Abstract
A detailed structural analysis of the entire human adenovirus capsid has been stymied by the complexity and size of this 150 MDa macromolecular complex. Over the past 10 years, the steady improvements in viral genome manipulation concomitant with advances in crystallographic techniques and data processing software has allowed structure determination of this virus by X-ray diffraction at 3.5 Å resolution. The virus structure revealed the location, folds, and interactions of major and minor (cement proteins) on the inner and outer capsid surface. This new structural information sheds further light on the process of adenovirus capsid assembly and virus-host cell interactions.
Introduction
Adenoviruses (HAdVs) are large, non-enveloped viruses containing a dsDNA genome of approximately 36 KD [1]. A significant percentage of the 56 different HAdV types cause acute respiratory, gastrointestinal and ocular infections [2,3]. One of these virus types, HAdV-5, is also being employed as a gene delivery vector for cardiovascular diseases and cancer [4,5] as well as a vaccine vehicle to protect against infection by diverse microbial agents [6,7]. A detailed structural analysis of HAdV may foster the development of antivirals to thwart virus infections as well as facilitate the generation of improved HAdV vectors for clinical applications. However, the adenovirus capsid contains nearly 1 million amino acids and is over 900 Å in diameter, and therefore presents a daunting task for structural analyses. Despite this impediment, early structural studies of HAdV have a rich and productive history. In fact, the HAdV hexon, the major outer capsid protein of the virus, was the first animal virus protein to be crystallized [8], allowing subsequent determination of its X-ray structure [9,10]. These early hexon X-ray diffraction studies set the stage for more complex structural analyses of the entire virus particle by cryoelectron microscopy and image reconstruction [11,12]. The continuing improvements in cryoEM methods allowed visualization of the locations, folds, and interactions of some of the major and minor (cement) proteins in the outer HAdV capsid [13,14]. Very recently, cryoEM techniques were used to visualize HAdV-5 at nearly atomic resolution, 3.6 Å, providing one of the most detailed analyses of the entire virus to date [15]. In addition, the X-ray structures of the isolated penton base [16] and fiber proteins [17] have provided insights on the early events in virus infection. Despite these major advances, the precise locations, folds and interactions between the hexon and several capsid proteins within the virus particle have yet to be completely resolved.
The challenges and solutions to solving the X-ray structure of adenovirus
Adenovirus, along with the bluetongue virus core [18], PRD1 [19], and reovirus core [20], are among the largest non-enveloped viruses to be studied by EM or X-ray diffraction techniques. The complexity of HAdV that displays pseudo T=25 icosahedral symmetry as well as its instability at non-neutral pH were factors that hindered the growth of X-ray diffraction quality virus crystals. In particular, the highly elongated fiber protein (~30 nm) that is present at each of the twelve 5-fold axes in most HAdV types, interfered with crystal lattice formation. Fiberless HAdV-5 particles [21,22] were considered as an alternative for virus crystallization, however these “bald” particles were difficult to propagate and were less stable than normal HAdV particles. As an alternative approach, an HAdV-5 vector bearing the short (~12 nm) flexible fiber of HAdV-35 (designated Ad35F) could be produced at high titer and was relatively stable. We were able to grow diffraction quality crystals of Ad35F (space group P1; a=853.9 Å, b=855.6 Å, c=865.3 Å, =119.6°, =91.7°, =118.1°) at near neutral pH conditions identified in a robotic screening approach [23]. However, the Ad35F crystals were weakly diffracting as determined at several different synchrotron X-ray beamlines. Eventually, a relatively new synchrotron beamline (GM/CA CAT 23-ID-D, APS/Chicago) with a high flux X-ray source was found to be capable of achieving optimal exposure of the virus crystals. In addition, we were able to identify appropriate freezing conditions of virus crystals using Paratone N as a cryoprotectant. This allowed the collection of multiple images from each crystal, a situation that was crucial for accumulating a large diffraction data set. The structure was determined using conventional molecular replacement methods employed in virus crystallography [23,24].
X-ray structure of Ad35F revealed inter-hexon associations
Ad35F crystals diffracted to a resolution of 3.5 Å resolution, revealing significant new structural features on the outer and inner surfaces of the virus capsid [24]. As the inner DNA core of the virus is not icosahedrally-ordered it could not be visualized. On the outer surface of the capsid, the 240 trimeric hexons, that have 4 unique locations, were revealed as expected from earlier cryoEM studies (Fig. 1a). However, the X-ray structure of Ad35F revealed much greater detail of the hexon, including the locations and orientations of several hypervariable region loops that were missing in previous structural analyses of the isolated hexon. It is likely that the greater structural information gleaned from X-ray structure is due in part to the fact that hexons provide the major contact sites between adjacent virus particles in the crystal lattice [23]. In addition, certain HVR loops interact with neighboring loops and/or neighboring cement proteins (Fig. 1b).
Figure 1.
