Abstract
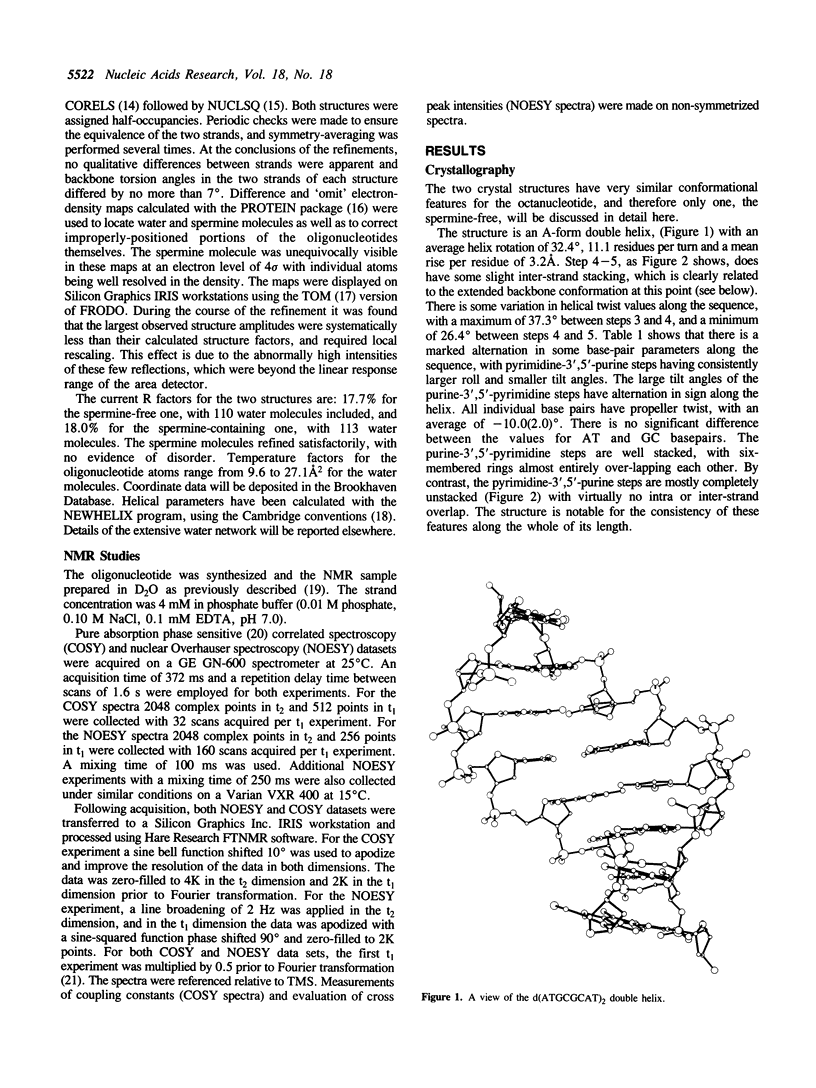
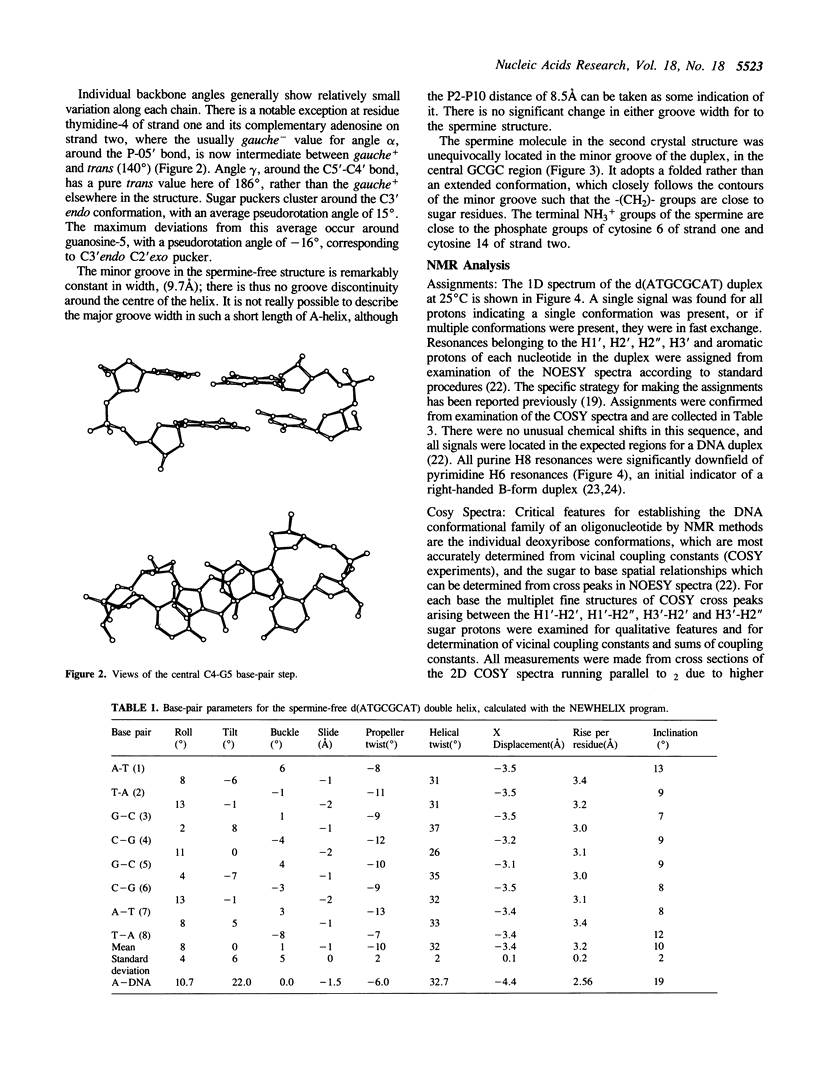
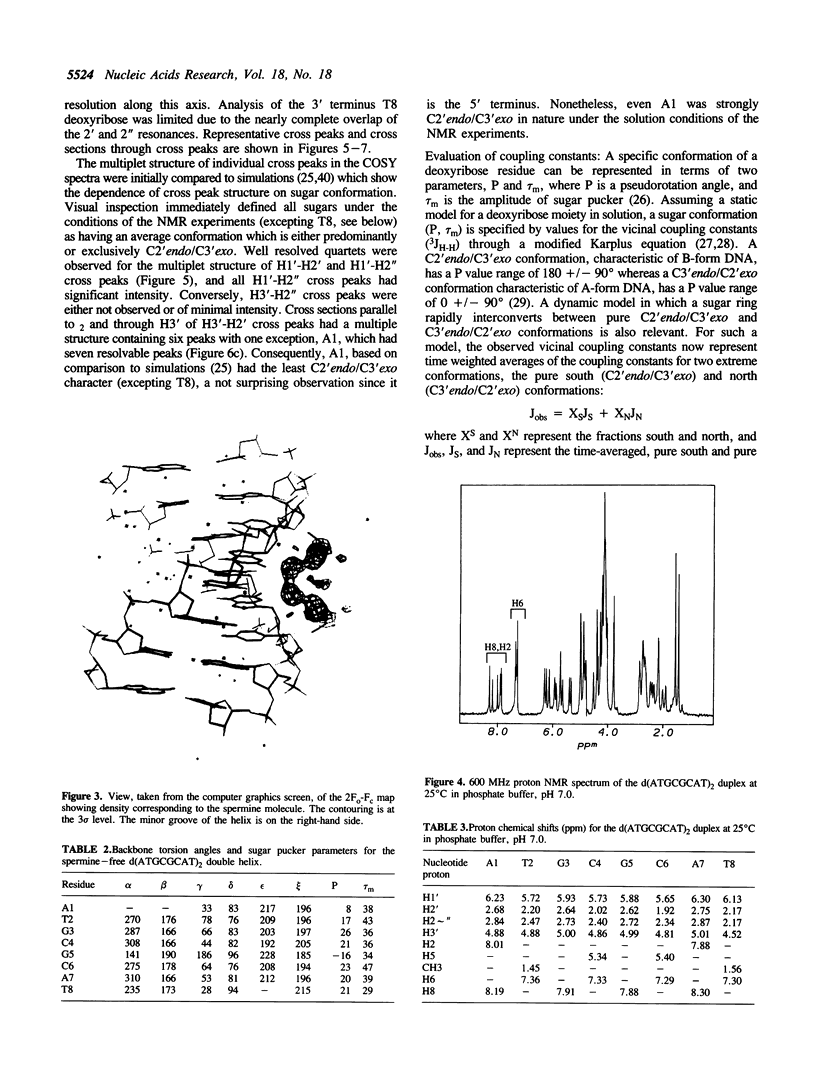
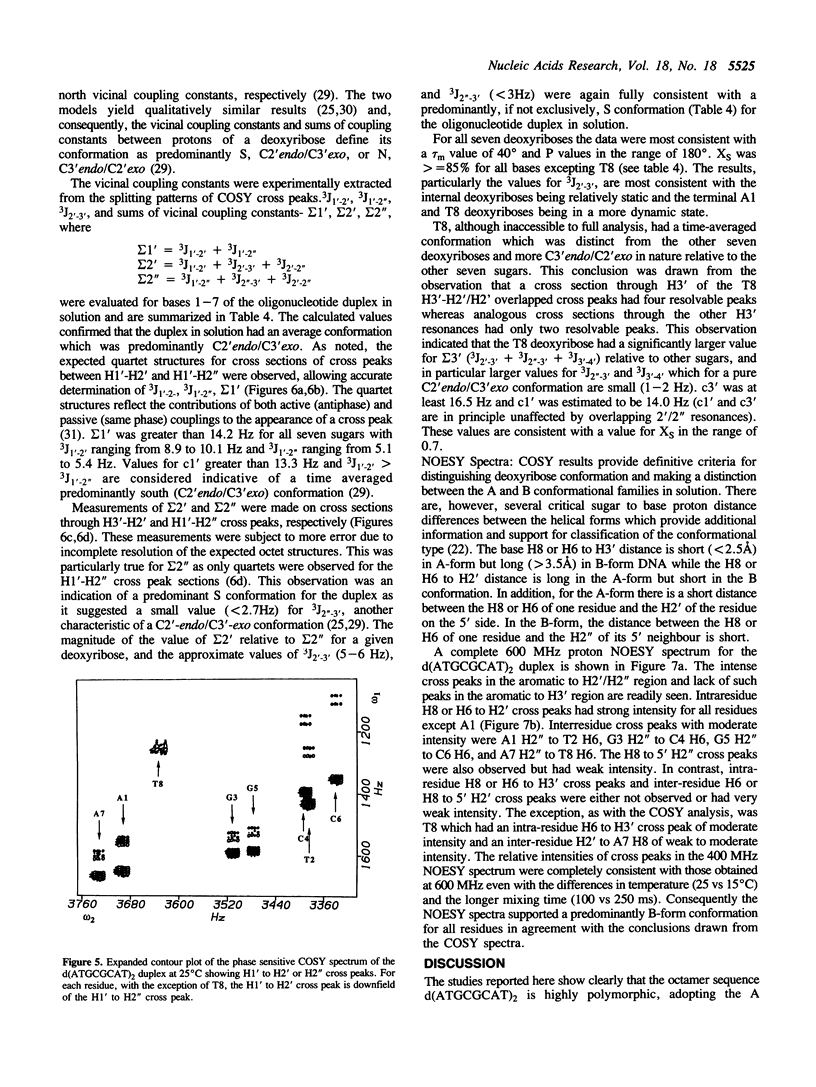
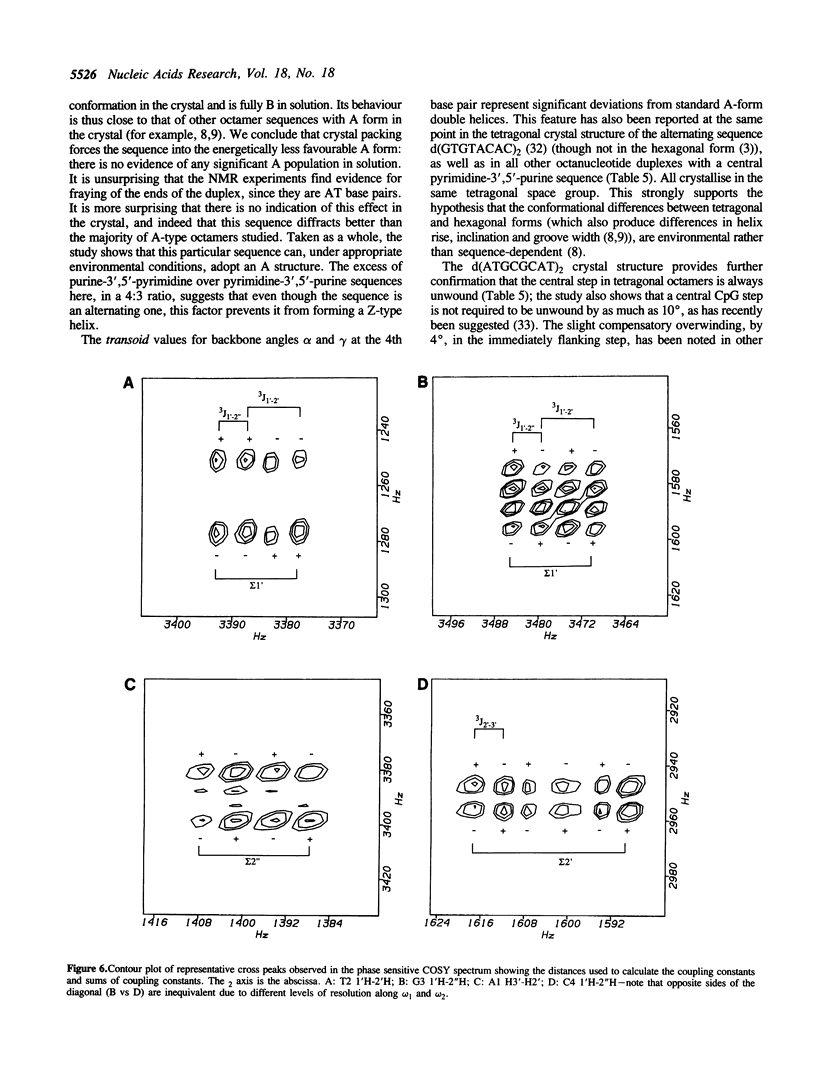
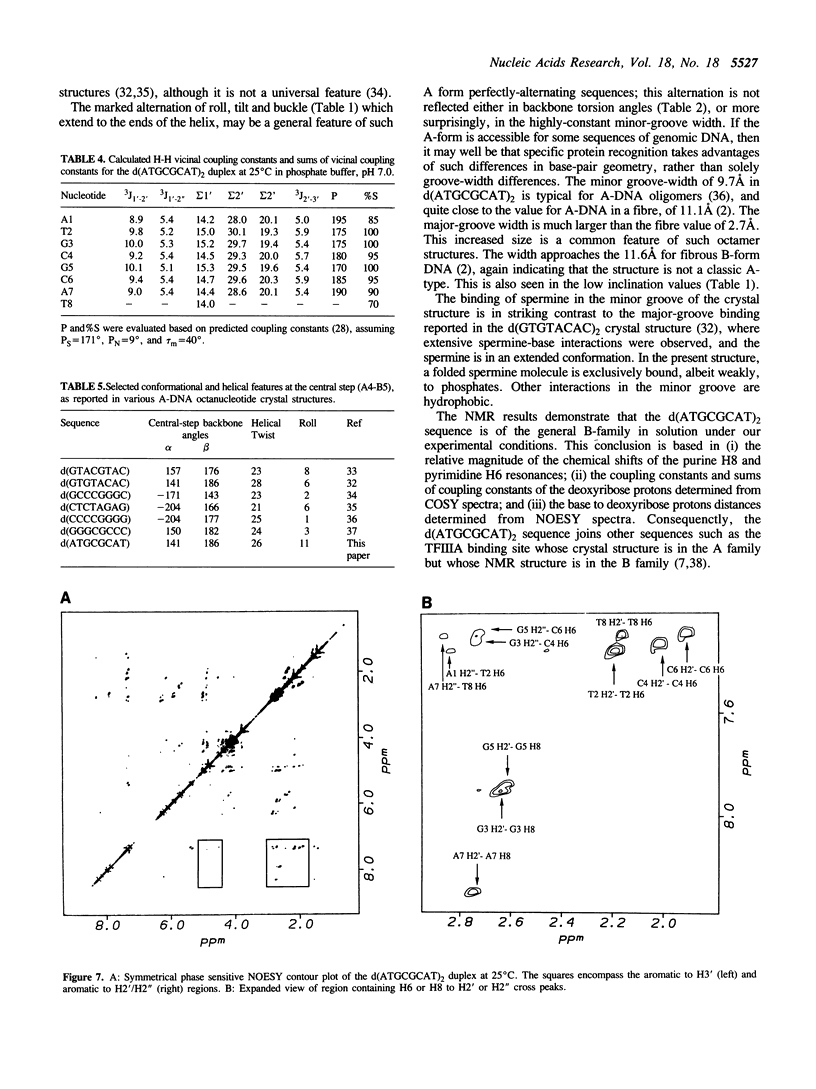
A combined crystal-structure determination and NMR analysis of the octanucleotide d(ATGCGCAT)2 is reported. The X-ray analysis shows that the structure is A-form duplex in crystal state. The NMR study shows that in solution this sequence is B-type. The conformational results from each technique are presented in detail. The implications of these findings in terms of conformational flexibility and ligand binding are discussed.
Full text
PDF

Selected References
These references are in PubMed. This may not be the complete list of references from this article.
- Aboul-ela F., Varani G., Walker G. T., Tinoco I., Jr The TFIIIA recognition fragment d(GGATGGGAG).d(CTCCCATCC) is B-form in solution. Nucleic Acids Res. 1988 Apr 25;16(8):3559–3572. doi: 10.1093/nar/16.8.3559. