Abstract
A protocol to generate parameters for the AMOEBA polarizable force field for small organic molecules has been established, and polarizable atomic typing utility, Poltype, which fully automates this process, has been implemented. For validation, we have compared with quantum mechanical calculations of molecular dipole moments, optimized geometry, electrostatic potential, and conformational energy for a variety of neutral and charged organic molecules, as well as dimer interaction energies of a set of amino acid side chain model compounds. Furthermore, parameters obtained in gas phase are substantiated in liquid-phase simulations. The hydration free energy (HFE) of neutral and charged molecules have been calculated and compared with experimental values. The RMS error for the HFE of neutral molecules is less than 1 kcal/mol. Meanwhile, the relative error in the predicted HFE of salts (cations and anions) is less than 3% with a correlation coefficient of 0.95. Overall, the performance of Poltype is satisfactory and provides a convenient utility for applications such as drug discovery. Further improvement can be achieved by the systematic study of various organic compounds, particularly ionic molecules, and refinement and expansion of the parameter database.
Keywords: AMOEBA, Polarizable force field, Small molecule modeling, Poltype, Atomic typer, Molecular dynamics
1 Introduction
The classical fixed-charge molecular mechanics force field has been the conventional model to study biological macromolecular systems. However, Buckingham [1] pointed out as early as 1967 that the intermolecular forces are electrostatic in nature and can be modeled with electric multipole moments and induction. Despite investigations of various polarizable force fields [2–8], few large-scale macromolecular simulations have taken advantage of these models. Furthermore, the need for a polarizable force field has been widely acknowledged [9, 10]. Studies identify that fixed-charge force fields have difficulties with calculating the solvation free energy of polar small molecules, particular those containing hydroxyl groups [11], which are common in carbohydrates. Patel et al. [12] studied polarization in further detail and proposed the TIP4P-QDP charge-dependent polarizability water model. This work revealed the effects of polarization variability on the enhanced structure at liquid–vapor interface. Additionally, the energetics [13] as well as thermodynamics of single-atom monovalent [14, 15] and divalent [16] ions in solvent have been studied with polarizable models. The importance of polarization in protein–ligand recognition [17–19] has been identified. Although other classical force fields have been extended to include polarization interactions, such as PIPF-CHARMM [20], most polarizable force fields lack higher-order electronic moments.
The AMOEBA (Atomic multipole optimized energetics for biomolecular applications) polarizable force field is an effort to achieve chemical accuracy of conformational and interaction energies to quantum mechanical models. Moreover, AMOEBA addresses differences in systems where assuming an averaged polarization is not adequate. Its permanent point multipole up to quadrupole is capable of describing the intricate electrostatic potential surfaces. Polarization is represented with polarizable point dipoles that can fully describe the directionality of electron redistribution without the need for fictitious particles. While small molecule parameters can be obtained relatively easily for fixed-charge force fields [21, 22], adoption of a multi-pole-based force field has been limited due to the lack of automation for parameterization. Higher-order multipole moments require the choice of a local frame, and the assignment of atomic polarizabilities is necessary as well. This work articulates a procedure to generate the AMOEBA force field parameters for small molecules such as protein ligands. Additionally, a utility, Poltype, has been implemented to fully automate this procedure and is available at http://water.bme.utexas.edu/wiki/index.php/Software:Poltype. The parameters obtained from this procedure are substantiated via comparisons with quantum mechanics calculations, other molecular mechanics simulations, and experimental measurements for a range of properties.
2 Methods
2.1 AMOEBA force field
The AMOEBA force field is a polarizable molecular mechanics model that treats electrostatic interactions with higher-order moments up to quadrupoles. The potential energy model has been described previously [23–25] and is briefly explained here for reference. The potential energy function comprises bonded and non-bonded interactions. Bonded interactions include bond stretching, angle-bending, bond-angle stretch-bending, out-of-plane bending, and rotation about torsion. Non-bonded interactions include van der Waals, permanent and induced electrostatics.
Bonded interactions in the AMOEBA force field adopt non-harmonic functional forms. Bond stretch energies utilize the fourth-order Taylor expansion of the Morse potential. Bond-angle bend and torsion energies utilize a sixth-order potential and a six-term Fourier series expansion, respectively. These valence functional forms are the same as those used by the MM3 [26] classical molecular mechanics potential. Additionally, out-of-plane bending was restrained at sp2-hybridized trigonal centers with a Wilson–Decius cross-function [27].
Bond lengths, bond/torsion phase angles, and energies are in units of Å, degrees, and kcal/mol, respectively. The repulsion–dispersion interactions are represented with a buffered 14-7 potential [28].
The potential is a function of separation distance, Rij, between atoms i and j where is the minimum energy distance and is combined for heterogeneous atom pairs . In addition, the potential minimum in kcal/mol is combined for heterogeneous atom pairs .
Permanent electrostatic interactions are computed with higher-order moments where
is a multipole composed of charge, qi, dipoles, diα, and quadrupoles, Qiβγ. The interaction energy between two multipole sites is
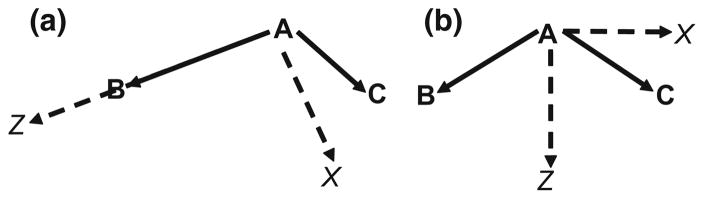
Multipoles are defined at atomic centers in relation to a local frame defined by other atoms that are bonded to it. A triplet of atoms is used to specify a local frame following the z-then-x convention. Figure 1a illustrates an example of an asymmetric local frame for atom A defined by atoms A, B, and C. The vector created by AB is the direction of the positive z-axis. The positive x-axis lies on the plane created by ABC and creates an acute angle with AC. The positive y-axis is defined to create a cubic right-handed coordinate system. Figure 1b illustrates an example of a local frame in which B and C are symmetric with respect to A, such as a water molecule. The z-axis is defined as the bisector of ∠BAC. The x-axis is defined as the vector along the aforementioned plane that creates an acute angle with AB and is orthogonal to the z-axis. As with the former case, the y-axis is defined to create a right-handed coordinate system.
Fig. 1.
Given atoms A, B, and C, the local frame is identified by the z-axis and x-axis as shown. The y-axis is defined to create a right-handed coordinate system with the existing axes
Electronic polarization describes the redistribution of electron density due to an external field. Polarizable point dipoles are utilized by AMOEBA at atomic centers to describe this effect. An iterative induction approach originally developed by Thole [29] is adopted in which an induced dipole at site i continues to polarize all other sites until convergence is achieved at all induced dipole sites. This method imposes a damped polarization interaction at very short range in order to avoid a well-known artifact of point polarizability models by smearing one of the atomic multipole moments in each pair of interaction sites [30]. The damping functions for charge, dipole, and quadrupole interactions have been derived previously [24]. The smearing function of a charge has the form
and u = rij/(αiαj)1/6 where rij is the linear separation between sites i, j and αi, αj are their corresponding atomic polarizabilities. The factor “a” is a dimensionless width parameter that determines the damping strength.