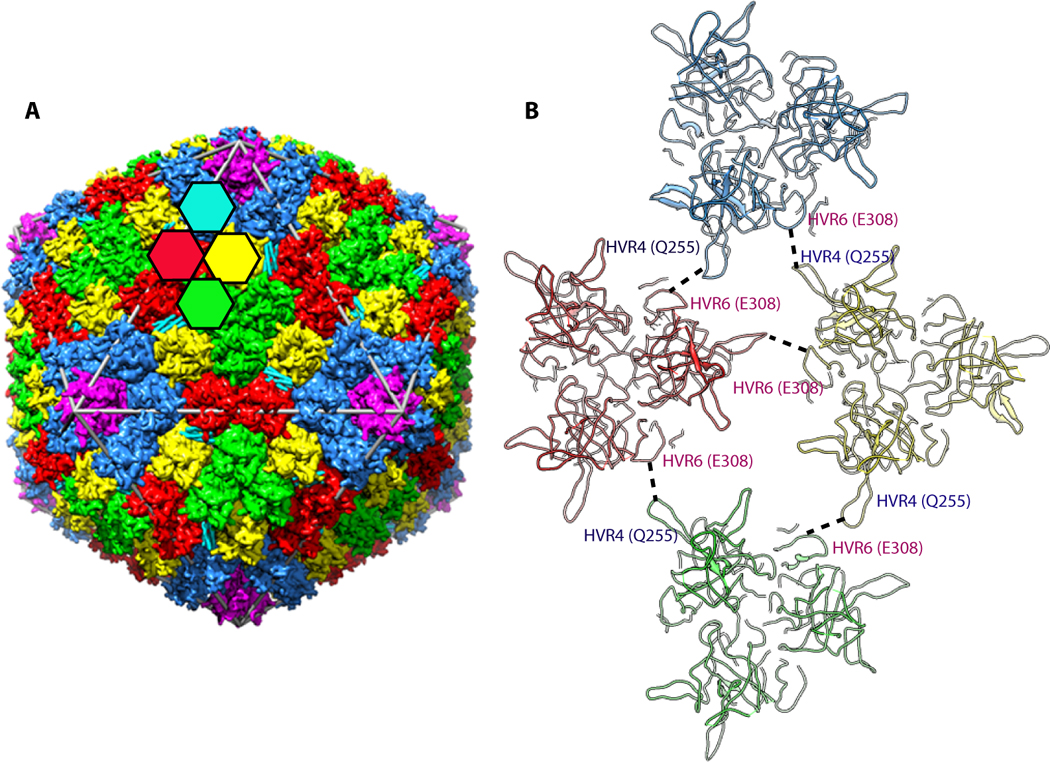
A. A schematic representation illustrating the arrangement of major capsid proteins (hexon and penton base) in the adenovirus capsid. The colored hexagons represent the 4 unique hexon trimers of the pseudo T=25 icosahedral lattice. Five-fold related penton base subunits shown in magenta are located at each of the 12 vertices of the capsid. B. Interaction of hypervariable (HVR) loops HVR4 and HVR6 that help stabilize the Ad capsid. These interactions result in the ordering of HVR4 loop, which was disordered in the isolated hexon structure.
Penton base association with the fiber protein at the 5-fold vertices
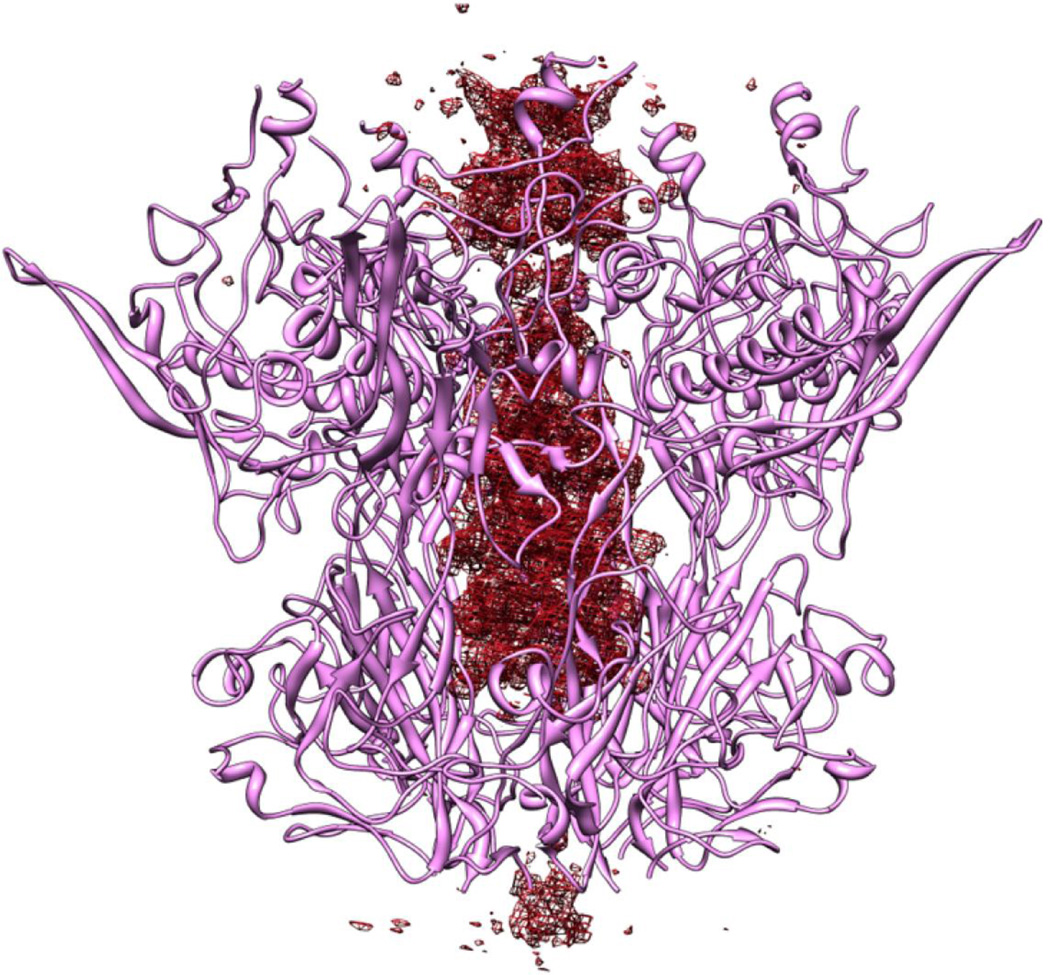
The penton base protein, present at each vertex in Ad35F, serves as the attachment site for cell surface integrins αvβ3 and αvβ5 that serve as receptors for virus internalization into host cells [25]. Interestingly, the penton base in the virus crystal structure adopted a different conformation than that previously observed in the isolated protein [16]. In the virus the central pore of the penton base is wider than that of the isolated penton. Moreover, the Ad35F fiber shaft appears to penetrate the pore of the virus penton base, inserting well into the center of the pentamer (Fig. 2). These findings suggest that the penton base is a rather flexible molecule that can undergo conformational changes. These structural changes might have been induced by the high calcium conditions used to produce virus crystals [23]. Conformational changes in the penton base were indicated by a cryoEM study of adenovirus in association with integrin αvβ5 [26]. However, the integrin-induced changes that were hypothesized were distinct from those observed in the virus crystal structure. Thus, further structural and functional studies will be needed to identify the triggers for penton base conformational change and the specific conformations adopted at particular points in the virus life cycle.
