Background: Early events of protein folding are difficult to follow.
Results: The unfolded state may have extended and compact conformers, which interconvert at early microsecond.
Conclusion: Hydrophobic compaction and secondary structure formation do not occur simultaneously in aqueous solution. They occur simultaneously in the presence of urea.
Significance: Hydrophobic collapse has been monitored at single molecular resolution.
Keywords: Circular Dichroism (CD), Cytochrome c, Fluorescence Correlation Spectroscopy, Protein Folding, Single Molecule Biophysics, Hydrophobic Collapse, Unfolded State
Abstract
What happens in the early stage of protein folding remains an interesting unsolved problem. Rapid kinetics measurements with cytochrome c using submillisecond continuous flow mixing devices suggest simultaneous formation of a compact collapsed state and secondary structure. These data seem to indicate that collapse formation is guided by specific short and long range interactions (heteropolymer collapse). A contrasting interpretation also has been proposed, which suggests that the collapse formation is rapid, nonspecific, and a trivial solvent related compaction, which could as well be observed by a homopolymer (homopolymer collapse). We address this controversy using fluorescence correlation spectroscopy (FCS), which enables us to monitor the salt-induced compaction accompanying collapse formation and the associated time constant directly at single molecule resolution. In addition, we follow the formation of secondary structure using far UV CD. The data presented here suggest that both these models (homopolymer and heteropolymer) could be applicable depending on the solution conditions. For example, the formation of secondary structure and compact state is not simultaneous in aqueous buffer. In aqueous buffer, formation of the compact state occurs through a two-state co-operative transition following heteropolymer formalism, whereas secondary structure formation takes place gradually. In contrast, in the presence of urea, a compaction of the protein radius occurs gradually over an extended range of salt concentration following homopolymer formalism. The salt-induced compaction and the formation of secondary structure take place simultaneously in the presence of urea.
Introduction
Several experimental and theoretical techniques have been devised to study dynamics of the unfolded state and to monitor early events of protein folding (1–4). Many of these studies involve measurements of rapid kinetics using mixing techniques with sub-millisecond and μs time resolutions (5–8). Cytochrome c has been found to be an attractive model system for these experiments (9).
Two long-standing controversial questions related to the nature and time scale of the initial events of protein folding have been addressed by many of these rapid kinetics measurements. First, is it the hydrophobic collapse, which occurs first, followed by the formation of the secondary structure, or do they occur simultaneously? Second, is the initial event of protein folding directed by specific interactions between the participating amino acids, or is it nonspecific? Rapid kinetics measurements show barrier limited formation of a native-like collapsed states in the early time scale (6, 10). These experiments and other data seem to suggest that hydrophobic collapse and secondary structure formation take place simultaneously (11). These studies indicated that collapse is guided by specific short and long range interactions and barrier-limited (heteropolymer collapse) (6, 9–10). A contrasting interpretation was brought up by Sosnick et al. (12, 13), who suggested that the initial collapse is nonspecific. According to this interpretation, the initial collapse is a trivial compaction, which any homopolymer would experience when the denaturant gets diluted by aqueous buffer (homopolymer collapse). The controversy has been found difficult to resolve because of several reasons. First, the number of experimental techniques, which could probe early change in conformation and μsec dynamics in the unfolded state, is limited. Second, pathways of protein folding are heterogeneous with complications arising from parallel pathways, trapped intermediates, and misfolding.
We addressed these questions by making use of fluorescence correlation spectroscopy (FCS)2 and far UV CD to study the salt-induced collapse and secondary structure formation of cytochrome c from Saccharomyces cerevisiae. Bovine serum albumin also was used to verify some of the key experimental results on a different protein system. FCS is a unique experimental technique that can measure conformation of the unfolded state and its fluctuation dynamics in the μs time scale using the same experimental data. Additionally, FCS offers single molecule sensitivity, which could efficiently isolate folding events from any competing undesired processes such as aggregation.
Using FCS, we observed the presence of a fluctuation dynamics between an extended and a collapsed conformer of cytochrome c unfolded at pH 2. The time constant of their interconversion was 50 μs. The equilibrium population of collapsed conformer increased upon the addition of salt as monitored by a decrease in the hydrodynamic radius (rH). Using far UV CD and FCS, we independently monitored the formation of secondary structure and that of the collapsed state, respectively. In aqueous buffer, the formation of collapsed state followed a cooperative two-state transition occurring at low salt concentration (heteropolymer collapse), although secondary structure forms gradually over a large range of salt concentration. In aqueous buffer, these two events are not simultaneous. A salt-induced compaction of the hydrodynamic radius has been observed in the presence of urea. However, the contraction of the unfolded chain and secondary structure formation occurred simultaneously in urea. In the presence of urea, the contraction followed homopolymer formalism.
EXPERIMENTAL PROCEDURES
Iso-1-cytochrome c (C2436), sodium perchlorate (CAS-7601-89-0), and urea (CAS-13-6) were obtained from Sigma. Alexa Fluor 488-maleimide and tetramethyl rodamine 5 maleimide (TMR) were obtained from Molecular Probes (Eugene, OR). All of the other reagents used were of the highest available grade.
Far UV CD spectra were carried out using a Jasco J715 spectropolarimeter. Experiments were performed typically with 9 μm TMR-maleimide-labeled cytochrome c-TMR. Far-UV CD spectra were recorded over a 200–250-nm range, and each spectrum was averaged using 10 repeat scans. Steady state fluorescence experiments were carried out with TMR-labeled protein using a PTI fluorescence spectrometer (Photon Technology Intl.). The fluorescence emission spectra were recorded between 560–650 nm using an excitation wavelength of 540 nm. Final concentration of the labeled cytochrome c used for the steady-state fluorescence was 1 μm.
MALDI mass spectrometry was carried out using a 4800 MALDI-TOF-TOF mass spectrometer (Applied Biosystems) equipped with a microchannel plate detector. The delay time used was 500 ns. Mass spectra were acquired by averaging 600 to 800 shots. Labeled cytochrome c fragment of 1 μm concentration was used for the mass spectrometry measurements.
Cytochrome c from S. cerevisiae contains one free cysteine (Cys-102), which was labeled with TMR using published procedures (14). Labeling of BSA with Alexa Fluor 488-maleimide was carried out using an identical method. Labeling of cytochrome c with TMR does not result in any significant effect on the structure, conformation, and folding of the protein (14). FCS experiments were carried out with a ConfoCor 3 LSM setup (Carl Zeiss, Evotec, Jena, Germany) using a 40× water immersion objective. The detailed experimental procedure has been published previously (15, 16).
For FCS data analysis, correlation function curves observed with the FCS experiments were analyzed using different models as described below.
Model 1 (Simple Diffusion Model)
For a single component system without any conformational event, the diffusion time (τD) of a fluorophore (for example, the free dye, TMR or Alexa Fluor 488-maleimide-labeled bovine serum albumin) and the number of particles in the observation volume (N) can be calculated by fitting the correlation function (G(τ)) to Equation 1 (17),
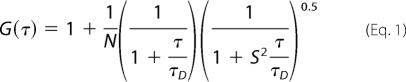 |
where S is the structure parameter, the depth-to-diameter ratio of the Gaussian observation volume.
