Background: The function of the CutA divalent cation tolerance homolog (Escherichia coli) (CUTA) is largely unknown.
Results: CUTA has several isoforms, and the longest interacts with BACE1, regulates intracellular trafficking of BACE1, and affects Aβ generation.
Conclusion: CUTA is a novel BACE1-interacting protein and regulates the β-cleavage of APP.
Significance: CUTA may play a role in Alzheimer disease.
Keywords: Alzheimer Disease, Amyloid, Amyloid Precursor Protein, Secretases, Trafficking, β-Site APP Cleaving Enzyme 1, CutA Divalent Cation Tolerance Homolog (E. coli)
Abstract
Accumulation of the neurotoxic β-amyloid (Aβ) peptide in the brain is central to the pathogenesis of Alzheimer disease. Aβ is derived from the β-amyloid precursor protein (APP) through sequential cleavages by β- and γ-secretases, and the production of Aβ is greatly affected by the subcellular localization of these factors. CUTA, the mammalian CutA divalent cation tolerance homolog (E. coli), has been proposed to mediate acetylcholinesterase activity and copper homeostasis, which are important in Alzheimer disease pathology. However, the exact function of CUTA remains largely unclear. Here we show that human CUTA has several variants that differ in their N-terminal length and are separated as heavy (H) and light (L) components. The H component has the longest N terminus and is membrane-associated, whereas the L component is N-terminally truncated at various sites and localized in the cytosol. Importantly, we demonstrate that the H component of CUTA interacts through its N terminus with the transmembrane domain of β-site APP cleaving enzyme 1 (BACE1), the putative β-secretase, mainly in the Golgi/trans-Golgi network. Overexpression and RNA interference knockdown of CUTA can reduce and increase BACE1-mediated APP processing/Aβ secretion, respectively. RNA interference of CUTA decelerates intracellular trafficking of BACE1 from the Golgi/trans-Golgi network to the cell surface and reduces the steady-state level of cell surface BACE1. Our results identify the H component of CUTA as a novel BACE1-interacting protein that mediates the intracellular trafficking of BACE1 and the processing of APP to Aβ.
Introduction
The formation of senile plaques and neurofibrillary tangles are two major pathological hallmarks of Alzheimer disease (AD).2 Senile plaques are extracellular deposits of fibrillar β-amyloid (Aβ) peptide, a proteolytic product of β-amyloid precursor protein (APP) (1). Neurofibrillary tangles are intracellular bundles of self-assembled paired helical filaments composed of hyperphosphorylated microtubule-associated protein Tau (2). Multiple lines of evidence suggest that a primary cause of AD pathogenesis is the overproduction/excessive accumulation of Aβ, which triggers a cascade of neurodegenerative steps, including the formation of senile plaques and intraneuronal fibrillary tangles and neuronal loss in susceptible brain regions (3). Amyloidogenic Aβ peptide is proteolytically derived from APP within the secretory pathway by distinct enzymatic activities known as β- and γ-secretase cleavages (3). Therefore, regulation of these enzymes through their expression level, enzymatic activity, and/or subcellular localization may affect the level of Aβ and be useful in potential therapeutics for AD.
The putative β-secretase, β-site APP cleaving enzyme 1 (BACE1), is a novel membrane-bound aspartyl protease containing a single type I transmembrane domain near its C terminus (4–7). Cleavage of APP by BACE1 is the first step leading to Aβ generation, and BACE1 activity is thought to be the rate-limiting factor in Aβ production. BACE1 deficiency can rescue memory deficits and cholinergic dysfunction in an AD mouse model (Tg2576), correlating with a dramatic reduction in Aβ40/42 levels (8–10). Significantly, several studies show that BACE1 levels and activity are elevated in the brain regions affected by AD (11, 12), suggesting that BACE1 may be a good therapeutic target. Optimal BACE1 activity requires an acidic environment; as expected, BACE1 is primarily localized in the Golgi, trans-Golgi network (TGN), and endosomes. BACE1 is also found on the cell surface (5, 13–15). However, the detailed mechanisms regulating BACE1 trafficking and activity are not well understood.
The mammalian CutA divalent cation tolerance homolog (Escherichia coli), CUTA, was discovered in a search for the membrane anchor of acetylcholinesterase (AChE), an enzyme that degrades acetylcholine and an important target for inhibition during AD treatment (16). Although it was later determined that CUTA does not directly interact with AChE and that the real membrane anchor protein of AChE is PRiMA (17), mouse CUTA was recently found to affect the processing and trafficking of AChE (18). The CUTA protein forms trimers through a region of ∼100 residues that is strongly conserved from bacteria to vertebrates. The trimeric core is preceded by a region of variable length with several variants in mammals (19). In bacteria, CutA is involved in copper tolerance, and some mutations in the cutA gene lead to copper sensitivity due to its increased uptake (20). Additional studies showed that many CutA proteins have a copper binding capacity and that copper could induce reversible aggregation of the CutA protein (21, 22). Furthermore, the trimeric core of CutA is also homologous to that of the intracellular signal transduction protein P-II, implying a role in signal transduction (21). However, the exact functions of the mammalian CUTA protein, especially human CUTA, remain largely unclear.
In this study we show that human CUTA is expressed in various forms. Importantly, we find that the longest CUTA isoform can interact with BACE1 and regulate BACE1 intracellular trafficking, thereby mediating APP processing and Aβ generation.