Figure 2.
Fo-Fc difference density (colored red) revealed along the 5-fold axis that penetrates the pore formed by the 5-fold related penton base subunits. This density is suggested to correspond to the shaft region of the trimeric fiber molecule, although sidechain density is not clearly identifiable presumably because of the symmetry mismatch between the fiber and the 5-fold capsid vertex.
Cement proteins on the outside and inside surface of the virus capsid
A prominent structural element on the outside of the virus capsid is a well-ordered four-helix bundle (4HB) that is present at three copies per facet. The longest helix is approximately 80 Å in length and the entire 4HB corresponds to a ~200 amino acid domain of a cement protein. Earlier cryoEM studies [11,14] assigned this structure to protein IIIa, a 63 KDa cement protein that is required for the late stage of virus maturation [27]. However, more recent cryoEM analyses at ~ 6Å [13] and at 3.6Å [15] resolution assigned this protein module to the C-termini of protein IX. Our X-ray structure of Ad35F 3.5 Å resolution revealed that two of the helices in the 4HB were clearly connected, raising doubt as to whether this module could be the C-termini of protein IX. The principles of quasi-equivalence as well as the established protein IIIa copy number (60 per capsid), are consistent with the idea that the 4HB is a domain of protein IIIa. Further improvements in the X-ray maps of HAdV are needed to definitively assign this domain to a particular cement protein. The accurate identification of this cement protein should aid our understanding of capsid assembly, as there are close contacts between the 4HB and the HVR4 loop of hexon trimers.