Model 2 (Simple Diffusion with Conformation Fluctuation Model)
Because the correlation functions obtained with cytochrome c-TMR at pH 2 could not be analyzed using model 1 described above, we needed to use model 2 for the successful representation of the data. This model (model 2) assumes a single diffusing species (with the diffusion time of τD), which undergoes a chemical reaction or conformational change. The correlation function of the system can be represented by Equation 2 (17).
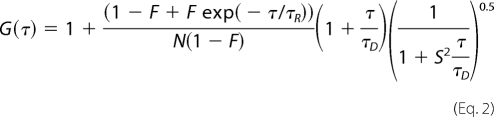 |
The parameter τR represents the time constant of a conformational fluctuation between a fluorescent and a non-fluorescent conformer (for example, A ⇄ B, and both of the conformers, A and B, have the same τD) provided that the said event occurs faster than the diffusion time (τR ≪ τD). F is the amplitude of τR, which corresponds to the average fraction of the molecules in the non-fluorescent (dark) state (17). For both Equations 1 and 2, a rapid exponential component with amplitude 20–30% and the time constant of 1–3 μs for the triplet state photophysics also is present.
The diffusion coefficient (D) of the molecule can be calculated from τD using Equation 3,
 |
where ω is the beam radius of the observation volume, which can be obtained by measuring τD of a fluorophore of known D.
The hydrodynamic radius (rH) of a labeled molecule can be calculated from D using the Stokes Einstein equation (Equation 4),
 |
where k is the Boltzmann constant, T is the temperature, and η corresponds to the viscosity of the solution.
FCS data were further analyzed by maximum entropy method (MEM) (18). This is a model-free method in which the multicomponent correlation function can be represented by n, the number of non-interacting fluorescent species. These species can have diffusion time values between 0.001 and 500 ms. MEM minimizes the parameter χ2 and maximizes the entropic quantity, S = Σipilnpi (where pi = aiΣiaj) to obtain an optimized fit (15, 18).
RESULTS
FCS has been used extensively to measure hydrodynamic radius (rH) of fluorescently labeled molecules at a single molecule resolution (19, 20). Additionally, FCS has been used to monitor conformational dynamics of biological molecules at μs time scale (3, 4). As a matter of fact, FCS is one of the very few experimental techniques that can measure protein dynamics in the μs time scale. This is because NMR and stopped flow methods are limited by their time resolution typically at ms; time resolved fluorescence decay experiments are effective only at the ns or ps time range. Computational studies including molecular dynamics simulations are frequent at the ns or ps time scale because of computational constraints one would encounter to perform simulations at μs time scale.
In a typical FCS experiment, intensity fluctuations of a labeled molecule inside a small observation volume are monitored with time. Intensity fluctuations may arise due to the diffusion of the molecule (with a diffusion time of τD) in or out of the observation volume (Equation 1) or due to the kinetics of a conformational change (with the time constant of τR) whose rate is faster than the molecular diffusion (τR ≪ τD, Equation 2). Analysis of the diffusion component (τD) using Equations 3 and 4 provides hydrodynamic radius (rH). Analysis of the kinetic component provides information about the time constant (τR) of the conformational fluctuations in the μs time scale.
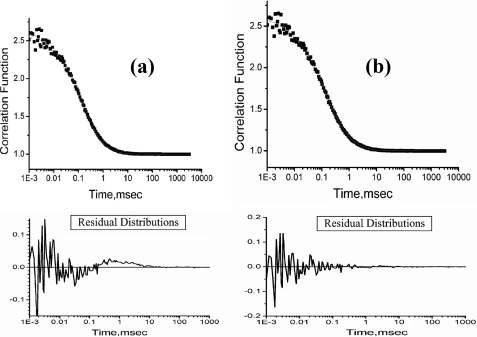
We have used FCS and far UV CD to study salt-induced collapse and secondary structure formation of TMR-labeled cytochrome c (cytochrome c-TMR) at pH 2. Labeling with TMR does not affect the conformation and folding of cytochrome c (14, 21). At low pH (for example, at pH 2), the protein becomes positively charged. To avoid the repulsive interactions between the positively charged residues at acidic pH, it unfolds and becomes extended. Unfolding of cytochrome c-TMR at pH 2 leads to a large decrease in the secondary structure as observed by far UV CD experiments (supplemental Fig. S1). The correlation functions obtained by the FCS experiments with cytochrome c-TMR at pH 2 could not be fit to Equation 1 resulting in non-random residual distributions (Fig. 1a). The data instead have been fit to Equation 2 (Fig. 1b), which contains an exponential time constant (τR of 50 μs) in addition to a diffusion component (of diffusion time of τD). Using the τD value obtained from the diffusion component, we have calculated the hydrodynamic radius (rH) of the protein at pH 2 to be 31 Å (using Equations 3 and 4, “Experimental Procedures”). The parameter τR denotes the time constant of a conformational fluctuation occurring between a non-fluorescent and a fluorescent conformer (also see “Experimental Procedures” for explanations of Equations 1 and 2). The parameter F denotes the amplitude of τR describing the relative population of the compact non-fluorescent state in equilibrium. Fitting of the FCS data using two exponential components (1 diffusion + 2 exponential components) does not improve the quality of the fit.
FIGURE 1.
a, representative correlation function obtained by FCS experiments with on average a single cytochrome c-TMR molecule at pH 2. The fit of this data to Equation 1 yields non-random behavior of the residual distribution (shown at the bottom of the figure) suggesting that Equation 1 may not be appropriate. b, correlation function obtained with cytochrome c-TMR at pH 2 could be fit successfully to Equation 2. Residual distribution calculated using Equation 2 (shown at the bottom of the figure) is completely random, indicating the goodness of the fit.