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Aggarwal A. K., Rodgers D. W., Drottar M., Ptashne M., Harrison S. C. Recognition of a DNA operator by the repressor of phage 434: a view at high resolution. Science. 1988 Nov 11;242(4880):899–907. doi: 10.1126/science.3187531. [DOI] [PubMed] [Google Scholar]
- Altona C., Sundaralingam M. Conformational analysis of the sugar ring in nucleosides and nucleotides. A new description using the concept of pseudorotation. J Am Chem Soc. 1972 Nov 15;94(23):8205–8212. doi: 10.1021/ja00778a043. [DOI] [PubMed] [Google Scholar]
- Celda B., Widmer H., Leupin W., Chazin W. J., Denny W. A., Wüthrich K. Conformational studies of d-(AAAAATTTTT)2 using constraints from nuclear overhauser effects and from quantitative analysis of the cross-peak fine structures in two-dimensional 1H nuclear magnetic resonance spectra. Biochemistry. 1989 Feb 21;28(4):1462–1471. doi: 10.1021/bi00430a006. [DOI] [PubMed] [Google Scholar]
- Chou S. H., Flynn P., Reid B. Solid-phase synthesis and high-resolution NMR studies of two synthetic double-helical RNA dodecamers: r(CGCGAAUUCGCG) and r(CGCGUAUACGCG). Biochemistry. 1989 Mar 21;28(6):2422–2435. doi: 10.1021/bi00432a013. [DOI] [PubMed] [Google Scholar]