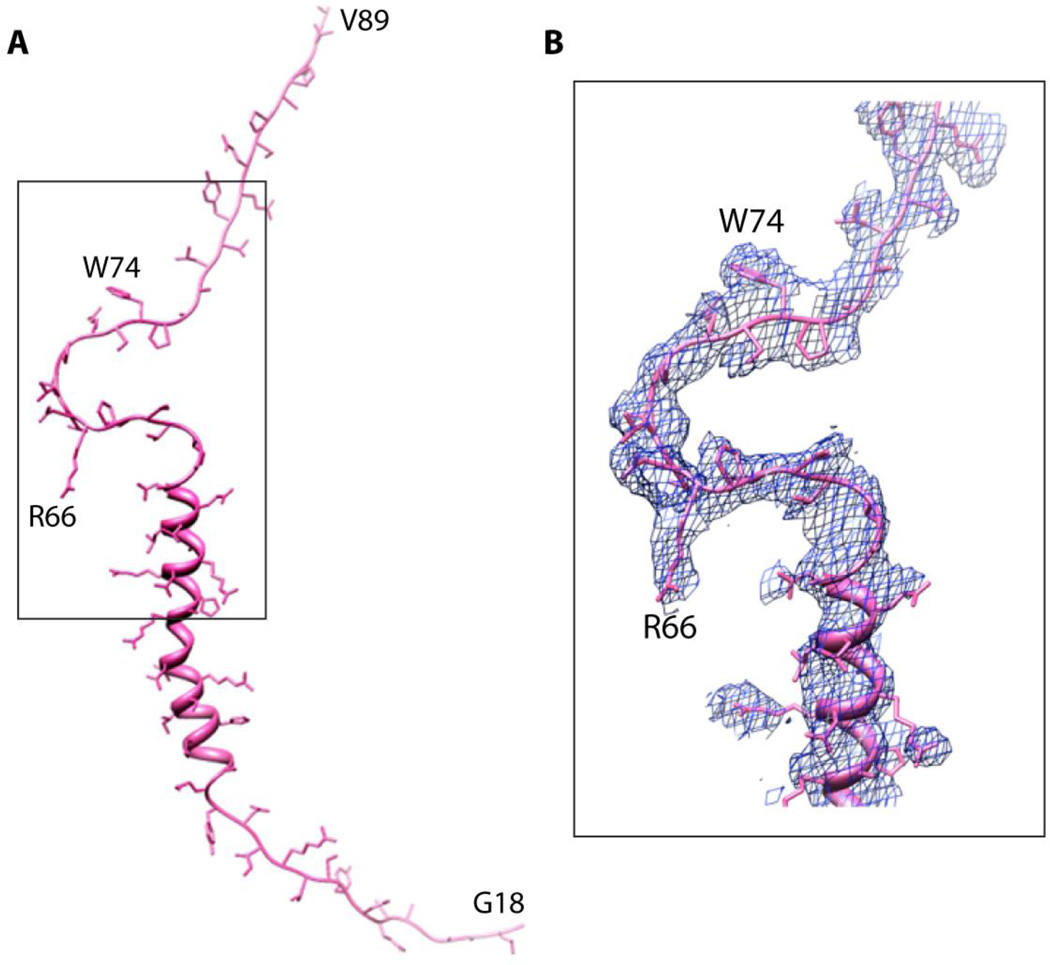
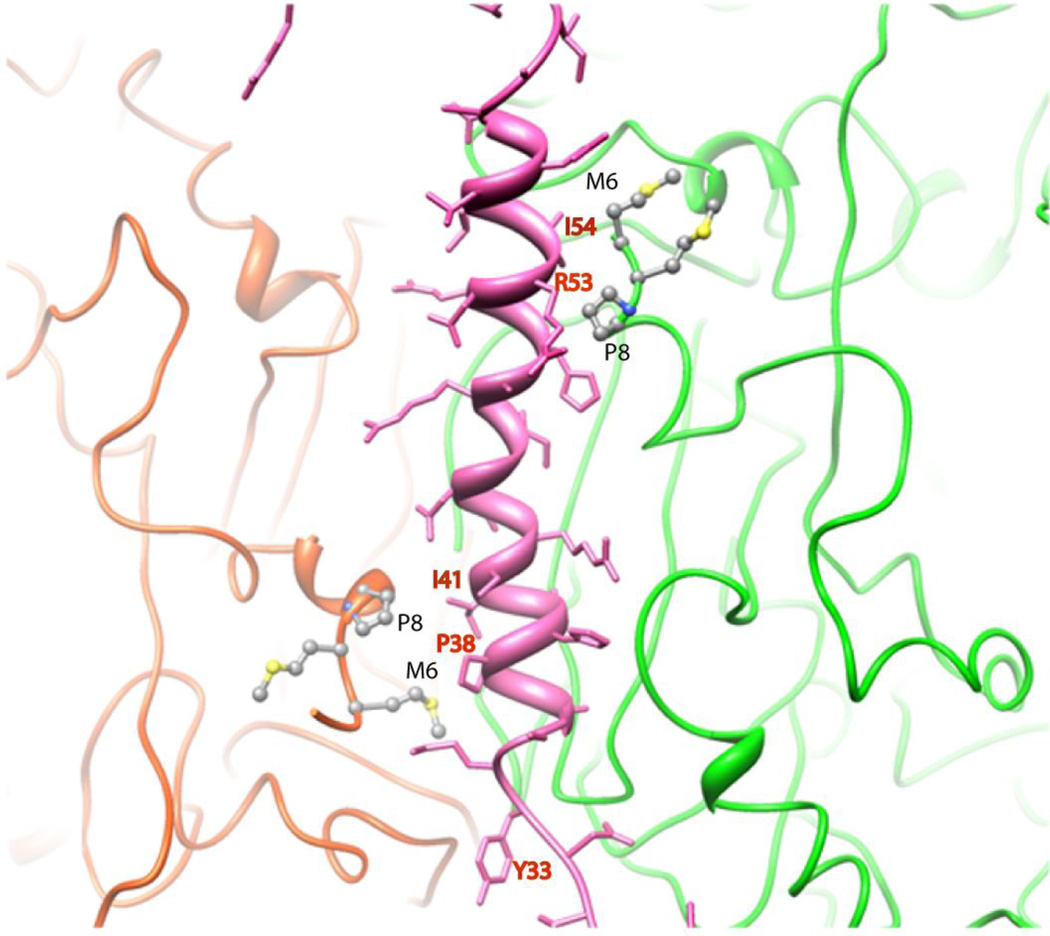
The X-ray structure of Ad35F also revealed well-ordered density, corresponding to cement protein VIII that lies underneath the capsid. Approximately 70 amino acid residues could be fitted into the X-ray density of pVIII that included an N-terminal alpha helical domain of ~ 20 residues followed by a prominent U-shaped bend that contained several proline residues and ending with an extended polypeptide region (Fig. 3). Interestingly, pVIII is present at two distinct locations on the inner surface of the capsid and these sites closely trace the outline of hexons that comprise the group of nine hexons (GON). The group of nine hexons, which can be dissociated from intact virions by treatment with 10% pyridine, are thought to represent an assembly intermediate during the capsid maturation [28]. Two isoleucine residues, Ile 41 and Ile54, located on the N-terminal helical domain of pVIII mediate non-polar associations with two neighboring hexon N-termini (Fig. 4) while a third hexon association is mediated in a similar fashion via the proline-containing U-shaped domain. Thus, pVIII appears to play a significant role in stabilizing the inside surface of the virus capsid by interactions with the N-termini of multiple hexon trimers.
Figure 3.
A. Model showing the extended structure of a PVIII molecule. Nearly 70 residues of VIII are ordered in one of the two independent molecules present in the adenovirus structure. B. Electron density corresponding to the inverted U shaped structure highlighted by the rectangle in panel A.
Figure 4.
Interactions mediated by the protein VIII helix located between two hexon subunits. The VIII helix, shown in pink, helps to stabilize the N-termini of two hexon subunits through non-polar interactions.
Additional alpha helical domains, organized as pairs, were revealed underneath the vertex region. We were not able to definitively identify these protein segments in the X-ray structure, although they have been assigned to protein IIIa in cryoEM studies of the virus [13,15]. However, it remains possible that they correspond to one or more domains of a different cement protein pVI that is released along with other vertex proteins (i.e. penton base and fiber) upon heating. Protein VI plays multiple roles in the virus life cycle including endosome disruption [29], acting as a cofactor for protease cleavage [30] and serving as a chaperone for hexon import into the nucleus prior virus assembly [31]. Recent studies have demonstrated that