EXPERIMENTAL PROCEDURES
Cell Culture, Vectors, Small Interfering RNA (siRNA), and Transfection
Human embryonic kidney HEK 293T cells, HeLa cells, and human neuroblastoma SH-SY5Y cells were maintained in DMEM (Mediatech, Manassas, VA) supplemented with 10% fetal bovine serum (Hyclone, Logan, CT). HEK cells stably expressing human APP Swedish mutants (HEK-Swe) were maintained in DMEM (Mediatech) supplemented with 10% fetal bovine serum and 0.4 mg/ml G418 (Omega Scientific, Tarzana, CA). Cells were transiently transfected with pcDNA, BACE1, CUTA, APP, Nicastrin (NCT), and various truncated/mutated BACE1 and CUTA vectors (for more information on vectors used in this study please see supplemental Fig. 1) using TurbofectTM (Fermentas Inc., Glen Burnie, MD) following the manufacturer's protocol. For RNA interference (RNAi), to down-regulate human CUTA expression, three CUTA targeting siRNAs (1, 5′-TGAGGTGCTGATGATGATTAA-3′; 2, 5′-GCGTCAACCTCATCCCTCAGATTAC-3′; 3, 5′-GTAATCTGAGGGATGAGGTTGACGC-3′) and a scrambled control siRNA (Invitrogen) were transfected into cells using Lipofectamine RNAiMAX reagent (Invitrogen) following the manufacturer's protocol.
Western Blot and Antibodies
Treated cells were lysed in a lysis buffer (50 mm Tris-HCl, pH 7.4, 150 mm sodium chloride, 5 mm EDTA, 1% Nonidet P-40, supplemented with a protease inhibitor mixture). Equal protein amounts of cell lysates were subjected to sodium dodecyl sulfate-polyacrylamide gel electrophoresis (SDS-PAGE) followed by Western blot as indicated. Antibodies used here include a rabbit polyclonal antibody against human CUTA (R-CUTA) that was developed by immunizing the rabbit with a recombinant CUTA protein lacking the first 42 amino acids (based on CUTA isoform 1 numbering) and rabbit polyclonal antibodies 689 against BACE1 (23), Ab14 against PS1 N-terminal fragment (NTF) (24), 369 against APP C-terminal fragment (CTF) (25), and 716 against Nicastrin (26). The mouse monoclonal antibody 3D5 against BACE1 was a gift from Dr. Robert Vassar. Mouse anti-α-tubulin, mouse anti-β-actin, and mouse anti-HA antibodies were from Sigma. The mouse monoclonal anti-Myc antibody 9E10 was from Invitrogen. The mouse monoclonal antibody 6E10 against human Aβ and a rabbit polyclonal antibody against sAPPβ were from Covance (Princeton, NJ). Anti-ADAM10, anti-TACE and anti-GAPDH antibodies were from Santa Cruz Biotechnology (Santa Cruz, CA).
RNA Isolation and Quantitative Real-time Polymerase Chain Reaction (RT-PCR)
Total RNA was extracted using TRIzol reagent (Invitrogen). SuperScript First-Strand kit (Invitrogen) was used to synthesize cDNAs. RT-PCR was carried out with an ICycler instrument (Bio-Rad) using the IQTM SYBR Green supermix (Bio-Rad). Primers used for human CUTA were 5′-CCTGCGTCAACCTCATCCC-3′ and 5′-GCAATTACCTCGGCCACTTC-3′. A pair of GAPDH primers was used for control (27).
Co-immunoprecipitation
HEK 293T cells transfected with various vectors as well as human brain tissues were lysed in lysis buffer. Equal protein amounts of cell lysates were incubated with normal rabbit IgG or indicated antibodies together with Trueblot IPTM beads (eBioscience, San Diego, CA) at 4 °C overnight. Immunoprecipitated proteins were analyzed by Western blot.
Cell Surface Biotinylation
Treated cells were washed with ice-cold phosphate-buffered saline containing 1 mm each of CaCl2 and MgCl2 and incubated at 4 °C with 0.5 mg/ml Sulfo-NHS-LC-biotin (Thermo Scientific, Rockford, IL). Cells were then lysed, and lysates were affinity-precipitated with streptavidin-agarose beads (Thermo Scientific). Biotinylated proteins were subjected to Western blot analysis.
BACE1 Activity Assay
The in vitro activity of BACE1 was assayed using a commercial kit (Sigma) following the manufacturer's protocol.
Pulse-Chase and Biotinylation Analysis
293T cells transfected first with BACE1-HA and then with CUTA siRNA or scrambled control siRNA were starved for 30 min and labeled by [35S]methionine (100 μCi/ml) for 15 min at 37 °C. After washing with phosphate-buffered saline, cells were chased in normal growth medium at 20 °C for 2 h to accumulate labeled proteins in TGN. Cells were then incubated for various times at 37 °C. At the end of each chase time, cells were biotinylated at 4 °C. Cell lysates were affinity-precipitated by streptavidin-agarose beads (Thermo Scientific), and biotinylated cell surface proteins were eluted with 2% SDS. After dilution with a Triton X-100 containing buffer, eluted proteins were immunoprecipitated using an anti-HA antibody, separated on SDS-PAGE gels, and analyzed by autoradiography.
Membrane Fractionation
Membrane fractionation assays were carried out as described previously with some modifications (28). Briefly, treated 293T cells were washed with phosphate-buffered saline, collected with homogenization buffer (10 mm Tris, pH 7.4, 1 mm EDTA, 200 mm sucrose, 1 mm phenylmethylsulfonyl fluoride), and homogenized by 10 passages through a 25G 7/8 needle (BD Biosciences). Samples were centrifuged at 900 × g for 10 min to remove cell debris and nuclei. Supernatants were centrifuged at 100,000 × g for 60 min at 4 °C. After transferring the supernatant (cytosol) to a new tube, the pellet was resuspended in 100 mm NaHCO3 buffer, pH 11.3, incubated on ice for 30 min, and centrifuged at 100,000 × g for 60 min at 4 °C. The pellet was washed and resuspended with lysis buffer.