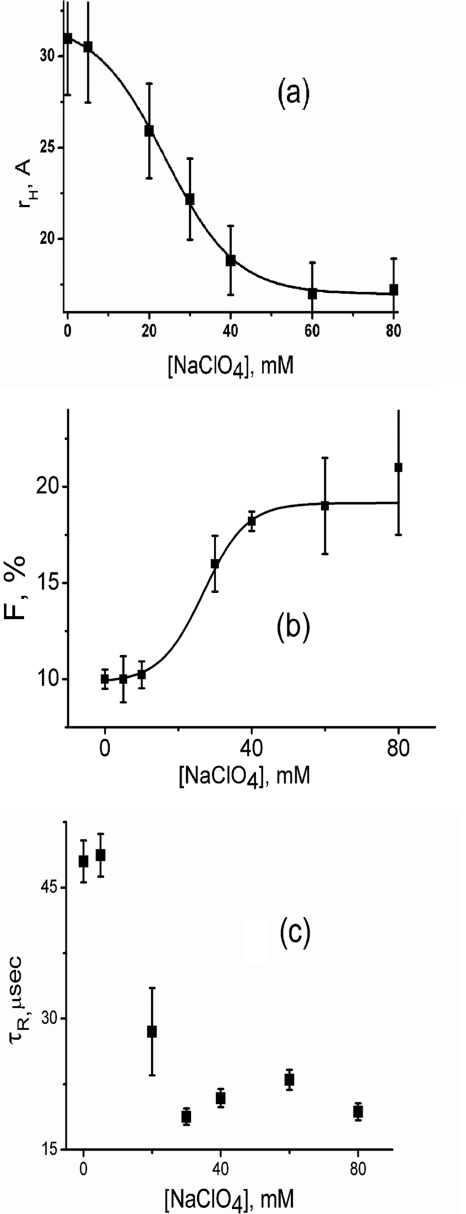
The addition of salts (for example, sodium perchlorate) increases the dielectric constant of the aqueous solution. Because the repulsive interactions between the positive charged residues are inversely proportional to the dielectric constant of the medium, the hydrophobic interactions predominate resulting in the compaction of the unfolded state. The value of rH, as observed by FCS experiments with cytochrome c-TMR, decreases with the addition of sodium perchlorate at pH 2 (Fig. 2a), suggesting the formation of a compact collapsed state. The value of rH in the presence of 120 mm sodium perchlorate at pH 2 has been found to be 18 Å. The parameter F increases with the increase in the sodium perchlorate concentration (Fig. 2b), which occurs in concert with the decrease in rH. The value of τR remains at 50 μs at low salt concentration and decreases to 20 μs as the salt induced transition occurs (Fig. 2c). In addition, the presence of 120 mm sodium perchlorate leads to partial refolding of cytochrome c-TMR as observed by an increase in the ellipticity measured by far UV CD (supplemental Fig. S1).
FIGURE 2.
The variation of rH (a), F (b), and τR (c) with sodium perchlorate concentrations at pH 2. These parameters have been calculated by fitting the correlation functions using Equation 2. The lines drawn through rH and F data show their fit to a two-state unfolding transition model. The data suggests the presence of a conformational fluctuation between equilibrium distributions of U (rH of 31 Å) and IC (rH of 18 Å) at pH 2. The presence of sodium perchlorate shifts the equilibrium toward IC. The equilibrium between U and IC could be defined successfully using a two-state transition hypothesis. The error bars shown in this and other figures have been calculated using at least three independent measurements.
The results discussed above could be explained by assuming the presence of a conformational equilibrium between an extended (U, 31 Å) and a compact collapsed (IC, 18 Å) conformer in the unfolded state of cytochrome c-TMR at pH 2.
The presence of an equilibrium containing three states (for example, one described in Equation 6) can be ruled out. This is because, the use of Equation 2 (which represents a two-state equilibrium between U and IC, Equation 5 above) is sufficient to successfully fit the correlation functions data of cytochrome c-TMR at pH 2. The use of an equation containing two exponential components (with time constants of τR1 and τR2) in addition to a diffusion component (of diffusion time of τD), which would define a three states equilibrium (Equation 6), does not improve the quality of the fit.
 |
It has been shown that the presence of heme quenches TMR fluorescence of the protein (22). The compact collapsed conformer (IC) is non-fluorescent because heme (the quencher) is close to the attached fluorophore (TMR) ensuring efficient quenching of TMR fluorescence. Alternatively, the extended conformer (U) is strongly fluorescent because heme (the quencher) and TMR (the fluorophore) are distant from each other. The parameter F, which corresponds to the population of the non-fluorescent conformer (see “Experimental Procedures”), would represent the population of IC (the compact conformer) in the equilibrium between U and IC. The time constant (τR) of the interconversion between U and IC has been found to be 50 μs. The population of IC increases with sodium perchlorate concentration as observed by a decrease in rH (Fig. 2a) and an increase in F (Fig. 2b). The decrease in rH with sodium perchlorate occurs in a single co-operative step that could be fit successfully using a two-state transition model, and the midpoint is 23 mm. The increase in F with the sodium perchlorate concentration occurs simultaneously (Fig. 2b), and a fit using the two state transition hypotheses leads to a midpoint of 20 mm.
The presence of an equilibrium between an extended and a compact conformer in the unfolded state of cytochrome c has been shown before by fluorescence spectroscopy and small angle x-ray scattering experiments (14, 23, 24). The formation of the collapsed unfolded states for multiple other proteins has also been demonstrated (25–29). Shastry and Roder (6) observed a 60-μs time kinetics of the formation of the compact state of cytochrome c using a continuous flow capillary mixing technique. Pascher et al. (29) observed a 40-μs time kinetics for the early events of cytochrome c folding. These results are comparable with the 50-μs time constant observed in the present study.
Several control experiments have been carried out to establish that the parameter τR measures a true physical event and is not a fitting artifact. First, experiments have been carried out with TMR (the free dye) using the same setup and experimental conditions. Supplemental Fig. S2 shows the correlation functions obtained by the FCS experiments with TMR at pH 2. The data are fit successfully to Equation 1 and the use of τR is not needed. Second, FCS experiments have been carried out with cytochrome c-TMR using different laser power and pinhole diameters. The possibility of an artifactual extra component at large pinhole diameter has been reported previously (30). We see no systematic variation in the value of τR and F with the variation of laser power and pinhole diameters studied at pH 2 in the absence or presence of sodium perchlorate (supplemental Figs. S3 and S4). This experiment suggests that τR represents a true conformational event and not an erroneous component arising from the imperfect Gaussian approximation of the confocal volume element.
In the third control experiment, we used cyanogen bromide-treated labeled cytochrome c-TMR as the control. Cyanogen bromide treatment with the protein yields a fragment with molecular weight of 3139 Da (as judged by mass spectrometry, supplemental Fig. S5). Although the fragment was still labeled with TMR, it did not contain the heme group as judged by absorption spectroscopy (supplemental Fig. S6). The absence of heme in the cyanogen bromide fragment resulted in a complete absence of fluorescence quenching observed in the full-length protein (supplemental Fig. S7 shows the fluorescence spectra of native and cyanogen bromide-treated fragment of cytochrome c with the same TMR concentration). Because the fragment lacks the non-fluorescent state and the parameter F in Equation 2 denotes the relative amplitude of the non-fluorescent state, the correlation function of the cyanogen bromide-treated fragment is not expected to contain the τR component. Alternatively, the presence of any τR component in the correlation function of the cyanogen bromide fragment would represent an artifact and not a physical motion. The correlation function data of the cyanogen bromide-treated protein can be fit successfully to Equation 1 and the use of τR is not needed (supplemental Fig. S8).