2.2 Protocol
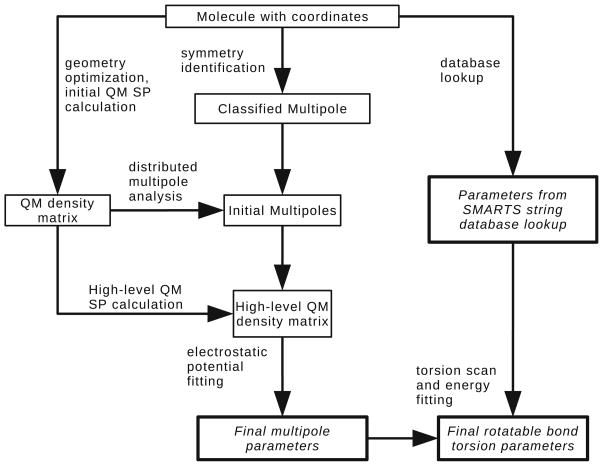
Given the structure, net charge, and multiplicity of a molecule, all parameters can be systematically determined. The AMOEBA force field requires parameters for atomic charges, dipoles, quadrupoles, polarizabilities, damping coefficients (for high valence ions only), van der Waals diameters, and well depths. Valence parameters include force constants and equilibrium values for bond lengths, angles, and torsion force constants of up to sixfold. Figure 2 depicts an overview of the parameterization process. This procedure is implemented by the Poltype polarizable atomic typing utility.
Fig. 2.
Overview of the parameterization procedure for Poltype
Prior to parameterization, the molecule with coordinates (Fig. 2) is needed. Rotatable bonds are identified about four heavy atoms in which the second and third atoms share a single bond. Bond types can be provided in the structure given to Poltype. If not assigned, atom and bond perception are performed by taking advantage of the mechanism offered by The Open Babel Package, version 2.3.0. available at http://openbabel.sourceforge.net. Symmetric multipoles are classified (Fig. 2) based on an iterative algorithm to identify graph invariant indices [31, 32] based on the maximum graph theoretical distance, heavy valence, aromaticity, ring atom, atomic number, heavy bond sum, and formal charge of an atom.
Multipoles are obtained with Stone’s distributed multi-pole analysis [33] and then refined via electrostatic potential fitting. All quantum mechanics (QM) calculations are performed with Gaussian 09 [34]. The structure is first optimized at the HF/6-31G* level. The initial QM single-point calculation (Fig. 2) then computes the electron density matrix using the MP2/6-311G** level of theory and basis set. Additionally, a grid of electrostatic potentials is populated from high-level basis set single-point calculations (Fig. 2), which may be either the MP2/6-311++G(2d, 2p) or MP2/aug-cc-pVTZ basis sets. The electrostatic potential is computed for a grid around each molecule. Grid points of four shells of increasing distance around a molecule with an offset of 1 and 0.35 Å apart are generated. The GDMA program [35] implements distributed multipole analysis [33]. It arranges multipole sites at atomic centers and analytically assigns initial multipoles (Fig. 2) based on the density matrix. In GMDA v2.2, “Switch 0” and “Radius H 0.65” are set to access the original DMA procedure. Atomic polarizabilities are assigned based solely on the element type of each atom. Polarization groups are partitioned between rotatable bonds. The final multipole parameters (Fig. 2) are further optimized by fitting to electrostatic potentials with a 0.1 kcal mol−1 electron−2 gradient convergence criteria. When there is intramolecular polarization, the electrostatic potential around the molecule is calculated from the permanent multipoles with fully induced dipoles added. Potential fitting and multipole assignment are currently based on modified utilities available in TINKER 5.1, and standalone versions of Poltype will be developed within the Force Field X (FFX) platform available at http://ffx.kenai.com/. In accordance with the solvation study conducted by Shi et al. [36], quadrupoles of hydroxyl groups are scaled by 60% after electrostatic potential fitting.
Diameter and well-depth values for van der Waals are assigned based on elements and their valence orbitals. A SMARTS string pattern was used to search for bond orders with its neighbors and assigned after a database lookup (Fig. 2). Hydrogen atoms also have a reduction factor that is based on the valence orbital of the atom to which it is bonded. Force constants for bond length, angle-bend, stretch-bend, out-of-plane bend, and torsions about non-rotatable bonds are similarly obtained from a database lookup (Fig. 2). Equilibrium values are taken from the QM optimized geometry.
Torsional parameters about rotatable bonds (Fig. 2) are obtained by comparing the conformational energy profile calculated from QM with the AMOEBA model that includes electrostatics, vdW, bonds, angles, parameters. A set of torsion parameters is identified by 4 atom classes that surround the rotatable bond and are composed of force constants for each periodicity (1–6). The dihedral angle is scanned by minimizing all torsions about the rotatable bond of interest at 30° intervals with restraints. The sixth-order Fourier series is then fit to the difference between the QM conformational energy and AMOEBA’s energy without the rotatable-bond torsion term. The QM conformational energy was obtained at the M06L/6-31G** level. Since torsion scanning gives 12 data points, no more than 8 parameters may be used to fit to the conformational energy profile. Torsions about the same central bond that are also in-phase are collapsed into one set of parameters for the fitting, and the contributions are distributed evenly among the parameters. Additionally, if a torsional parameter is greater than the difference between the maximum and minimum energy, then that parameter was omitted, and the rest of the parameters were fit again. However, if all parameters are removed after the magnitude test, the torsion parameters of only the atoms used to restrain the torsions were fitted. If more than one rotatable bond contains the same classes, the force constants of all classes are averaged.