Immunostaining
HeLa cells were transfected with the indicated vectors for 24 h. Cells were fixed in 3.7% paraformaldehyde, permeabilized, blocked in 5% bovine serum albumin, and then incubated with antibodies against Myc, HA, TGN46 (Novus Biologicals, Littleton, CO), EEA1 (BD Biosciences), and/or LAMP2 (Santa Cruz Biotechnology) for 1 h. Cells were then incubated with Alexa Fluor 350, 488, and 594-conjugated secondary antibodies. Specimens were examined under a Decon microscope.
RESULTS
Human CUTA Has Various Forms That Localize to Different Subcellular Compartments
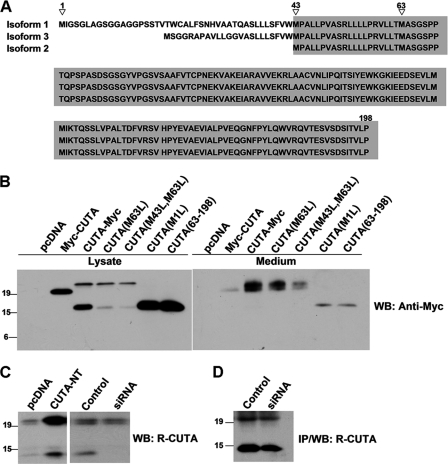
The human CUTA gene is reported to have five variant transcripts that encode three protein isoforms (Fig. 1A). The longest CUTA isoform (isoform 1) has 198 amino acids. CUTA isoform 3 has a different N terminus than that of isoform 1, but the two share an identical sequence at the C terminus. CUTA isoform 2 is completely contained within isoform 1 and isoform 3 and starts at a downstream methionine at position 43 (numbered according to isoform 1). In addition, another methionine codon at position 63 (Met-63), shared by all three CUTA isoforms, possibly serves as another translation start site, as its corresponding codon in the mouse cuta gene has been found to be a translation start site (18). It has previously been shown that mouse cuta variants are initiated at three in-frame ATG codons, and their encoded proteins can be separated as heavy and light components by SDS-PAGE (18).
FIGURE 1.
Human CUTA has several variants that can be separated into heavy and light components. A, alignment of the three known human CUTA isoforms is shown. Identical amino acids shared by the three isoforms are highlighted in gray. Potential translation initiation methionine sites are indicated by triangles. B, HEK 293T cells were transfected with control pcDNA, N-terminal tagged CUTA (Myc-CUTA), C-terminal tagged CUTA (CUTA-Myc), C-terminal Myc-tagged CUTA with methionine 1 mutated to leucine (M1L), methionine 63 mutated to leucine (M63L), or methionines 43 and 63 mutated to leucines (M43L, M63L), and C-terminal Myc-tagged CUTA that had amino acids 1–62 of its N terminus truncated (63–198). After 44 h, culture media was changed to fresh media. After another 4 h, conditioned media was collected, and cells were lysed. Equal protein amounts of cell lysates and equal volume amounts of conditioned media were subjected to SDS-PAGE and Western blot (WB) using the Myc antibody 9E10. C, cells were transfected with pcDNA or non-tagged CUTA (CUTA-NT) (left panel) or transfected with control siRNA or CUTA siRNA 3 (right panel). Equal protein amounts of cell lysates were subjected to SDS-PAGE and Western blot using the R-CUTA antibody. D, lysates containing equalized amounts of protein from cells transfected with control siRNA or CUTA siRNA 3 were incubated with the R-CUTA antibody for immunoprecipitation (IP). Immunoprecipitated proteins were analyzed by Western blot using the same antibody.
To characterize human CUTA, we overexpressed human CUTA isoform 1 with a Myc tag at either the N or the C terminus in HEK 293T cells. SDS-PAGE and Western blots with a Myc antibody detected only one band in cells transfected with Myc-CUTA. This band has a molecular mass of about 22 kDa that is consistent with full-length CUTA (including the Myc tag and linker sequence) (Fig. 1B). In cells transfected with CUTA-Myc, there were two bands detected at 24 kDa (heavy component (H)) and 17 kDa (light component (L)) (Fig. 1B); the molecular mass of the H component is consistent with that of full-length CUTA (including the Myc tag and linker sequence).
Because the translation of mouse cuta can be initiated at different methionine codons (18), we also assessed the use of various methionine codons for translation in human CUTA by mutagenesis studies. As shown in Fig. 1B, when methionine 43 was mutated into leucine (M43L), or both methionines 43 and 63 were mutated into leucines (M43L, M63L), the generation of the L component of CUTA was greatly reduced. When methionine 1 was mutated into leucine (M1L), there was no generation of the H component of CUTA but a strong generation of the L component whose molecular mass was the same as that of CUTA-(63–198) (Fig. 1B and supplemental Fig. 2). These results are consistent with the findings in mouse cuta and suggest that human CUTA translation can be initiated at different sites, methionines 1, 43, and 63. Furthermore, in cells overexpressing CUTA-(43–198)/isoform 2, the isoform thought to represent CUTA because of its conservation among the three known isoforms (29), both CUTA-(43–198) and CUTA-(63–198) were detected, with the latter showing a higher expression level (supplemental Fig. 2), suggesting that methionine 63 may be a major initiation site for the L component of CUTA. Alternatively, it is possible that the L component of CUTA may also be derived through cleavage of the H component, as proposed previously (18, 29). We also found that mutations of M43L and M63L did not completely abolish the generation of the L component (Fig. 1B). However, based on the molecular mass difference between the H and the L components, cleavage of CUTA should also generate an N-terminal fragment with a molecular mass of about 7 kDa, and this was not observed in cells transfected with Myc-CUTA (Fig. 1B). One possible explanation is that the generated N-terminal fragment may be quickly degraded and/or cleaved into smaller fragments and thus cannot be detected. Nevertheless, our results demonstrate that human CUTA has various forms that can be separated as H and L components. The H component represents CUTA that is translated at methionine 1, and the L component includes mostly CUTA isoforms translated at methionines 43 (i.e. CUTA isoform 2) and 63, and a small fraction of CUTA cleavage products.