the N-terminal amphipathic helix (AH) of pVI plays a crucial role in endosome disruption [32]. Specifically, a single amino acid substitution, L40Q, in the AH of pVI attenuates virus-mediated membrane association and disruption, leading to accumulation of virus particles in endosomes. One unresolved question related to HAdV-mediated membrane disruption is the precise pVI copy number present in the mature virion. The answer to this question is not only an important for understanding the process of virus assembly but it is also relevant for obtaining a complete picture of capsid disassembly and membrane disruption. Previous estimates based on biochemical or mass spectrometry indicated that there are 340–360 copies of pVI per virion [33,34]. As this copy number is higher than that of the 240 hexon trimers in the virion, this raises the possibility that some of the hexons in the virion are associated with more than one pVI molecule. It is also possible that previous estimates of the pVI copy number are too high as suggested by recent cryoEM analyses revealing the association of single densities with each hexon [35]. Further biochemical and structural analyses are needed to accurately determine the pVI-hexon stoichiometry in the HAdV capsid.
Conclusions and Future Directions
The crystal structure of HAdV at 3.5 Å resolution represents one of the largest macromolecular structures determined by X-ray diffraction. The HAdV crystal structure revealed the locations and associations of the four different hexon trimers as well as a surprising conformational change in the penton base and fiber on the outer surface of the capsid. Although the cause for the penton alteration has not been established, these findings emphasize the plasticity of the vertex region that is known to undergo release during capsid disassembly. Further functional and biochemical studies are needed to establish the basis for these structural observations and to determine their relationship to the structural information obtained by cryoEM analyses [15,36]. The crystal structure of HAdV also revealed the locations, folds and associations of several cement proteins, although only one of these, protein VIII, was definitely identified by fitting the sequence (~ 70 residues) into the X-ray density. CryoEM studies have tentatively assigned the locations of the other cement proteins in the virion, IIIa, VI and IX. We are currently exploring the incorporation of heavy atoms into the virus crystals to positively assign the sequences of these cement molecules by X-ray diffraction.