- Drew H. R., Dickerson R. E. Structure of a B-DNA dodecamer. III. Geometry of hydration. J Mol Biol. 1981 Sep 25;151(3):535–556. doi: 10.1016/0022-2836(81)90009-7. [DOI] [PubMed] [Google Scholar]
- Gao X. L., Patel D. J. NMR studies of echinomycin bisintercalation complexes with d(A1-C2-G3-T4) and d(T1-C2-G3-A4) duplexes in aqueous solution: sequence-dependent formation of Hoogsteen A1.T4 and Watson--Crick T1.A4 base pairs flanking the bisintercalation site. Biochemistry. 1988 Mar 8;27(5):1744–1751. doi: 10.1021/bi00405a054. [DOI] [PubMed] [Google Scholar]
- Haran T. E., Shakked Z., Wang A. H., Rich A. The crystal structure of d(CCCCGGGG): a new A-form variant with an extended backbone conformation. J Biomol Struct Dyn. 1987 Oct;5(2):199–217. doi: 10.1080/07391102.1987.10506390. [DOI] [PubMed] [Google Scholar]
- Hare D. R., Wemmer D. E., Chou S. H., Drobny G., Reid B. R. Assignment of the non-exchangeable proton resonances of d(C-G-C-G-A-A-T-T-C-G-C-G) using two-dimensional nuclear magnetic resonance methods. J Mol Biol. 1983 Dec 15;171(3):319–336. doi: 10.1016/0022-2836(83)90096-7. [DOI] [PubMed] [Google Scholar]