3 Results and discussion
3.1 Monomeric comparisons
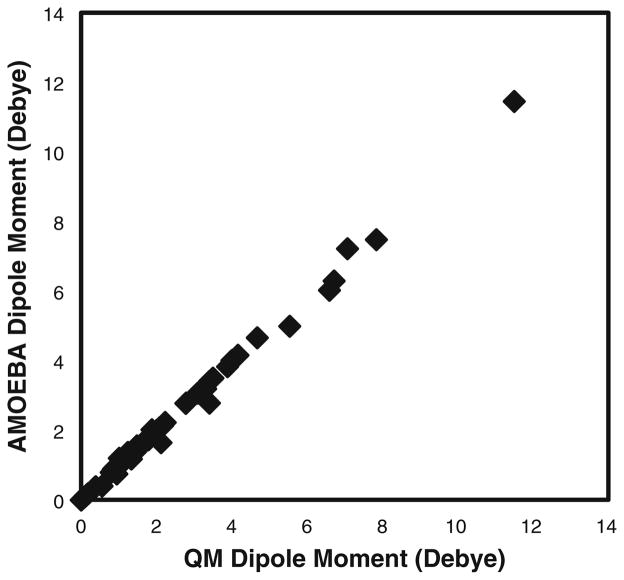
Quantum mechanics calculations provide molecular properties such as dipole moments, optimized structures, conformational energies, and electrostatic potentials of a grid around a molecule. The parameters of a diverse set of small organic molecules have been obtained using the Poltype polarizable atomic typing utility. A representative set of amino acid side-chain model compounds obtained from the Atlas of Protein Side-Chain Interactions [37, 38] was parameterized. Additionally, the parameters for a subset of small molecules, for which experimental hydration free energies are available [39, 40], were obtained as well. The full listing of dipole moments, electrostatic potential room mean square deviation, optimized structure, and conformation energies computed with Poltype/AMOEBA parameters and quantum mechanics are provided in the supplementary material. The molecular dipole moment of the optimized geometry computed using the Poltype/AMOEBA parameters compared with quantum mechanics calculations is shown in Fig. 3. The RMS error of all molecular dipoles is 0.16 Debye, and the correlation coefficient is 0.998. Molecules with particularly large dipole moment errors are anionic molecules such as CH3S-and C6H5S- with errors of 0.62 and 0.42 Debye, respectively. The molecular dipole moment from quantum mechanics calculations are 6.71 and 3.40 Debye, respectively. Although C6H5S- may not have a large relative error, it poses one of the largest absolute errors. The electrostatic potential (ESP) RMS difference of a grid of point charges around a molecule is 0.16 kcal/mol, and the molecules with the largest errors follow the same trend as that for dipoles. The average RMS distance between optimized geometries from Poltype/AMOEBA molecules and those optimized from quantum mechanics is 0.08 Å. For conformational energies, the correlation of all conformations prior to torsional fitting about rotatable bonds yielded a 3.6 kcal/mol RMS deviation from QM and a 0.13 correlation coefficient. After fitting, the RMS deviation decreased to 1.24 kcal/mol with a 0.91 correlation coefficient.
Fig. 3.
Molecular dipole moment computed from AMOEBA parameters and quantum mechanical calculations
3.2 Dimer calculations
The packing of side chains makes significant contribution to protein stability. Investigation of interactions between side-chain model compounds is commonly used for evaluating the potential energy models. Typically, fixed-charge potential energy models are not able to accurately reproduce both gas-phase and solution-phase properties. However, AMOEBA aims to capture the energetics in different environments by including explicit polarization effects. The aforementioned Atlas of Protein Side-Chain Interactions [37, 38] were compiled by clustering interacting side chain pair conformations in 2,548 non-homologous protein structures from the Protein Data Bank. The geometry of the top cluster from the side chain pairs was used for dimer calculations. As the atlas only contains the conformation of heavy atoms, the systems were prepared [41] by adding hydrogen atoms to each model compound, and then each pair was optimized at the DFT/TZVP level. Heavy atoms were held fixed during optimization. Table 1 shows the interaction energy of the most common dimer configurations calculated with AMOEBA compared with other QM and molecular mechanics methods. Amino acids are identified by their standard abbreviations. Charged residues that are neutralized have an “(N)” designation. Interactions calculated with CCSD(T)/CBS [42–44] are considered reference energies. The OPLS-AA/L [45] and Amber parm03 [46] are energies computed with fixed-charge force fields and were taken directly from a study by Berka and coworkers [41]. Note that typically fixed-charge force fields use “enhanced” atomic charges for condensed-phase modeling such that comparison with gas-phase QM is not entirely useful. The DFT/TZVP and RI-MP2/aVTZ are quantum mechanics calculations used for comparison.
Table 1.
Interaction energies (kcal/mol) for amino acid pairs (identified by their standard abbreviations) calculated using several approaches in the gas phase
| Dimera | CCSD(T) CBSb | Poltype AMOEBA | OPLS-AA/Lc | parm03d | DFT TZVP | RI-MP2 aVTZ |
|---|---|---|---|---|---|---|
| RD | −110.80 | −100.33 | −105.71 | −90.37 | −110.60 | −110.21 |
| KE | −108.40 | −104.86 | −106.02 | −103.57 | −108.27 | −107.75 |
| DH(N) | −30.64 | −28.15 | −12.20 | −22.36 | −28.83 | −30.91 |
| D(N)H(N) | −17.97 | −15.10 | −10.90 | −7.80 | −16.26 | −17.94 |
| R(N)D(N) | −16.32 | −12.38 | −8.94 | – | −14.71 | −15.92 |
| K(N)E(N) | −10.76 | −9.54 | −8.80 | −9.11 | −9.81 | −10.65 |
| QN | −7.37 | −4.86 | −8.61 | −8.84 | −5.66 | −6.92 |
| TT | −6.50 | −7.99 | −7.96 | −6.83 | −4.81 | −6.28 |
| YY | −4.66 | −5.41 | −3.84 | −3.62 | 1.35 | −5.51 |
| TS | −4.50 | −5.15 | −4.38 | −4.40 | −3.36 | −4.30 |
| LW | −4.04 | −4.16 | −3.46 | −3.46 | 1.00 | −4.74 |
| YP | −3.79 | −3.83 | −3.05 | −3.09 | 0.44 | −4.11 |
| FF | −2.33 | −2.41 | −1.97 | −2.26 | 1.11 | −3.04 |
| MM | −2.03 | −1.95 | −3.14 | −2.35 | 1.22 | −2.01 |
| LY | −1.72 | −1.72 | −1.86 | −1.52 | 0.96 | −1.66 |
| LL | −1.62 | −1.60 | −1.40 | −1.66 | 0.00 | −1.60 |
| MC | −1.46 | −1.28 | −2.01 | −1.20 | 0.25 | −1.43 |
| VV | −1.39 | −1.52 | −1.36 | −1.43 | 0.44 | −1.28 |
| IL | −1.39 | −1.36 | −1.19 | −1.41 | 0.06 | −1.35 |
| II | −1.24 | −1.22 | −1.13 | −1.20 | 0.62 | −1.11 |
| LT | −1.09 | −1.11 | −0.91 | −1.05 | 0.02 | −1.02 |
| VL | −1.08 | −1.06 | −0.81 | −1.11 | 0.11 | −1.01 |
| AL | −1.07 | −1.11 | −1.00 | −0.94 | 0.71 | −0.93 |
| LG | −0.77 | −0.75 | −0.75 | −0.53 | −0.09 | −0.71 |
| MRE [%] | 8.69 | 19.54 | 13.55 | 83.61 | 6.52 | |
| MRX [%] | 34.01 | 60.19 | 56.58 | 166.28 | −30.62 | |
| MAE | 1.28 | 2.11 | 2.22 | 2.03 | 0.26 | |
| MAX | 10.47 | 18.44 | 20.43 | 6.01 | −0.85 | |
| RMS | 2.61 | 4.16 | 4.78 | 1.4 | 0.36 |
Interaction energies computed with the CCSD(T) level of theory extrapolated to the complete basis set limit (CBS) is used as the reference method. Interaction energies calculated with the AMOEBA force field parameterized with Poltype are performed as a part of this work. The DFT method was carried out with the TPSS functional and TZVP basis. The aug-cc-pVTZ basis set and resolution of identity approximation was used for the MP2 method. MRE is the unsigned mean relative error (%), MRX is the signed maximal relative error (%), MAE is the unsigned mean absolute error, MAX is the signed maximal absolute error, and RMS is the signed root mean square error
Other than Poltype/AMOEBA, interaction energies were calculated by (Berka [41])
Interaction energy computed using OPLS-AA/L force field (Kaminski [45])
Interaction energy computed using parm03 force field (Duan [46])
Overall, the mean relative error (MRE) and maximal relative error (MRX) of interaction energies computed with parameters from Poltype for the AMOEBA force field are lower than errors of other force fields as well as DFT, but comparable to RI-MP2 results. The mean absolute error (MAE), maximal absolute error (MAX), and root mean square error (RMS) are also lower than other molecular mechanics methods, but worse than DFT. Interestingly, the MRX of all molecular mechanical methods perform better than DFT, as the latter shows significant “relative” errors for weak associating dimers. Similarly, AMOEBA and the fixed-charge force fields yield a lower MAE compared to DFT. However, when an empirical dispersion function was incorporated in the DFT method [47, 48], the interaction energy prediction improves significantly [41]. RI-MP2 performs remarkably well in producing accurate interaction energies when compared to CCSD(T)/CBS.