All of the structural information obtained by X-ray diffraction analysis of HAdV corresponds to the outer and inner surfaces of the virion capsid. We still lack structural information on the dsDNA core of the virion that contains several nucleic acid associated proteins, V, VII and Mu. Unlike some other dsDNA viruses, the HAdV core lacks icosahedral symmetry thus impeding progress in structural determination by X-ray diffraction. It might be possible to obtain additional structural information on HAdV core by collecting very low resolution X-ray diffraction data [37,38] and/or by fitting the crystal structures of isolated protein V or VII into the virus core electron density. The knowledge and experience gained in solving the 3D structure of mature HAdV-5 may also facilitate structural determination of distinct HAdV serotypes as well as immature virus particles that contain unprocessed pre-proteins in their outer capsids [39,40]. Such knowledge should improve our understanding of adenovirus maturation and assembly as well as viral pathogenesis and may provide the basis for the development of safer and more efficient viral vectors.
Highlights.
X-ray structure of the entire adenovirus particle determined at 3.5 Å resolution
The penton base and fiber adopt an altered conformation in the virus.
The locations, associations and structural features of several cement proteins were revealed.
Acknowledgement
This work was supported by NIH grants AI070771 to V.S.R., HL054352 and EY011431 to G.R.N. and AI042929 to P.L.S. GM/CA CAT 23 ID-D, Argonne National Laboratories, Chicago Illinois, GUP beam time was used. GM/CA CAT has been funded in whole or partly with Federal funds from the National Cancer Institute (Y1-CO-1020) and the National Institute of General Medical Sciences (Y1-GM-1104). Use of the Advanced Photon Source was supported by the U.S. Department of Energy, Basic Energy Sciences, Office of Science, under contract No. DEAC02-06CH11357.
Footnotes
Publisher's Disclaimer: This is a PDF file of an unedited manuscript that has been accepted for publication. As a service to our customers we are providing this early version of the manuscript. The manuscript will undergo copyediting, typesetting, and review of the resulting proof before it is published in its final citable form. Please note that during the production process errors may be discovered which could affect the content, and all legal disclaimers that apply to the journal pertain.
References and recommended reading
Papers of particular interest, published within the annual period of review, have been highlighted as:
* of special interest
** of outstanding interest
- 1.Davison AJ, Benko M, Harrach B. Genetic content and evolution of adenovirus. Journal of General Virology. 2003;84:2895–2908. doi: 10.1099/vir.0.19497-0. [DOI] [PubMed] [Google Scholar]
- 2.Walsh MP, Seto J, Jones MS, Chodosh J, Xu W, Seto D. Computational analysis identifies human adenovirus type 55 as a re-emergent acute respiratory disease pathogen. J Clin Microbiol. 2010;48:991–993. doi: 10.1128/JCM.01694-09. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Horwitz MS. In: Adenoviruses. edn 3rd. Fields DMK BN, Howley PM, Chanock RM, Melnick JL, Monath TP, Roizman B, Straus SE, editors. Philadelphia: Lippincott-Raven; 1996. [Google Scholar]
- 4.Rosengart TK, Lee LY, Patel SR, Kligfield PD, Okin PM, Hackett NR, Isom OW, Crystal RG. Six-month assessment of a phase I trial of angiogenic gene therapy for the treatment of coronary artery disease using direct intramyocardial administration of an adenovirus vector expressing the VEGF121 cDNA. Ann Surg. 1999;230:466–470. doi: 10.1097/00000658-199910000-00002. discussion 470-462. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Kaplan JM. Adenovirus-based cancer gene therapy. Curr Gene Ther. 2005;5:595–605. doi: 10.2174/156652305774964677. [DOI] [PubMed] [Google Scholar]
- 6.Sullivan NJ, Geisbert TW, Geisbert JB, Xu L, Yang Zy, Roederer M, Koup RA, Jahrling PB, Nabel GJ. Accelerated vaccination for Ebola virus haemorrhagic fever in non-human primates. Nature. 2003;424:681–684. doi: 10.1038/nature01876. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Shiver JW, Emini EA. Recent advances in the development of HIV-1 vaccines using replication-incompetent adenovirus vectors. Annual Review of Immunology. 2004;55:355–372. doi: 10.1146/annurev.med.55.091902.104344. [DOI] [PubMed] [Google Scholar]
- 8.Franklin RM, Pettersson U, Akervall K, Strandberg B, Philipson L. Structural proteins of adenovirus. V. Size and structure of the adenovirus type 2 hexon. J Mol Biol. 1971;57:383–395. doi: 10.1016/0022-2836(71)90098-2. [DOI] [PubMed] [Google Scholar]
