membrane Proteins. J. Mol. Biol. 2001;311:891–907. doi: 10.1006/jmbi.2001.4908. [DOI] [PubMed] [Google Scholar]
- 15.Eilers M, Patel AB, Liu W, Smith SO. Comparison of Helix Interactions in Membrane and Soluble Alpha-Bundle Proteins. Biophys. J. 2002;82:2720–2736. doi: 10.1016/S0006-3495(02)75613-0. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Eilers M, Shekar SC, Shieh T, Smith SO, Fleming PJ. Internal Packing of Helical Membrane Proteins. Proc. Natl. Acad. Sci. U.S.A. 2000;97:5796–5801. doi: 10.1073/pnas.97.11.5796. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Fleishman SJ, Ben-Tal N. A Novel Scoring Function for Predicting the Conformations of Tightly Packed Pairs of Transmembrane Alpha-Helices. J. Mol. Biol. 2002;321:363–378. doi: 10.1016/s0022-2836(02)00590-9. [DOI] [PubMed] [Google Scholar]
- 18.Park Y, Helms V. Assembly of Transmembrane Helices of Simple Polytopic Membrane Proteins from Sequence Conservation Patterns. Proteins. 2006;64:895–905. doi: 10.1002/prot.21025. [DOI] [PubMed] [Google Scholar]
- 19.Pabuwal V, Li Z. Network Pattern of Residue Packing in Helical Membrane Proteins and its Application in Membrane Protein Structure Prediction. Protein Eng. Des. Sel. 2008;21:55–64. doi: 10.1093/protein/gzm059. [DOI] [PubMed] [Google Scholar]
- 20.Gao J, Li Z. Comparing Four Different Approaches for the Determination of Inter-Residue Interactions Provides Insight for the Structure Prediction of Helical Membrane Proteins. Biopolymers. 2009;91:547–556. doi: 10.1002/bip.21175. [DOI] [PubMed] [Google Scholar]
- 21.Gao J, Li Z. Conserved Network Properties of Helical Membrane Protein Structures and its Implication for Improving Membrane Protein Homology Modeling at the Twilight Zone. J. Comput. Aided Mol. Des. 2009;23:755–763. doi: 10.1007/s10822-008-9220-9. [DOI] [PubMed] [Google Scholar]
- 22.Gromiha MM, Selvaraj S. Role of Medium- and Long-Range Interactions in Discriminating Globular and Membrane Proteins. Int. J. Biol. Macromol. 2001;29:25–34. doi: 10.1016/s0141-8130(01)00150-7. [DOI] [PubMed] [Google Scholar]
- 23.Michino M, Abola E, GPCR Dock 2008 participants. Brooks CL, 3rd, Dixon JS, Moult J, Stevens RC. Community-Wide Assessment of GPCR Structure Modelling and Ligand Docking: GPCR Dock 2008. Nat. Rev. Drug Discov. 2009;8:455–463. doi: 10.1038/nrd2877. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Kufareva I, Rueda M, Katritch V, GPCR Dock 2010 participants. Stevens RC, Abagyan R. Status of GPCR Modeling and Docking as Reflected by Community Wide GPCR Dock 2010 Assessment. Structure. 2011;19:1108–1126. doi: 10.1016/j.str.2011.05.012. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Forrest LR, Tang CL, Honig B. On the Accuracy of Homology Modeling and Sequence Alignment Methods Applied to Membrane Proteins. Biophys. J. 2006;91:508–517. doi: 10.1529/biophysj.106.082313. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Larkin MA, Blackshields G, Brown NP, Chenna R, McGettigan PA, McWilliam H, Valentin F, Wallace IM, Wilm A, Lopez R, Thompson JD, Gibson TJ, Higgins DG. Clustal W and Clustal X Version 2.0. Bioinformatics. 2007;23:2947–2948. doi: 10.1093/bioinformatics/btm404. [DOI] [PubMed] [Google Scholar]
- 27.Pearl FM, Bennett CF, Bray JE, Harrison AP, Martin N, Shepherd A, Sillitoe I, Thornton J, Orengo CA. The CATH Database: An Extended Protein Family Resource for Structural and Functional Genomics. Nucleic Acids Res. 2003;31:452–455. doi: 10.1093/nar/gkg062. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Tusnady GE, Dosztanyi Z, Simon I. PDB_TM: Selection and Membrane Localization of Transmembrane Proteins in the Protein Data Bank. Nucleic Acids Res. 2005;33:D275–D278. doi: 10.1093/nar/gki002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Sali A, Blundell TL. Comparative Protein Modelling by Satisfaction of Spatial Restraints. J. Mol. Biol. 1993;234:779–815. doi: 10.1006/jmbi.1993.1626. [DOI] [PubMed] [Google Scholar]
- 30.Berman HM, Westbrook J, Feng Z, Gilliland G, Bhat TN, Weissig H, Shindyalov IN, Bourne PE. The Protein Data Bank. Nucleic Acids Res. 2000;28:235–242. doi: 10.1093/nar/28.1.235. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Stickle DF, Presta LG, Dill KA, Rose GD. Hydrogen Bonding in Globular Proteins. J. Mol. Biol. 1992;226:1143–1159. doi: 10.1016/0022-2836(92)91058-w. [DOI] [PubMed] [Google Scholar]
- 32.Muppirala UK, Li Z. A Simple Approach for Protein Structure Discrimination Based on the Network Pattern of Conserved Hydrophobic Residues. Protein Eng. Des. Sel. 2006;19:265–275. doi: 10.1093/protein/gzl009. [DOI] [PubMed] [Google Scholar]
- 33.Wallner B, Elofsson A. Can Correct Protein Models be Identified? Protein Science. 2003;12:1073–1086. doi: 10.1110/ps.0236803. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Gromiha MM, Selvaraj S. Inter-Residue Interactions in Protein Folding and Stability. Prog. Biophys. Mol. Biol. 2004;86:235–277. doi: 10.1016/j.pbiomolbio.2003.09.003. [DOI] [PubMed] [Google Scholar]
- 35.Handl J, Knowles J, Lovell SC. Artefacts and Biases Affecting the Evaluation of Scoring Functions on Decoy Sets for Protein Structure Prediction. Bioinformatics. 2009;25:1271–1279. doi: 10.1093/bioinformatics/btp150. [DOI] [PMC free article] [PubMed] [Google Scholar]