The charged pairs arginine–aspartate and lysine–glutamate (RD and KE) seem to be the source of the largest absolute error for all molecular mechanics methods. However, the relative error of the RD pair was less than 10% for Poltype/AMOEBA and OPLS-AA/L with a larger error for Amber parm03. The relative error for the KE pair was less than 5% for AMOEBA and OPLS force fields. Additionally, an SAPT decomposition of the KE interaction reveals that higher-order energy beyond first-order electrostatics and repulsion and second-order induction and dispersion stabilizes the pair by about 3 kcal/mol. Conversely, higher-order energy stabilizes the RD pair by more than 6 kcal/mol and suggests that the difficulty with this pair may be due to interactions not captured by the energy function of molecular mechanics models.
Pairs with polar residues yield lower absolute error for Poltype/AMOEBA. However, it should be noted that the conformation of residues such as aspartic acid may be artificial due to the system preparation described above. Since all geometries chosen for the aspartic acid have C–O bond lengths in a narrow range between 1.24 and 1.25 Å, this geometry only corresponds to the COO–carboxylate ion [49, 50]. Typically, protonated carboxylic acid exhibits asymmetric bond lengths of 1.31 and 1.2 Å. Since geometries in the current test set are obtained from PDB structures and then minimized with heavy atoms fixed and hydrogen atoms added, pairs with artificially protonated carboxylic acid such as D(N)H(N) do not accurately describe electron distributions of charged carboxylate nor neutral carboxylic acid. Reassignment of multipoles with the artificial structure indeed yields an interaction energy of the D(N)H(N) pair that more closely matches the reference energy of the structure. The R(N)D(N) pair exhibits a similar sensitivity to geometry in which an assignment of multipoles with the given structure yields the error in Table 1, but the assignment of multipoles with a monomer-optimized structure further underestimated the interaction energy by ~3 kcal/mol. These examples suggest that the electron distribution of unphysical structures, particularly protonation states that are incompatible with its heavy atom conformation, cannot be captured by molecular mechanical models including AMOEBA. The parameterization of molecular mechanics models is based on minimum energy structures as it is unlikely for simple classical mechanical model to capture the complete potential energy surface especially when the structures deviate significantly from the local minima and “chemical” changes are involved. Nonetheless, dimer interaction energy calculations provide insight into the non-covalent interactions of a system and are conducive to the development of a force field. This is particularly true for AMOEBA since polarization allows parameters to be transferable between gas- and condensed-phase without the need to “pre-polarize” and scale up partial charges. Moreover, other workers [51] support the proficiency of AMOEBA in predicting interaction energies of fragment pairs decomposed from the HIV-II protease crystal structure and show improvement over other classical molecular mechanics models.
3.3 Solvation
The thermodynamic properties of molecules developed with Poltype are studied and compared with experimental values. The parameters of several families of small molecules containing functional groups in drug-like molecules were obtained for AMOEBA using Poltype, and their hydration free energies (HFE) are computed with the Bennett acceptance ratio (BAR) [52]. In a similar procedure as a previous AMOEBA HFE study [36], perturbations of the solute required the decoupling of electrostatic and van der Waals interactions. The perturbation of electrostatic atomic multipoles and polarizabilities was scaled down linearly with λ = (1.0, 0.9, 0.8, 0.7, 0.6, 0.5, 0.4, 0.3, 0.2, 0.1, and 0.0). We also scale down the radius and well depth of vdW interactions linearly with λ = (1.0, 0.9, 0.8, 0.75, 0.7, 0.65, 0.6, 0.5, 0.4, 0.2, and 0.0). Molecular dynamics in solvent were carried out by placing the solute molecule at the origin of a cubic, pre-equilibrated, 28.78-Å periodic box containing 800 water molecules. The system was then equilibrated for 50 ps at 298 K. For each perturbation step, 500-ps molecular dynamics simulations were performed with 1 fs time steps and vdW cutoff of 12 Å at 298 K constant temperature using the Berendsen thermostat [53]. The long-range electrostatics for all the systems were treated using particle mesh Ewald (PME) summation [54–56]. The atomic coordinates at every 500 fs were used for post-analysis except for first 100 ps simulation. Gas-phase simulations were run on the single solute molecule for 50 ps with a time step of 0.1 ps at 298 K using a stochastic thermostat. Atomic coordinates at every 100 fs were used for post-analysis. Previously, Mobley et al. [57] conducted a study to compute HFE for a larger set of molecules with the fixed-charge general Amber force field (GAFF) [58] by assigning AM1-BCC partial charges [8, 59]. Hydration free energies with the AMOEBA force field, GAFF, and experimental results [39, 40] are listed in Table 2. Included in the table is also the free energy difference observed, while electrostatics and van der Waals interactions are perturbed. The RMS error of HFE with Poltype/AMOEBA for the set of molecules in this study is 0.75 kcal/mol. Although previous work with a larger set of molecules with GAFF yielded a lower RMS error, the error for the set in this study is 1.56 kcal/mol. When families of molecules are considered, alkenes have errors of ~0.6 kcal/mol, while the errors for GAFF are ~1 kcal/mol. Similarly, HFE predicted by Poltype/AMOEBA of nitro-containing molecules consistently yield lower errors than GAFF.
Table 2.