It was previously found that both the H and the L component of mouse CUTA can be secreted into the media (18). Here we found that both the H and L component of human CUTA were also secreted (Fig. 1B and supplemental Fig. 9). In CUTA-Myc-transfected cells that express both the H and the L component, the dominantly secreted form is the H component (Fig. 1B and supplemental Fig. 9), suggesting that the H component is preferentially secreted over the L component. Moreover, although the expression level of Myc-CUTA was higher than that of the H component of CUTA-Myc, the secretion of the former was lower than that of the latter (Fig. 1B), implying that the Myc modification of the N terminus may affect its secretion.
To study endogenous human CUTA, we developed a rabbit polyclonal antibody against CUTA-(43–198). In Western blot experiments, this R-CUTA antibody recognized two bands with molecular masses of 20 and 13 kDa in human SH-SY5Y cells transfected with control pcDNA and with non-tagged CUTA (Fig. 1C, left panel). The intensity of the two bands in cells transfected with non-tagged CUTA was much stronger than in cells transfected with pcDNA, suggesting that the two bands represent endogenous CUTA H and L components. However, when cells were subjected to RNA interference (RNAi) to down-regulate endogenous CUTA, a Western blot using this antibody only showed a significant down-regulation of the L component but not the H component of CUTA (Fig. 1C, right panel). We believe that the R-CUTA antibody recognizes some nonspecific band right at the same molecular mass as the H component of CUTA. This is supported by the finding that the H component band intensity was much stronger than that of the L component when detected by Western blot using this antibody (Fig. 1C), whereas the expression of the H component seemed to be much less than that of the L component when detected using the Myc antibody in cells transfected with CUTA-Myc (Fig. 1B).
To exclude potential interference from nonspecific band recognized by the R-CUTA antibody, we carried out an immunoprecipitation-Western blot using R-CUTA and indeed found that this antibody immunoprecipitated the H component of CUTA as lower levels than the L component (Fig. 1D). In addition, in cells transfected with different CUTA siRNAs, this antibody immunoprecipitated much less CUTA (both H and L components) than that in control cells, confirming the efficacy of RNAi in down-regulating both H and L components of CUTA (Fig. 1D). Together, these results show that endogenous human CUTA is also expressed as heavy and light components.
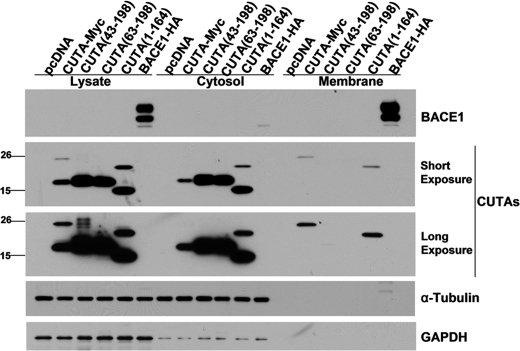
Although it was previously found in stably transfected cells that CUTA isoform 2, part of the L component, was localized in the mitochondria (29), another study showed that the vast majority of the mouse CUTA L component was in the cytosol, whereas the H component was in microsomes (18). Herein, we also found that the H component of human CUTA was associated with the membrane fraction, whereas the L component of human CUTA was in the cytosol (Fig. 2). As expected, CUTA-(43–198)/isoform 2 and CUTA-(63–198) of the L component were found only in the cytosol (Fig. 2). However, the H component of a C terminus-truncated CUTA form, CUTA-(1–164), was found both in the membrane and the cytosol, whereas the L component of CUTA-(1–164) was only found in the cytosol (Fig. 2). Therefore, our results suggest that the H component of human CUTA associates with membranes through its N terminus at amino acids 1–42, and the C terminus of CUTA might also contribute to its association with membranes, depending on the presence of the N terminus. Moreover, when the membrane fractions were treated with NaHCO3, pH 11.3, to detach peripheral membrane proteins from membranes, only the membrane bound GAPDH (30), but not the CUTA H component, was dissociated from the membrane fractions (Fig. 2 and supplemental Fig. 3), suggesting that the H component of CUTA is probably integrated into the membrane through its N terminus. The small amount of CUTA L component found in the NaHCO3 washing buffer (supplemental Fig. 3) may imply that some CUTA L component in the cytosol is loosely associated with membranes.
FIGURE 2.
The heavy component of CUTA containing the N terminus is associated with the membrane, whereas the light component of CUTA is located in the cytosol. Cells were transfected with pcDNA, full-length CUTA, CUTA with N-terminal amino acids 1–42 truncated (43–198), CUTA with N-terminal amino acids 1–62 truncated (63–198), CUTA with C-terminal amino acids 165–198 truncated (1–164), or BACE1-HA. After homogenization of cells, a small sample was used to analyze total lysates, and the rest was subjected to centrifuge to separate the cytosolic fraction and the membrane. The membrane pellet was rinsed with NaHCO3, pH 11.3, to dissociate peripheral membrane proteins and then resuspended in lysis buffer in a volume equal to that of the cytosolic fraction. Equal volume amounts of samples were subjected to SDS-PAGE and Western blot to detect CUTA (using the Myc antibody 9E10), GAPDH (to indicate membrane-bound protein), BACE1 (using an HA antibody to indicate the membrane fraction) and α-tubulin (to indicate the cytosolic fraction).