- 9.Roberts MM, White JL, GrÅtter MG, Burnett RM. Three-dimensional structure of the adenovirus major coat protein hexon. Science. 1986;232:1148–1151. doi: 10.1126/science.3704642. [DOI] [PubMed] [Google Scholar]
- 10.Rux JJ, Burnett RM. Type-specific epitope locations revealed by X-ray crystallographic study of adenovirus type 5 hexon. Mol Ther. 2000;1:18–30. doi: 10.1006/mthe.1999.0001. [DOI] [PubMed] [Google Scholar]
- 11.Stewart PL, Burnett RM, Cyrklaff M, Fuller SD. Image reconstruction reveals the complex molecular organization of adenovirus. Cell. 1991;67:145–154. doi: 10.1016/0092-8674(91)90578-m. [DOI] [PubMed] [Google Scholar]
- 12.Stewart PL, Fuller SD, Burnett RM. Difference imaging of adenovirus: Bridging the resolution gap between X-ray crystallography and electron microscopy. The EMBO Journal. 1993;12:2589–2599. doi: 10.1002/j.1460-2075.1993.tb05919.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Saban SD, Silvestry M, Nemerow GR, Stewart PL. Visualization of alpha-helices in a 6-angstrom resolution cryoelectron microscopy structure of adenovirus allows refinement of capsid protein assignments. J Virol. 2006;80:12049–12059. doi: 10.1128/JVI.01652-06. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Fabry CM, Rosa-Calatrava M, Conway JF, Zubieta C, Cusack S, Ruigrok RW, Schoehn G. A quasi-atomic model of human adenovirus type 5 capsid. The EMBO Journal. 2005;24:1645–1654. doi: 10.1038/sj.emboj.7600653. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15. Liu H, Jin L, Koh SB, Atanasov I, Schein S, Wu L, Zhou ZH. Atomic structure of human adenovirus by cryo-EM reveals interactions among protein networks. Science. 2010;329:1038–1043. doi: 10.1126/science.1187433. ** Liu et al report the the structure of adenovirus at 3.6 Å resolution using cryoelectron microscopy. The structure reveals the locations of the major capsid proteins as well as the cement proteins IIIa, VIII, and IX. Although there are differences in the virus models obtained by X-ray and cryoEM that need to be resolved, these cryoEM studies represent a tour de force in the field of virus structural biology.
- 16. Zubieta C, Schoehn G, Chroboczek J, Cusack S. The structure of the human adenovirus 2 penton. Mol Cell. 2005;17:121–135. doi: 10.1016/j.molcel.2004.11.041. * Zubieta et al report the first crystal structure of the penton base protein as an isolated protein. Removal of the N-terminal 50 residues of this protein enabled crystallization and subsequent structural analyses of its interaaction with a N-terminal fiber peptide. This paper provided a molecular description of how the fiber protein that has three fold symmetry is able to associate with the five-fold symmetrical penton base base.
- 17.van Raaij MJ, Mitraki A, Lavigne G, Cusack S. A triple beta-spiral in the adenovirus fibre shaft reveals a new structural motif for a fibrous protein. Nature. 1999;401:935–938. doi: 10.1038/44880. [DOI] [PubMed] [Google Scholar]
- 18.Grimes JM, Burroughs JN, Gouet P, Diprose JM, Malby R, Ziéntara S, Mertens PP, Stuart DI. The atomic structure of the bluetongue virus core. Nature. 1998;395:470–478. doi: 10.1038/26694. [DOI] [PubMed] [Google Scholar]
- 19.Abrescia NG, Cockburn JJ, Grimes JM, Sutton GC, Diprose JM, Butcher SJ, Fuller SD, San Martin C, Burnett RM, Stuart DI, et al. Insights into assembly from structural analysis of bacteriophage PRD1. Nature. 2004;432:68–74. doi: 10.1038/nature03056. [DOI] [PubMed] [Google Scholar]
- 20.Reinisch KM, Nibert ML, Harrison SC. Structure of the reovirus core at 3.6 A resolution. Nature. 2000;404:960–967. doi: 10.1038/35010041. [DOI] [PubMed] [Google Scholar]
- 21.Von Seggern DJ, Huang S, Fleck SK, Stevenson SC, Nemerow GR. Adenovirus vector pseudotyping in fiber-expressing cell lines: improved transduction of Epstein-Barr virus-transformed B cells. J Virol. 2000;74:354–362. doi: 10.1128/jvi.74.1.354-362.2000. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Chiu CY, Wu E, Brown SL, Von Seggern DJ, Nemerow GR, Stewart PL. Structural analysis of a fiber-pseudotyped adenovirus with ocular tropism suggests differential modes of cell receptor interactions. J Virol. 2001;75:5375–5380. doi: 10.1128/JVI.75.11.5375-5380.2001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Reddy VS, Natchiar SK, Gritton L, Mullen TM, Stewart PL, Nemerow GR. Crystallization and preliminary X-ray diffraction analysis of human adenovirus. Virology. 2010;402:209–214. doi: 10.1016/j.virol.2010.03.028. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Reddy VS, Natchiar SK, Stewart PL, Nemerow GR. Crystal structure of human adenovirus at 3.5 A resolution. Science. 2010;329:1071–1075. doi: 10.1126/science.1187292. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Wickham TJ, Mathias P, Cheresh DA, Nemerow GR. Integrins alpha v beta 3 and alpha v beta 5 promote adenovirus internalization but not virus attachment. Cell. 1993;73:309–319. doi: 10.1016/0092-8674(93)90231-e. [DOI] [PubMed] [Google Scholar]