Hydration free energies (kcal/mol) of small molecules obtained from experiment, Poltype/AMOEBA, and general Amber force field (GAFF) are shown in bold. The free energy differences as a result of scaling electrostatics and van der Waals, and the errors from the experimental value for Poltype/AMOEBA and GAFF are shown as well
| Molecule name | Expa | Poltype/AMOEBA
|
GAFFb |
||||||
|---|---|---|---|---|---|---|---|---|---|
| ΔGexp | ΔGele | ΔGvdw | ΔGAMOEBA | ErrorAMOEBA | ΔGele | ΔGvdw | ΔGGAFF | Error GAFF | |
| 2 Methylbut-2–ene | 1.31 | −1.78 | 2.51 | 0.72 | −0.59 | −0.55 | 2.83 | 2.28 | 0.97 |
| But-1-ene | 1.38 | −1.65 | 2.48 | 0.83 | −0.55 | −0.37 | 2.85 | 2.48 | 1.10 |
| 1-Nitrobutane | −3.09 | −4.50 | 1.95 | −2.55 | 0.54 | −2.43 | 0.92 | −1.51 | 1.58 |
| 2-Nitrophenol | −4.58 | −5.43 | 1.24 | −4.19 | 0.39 | −5.40 | 0.06 | −5.34 | −0.76 |
| 4-Nitrophenol | −10.64 | −11.09 | 1.49 | −9.61 | 1.03 | −8.04 | −0.18 | −8.22 | 2.42 |
| 22-Dimethylbutane | 2.51 | −0.75 | 2.44 | 1.70 | −0.81 | 0.01 | 2.52 | 2.53 | 0.02 |
| n-Octane | 2.88 | −1.37 | 3.11 | 1.74 | −1.14 | 0.01 | 3.12 | 3.13 | 0.25 |
| 23-Dimethylphenol | −6.16 | −7.24 | 2.28 | −4.96 | 1.20 | −6.49 | 1.82 | −4.67 | 1.49 |
| Tert-butylbenzene | −0.44 | −3.54 | 2.21 | −1.33 | −0.89 | −2.98 | 2.56 | −0.42 | 0.02 |
| 3-Chloropyridine | −4.01 | −5.42 | 1.40 | −4.02 | −0.01 | −3.78 | 1.28 | −2.50 | 1.51 |
| Di-n-butylamine | −3.24 | −6.86 | 3.28 | −3.58 | −0.34 | −4.71 | 3.08 | −1.63 | 1.61 |
| Di-n-propyl ether | −1.16 | −5.41 | 2.71 | −2.70 | −1.54 | −2.67 | 2.88 | 0.21 | 1.37 |
| Methyl isopropyl ether | −2.01 | −5.37 | 2.62 | −2.75 | −0.74 | −2.89 | 2.14 | −0.75 | 1.26 |
| Di-n-propyl sulfide | −1.28 | −4.16 | 2.35 | −1.81 | −0.53 | −2.15 | 2.64 | 0.49 | 1.77 |
| Dimethyl disulfide | −1.83 | −3.22 | 1.99 | −1.23 | 0.60 | −0.72 | 2.20 | 1.48 | 3.31 |
| Isobutyraldehyde | −2.86 | −5.29 | 2.17 | −3.12 | −0.26 | −4.98 | 2.05 | −2.93 | −0.07 |
| Propionaldehyde | −3.43 | −5.26 | 1.85 | −3.41 | 0.02 | −5.06 | 1.98 | −3.08 | 0.35 |
| Methanethiol | −1.24 | −2.85 | 2.02 | −0.83 | 0.41 | −2.25 | 1.99 | −0.26 | 0.98 |
| n-Butanethiol | 20.99 | −3.77 | 2.10 | −1.68 | −0.69 | −2.39 | 2.27 | −0.12 | 0.87 |
| Methyl acetate | −3.13 | −6.20 | 2.01 | −4.19 | −1.06 | −5.44 | 1.71 | −3.73 | −0.60 |
| Oct-1-yne | 0.71 | −2.40 | 2.94 | 0.54 | −0.17 | −0.83 | 3.29 | 2.46 | 1.75 |
| Pent-1-yne | 0.01 | −2.37 | 2.39 | 0.02 | 0.01 | −0.81 | 2.74 | 1.93 | 1.92 |
| Octan-1-ol | −4.09 | −7.87 | 2.68 | −5.20 | −1.11 | −5.13 | 2.48 | −2.65 | 1.44 |
| p-Dibromobenzene | −2.30 | −3.7 | 2.22 | −1.48 | 0.82 | −1.70 | 1.69 | −0.01 | 2.29 |
| Tribromomethane | −2.13 | −3.82 | 2.45 | −1.37 | 0.76 | −0.70 | 1.58 | 0.88 | 3.01 |
However, GAFF had errors of lower magnitude than Poltype/AMOEBA for two alkanes (22-dimethylbutane and n-octane). It should be noted that the free energy differences observed for these molecules due to van der Waals perturbations are consistent between AMOEBA and GAFF. It seems that electrostatics is too “attractive” in AMOEBA. The source of error in AMOEBA electrostatics is likely due to the ESP optimization procedure. For example, in the fitting process of n-octane, there are significantly changes in the quadrupoles of the terminal hydrogen atoms, which are shared by 6 atoms. Large deviations from those obtained from DMA may result in unphysical electrostatic parameters. Relaxing the convergence criteria of ESP fit-ting from 0.1 to 0.5 kcal mol−1 electron−2 gradient convergence criteria or moving the grid points away from the vdW surface may prevent unphysical multipoles resulting from the optimization. We are currently investigating this procedure especially for large linear molecules. Additionally, care must be taken when defining polarization groups. Some families such as aldehydes cannot be partitioned between carbonyl C=O and its neighboring heaving atom. When the groups are inappropriately partitioned across the bond, errors in HFE prediction increase to 1.53 and 1.55 kcal/mol for isobutyraldehyde and propionaldehyde, respectively.
Additionally, the hydration of ionic molecules is studied. For this preliminary study, a generic scaling factor for formally charged atoms was applied. For hydrogen atoms bonded to atoms with positive formal charge, their vdW diameters were scaled down by 10% from their original parameters. Conversely, the vdW diameters of atoms with a formal negative charge were scaled up by 10%. These scaling methods will be further investigated and refined. Simulations details are the same as those of neutral solutes. It should be noted, though, that difficulties arise in the comparison with experiments of homogenous ions as they are not directly accessible and must be conducted in salt solutions. The contributions of anions and cations then must be determined through various schemes such as self-consistent thermodynamic analysis [60], the TATB assumption [61], or the cluster-pair approximation [62]. The experimental hydration data that we compare with here apply the cluster-pair approximation, which is based on the correlation between ion–water clustering data and aqueous solvation free energies of neutral ion pairs. However, when comparing ion solvation quantities, differences between experimental methods and measurements should be taken into consideration. Therefore, the comparison of an anion–cation salt pair is more appropriate than single ions alone, if the HFE of the pair has been determined in a consistent manner. Experimental hydration free energy of single ions and salt and corresponding energies using Poltype/AMOEBA are shown in Table 3. The salt HFE of an anionic molecule is taken here to be the sum of HFE of the molecule and the sodium cation [63, 64]. Similarly, the salt HFE of a cationic molecule is taken to be the sum of the HFE of the molecule and the chlorine anion [63, 64]. The correlation coefficient between salt HFE obtained from experimental and Poltype/AMOEBA is 0.95, and the unsigned mean relative error is less than 3%. Phosphor and sulfur containing cationic molecules produced salt HFE that agreed well with experiment. Some of the largest errors come from the oxonium cations. As mentioned previously, a simple scaling has been applied to all atoms with a formal negative charge or hydrogen atoms bonded to atoms with a positive charge. Further investigation of accurate ion parameters is required.
Table 3.