CUTA Regulates BACE1-mediated APP Processing
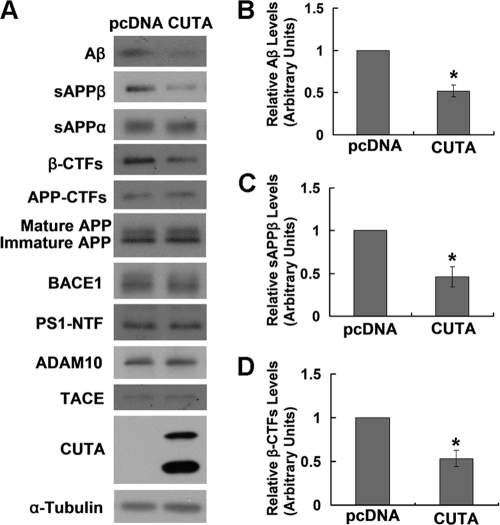
Because both copper and AChE are potentially affected by CUTA and play a role in AD, we next explored the possible involvement of CUTA in AD by studying its effect on APP processing/Aβ production, one of the most important events in AD pathogenesis. Notably, when CUTA isoform 1 was overexpressed in HEK-Swe cells, there was a significant decrease (∼50%) in the level of secreted Aβ (Fig. 3 A and B), accompanied by a dramatic decrease in the level of sAPPβ (Fig. 3, A and C) and APP β-C-terminal fragments (β-CTFs) (Fig. 3, A and D), two products derived from APP through BACE1 cleavage. These data strongly suggest that CUTA regulates BACE1-mediated APP processing. However, overexpression of CUTA had no effect on the protein levels of full-length APP (both immature and mature forms), BACE1, the NTF of PS1 that is an important γ-secretase component, and ADAM10 and TACE, two major α-secretases (Fig. 3A).
FIGURE 3.
Overexpression of CUTA reduces β-processing of APP. A, HEK-Swe cells were transfected with control pcDNA and CUTA-Myc. The sAPPα and sAPPβ in conditioned media were Western-blotted with respective antibodies. Aβ secreted in conditioned media was precipitated by trichloroacetic acid and Western-blotted with 6E10. Cell lysates were Western-blotted with antibodies against total APP (mature and immature forms) and APP-CTFs (369), β-CTFs (6E10), BACE1 (3D5), PS1 NTF (Ab14), ADAM10, TACE, CUTA (Myc), and α-tubulin. The levels of Aβ (B), sAPPβ (C), and APP β-CTFs (D) were quantified by densitometry and normalized to those of controls for comparison (set as one arbitrary unit). *, p < 0.05, n = 3.
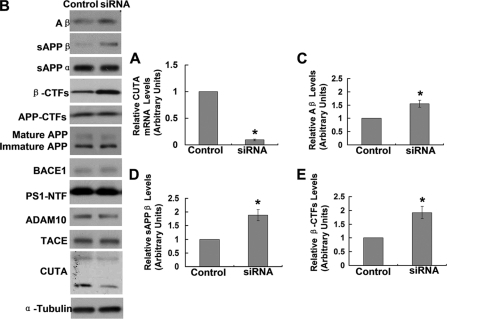
To further confirm the regulation of BACE1-mediated APP proteolysis by CUTA, we down-regulated the mRNA (Fig. 4A and supplemental Fig. 4A) and protein (Fig. 4B and supplemental Fig. 4B) levels of endogenous CUTA by RNAi. We found that the levels of Aβ (Fig. 4, B and C, and supplemental Fig. 4B), sAPPβ (Fig. 4, B and D, and supplemental Fig. 4B), and β-C-terminal fragments (β-CTFs) (Fig. 4, B and E) were markedly increased upon down-regulation of CUTA, whereas the protein levels of BACE1, full-length APP (both immature and mature forms), total APP CTFs, PS1-NTF, ADAM10, and TACE were unaffected (Fig. 4B).
FIGURE 4.
Down-regulation of CUTA increases β-processing of APP. HEK-Swe cells were transfected with control siRNA and CUTA siRNA 3. A, cells were subjected to RNA extraction and gene analysis by RT-PCR. The mRNA level of CUTA was normalized to that of GAPDH for comparison. *, p < 0.05, n = 3. B, the sAPPα and sAPPβ in conditioned media were Western-blotted with respective antibodies. Aβ secreted in conditioned media was precipitated by trichloroacetic acid and Western-blotted with 6E10. Cell lysates were Western-blotted with antibodies against total APP (mature and immature forms) and APP-CTFs (369), BACE1 (3D5), PS1 NTF (Ab14), ADAM10, TACE, CUTA (Myc), and α-tubulin. The levels of Aβ (C), sAPPβ (D), and APP β-CTFs (E) were quantified by densitometry and normalized to those of controls for comparison (set as one arbitrary unit). *, p < 0.05, n = 3.