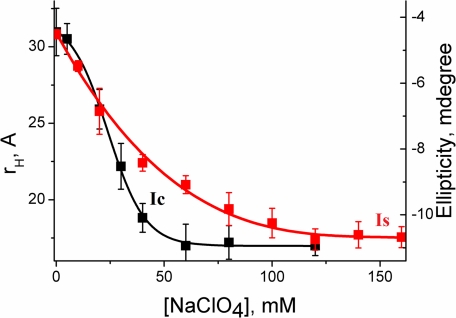
The addition of sodium perchlorate at pH 2 results in the formation of a molten globule-like intermediate (defined in the present work as IS). The formation of IS is accompanied by the formation of large secondary structure (supplemental Fig. S1). The salt-induced formation of secondary structure has been monitored independently by the increase in the ellipticity at 222 nm using far UV CD. Fig. 3 shows a comparison between the formation of IC (monitored by the decrease in rH and an increase in F) and that of IS (monitored by the increase in the ellipticity at 222 nm). Although the formation of IC is a two-state cooperative transition, the formation of IS occurs through a shallow transition and completes at higher salt concentration. Moreover, the formation of IS is significantly less cooperative. A comparison between these two processes (the formation of IC and IS) suggests that IC contains ∼60% of the secondary structure, whereas the remaining secondary structure forms gradually as the IS state forms (Fig. 3). Although the kinetics of the formation of IS is not measured directly, we could unambiguously show that the formation of IC and IS are not simultaneous and they correspond to two separate thermodynamic events. As mentioned before, IS is similar to the molten globule-like intermediate states studied extensively for multiple proteins including cytochrome c (31). The formation of the native state (N) occurs in subsequent slow steps requiring further refolding (such as using a buffer with a pH of 7.4). It may be important to point out that extreme care has been exercised to rule out any aggregation related complications in these measurements with the protein at low pH and in the presence of salt. This is particularly important for the bulk CD experiments, which require relatively high concentrations. For every experiment, the reversibility of the CD measurements has been established.
FIGURE 3.
The comparison between the formation of collapsed state (IC) and that of secondary structure (IS). The formation of IC has been monitored by rH using FCS data (black squares), whereas far UV CD at 222 nm (red squares) has been used to monitor the formation of IS. The data containing the variation of rH with sodium perchlorate concentration has been fit to a two-state unfolding transition as shown by the black line. The line through the CD data (red line), on the other hand, is for the presentation only and not a fit using a physically validated equation. All measurements have been carried out at pH 2.
Although urea has been widely used as a protein denaturant, the mechanism of its action has been debated extensively (32–36). Although an “indirect” mechanism has been proposed, it is now generally believed that urea unfolding occurs through “direct” binding to protein backbone. Interestingly enough, there are controversies regarding the nature of the direct binding of urea: either it is hydrophobic and directed toward the apolar residues (36), or it is performed through hydrogen bonding and directed toward the polar residues of the protein (37). It has been shown using hydrogen deuterium exchange that urea unfolds protein by forming direct hydrogen bonds to the peptide groups (38).
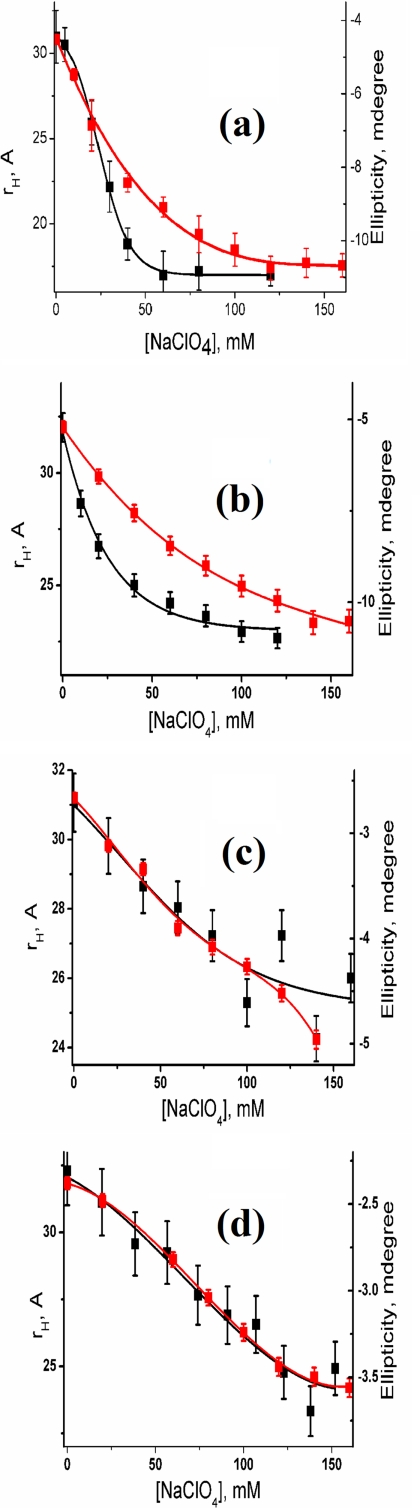
In a separate experiment, we use FCS and far UV CD to monitor the formation of IC and IS induced by sodium perchlorate in the presence of increasing concentration of urea at pH 2. The interaction of urea with the protein would oppose the formation of compact collapsed state, and thus, these experiments should monitor the competing effects of urea and sodium perchlorate on the formation of IC and IS. Fig. 4 shows the variation of rH and the ellipticity at 222 nm with sodium perchlorate concentration in the absence and presence of different concentrations of urea at pH 2. At low urea concentration (0 and 1 m urea), the formation of IC and IS is not simultaneous. The formation of IC occurs through a sharp two-state transition with a large decrease in rH and increase in F, whereas IS forms gradually through a shallow transition (see above).
FIGURE 4.
Variation of rH (black) and ellipticity at 222 nm (red) with sodium perchlorate concentration in the presence of 0 m urea (a), 1 m urea (b), 3 m urea (c), and 4 m urea (d). At low urea concentration (0 and 1 m urea), the formation of IC and IS are not simultaneous. In the presence of high concentration of urea (3 and 4 m urea), however, IC and IS form simultaneously. The variation of rH with sodium perchlorate concentration in the absence of urea (a) could be fit successfully using a two-state unfolding transition. The data deviate from this model in the presence of urea and the drawn lines are only for presentation purposes (b–d).
In the presence of 3 m urea and beyond at pH 2, F has been found to be very low (∼5%) or it may even be absent. In addition, no change in F (which is inherently less) is observed in the presence of added sodium perchlorate (Figure 5). However, a small but significant decrease in rH has been observed, which occurs along with an increase in the secondary structure (Fig. 4d). The absence of any significant amplitude of F in the absence or presence of sodium perchlorate concentrations suggests that there is no rapid equilibrium between the extended (U) and any compact (IC-like) state. Thus, the interconversion between U an IC must be slow with time constant higher than the diffusion time (τD). This conclusion comes directly from Equation 2 (see “Experimental Procedures”), which assumes τR ≪ τD for τR and F to be observed in the experimental correlation functions. This inference is supported also by Fig. 4, c and d, which show non-two state formation of IC, which occurs simultaneously with the formation of IS. The values of ellipticity at 222 nm and rH in the absence and presence of high concentration of sodium perchlorate for all urea concentrations are shown in Table 1. These values were used as the start and end points of all the experiments described above. Supplemental Fig. S9 shows the far UV CD spectra of the protein in the absence and presence of sodium perchlorate in the presence of different concentrations of urea.
FIGURE 5.