Hydration free energy (kcal/mol) of ionic molecules and their corresponding salt
| Molecule | ΔGexp | ΔGsalt exp | ΔGAMOEBA | ΔGsalt AMOEBA |
|---|---|---|---|---|
| (CH3)2PH2+ | −57 | −131.5 | −47.87 | −134.37 |
| CH3PH3+ | −63 | −137.5 | −51.80 | −138.30 |
| (CH3)2SH+ | −64.5 | −139 | −51.53 | −138.03 |
| CH3SH2+ | −74 | −148.5 | −57.54 | −144.04 |
| CH3NH3+ | −76.4 | −150.9 | −68.16 | −154.66 |
| (CH3)2NH2+ | −68.6 | −143.1 | −63.01 | −149.51 |
| HC(OH)NH2+ | −78 | −152.5 | −67.31 | −153.81 |
| C6H5NH3+ | −72.4 | −146.9 | −63.15 | −149.65 |
| C5H5NH+ | −58 | −132.5 | −52.19 | −138.69 |
| imidazoleH+ | −64 | −138.5 | −54.45 | −140.95 |
| CH3C(OH)CH3+ | −77.1 | −151.6 | −53.57 | −140.07 |
| (CH3)2OH+ | −79.7 | −154.2 | −59.01 | −145.51 |
| CH3CH2OH2+ | −88.4 | −162.9 | −68.32 | −154.82 |
| CH3OH2+ | −93 | −167.5 | −73.75 | −160.25 |
| CH3O− | −95 | −198.2 | −105.67 | −197.47 |
| HCO2− | −76.2 | −179.4 | −91.27 | −183.07 |
| CH3S− | −73.8 | −177 | −85.13 | −176.93 |
| C6H5S− | −63.4 | −166.6 | −67.25 | −159.05 |
4 Conclusions
A protocol to develop AMOEBA models for small molecules has been established. In this work, we have described a standard approach to generate the AMOEBA force field for small and drug-like molecules. Although the parameterization process for the AMOEBA polarizable force field requires more sophistication compared to the process for fixed-charge force fields, a straightforward procedure is described here. Additionally, the Poltype utility allows one to generate the AMOEBA polarizable force field for a small molecule in a fully automated manner. We have shown good agreement with quantum mechanics measurements in gas phase for monomers and dimers for neutral as well as charged molecules. Furthermore, parameters obtained in gas phase are transferred directly to liquid-phase systems without modification. The hydration free energy (HFE) of neutral and charged molecules have been calculated with the Bennett acceptance ratio and compared with experimental values. The RMS error for the HFE of neutral molecules is less than 1 kcal/mol, while the unsigned mean relative error is less than 3% and a correlation coefficient is 0.95 for the HFE of salts containing charged molecules. Although some assignments such as the van der Waals diameters of atoms with formal charge of ionic molecules need further investigation, Poltype readily facilitates the systematic study of these chemical functional groups. Since the requirement to perform quantum mechanical may be computationally demanding, future work of parameterization for the AMOEBA force field would be to develop a semi-empirical method such as AM1-BCC [8, 59] to assign atomic multipoles. Although the HFE RMS error of the neutral molecules in this study demonstrate improved solvation energies than fixed-charge force fields, confirmation with a more extensive dataset is still necessary. We believe the advantage of polarizable force fields such as AMOEBA will be further illustrated in processes where environmental changes are involved.
Supplementary Material
Acknowledgments
This research was supported by grants from the National Institute of General Medical Sciences (R01GM079686) and the Robert A. Welch Foundation (F-1691) to P.R.
Abbreviations
- AMOEBA
Atomic multipole optimized energetics for biomolecular applications
- DMA
Distributed multipole analysis
- HFE
Hydration free energy
- ESP
Electrostatic potential
Footnotes
Electronic supplementary material The online version of this article (doi:10.1007/s00214-012-1138-6) contains supplementary material, which is available to authorized users.
References
- 1.Buckingham AD. Permanent and induced molecular moments and long-range intermolecular forces. Adv Chem Phys. 1967;12:107–142. [Google Scholar]
- 2.Jensen L, Astrand PO, Osted A, Kongsted J, Mikkelsen KV. Polarizability of molecular clusters as calculated by a dipole interaction model. J Chem Phys. 2002;116(10):4001–4010. [Google Scholar]
- 3.Gresh N, Cisneros GA, Darden TA, Piquemal JP. Anisotropic, polarizable molecular mechanics studies of inter- and intramoecular interactions and ligand-macromolecule complexes. A bottom-up strategy. J Chem Theory Comput. 2007;3(6):1960–1986. doi: 10.1021/ct700134r. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Piquemal JP, Williams-Hubbard B, Fey N, Deeth RJ, Gresh N, Giessner-Prettre C. Inclusion of the ligand field contribution in a polarizable molecular mechanics: SIBFA-LF. J Comput Chem. 2003;24(16):1963–1970. doi: 10.1002/jcc.10354. [DOI] [PubMed] [Google Scholar]
- 5.Rick SW, Stuart SJ, Berne BJ. Dynamical fluctuating charge force-fields: application to liquid water. J Chem Phys. 1994;101(7):6141–6156. [Google Scholar]
- 6.Xie W, Orozco M, Truhlar DG, Gao J. X-Pol potential: an electronic structure-based force field for molecular dynamics simulation of a solvated protein in water. J Chem Theory Comput. 2009;5(3):459–467. doi: 10.1021/ct800239q. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Senn HM, Thiel W. QM/MM methods for biomolecular systems. Angewandte Chemie-Int Ed. 2009;48(7):1198–1229. doi: 10.1002/anie.200802019. [DOI] [PubMed] [Google Scholar]
- 8.Jakalian A, Jack DB, Bayly CI. Fast, efficient generation of high-quality atomic charges. AM1-BCC model: II. Parameterization and validation. J Comput Chem. 2002;23(16):1623–1641. doi: 10.1002/jcc.10128. [DOI] [PubMed] [Google Scholar]
- 9.Cieplak P, Dupradeau FY, Duan Y, Wang JM. Polarization effects in molecular mechanical force fields. J Phys Condens Matter. 2009;21(33):333102. doi: 10.1088/0953-8984/21/33/333102. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Lopes PEM, Roux B, MacKerell AD. Molecular modeling and dynamics studies with explicit inclusion of electronic polarizability: theory and applications. Theor Chem Acc. 2009;124(1–2):11–28. doi: 10.1007/s00214-009-0617-x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Klimovich PV, Mobley DL. Predicting hydration free energies using all-atom molecular dynamics simulations and multiple starting conformations. J Comput-Aided Mol Des. 2010;24(4):307–316. doi: 10.1007/s10822-010-9343-7. [DOI] [PubMed] [Google Scholar]