H Component of CUTA Isoform 1 Interacts with BACE1
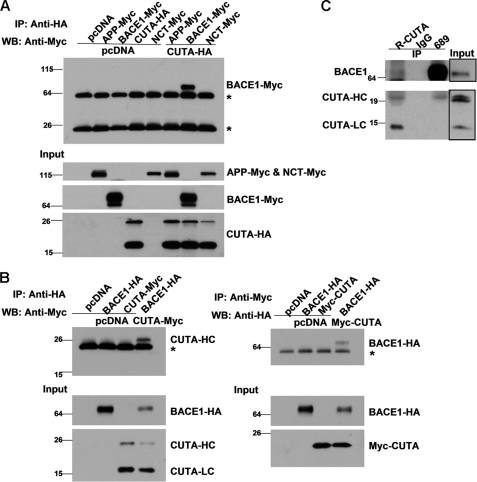
Because overexpression of CUTA had no effect on the protein levels of BACE1, it is possible that the enzymatic activity of BACE1 is not affected by CUTA. Indeed, our results showed that overexpression of CUTA did not alter in vitro enzymatic activity of BACE1 (supplemental Fig. 5). So we postulated that CUTA may affect BACE1-mediated APP processing through another mechanism and tested whether CUTA can interact with BACE1. When full-length CUTA-HA was co-expressed with BACE1, APP, or Nicastrin (NCT, an important γ-secretase component) that were all Myc-tagged at the C terminus, immunoprecipitation with an HA antibody only pulled down BACE1 but not APP or NCT (Fig. 5A). When full-length CUTA-Myc and BACE1-HA were co-expressed, the HA antibody pulled down the H component but not the L component of CUTA (Fig. 5B, left panels). A Myc antibody also pulled down BACE1 in cells co-expressing Myc-CUTA and BACE1-HA (Fig. 5B, right panels). Moreover, we carried out a co-immunoprecipitation study in human brain lysates and found that the CUTA antibody R-CUTA immunoprecipitated endogenous BACE1, whereas the BACE1 antibody 689 immunoprecipitated the endogenous CUTA H component (Fig. 5C). Together, these results demonstrate that the H but not the L component of CUTA isoform 1 can interact with BACE1. Because the N terminus of CUTA isoform 3 is different from and shorter than that of CUTA isoform 1, we also tested it and found that CUTA isoform 3 did not interact with BACE1 (supplemental Fig. 6), suggesting that the N terminus of CUTA isoform 1 mediates its unique interaction with BACE1.
FIGURE 5.
The H component of CUTA interacts with BACE1. A, CUTA with an HA tag at the C terminus (CUTA-HA) was co-transfected with APP, BACE1, or NCT, all of which were Myc-tagged at the C terminus into HEK 293T cells. Cells co-transfected with pcDNA, and the indicated vectors were used as controls. Equal protein amounts of cell lysates were used for immunoprecipitation (IP) with an HA antibody and Western blot (WB) with the Myc antibody 9E10 to detect CUTA-interacting proteins. Ten percent of cell lysates were used as input to detect the expression of transfected proteins. B, pcDNA, BACE1-HA, and CUTA-Myc (left panel) or Myc-CUTA (right panel) were pairwise co-transfected into HEK 293T cells. Equal protein amounts of cell lysates were immunoprecipitated with an HA antibody and Western-blotted with a Myc antibody (left panels) or immunoprecipitated with the Myc antibody and Western-blotted with the HA antibody (right panels). Ten percent of cell lysates were used as input. *, nonspecific bands. C, the lysates of human brain tissues were immunoprecipitated with normal rabbit IgG, the R-CUTA antibody, or the BACE1 antibody 689. Immunoprecipitated proteins and input (5% of cell lysates) were subjected to SDS-PAGE and Western blot using another anti-BACE1 antibody (3D5) or R-CUTA.
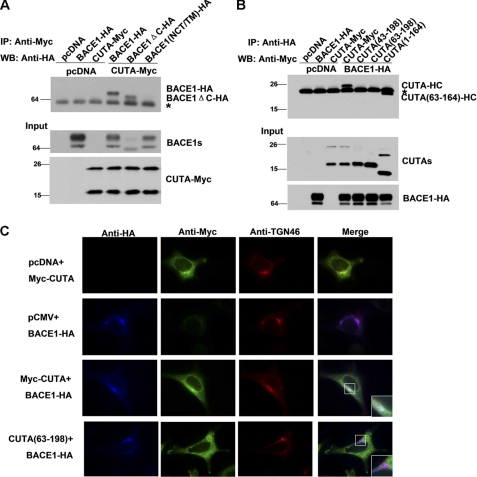
Because BACE1 is a type I transmembrane protein and the H component of CUTA isoform 1 is associated with the membrane through its N terminus, we investigated the domains important for BACE1 interaction with CUTA. We first generated a BACE1 chimera mutant (BACE1(NCT/TM)) that has its transmembrane domain substituted with the NCT transmembrane domain. We found that BACE1(NCT/TM) was delivered to the cell surface similarly to that of wild type BACE1 (supplemental Fig. 7A) and that overexpression of BACE1(NCT/TM) dramatically increased the levels of APP β-CTFs and sAPPβ as does wild type BACE1 (supplemental Fig. 7B), indicating that such a transmembrane domain replacement does not affect the trafficking or enzymatic activity of BACE1. However, we found that this BACE1 chimera mutant did not interact with CUTA anymore (Fig. 6A). In addition, we found that overexpression of CUTA only reduced the levels of Aβ, sAPPβ, and APP β-CTFs in cells overexpressing wild type BACE1 but not in cells overexpressing BACE1(NCT/TM) (supplemental Fig. 7C). A deletion of the C terminus domain of BACE1 had no effect on its interaction with CUTA (Fig. 6A). In addition, in cells co-expressing BACE1-HA and various CUTA forms, the HA antibody only pulled down the H components of CUTA isoform 1 and CUTA-(1–164) but not their L components, CUTA-(43–198) or CUTA-(63–198) (Fig. 6B). These results suggest that the transmembrane domain of BACE1 mediates BACE1 interaction with the N terminus of CUTA isoform 1.
FIGURE 6.