- Heinemann U., Lauble H., Frank R., Blöcker H. Crystal structure analysis of an A-DNA fragment at 1.8 A resolution: d(GCCCGGGC). Nucleic Acids Res. 1987 Nov 25;15(22):9531–9550. doi: 10.1093/nar/15.22.9531. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hunter W. N., D'Estaintot B. L., Kennard O. Structural variation in d(CTCTAGAG). Implications for protein-DNA interactions. Biochemistry. 1989 Mar 21;28(6):2444–2451. doi: 10.1021/bi00432a015. [DOI] [PubMed] [Google Scholar]
- Jain S., Sundaralingam M. Effect of crystal packing environment on conformation of the DNA duplex. Molecular structure of the A-DNA octamer d(G-T-G-T-A-C-A-C) in two crystal forms. J Biol Chem. 1989 Aug 5;264(22):12780–12784. [PubMed] [Google Scholar]
- Jain S., Zon G., Sundaralingam M. Base only binding of spermine in the deep groove of the A-DNA octamer d(GTGTACAC). Biochemistry. 1989 Mar 21;28(6):2360–2364. doi: 10.1021/bi00432a002. [DOI] [PubMed] [Google Scholar]
- McCall M., Brown T., Hunter W. N., Kennard O. The crystal structure of d(GGATGGGAG): an essential part of the binding site for transcription factor IIIA. Nature. 1986 Aug 14;322(6080):661–664. doi: 10.1038/322661a0. [DOI] [PubMed] [Google Scholar]