- 12.Bauer BA, Warren GL, Patel S. Incorporating phase-dependent polarizability in nonadditive electrostatic models for molecular dynamics simulations of the aqueous liquid-vapor interface. J Chem Theory Comput. 2009;5(2):359–373. doi: 10.1021/ct800320f. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Cezard C, Bouvier B, Brenner V, Defranceschi M, Millie P, Soudan JM, Dognon JP. Theoretical investigation of small alkali cation-molecule: a model potential approach. J Phys Chem B. 2004;108(4):1497–1506. [Google Scholar]
- 14.Jiao D, King C, Grossfield A, Darden TA, Ren PY. Simulation of Ca2+ and Mg2+ solvation using polarizable atomic multipole potential. J Phys Chem B. 2006;110(37):18553–18559. doi: 10.1021/jp062230r. [DOI] [PubMed] [Google Scholar]
- 15.Yu HB, Whitfield TW, Harder E, Lamoureux G, Vorobyov I, Anisimov VM, MacKerell AD, Roux B. Simulating monovalent and divalent ions in aqueous solution using a drude polarizable force field. J Chem Theory Comput. 2010;6(3):774–786. doi: 10.1021/ct900576a. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Wu JC, Piquemal JP, Chaudret R, Reinhardt P, Ren PY. Polarizable molecular dynamics simulation of Zn(II) in water using the AMOEBA force field. J Chem Theory Comput. 2010;6(7):2059–2070. doi: 10.1021/ct100091j. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Jiao D, Golubkov PA, Darden TA, Ren P. Calculation of protein-ligand binding free energy by using a polarizable potential. Proc Natl Acad Sci USA. 2008;105(17):6290–6295. doi: 10.1073/pnas.0711686105. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Jiao D, Zhang JJ, Duke RE, Li GH, Schnieders MJ, Ren PY. Trypsin-ligand binding free energies from explicit and implicit solvent simulations with polarizable potential. J Comput Chem. 2009;30(11):1701–1711. doi: 10.1002/jcc.21268. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Shi Y, Jiao DA, Schnieders MJ, Ren PY. Trypsin-ligand binding free energy calculation with AMOEBA. EMBC 2009 Ann Int Conf IEEE Eng Med Biol Soc. 2009;1–20:2328–2331. doi: 10.1109/IEMBS.2009.5335108. http://dx.crossref.org/10.1109%2FIEMBS.2009.5335108. [DOI] [PMC free article] [PubMed]
- 20.Xie WS, Pu JZ, MacKerell AD, Gao JL. Development of a polarizable intermolecular potential function (PIPF) for liquid amides and alkanes. J Chem Theory Comput. 2007;3(6):1878–1889. doi: 10.1021/ct700146x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Wang JM, Wang W, Kollman PA, Case DA. Automatic atom type and bond type perception in molecular mechanical calculations. J Mol Graphics Model. 2006;25(2):247–260. doi: 10.1016/j.jmgm.2005.12.005. [DOI] [PubMed] [Google Scholar]
- 22.Moriarty NW, Grosse-Kunstleve RW, Adams PD. Electronic ligand builder and optimization workbench (eLBOW): a tool for ligand coordinate and restraint generation. Acta Crystallogr Sect D Biol Crystallogr. 2009;65:1074–1080. doi: 10.1107/S0907444909029436. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Ren PY, Ponder JW. Consistent treatment of inter- and intramolecular polarization in molecular mechanics calculations. J Comput Chem. 2002;23(16):1497–1506. doi: 10.1002/jcc.10127. [DOI] [PubMed] [Google Scholar]
- 24.Ren PY, Ponder JW. Polarizable atomic multipole water model for molecular mechanics simulation. J Phys Chem B. 2003;107(24):5933–5947. [Google Scholar]
- 25.Ponder JW, Wu CJ, Ren PY, Pande VS, Chodera JD, Schnieders MJ, Haque I, Mobley DL, Lambrecht DS, DiStasio RA, Head-Gordon M, Clark GNI, Johnson ME, Head-Gordon T. Current Status of the AMOEBA polarizable force field. J Phys Chem B. 2010;114(8):2549–2564. doi: 10.1021/jp910674d. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Allinger NL, Yuh YH, Lii JH. Molecular mechanics: the MM3 force-field for hydrocarbons.1. J Am Chem Soc. 1989;111(23):8551–8566. [Google Scholar]
- 27.Wilson EB, Decius JC, Cross PC. Molecular vibrations: the theory of infrared and raman vibrational spectra. McGraw-Hill; New York: 1955. [Google Scholar]
- 28.Halgren TA. Representation of vanderwaals (Vdw) interactions in molecular mechanics force-fields: potential form, combination rules, and Vdw parameters. J Am Chem Soc. 1992;114(20):7827–7843. [Google Scholar]
- 29.Thole BT. Molecular polarizabilities calculated with a modified dipole interaction. Chem Phys. 1981;59(3):341–350. [Google Scholar]
- 30.Burnham CJ, Li JC, Xantheas SS, Leslie M. The parametrization of a Thole-type all-atom polarizable water model from first principles and its application to the study of water clusters (n = 2–21) and the phonon spectrum of ice Ih. J Chem Phys. 1999;110(9):4566–4581. [Google Scholar]
- 31.Weininger D. Smiles, a chemical language and information-system.1. Introduction to methodology and encoding rules. J Chem Inf Comput Sci. 1988;28(1):31–36. [Google Scholar]
- 32.Morgan HL. The generation of a unique machine description for chemical structures-a technique developed at chemical abstracts service. J Chem Document. 1965;5(2):107–113. [Google Scholar]
- 33.Stone AJ, Alderton M. Distributed multipole analysis: methods and applications. Mol Phys. 1985;56(5):1047–1064. [Google Scholar]
- 34.Frisch MJ, Trucks GW, Schlegel HB, Scuseria GE, Robb MA, Cheeseman JR, Scalmani G, Barone V, Mennucci B, Petersson GA, Nakatsuji H, Caricato M, Li X, Hratchian HP, Izmaylov AF, Bloino J, Zheng G, Sonnenberg JL, Hada M, Ehara M, Toyota K, Fukuda R, Hasegawa J, Ishida MTN, Honda Y, Kitao HNO, Vreven T, Montgomery JAJ, Peralta JE, Ogliaro F, Bearpark M, Heyd JJ, Brothers E, Kudin KN, Staroverov VN, Kobayashi R, Normand J, Raghavachari K, Rendell A, Burant JC, Iyengar SS, Tomasi J, Cossi M, Rega N, Millam JM, Klene M, Knox JE, Cross JB, Bakken V, Adamo C, Jaramillo J, Gomperts R, Stratmann RE, Yazyev O, Austin AJ, Cammi R, Pomelli C, Ochterski JW, Martin RL, Morokuma K, Zakrzewski VG, Voth GA, Sal-vador P, Dannenberg JJ, Dapprich S, Daniels AD, Farkas Ö, Foresman JB, Ortiz JV, Cioslowski J, Fox DJ. Gaussian 09. Revision D.1 edn. Gaussian, Inc; Wallingford CT: 2009. [Google Scholar]
- 35.Stone AJ. Distributed multipole analysis: stability for large basis sets. J Chem Theory Comput. 2005;1(6):1128–1132. doi: 10.1021/ct050190+. [DOI] [PubMed] [Google Scholar]
- 36.Shi Y, Wu CJ, Ponder JW, Ren PY. Multipole electrostatics in hydration free energy calculations. J Comput Chem. 2011;32(5):967–977. doi: 10.1002/jcc.21681. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Singh J, Thornton JM. Atlas of protein side-chain interactions. I & II. IRL press; Oxford: 1992. [Google Scholar]