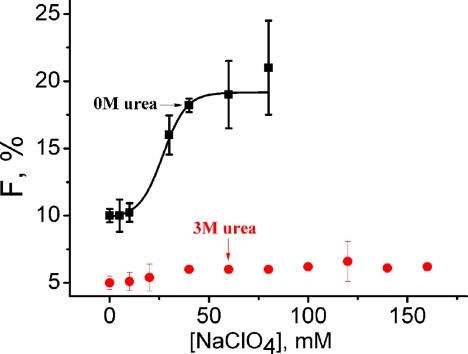
The variation of amplitude of τR (F, %) of cytochrome c at pH 2 with sodium perchlorate concentration in the absence (black) and presence (red) of 3 m urea. A large increase in F with the salt concentration has been observed in the absence of urea, whereas no significant change is present in the presence of 3 m urea.
TABLE 1.
The values of hydrodynamic radius (rH) and ellipticity at 222 nm of U, IC, IS, and N in the presence of different urea concentration
N corresponds to the native state of TMR-cytochrome c in 20 mm sodium phosphate buffer at pH 7.4. Ellipticity and rH of U change in the presence of different concentrations of urea, and the change depends on the extent of the residual structure present in U. deg, degree.
| Urea Concentration (m) |
U |
IC |
IS |
N |
||||
|---|---|---|---|---|---|---|---|---|
| Ellipticity at 222 nm | rH | Ellipticity at 222 nm | rH | Ellipticity at 222 nm | rH | Ellipticity at 222 nm | rH | |
| mol deg | Å | mol deg | Å | mol deg | Å | mol deg | Å | |
| 0 | −4.5 | 30.7 | −8.2 | 18 | −10.6 | 18 | −15.1 | 20 |
| 1 | −5.1 | 31.9 | −8 | 22.5 | −10.5 | 22.5 | −15.1 | 20 |
| 3 | −2.7 | 31.1 | −5 | 25.5 | −5 | 25.5 | −15.1 | 20 |
| 4 | −2.4 | 32.3 | −3.6 | 25 | −3.6 | 25 | −15.1 | 20 |
Because the above experiments have been carried out with only one protein (cytochrome c-TMR), we wanted to repeat some of these measurements with at least another protein system to find out whether any general trend could be established. FCS experiments with Alexa Fluor 488-maleimide-labeled bovine serum albumin (BSA-Alexa Fluor 488-maleimide) suggest a sharp collapse formation at pH 2 in the presence of increasing concentrations of sodium perchlorate (supplemental Fig. S10). Identical to what has been observed with cytochrome c-TMR, collapse formation follow a shallow transition in the presence of urea at pH 2 (Fig. 4). Although the results obtained with BSA-Alexa Fluor 488-maleimide are identical to that of cytochrome c-TMR, we are currently investigating several other protein systems containing different secondary structural elements (it should be noted that both cytochrome c-TMR and BSA-Alexa Fluor 488-maleimide are predominately α-helical) to determine whether the nature of secondary structure may have any influence on the interconnection between the hydrophobic collapse and secondary structure formation.
DISCUSSION
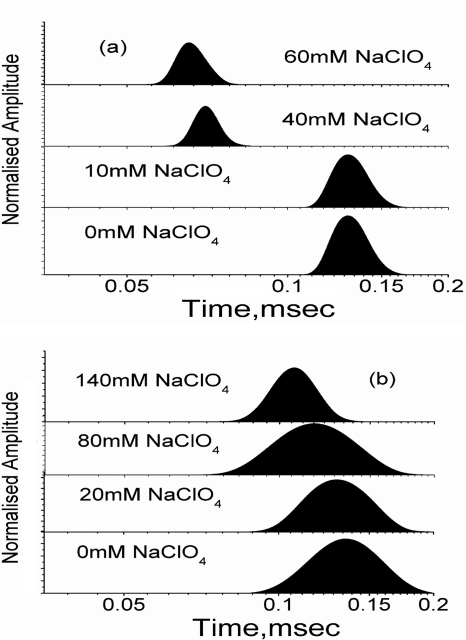
The nature of the collapse or whether it is a homopolymer (nonspecific) or heteropolymer (specific) has been the subjects of long-standing debates. A collapse process pertaining to a homopolymer is expected to be continuous and extended (39). Computational and experimental studies suggest that homopolymer collapse comprise multiple kinetic stages leading to extended process (39–41). The observed formation of IC in the absence of urea (Fig. 4a) could be fit successfully to a two-state transition. In contrast, the presence of high concentration of urea results in relatively extended transitions for IC, which deviates from the two-state behavior (Fig. 4d). To further clarify, we have shown the MEM distribution of τD values in the presence of different sodium perchlorate concentration at pH 2 in the absence (Fig. 6a) and presence of 3 m urea (Fig. 6b). In the absence of urea, the MEM profiles remain unchanged at a relatively large value of τD before the collapse and shift sharply to the lower value as the transition occurs (Fig. 6a). In the presence of 3 m urea, this is not the case, and the shift in the MEM profiles occurs in continuous manner (Fig. 6b). The FCS data presented in this paper thus suggest that the collapse in the absence of urea may not follow nonspecific homopolymer formalism. In the presence of urea, on the other hand, the data seem to suggest a homopolymer-like nonspecific compaction. The presence of “specific” and “nonspecific” components in the early folding of Barstar has been identified using multisite fluorescence resonance energy transfer measurements (42).
FIGURE 6.
The variation of the maximum entropy profiles of cytochrome c-TMR with sodium perchlorate concentration in the absence (a) and presence (b) of 3 m urea at pH 2. In the absence of urea, the maximum entropy profiles shift sharply toward lower diffusion time as IC forms. This behavior of maximum entropy profiles is not consistent with the homopolymer collapse hypothesis. In the presence of urea, however, IC forms slowly and gradually over an extended range of sodium perchlorate concentration, as expected for a homopolymer collapse.
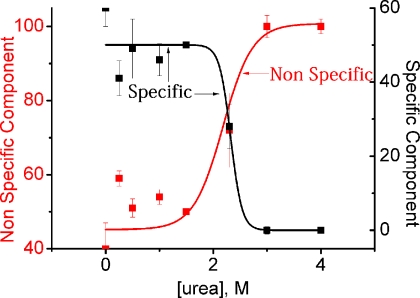
Using the considerations outlined in the previous paragraph, the specific and nonspecific contributions to the secondary structure formation have been calculated in the presence of different urea concentrations and shown in Fig. 7. Fig. 7 shows that the nonspecific component to the secondary structure of IS increases with the urea concentration in a sigmoidal fashion. For example, the nonspecific component of the secondary structure is ∼40% in the absence of urea, whereas it increases to 100% in the presence of 4 m urea. The specific component, on the other hand, decreases identically from 60% (in the absence of urea) to 0% (in the presence of 4 m urea). In the presence of high concentration of urea, the protein behaves like a homopolymer, resulting in simultaneous formation of collapse and secondary structure (which is entirely due to nonspecific contacts).
FIGURE 7.
The variation in the percentage of nonspecific (red) and specific contributions (black) to the secondary structure with urea concentration. A percentage of nonspecific component increases and that of specific component decreases with urea concentration. It is important to note that these percentage calculations are based on total secondary structure change between the U and IS and the native state (N) is not considered.