- Otwinowski Z., Schevitz R. W., Zhang R. G., Lawson C. L., Joachimiak A., Marmorstein R. Q., Luisi B. F., Sigler P. B. Crystal structure of trp repressor/operator complex at atomic resolution. Nature. 1988 Sep 22;335(6188):321–329. doi: 10.1038/335321a0. [DOI] [PubMed] [Google Scholar]
- Rabinovich D., Haran T., Eisenstein M., Shakked Z. Structures of the mismatched duplex d(GGGTGCCC) and one of its Watson-Crick analogues d(GGGCGCCC). J Mol Biol. 1988 Mar 5;200(1):151–161. doi: 10.1016/0022-2836(88)90340-3. [DOI] [PubMed] [Google Scholar]
- Rhodes D., Klug A. An underlying repeat in some transcriptional control sequences corresponding to half a double helical turn of DNA. Cell. 1986 Jul 4;46(1):123–132. doi: 10.1016/0092-8674(86)90866-4. [DOI] [PubMed] [Google Scholar]
- Rinkel L. J., Altona C. Conformational analysis of the deoxyribofuranose ring in DNA by means of sums of proton-proton coupling constants: a graphical method. J Biomol Struct Dyn. 1987 Feb;4(4):621–649. doi: 10.1080/07391102.1987.10507665. [DOI] [PubMed] [Google Scholar]