The N terminus of CUTA and the transmembrane domain of BACE1 are crucial for their interaction in the Golgi/TGN. A, CUTA-Myc was co-transfected with BACE1, BACE1 lacking the C terminus (BACE1-ΔC), or BACE1 that has its transmembrane domain substituted with that of Nicastrin (BACE1(NCT/TM)), all of which were HA-tagged at the C terminus, into HEK 293T cells. Cells co-transfected with pcDNA and the indicated vectors were used as controls. Equal protein amounts of cell lysates were used for immunoprecipitation (IP) with a Myc antibody and Western blot (WB) with an HA antibody. Ten percent of cell lysates were used as input. B, BACE1-HA was co-transfected with CUTA, CUTA-(43–198), CUTA-(63–198), or CUTA-(1–164), all of which were Myc-tagged at the C terminus into HEK 293T cells. Equal protein amounts of cell lysates were used for IP with an HA antibody and Western blot with a Myc antibody. Ten percent of cell lysates were used as input. *, nonspecific bands. C, HeLa cells were transfected with pcDNA, Myc-CUTA, CUTA-(63–198), and/or BACE1-HA, as indicated. Cells were then fixed, permeabilized, and immunostained with primary antibodies against HA (indicating BACE1, in blue), Myc (indicating CUTA, in green), and TGN46 (indicating the Golgi/TGN organelle, in red), and respective secondary antibodies conjugated with Alexa Fluor 350, 488, or 594. Samples were examined by immunofluorescence microscopy.
BACE1 is mainly localized in acidic environments such as the Golgi/TGN and endosomes. To determine the subcellular compartments in which CUTA and BACE1 interact, we co-expressed Myc-CUTA and BACE1-HA and carried out immunostaining with Myc and HA antibodies. Our results showed that a large amount of full-length CUTA and BACE1 colocalized with each other, and their colocalization is mainly at TGN46 (Golgi/TGN marker)-positive sites (Fig. 6C), not at EEA1 (early endosome marker)-positive or LAMP2 (late endosome marker)-positive sites (supplemental Fig. 8), indicating that the interaction between CUTA and BACE1 mainly occurs in the Golgi/TGN. CUTA-(63–198), an L component of CUTA, was found to be expressed throughout the cytosol and showed no colocalization with BACE1 or TGN46 (Fig. 6C).
We also explored whether the interaction between CUTA and BACE1 has any effect on CUTA secretion. However, our results showed that overexpression of BACE1 did not alter the secretion of CUTA (supplemental Fig. 9), suggesting that the secretion of CUTA may be mediated by other proteins than BACE1.
H Component of CUTA Isoform 1 Regulates APP Processing through Mediation of BACE1 Trafficking
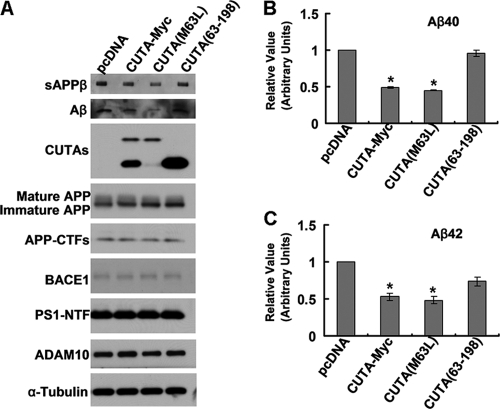
Because only the H component of CUTA isoform 1 interacts with BACE1, we speculate that only the H component, not the L component, is responsible for mediating BACE1-regulated APP processing. Indeed, only upon transfection of CUTA-Myc and CUTA(M63L) that overexpress the H component were the levels of sAPPβ and Aβ (Fig. 7A), including Aβ 40 and 42 (Fig. 7, B and C), dramatically reduced. Overexpression of CUTA-(63–198), which does not interact with BACE1, had no effect on the levels of sAPPβ and Aβ (Fig. 7A).
FIGURE 7.
Only BACE1-interacting CUTA forms regulate β-processing of APP and Aβ secretion. HEK-Swe cells were transfected with control pcDNA, CUTA-Myc, CUTA(M63L), or CUTA-(63–198). A, secreted Aβ and sAPPβ in conditioned media and mature/immature APP, APP-CTFs, BACE1, PS1-NTF, ADAM10, CUTA, and α-tubulin in cell lysates were detected by Western blot with respective antibodies. Secreted Aβ40 (B) and Aβ42 (C) in conditioned media were quantified by ELISA. The levels of Aβ40 and Aβ42 were normalized to those of controls for comparison (set as one arbitrary unit). *, p < 0.05, n = 4.
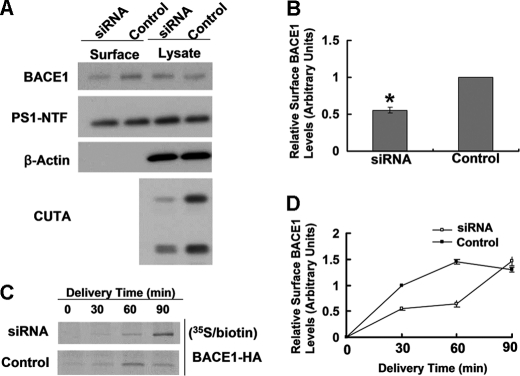
Because altered protein trafficking of BACE1 has been found to affect its cleavage of APP, we studied whether intracellular trafficking of BACE1 is affected by CUTA. When CUTA was down-regulated by RNAi, we found that the steady-state level of cell surface BACE1 was significantly decreased, whereas the level of cell surface PS1-NTF and the total levels of BACE1 and PS1-NTF were unaffected (Fig. 8, A and B). In addition, as expected, cell surface level of BACE1(NCT/TM), which does not interact with CUTA, was not affected by CUTA overexpression (supplemental Fig. 7D).
FIGURE 8.