- 26.Lindert S, Silvestry M, Mullen TM, Nemerow GR, Stewart PL. Cryo-electron microscopy structure of an adenovirus-integrin complex indicates conformational changes in both penton base and integrin. J Virol. 2009;83:11491–11501. doi: 10.1128/JVI.01214-09. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Cuillel M, Cortolezzis B, Chroboczek J, Langowski J, Ruigrok RW, Jacrot B. Purification and characterization of wild-type and ts 112 mutant protein IIIa of human adenovirus 2 expressed in Escherichia coli. Virology. 1990;175:222–231. doi: 10.1016/0042-6822(90)90202-3. [DOI] [PubMed] [Google Scholar]
- 28.Crowther RA, Franklin RM. The structure of the groups of nine hexons from adenovirus. J Mol Biol. 1972;68:181–184. doi: 10.1016/0022-2836(72)90273-2. [DOI] [PubMed] [Google Scholar]
- 29.Wiethoff CM, Wodrich H, Gerace L, Nemerow GR. Adenovirus protein VI mediates membrane disruption following capsid disassembly. J Virol. 2005;79:1992–2000. doi: 10.1128/JVI.79.4.1992-2000.2005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Mangel WF, McGrath WJ, Toledo DL, Anderson CW. Viral DNA and a viral peptide can act as cofactors of adenovirus virion proteinase activity. Nature. 1993;361:274–275. doi: 10.1038/361274a0. [DOI] [PubMed] [Google Scholar]
- 31.Wodrich H, Guan T, Cingolani G, Von Seggern D, Nemerow G, Gerace L. Switch from capsid protein import to adenovirus assembly by cleavage of nuclear transport signals. EMBO J. 2003;22:6245–6255. doi: 10.1093/emboj/cdg614. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Moyer CL, Wiethoff CM, Maier O, Smith JG, Nemerow GR. Functional genetic and biophysical analyses of membrane disruption by human adenovirus. J Virol. 2011;85:2631–2641. doi: 10.1128/JVI.02321-10. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Lehmberg E, Traina JA, Chakel JA, Chang RJ, Parkman M, McCaman MT, Murakami PK, Lahidji V, Nelson JW, Hancock WS, et al. Reversed-phase high-performance liquid chromatographic assay for the adenovirus type 5 proteome. J Chromatogr B Biomed Sci Appl. 1999;732:411–423. doi: 10.1016/s0378-4347(99)00316-3. [DOI] [PubMed] [Google Scholar]
- 34.van Oostrum J, Burnett RM. Molecular composition of the adenovirus type 2 virion. Journal of Virology. 1985;56:439–448. doi: 10.1128/jvi.56.2.439-448.1985. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Silvestry M, Lindert S, Smith JG, Maier O, Wiethoff CM, Nemerow GR, Stewart PL. Cryo-electron microscopy structure of adenovirus type 2 temperature-sensitive mutant 1 reveals insight into the cell entry defect. J Virol. 2009;83:7375–7383. doi: 10.1128/JVI.00331-09. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36. Liu H, Wu L, Zhou ZH. Model of the trimeric fiber and its interactions with the pentameric penton base of human adenovirus by cryo-electron microscopy. J Mol Biol. 2011;406:764–774. doi: 10.1016/j.jmb.2010.11.043. * Liu et al report the detailed structural analyses of the penton base fiber association in virus particles using cryoEM. These studies reveal the varied interactions between the top of the penton base and the N-terminal region of the fiber shaft domain.
- 37.Tsuruta H, Reddy VS, Wikoff WR, Johnson JE. Imaging RNA and dynamic protein segments with low-resolution virus crystallography: experimental design, data processing and implications of electron density maps. J Mol Biol. 1998;284:1439–1452. doi: 10.1006/jmbi.1998.2231. [DOI] [PubMed] [Google Scholar]
- 38.Venkataraman S, Reddy SP, Loo J, Idamakanti N, Hallenbeck PL, Reddy VS. Structure of Seneca Valley Virus-001: an oncolytic picornavirus representing a new genus. Structure. 2008;16:1555–1561. doi: 10.1016/j.str.2008.07.013. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39. Pérez-Berná AJ, Marabini R, Scheres SH, Menéndez-Conejero R, Dmitriev IP, Curiel DT, Mangel WF, Flint SJ, San Martín C. Structure and uncoating of immature adenovirus. J Mol Biol. 2009;392:547–557. doi: 10.1016/j.jmb.2009.06.057. * Perez-Berna et al reveal the structural features of a temperature sensitive adenovirus (ts1) by cryoEM that provides the tentative locations of unprocessed preproteins IIIa and VI. Interestingly, this immature form of the virus does not show significant alterations on the outer capsid surface.
- 40.Imelli N, Ruzsics Z, Puntener D, Gastaldelli M, Greber U. Genetic reconstitution of the human adenovirus type 2 temperature-sensitive 1 mutant defective in endosomal escape. Virol J. 2009;6:174. doi: 10.1186/1743-422X-6-174. [DOI] [PMC free article] [PubMed] [Google Scholar]