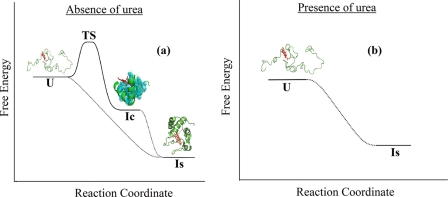
Fig. 8 summarizes the results discussed in the present work and shows a possible reaction coordinate diagram for the formation of IC and IS in the absence and presence of urea. In the absence of urea (Fig. 8a), U and IC are involved in a rapid equilibrium driven by specific contacts and thus, a defined transition state is expected. The present data are inadequate to determine whether IS forms sequentially through the formation of IC or they occur in parallel. Nevertheless, it is difficult to demonstrate IS formation (parallel or sequential) using defined transition states. The not-so-defined nature of IS formation (sequential or parallel) is thus demonstrated using dotted lines in Fig. 8a. Of course, it is possible that these pathways are composed of several steps requiring low transition state barriers. In the presence of urea (Fig. 8b), there is no defined equilibrium between U and IC (or IS) and these two processes (the formation of IC-like states and secondary structure) occur simultaneously through a homopolymer-like mechanism. This is to be noted that the formation of the native state (N) is not considered either in Fig. 8 or in any of the percentage calculations discussed in the previous paragraph (also see Fig. 7). Because the formation of N is not considered, a large change in the secondary structure may be excluded from the calculation. This is not significant in the absence of urea, because the extent of secondary structure of IS and that of N are not very different. However, in the presence of urea, the extent of secondary structure of IS would be significantly less (as it contains only the nonspecific component). The formation of the rest and majority of the secondary structure (IS to N transition) would take place slowly as the native state (N) form.
FIGURE 8.
A schematic diagram showing the sodium perchlorate induced formation of IC and IS in aqueous buffer (a) and in the presence (b) of 3 m urea. The unfolded extended conformer (U) does not have any secondary structure although the cofactor heme is bound to the protein. U has been found to be in rapid equilibrium with a compact conformer (IC) and the time constant of their interconversion is 50 μs. The addition of sodium perchlorate shifts the equilibrium toward IC, which is accompanied by a decrease in rH and an increase in F. The hydrophobic residues, which may be involved in the formation of IC, are shown by space-filling spheres. The cofactor heme, because it is hydrophobic, may also contribute toward the formation of IC. IC has been shown to contain partial secondary structure (presumably at the N and C-terminal helix regions). Sodium perchlorate induced formation of IC is a sharp cooperative transition with a defined transition state. The formation of the secondary structure (IS) takes place gradually over an extended range of sodium perchlorate concentration. This is an extended homopolymer-like transition and shown using dotted lines. The formation of the native protein is slow and is not shown. No rapid equilibrium exists between the extended (U) and any IC-like state in the presence of 3 m urea. A small extent of chain contraction (observed by the decrease in rH) and an increase in the secondary structure formation occur slowly and simultaneously in the presence of urea. The structure of IC (or IS) in the presence of urea is not defined, although these states are more compact than U and more extended than N (or IC observed without urea).
Thus, the long-standing specific versus nonspecific controversy of the initial protein folding events in the refolding buffer may now be addressed. The present data explains the apparent disagreement between distance measurements (light scattering or fluorescence data indicating specific nature) and secondary structure measurements (CD experiments suggesting the presence of nonspecific nature) for the early folding processes. We show that, if the collapse is probed by the contraction of the unfolded state, the initial collapse formation would seem to be specific. This conclusion comes directly from Fig. 4a because the change in rH with sodium perchlorate concentration in the absence of urea is cooperative and rapid, as expected for a specific transition. On the other hand, if the early formation of the secondary structure is monitored, it would show both nonspecific and specific contributions. The present data are also in agreement with the far UV circular dichroism measurements using microsecond continuous flow mixer, which show early formation of ∼20% of the native secondary structure at the earliest time point monitored (5).
It has been shown that pronounced biases toward the native or non-native structures exist in the unfolded states of a protein (43, 44). Because U and IC maintain an equilibrium in the unfolded state, the nature of contact formation in the unfolded state would dictate the nature of bias present in IC. The presence of non-native or misfolded bias in IC, for example, would be kinetically inefficient for the subsequent folding events because these misfolded contacts needed to be broken before the protein could be folded properly. These processes of breaking of misfolding contacts would slow down the formation of the secondary structure (formation of IS). Although it is difficult to monitor the nature of bias in the unfolded state of a protein experimentally, several successful attempts are available. The presence of high content of β-structures has been observed in the collapsed unfolded state of CspTm (45). The possibility of the formation of short and highly dynamic β-segments resulting in non-native hydrogen bond interaction has been postulated within the unfolded state, which could contribute to the chain contraction (46, 47). The use of NMR has been particularly useful to obtain molecular details about the local structures of the unfolded state (48–51).
The hydrodynamic radius (rH) of a protein in its completely unfolded state in good solvent (like urea or guanidium hydrochloride) can be defined by an empirical relationship as described below (52).
The value of rH of a native or native like protein in poor solvent (like aqueous buffer) is also defined well by Equation 8 (52).
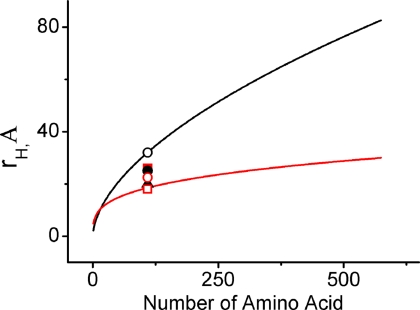
Fig. 9 shows the predicted values of rH of the completely unfolded (in good solvent) and collapsed (in poor solvent) proteins as a function of the number of amino acid residues. The predicted values of rH have been generated using Equations 7 and 8. The experimental values obtained by the FCS experiments at different solution conditions are also shown ion Fig. 9. The observed value of rH in the presence 4 m urea at pH 2 (the completely unfolded protein in good solvent) obtained by the present FCS experiments falls exactly on the predicted line (Fig. 9). Similarly, the value of rH in the presence of sodium perchlorate and no urea at pH 2 (the collapsed protein in poor solvent) also is in exact agreement with the prediction. Not surprisingly, the rH values of Ii in the presence of different urea concentrations deviates significantly from both the predicted lines: they are more compact than the completely unfolded state in urea, whereas they are more extended than the collapsed/native state in aqueous buffer. Depending on the concentration of urea, the rH values of IC would be either near the predicted line for the extended state in good solvent (n = 0.6), or near the predicted line for the collapsed/native state in poor solvent (n = 0.3) or distant from both. These partially folded/unfolded states of different compactness or secondary structure are not defined well, although they may well have significant implications in the mechanism of protein folding in general and in the field of the intrinsically disordered proteins in particular. The present data support Muller-Spath et al. (53) who show that the dimensions of an intrinsically disordered protein (or the unfolded state of a globular protein) would depend on several factors, including hydrophobicity, charge interactions, solution conditions, screening effects of charged denaturants such as guanidinium hydrochloride, and the binding of denaturants with the protein chain.
FIGURE 9.