- Schmitz U., Zon G., James T. L. Deoxyribose conformation in [d(GTATATAC)]2: evaluation of sugar pucker by simulation of double-quantum-filtered COSY cross-peaks. Biochemistry. 1990 Mar 6;29(9):2357–2368. doi: 10.1021/bi00461a021. [DOI] [PubMed] [Google Scholar]
- Scott E. V., Zon G., Marzilli L. G., Wilson W. D. 2D NMR investigation of the binding of the anticancer drug actinomycin D to duplexed dATGCGCAT: conformational features of the unique 2:1 adduct. Biochemistry. 1988 Oct 4;27(20):7940–7951. doi: 10.1021/bi00420a053. [DOI] [PubMed] [Google Scholar]
- Shakked Z., Guerstein-Guzikevich G., Eisenstein M., Frolow F., Rabinovich D. The conformation of the DNA double helix in the crystal is dependent on its environment. Nature. 1989 Nov 23;342(6248):456–460. doi: 10.1038/342456a0. [DOI] [PubMed] [Google Scholar]
- Shakked Z., Rabinovich D. The effect of the base sequence on the fine structure of the DNA double helix. Prog Biophys Mol Biol. 1986;47(3):159–195. doi: 10.1016/0079-6107(86)90013-1. [DOI] [PubMed] [Google Scholar]