- 38.Thornton JM. 2009 http://www.ebi.ac.uk/thornton-srv/databases/sidechains/
- 39.Abraham MH, Whiting GS, Fuchs R, Chambers EJ. Thermodynamics of solute transfer from water to hexadecane. J Chem Soc Perkin Trans. 1990;2(2):291–300. [Google Scholar]
- 40.Chambers CC, Hawkins GD, Cramer CJ, Truhlar DG. Model for aqueous solvation based on class IV atomic charges and first solvation shell effects. J Phys Chem. 1996;100(40):16385–16398. [Google Scholar]
- 41.Berka K, Laskowski R, Riley KE, Hobza P, Vondrasek J. Representative amino acid side chain interactions in proteins. A comparison of highly accurate correlated ab initio quantum chemical and empirical potential procedures. J Chem Theory Comput. 2009;5(4):982–992. doi: 10.1021/ct800508v. [DOI] [PubMed] [Google Scholar]
- 42.Tsuzuki S, Honda K, Uchimaru T, Mikami M. Ab initio calculations of structures and interaction energies of toluene dimers including CCSD(T) level electron correlation correction. J Chem Phys. 2005;122(14):144323. doi: 10.1063/1.1876092. [DOI] [PubMed] [Google Scholar]
- 43.Sinnokrot MO, Sherrill CD. Highly accurate coupled cluster potential energy curves for the benzene dimer: Sandwich, T-shaped, and parallel-displaced configurations. J Phys Chem A. 2004;108(46):10200–10207. [Google Scholar]
- 44.Hobza P, Sponer J. Structure, energetics, and dynamics of the nucleic Acid base pairs: nonempirical ab initio calculations. Chem Rev. 1999;99(11):3247–3276. doi: 10.1021/cr9800255. [DOI] [PubMed] [Google Scholar]
- 45.Kaminski GA, Friesner RA, Tirado-Rives J, Jorgensen WL. Evaluation and reparametrization of the OPLS-AA force field for proteins via comparison with accurate quantum chemical calculations on peptides. J Phys Chem B. 2001;105(28):6474–6487. [Google Scholar]
- 46.Duan Y, Wu C, Chowdhury S, Lee MC, Xiong GM, Zhang W, Yang R, Cieplak P, Luo R, Lee T, Caldwell J, Wang JM, Kollman P. A point-charge force field for molecular mechanics simulations of proteins based on condensed-phase quantum mechanical calculations. J Comput Chem. 2003;24(16):1999–2012. doi: 10.1002/jcc.10349. [DOI] [PubMed] [Google Scholar]
- 47.Rapcewicz K, Ashcroft NW. Fluctuation attraction in condensed matter: a nonlocal functional-approach. Phys Rev B. 1991;44(8):4032–4035. doi: 10.1103/physrevb.44.4032. [DOI] [PubMed] [Google Scholar]
- 48.Andersson Y, Hult E, Apell P, Langreth DC, Lundqvist BI. Density-functional account of van der Waals forces between parallel surfaces. Solid State Commun. 1998;106(5):235–238. [Google Scholar]
- 49.Ahmed HU, Blakeley MP, Cianci M, Cruickshank DWJ, Hubbard JA, Helliwell JR. The determination of protonation states in proteins. Acta Crystallog Sect D Biol Crystallogr. 2007;63:906–922. doi: 10.1107/S0907444907029976. [DOI] [PubMed] [Google Scholar]
- 50.Fenn TD, Schnieders MJ, Brunger AT, Pande VS. Polarizable atomic multipole X-Ray refinement: hydration geometry and application to macromolecules. Biophys J. 2010;98(12):2984–2992. doi: 10.1016/j.bpj.2010.02.057. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Faver JC, Benson ML, He XA, Roberts BP, Wang B, Marshall MS, Kennedy MR, Sherrill CD, Merz KM. Formal estimation of errors in computed absolute interaction energies of protein-ligand complexes. J Chem Theory Comput. 2011;7(3):790–797. doi: 10.1021/ct100563b. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Bennett CH. Efficient estimation of free-energy differences from montecarlo data. J Comput Phys. 1976;22(2):245–268. [Google Scholar]
- 53.Berendsen HJC, Postma JPM, Vangunsteren WF, Dinola A, Haak JR. Molecular-dynamics with coupling to an external bath. J Chem Phys. 1984;81(8):3684–3690. [Google Scholar]
- 54.Essmann U, Perera L, Berkowitz ML, Darden T, Lee H, Pedersen LG. A smooth particle mesh ewald method. J Chem Phys. 1995;103(19):8577–8593. [Google Scholar]
- 55.Darden T, York D, Pedersen L. Particle mesh ewald: an N.Log(N) method for ewald sums in large systems. J Chem Phys. 1993;98(12):10089–10092. [Google Scholar]
- 56.Sagui C, Darden TA. Molecular dynamics simulations of biomolecules: Long-range electrostatic effects. Annu Rev Biophys Biomol Struct. 1999;28:155–179. doi: 10.1146/annurev.biophys.28.1.155. [DOI] [PubMed] [Google Scholar]
- 57.Mobley DL, Bayly CI, Cooper MD, Shirts MR, Dill KA. Small molecule hydration free energies in explicit solvent: an extensive test of fixed-charge atomistic simulations. J Chem Theory Comput. 2009;5(2):350–358. doi: 10.1021/ct800409d. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58.Wang JM, Wolf RM, Caldwell JW, Kollman PA, Case DA. Development and testing of a general amber force field. J Comput Chem. 2004;25(9):1157–1174. doi: 10.1002/jcc.20035. [DOI] [PubMed] [Google Scholar]
- 59.Jakalian A, Bush BL, Jack DB, Bayly CI. Fast, efficient generation of high-quality atomic Charges. AM1-BCC model: I. Method. J Comput Chem. 2000;21(2):132–146. doi: 10.1002/jcc.10128. [DOI] [PubMed] [Google Scholar]
- 60.Schmid R, Miah AM, Sapunov VN. A new table of the thermodynamic quantities of ionic hydration: values and some applications (enthalpy-entropy compensation and Born radii) Phys Chem Chem Phys. 2000;2(1):97–102. [Google Scholar]
- 61.Krishnan CV, Friedman HL. Solvation enthalpies of various ions in water and heavy water. J Phys Chem. 1970;74(11):2356. [Google Scholar]
- 62.Tissandier MD, Cowen KA, Feng WY, Gundlach E, Cohen MH, Earhart AD, Tuttle TR, Coe JV. The proton’s absolute aqueous enthalpy and Gibbs free energy of solvation from cluster ion solvation data (vol 102A, pg 7791, 1998) J Phys Chem A. 1998;102(46):9308. [Google Scholar]
- 63.Grossfield A, Ren PY, Ponder JW. Ion solvation thermodynamics from simulation with a polarizable force field. J Am Chem Soc. 2003;125(50):15671–15682. doi: 10.1021/ja037005r. [DOI] [PubMed] [Google Scholar]
- 64.Kelly CP, Cramer CJ, Truhlar DG. Aqueous solvation free energies of ions and ion-water clusters based on an accurate value for the absolute aqueous solvation free energy of the proton. J Phys Chem B. 2006;110(32):16066–16081. doi: 10.1021/jp063552y. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.