The calculated variation of the hydrodynamic radii of different proteins with the number of residues for the extended unfolded states in the good solvent (black) and the native states in the poor solvent (red). The rH values determined from the present FCS measurements at different solution conditions are also shown. These are unfolded state (black open circles), native state (black filled triangles), IC in the absence (red open squares) and presence of 1 m urea (red open circles), 3 m urea (black filled circles) and 4 m urea (red filled squares).
Supplementary Material
Acknowledgments
We thank Professor S. Maiti of the Tata Institute of Fundamental Research for the Maximum Entropy Method analysis software. We also thank Professors Elliot Elson and Carl Frieden of the Washington University School of Medicine for critical comments on this work. Professor S. Roy of the Indian Institute of Chemical Biology is acknowledged for help and support.
This work was supported by Council for Scientific and Industrial Research Empower Project Grant OLP004.

This article contains supplemental Figs. S1–S10.
- FCS
- fluorescence correlation spectroscopy
- TMR
- tetramethyl rodamine 5 maleimide
- MEM
- maximum entropy method.
REFERENCES
- 1. Eaton W. A., Thompson P. A., Chan C. K., Hage S. J., Hofrichter J. (1996) Fast events in protein folding. Structure 4, 1133–1139 [DOI] [PubMed] [Google Scholar]
- 2. Hagen S. J., Hofrichter J., Szabo A., Eaton W. A. (1996) Diffusion-limited contact formation in unfolded cytochrome c: Estimating the maximum rate of protein folding. Proc. Natl. Acad. Sci. U.S.A. 93, 11615–11617 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3. Chattopadhyay K., Elson E. L., Frieden C. (2005) The kinetics of conformational fluctuations in an unfolded protein measured by fluorescence methods. Proc. Natl. Acad. Sci. U.S.A. 102, 2385–2389 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4. Chen H., Rhoades E., Butler J. S., Loh S. N., Webb W. W. (2007) Dynamics of equilibrium structural fluctuations of apomyoglobin measured by fluorescence correlation spectroscopy. Proc. Natl. Acad. Sci. U.S.A. 104, 10459–10464 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5. Akiyama S., Takahashi S., Ishimori K., Morishima I. (2000) Stepwise formation of α-helices during cytochrome c folding. Nat. Struct. Biol. 7, 514–520 [DOI] [PubMed] [Google Scholar]
- 6. Shastry M. C., Roder H. (1998) Evidence for barrier-limited protein folding kinetics on the microsecond time scale. Nat. Struct. Biol. 5, 385–392 [DOI] [PubMed] [Google Scholar]
- 7. Chattopadhyay K., Zhong S., Yeh S. R., Rousseau D. L., Frieden C. (2002) The intestinal fatty acid binding protein: The role of turns in fast and slow folding processes. Biochemistry 41, 4040–4047 [DOI] [PubMed] [Google Scholar]
- 8. Takahashi S., Yeh S. R., Das T. K., Chan C. K., Gottfried D. S., Rousseau D. L. (1997) Folding of cytochrome c initiated by submillisecond mixing. Nat. Struct. Biol. 4, 44–50 [DOI] [PubMed] [Google Scholar]
- 9. Goldbeck R. A., Chen E., Kliger D. S. (2009) Early events, kinetic intermediates, and the mechanism of protein folding in cytochrome c. Int. J. Mol. Sci. 10, 1476–1499 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10. Chen E., Goldbeck R. A., Kliger D. S. (2004) The earliest events in protein folding: A structural requirement for ultrafast folding in cytochrome c. J. Am. Chem. Soc. 126, 11175–11181 [DOI] [PubMed] [Google Scholar]
- 11. Uversky V. N., Fink A. L. (2002) The chicken-egg scenario of protein folding revisited. FEBS Lett. 515, 79–83 [DOI] [PubMed] [Google Scholar]
- 12. Sosnick T. R., Mayne L., Englander S. W. (1996) Molecular collapse: The rate-limiting step in two-state cytochrome c folding. Proteins 24, 413–426 [DOI] [PubMed] [Google Scholar]
- 13. Sosnick T. R., Shtilerman M. D., Mayne L., Englander S. W. (1997) Ultrafast signals in protein folding and the polypeptide contracted state. Proc. Natl. Acad. Sci. U.S.A. 94, 8545–8550 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14. Haldar S., Mitra S., Chattopadhyay K. (2010) Role of protein stabilizers on the conformation of the unfolded state of cytochrome c and its early folding kinetics: Investigation at single molecular resolution. J. Biol. Chem. 285, 25314–25323 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15. Ghosh R., Sharma S., Chattopadhyay K. (2009) Effect of arginine on protein aggregation studied by fluorescence correlation spectroscopy and other biophysical methods. Biochemistry 48, 1135–1143 [DOI] [PubMed] [Google Scholar]
- 16. Haldar S., Chattopadhyay K. (2011) Effects of arginine and other solution additives on the self-association of different surfactants: An investigation at single-molecule resolution. Langmuir 27, 5842–5849 [DOI] [PubMed] [Google Scholar]
- 17. Haupts U., Maiti S., Schwille P., Webb W. W. (1998) Dynamics of fluorescence fluctuations in green fluorescent protein observed by fluorescence correlation spectroscopy. Proc. Natl. Acad. Sci. U.S.A. 95, 13573–13578 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18. Sengupta P., Garai K., Balaji J., Periasamy N., Maiti S. (2003) Measuring size distribution in highly heterogeneous systems with fluorescence correlation spectroscopy. Biophys. J. 84, 1977–1984 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19. Elson E. L. (2004) Quick tour of fluorescence correlation spectroscopy from its inception. J. Biomed Opt 9, 857–864 [DOI] [PubMed] [Google Scholar]
- 20. Hess S. T., Huang S., Heikal A. A., Webb W. W. (2002) Biological and chemical applications of fluorescence correlation spectroscopy: A review. Biochemistry 41, 697–705 [DOI] [PubMed] [Google Scholar]
- 21. Werner J. H., Joggerst R., Dyer R. B., Goodwin P. M. (2006) A two-dimensional view of the folding energy landscape of cytochrome c. Proc. Natl. Acad. Sci. U.S.A. 103, 11130–11135 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22. Pletneva E. V., Gray H. B., Winkler J. R. (2005) Many faces of the unfolded state: Conformational heterogeneity in denatured yeast cytochrome c. J. Mol. Biol. 345, 855–867 [DOI] [PubMed] [Google Scholar]
- 23. Segel D. J., Fink A. L., Hodgson K. O., Doniach S. (1998) Protein denaturation: A small-angle x-ray scattering study of the ensemble of unfolded states of cytochrome c. Biochemistry 37, 12443–12451 [DOI] [PubMed] [Google Scholar]
- 24. Gast K., Zirwer D., Damaschun H., Hahn U., Müller-Frohne M., Wirth M., Damaschun G. (1997) Ribonuclease T1 has different dimensions in the thermally and chemically denatured states: A dynamic light scattering study. FEBS Lett. 403, 245–248 [DOI] [PubMed] [Google Scholar]