- Takusagawa F. The crystal structure of d(GTACGTAC) at 2.25 A resolution: are the A-DNA's always unwound approximately 10 degrees at the C-G steps? J Biomol Struct Dyn. 1990 Feb;7(4):795–809. doi: 10.1080/07391102.1990.10508524. [DOI] [PubMed] [Google Scholar]
- Wang A. H., Fujii S., van Boom J. H., Rich A. Molecular structure of the octamer d(G-G-C-C-G-G-C-C): modified A-DNA. Proc Natl Acad Sci U S A. 1982 Jul;79(13):3968–3972. doi: 10.1073/pnas.79.13.3968. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Westhof E., Dumas P., Moras D. Crystallographic refinement of yeast aspartic acid transfer RNA. J Mol Biol. 1985 Jul 5;184(1):119–145. doi: 10.1016/0022-2836(85)90048-8. [DOI] [PubMed] [Google Scholar]
- Wolk S., Thurmes W. N., Ross W. S., Hardin C. C., Tinoco I., Jr Conformational analysis of d(C3G3), a B-family duplex in solution. Biochemistry. 1989 Mar 21;28(6):2452–2459. doi: 10.1021/bi00432a016. [DOI] [PubMed] [Google Scholar]
- Yoon C., Privé G. G., Goodsell D. S., Dickerson R. E. Structure of an alternating-B DNA helix and its relationship to A-tract DNA. Proc Natl Acad Sci U S A. 1988 Sep;85(17):6332–6336. doi: 10.1073/pnas.85.17.6332. [DOI] [PMC free article] [PubMed] [Google Scholar]