- 25. Sadqi M., Lapidus L. J., Muñoz V. (2003) How fast is protein hydrophobic collapse? Proc. Natl. Acad. Sci. U.S.A. 100, 12117–12122 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26. Ziv G., Haran G. (2009) Protein folding, protein collapse, and tanford's transfer model: Lessons from single-molecule FRET. J. Am. Chem. Soc. 131, 2942–2947 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27. Kimura T., Uzawa T., Ishimori K., Morishima I., Takahashi S., Konno T., Akiyama S., Fujisawa T. (2005) Specific collapse followed by slow hydrogen-bond formation of β-sheet in the folding of single-chain monellin. Proc. Natl. Acad. Sci. U.S.A. 102, 2748–2753 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28. Ziv G., Thirumalai D., Haran G. (2009) Collapse transition in proteins. Phys. Chem. Chem. Phys. 11, 83–93 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29. Pascher T., Chesick J. P., Winkler J. R., Gray H. B. (1996) Protein folding triggered by electron transfer. Science 271, 1558–1560 [DOI] [PubMed] [Google Scholar]
- 30. Hess S. T., Webb W. W. (2002) Focal volume optics and experimental artifacts in confocal fluorescence correlation spectroscopy. Biophys. J. 83, 2300–2317 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31. Kataoka M., Hagihara Y., Mihara K., Goto Y. (1993) Molten globule of cytochrome c studied by small angle x-ray scattering. J. Mol. Biol. 229, 591–596 [DOI] [PubMed] [Google Scholar]
- 32. Bennion B. J., Daggett V. (2003) The molecular basis for the chemical denaturation of proteins by urea. Proc. Natl. Acad. Sci. U.S.A. 100, 5142–5147 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33. Caflisch A., Karplus M. (1999) Structural details of urea binding to barnase: A molecular dynamics analysis. Structure 7, 477–488 [DOI] [PubMed] [Google Scholar]
- 34. Oostenbrink C., van Gunsteren W. F. (2005) Methane clustering in explicit water: Effect of urea on hydrophobic interactions. Phys. Chem. Chem. Phys. 7, 53–58 [DOI] [PubMed] [Google Scholar]
- 35. Zou Q., Habermann-Rottinghaus S. M., Murphy K. P. (1998) Urea effects on protein stability: Hydrogen bonding and the hydrophobic effect. Proteins 31, 107–115 [PubMed] [Google Scholar]
- 36. Stumpe M. C., Grubmüller H. (2008) Polar or apolar–the role of polarity for urea-induced protein denaturation. PLoS Comput Biol. 4, e1000221. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37. O'Brien E. P., Dima R. I., Brooks B., Thirumalai D. (2007) Interactions between hydrophobic and ionic solutes in aqueous guanidinium chloride and urea solutions: Lessons for protein denaturation mechanism. J. Am. Chem. Soc. 129, 7346–7353 [DOI] [PubMed] [Google Scholar]
- 38. Lim W. K., Rösgen J., Englander S. W. (2009) Urea, but not guanidinium, destabilizes proteins by forming hydrogen bonds to the peptide group. Proc. Natl. Acad. Sci. U.S.A. 106, 2595–2600 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39. Hagen S. J., Eaton W. A. (2000) Two-state expansion and collapse of a polypeptide. J. Mol. Biol. 301, 1019–1027 [DOI] [PubMed] [Google Scholar]
- 40. Chu B., Ying Q., Grosberg Y. A. (1995) Two-stage kinetics of single-chain collapse polystyrene in cyclohexane. Macromolecules 28, 180–189 [Google Scholar]
- 41. Zhu P. W., Napper D. H. (1997) The longer time collapse kinetics of interfacial poly (N-isopropylacrylamide) in water. J. Chem. Phys. 106, 6492–6498 [Google Scholar]
- 42. Sinha K. K., Udgaonkar J. B. (2007) Dissecting the non-specific and specific components of the initial folding reaction of barstar by multi-site FRET measurements. J. Mol. Biol. 370, 385–405 [DOI] [PubMed] [Google Scholar]
- 43. Srinivasan R., Rose G. D. (1999) A physical basis for protein secondary structure. Proc. Natl. Acad. Sci. U.S.A. 96, 14258–14263 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44. Baldwin R. L., Rose G. D. (1999) Is protein folding hierarchic? II. Folding intermediates and transition states. Trends Biochem. Sci. 24, 77–83 [DOI] [PubMed] [Google Scholar]
- 45. Hoffmann A., Kane A., Nettels D., Hertzog D. E., Baumgärtel P., Lengefeld J., Reichardt G., Horsley D. A., Seckler R., Bakajin O., Schuler B. (2007) Mapping protein collapse with single-molecule fluorescence and kinetic synchrotron radiation circular dichroism spectroscopy. Proc. Natl. Acad. Sci. U.S.A. 104, 105–110 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46. Möglich A., Joder K., Kiefhaber T. (2006) End-to-end distance distributions and intrachain diffusion constants in unfolded polypeptide chains indicate intramolecular hydrogen bond formation. Proc. Natl. Acad. Sci. U.S.A. 103, 12394–12399 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47. Nettels D., Müller-Späth S., Küster F., Hofmann H., Haenni D., Rüegger S., Reymond L., Hoffmann A., Kubelka J., Heinz B., Gast K., Best R. B., Schuler B. (2009) Single-molecule spectroscopy of the temperature-induced collapse of unfolded proteins. Proc. Natl. Acad. Sci. U.S.A. 106, 20740–20745 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48. Kristjansdottir S., Lindorff-Larsen K., Fieber W., Dobson C. M., Vendruscolo M., Poulsen F. M. (2005) Formation of native and non-native interactions in ensembles of denatured ACBP molecules from paramagnetic relaxation enhancement studies. J. Mol. Biol. 347, 1053–1062 [DOI] [PubMed] [Google Scholar]
- 49. Mukrasch M. D., Markwick P., Biernat J., Bergen M., Bernadó P., Griesinger C., Mandelkow E., Zweckstetter M., Blackledge M. (2007) Highly populated turn conformations in natively unfolded Tau protein identified from residual dipolar couplings and molecular simulation. J. Am. Chem. Soc. 129, 5235–5243 [DOI] [PubMed] [Google Scholar]
- 50. Meng W., Raleigh D. P. (2011) Analysis of electrostatic interactions in the denatured state ensemble of the N-terminal domain of L9 under native conditions. Proteins 79, 3500–3510 [DOI] [PubMed] [Google Scholar]
- 51. Zhang O., Forman-Kay J. D. (1997) NMR studies of unfolded states of an SH3 domain in aqueous solution and denaturing conditions. Biochemistry 36, 3959–3970 [DOI] [PubMed] [Google Scholar]
- 52. Wilkins D. K., Grimshaw S. B., Receveur V., Dobson C. M., Jones J. A., Smith L. J. (1999) Hydrodynamic radii of native and denatured proteins measured by pulse field gradient NMR techniques. Biochemistry 38, 16424–16431 [DOI] [PubMed] [Google Scholar]
- 53. Müller-Späth S., Soranno A., Hirschfeld V., Hofmann H., Rüegger S., Reymond L., Nettels D., Schuler B. (2010) From the cover: Charge interactions can dominate the dimensions of intrinsically disordered proteins. Proc. Natl. Acad. Sci. U.S.A. 107, 14609–14614 [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.