Down-regulation of CUTA slows cell surface delivery of BACE1 and reduces the steady-state level of BACE1 at the cell surface. A, after RNAi down-regulation of CUTA, live SH-SY5Y cells were subjected to biotinylation. Cell lysates were affinity-precipitated with streptavidin-agarose beads. The levels of biotinylated BACE1 and PS1-NTF as well as their total protein levels were analyzed by Western blot. B, cell surface levels of BACE1 in A were quantified by densitometry and normalized to that of a control (set as one arbitrary unit) for comparison. *, p < 0.05, n = 3. C, upon down-regulation of CUTA by RNAi, HEK 293T cells transfected with BACE1-HA were pulse-labeled with [35S]methionine for 15 min and incubated at 20 °C to accumulate isotope-labeled proteins in the TGN. Cells were then incubated at 37 °C for the indicated periods followed by biotinylation at 4 °C. After affinity precipitation, biotinylated proteins were immunoprecipitated with an anti-HA antibody and then subjected to SDS-PAGE and autoradiography. D, the band intensity in C was quantified by densitometry for comparison.
The steady-state level of BACE1 at the cell surface is dependent on an equilibrium between the transport of BACE1 from the Golgi/TGN to the cell surface and the endocytosis of BACE1 from the cell surface to endosomes. Because CUTA and BACE1 mainly colocalize in the Golgi/TGN but not in endosomes (Fig. 6C and supplemental Fig. 5), CUTA probably affects the transport of BACE1 to the cell surface. To investigate this, we performed pulse-chase and biotinylation analyses. We found that in control cells the majority of [35S]methionine-labeled BACE1 arrived at the cell surface 60 min after chasing (Fig. 8, C and D). However, in CUTA-down-regulated cells, the time for the majority of [35S]methionine-labeled BACE1 to reach the cell surface was delayed to 90 min after chasing (Fig. 8, C and D), indicating that down-regulation of CUTA slows cell surface delivery of BACE1 from the Golgi/TGN.
DISCUSSION
Optimal activity of BACE1 requires an acidic environment. Consistently, BACE1 localizes to acidic subcellular compartments such as the Golgi/TGN and endosomes. In addition, BACE1 can also be detected at the cell surface during its intracellular trafficking through the secretory pathway (5, 13–15). Some previous studies have identified several BACE1-interacting proteins, including reticulon/Nogo (31–33), Golgi-localized γ-ear-containing ARF-binding protein (34–36), and sorting nexin 6 (37), all of whose disturbance will affect the normal intracellular trafficking of BACE1 and result in altered β-processing of APP for Aβ production. Herein, we have identified another BACE1-interacting protein, the full-length CUTA isoform 1 that regulates intracellular trafficking of BACE1 from the Golgi/TGN to the cell surface. Our results show that the N terminus of full-length CUTA isoform 1 can bind to the transmembrane domain of BACE1, mainly in the Golgi/TGN, and that a reduction in CUTA leads to delayed cell surface delivery of BACE1, leading BACE1 to cleave more APP for Aβ generation in the Golgi/TGN, which is the major subcellular localization of APP at steady state (25, 38, 39).
CUTA is conserved across a wide range of species from bacteria to mammals and ubiquitously expressed in all tissues, suggesting that CUTA exerts some fundamental function in the cell. Some studies have suggested that CUTA may play roles in copper tolerance/toxicity (20, 40), AChE activity (18), signal transduction (21), etc. However, so far the exact functions of CUTA, especially those of human CUTA, remain largely undetermined. In this study we find that human CUTA is expressed in various isoforms that differ by the length of the N terminus and can be separated into H and L components by SDS-PAGE. Notably, the H component, representing full-length CUTA or isoform 1, is membrane-associated and may have its N terminus integrated into the membrane, whereas the cytosolic L component contains other CUTA isoforms shortened at the N terminus and possibly a small fraction of CUTA cleavage products. Different subcellular localizations of the H and the L components of CUTA suggest that the two components may have different functions in the cell. Indeed, our results show that only full-length CUTA isoform 1 and not the other CUTA isoforms in the L component can interact with BACE1 and regulate β-cleavage of APP. In another study, full-length mouse CUTA was also found to mediate the processing and trafficking of AChE, possibly through intermediate proteins as CUTA does not directly interact with AChE (18). The L component of CUTA might be related to the cell response to copper, as one study found that overexpression of CUTA isoform 2 enhanced the cytotoxicity of copper (40).
Together, our results show that human CUTA can interact with BACE1 and thus mediate APP processing and Aβ generation. Because CUTA has also been proposed to mediate AChE activity and copper homeostasis, another two important events in AD pathology, CUTA may play an important role in AD through multiple pathways; this deserves further scrutiny.
Supplementary Material
Acknowledgments
We thank Dr. Robert Vassar for providing the 3D5 antibody, Dr. Eliezer Masliah for providing human brain tissue samples, and Dr. Yaomin Chen for providing the BACE1(NCT/TM)-HA construct.
This work was supported by grants from the Alzheimer Association (to Y.-W. Z.), National Natural Science Foundation of China (Grants 30973150 and 81161120496), 973 Prophase Project (Grant 2010CB535004), and Natural Science Foundation of Fujian Province of China Grants 2009J06022 (to Y.-W. Z.) and 2010J01235 (to X. Z.).

This article contains supplemental Figs. 1–9,
- AD
- Alzheimer disease
- Aβ
- β-amyloid
- AChE
- acetylcholinesterase
- APP
- amyloid precursor protein
- sAPP
- soluble APP
- BACE1
- β-site APP cleaving enzyme 1
- CTF
- C-terminal fragment
- NTF
- N-terminal fragment
- CUTA
- CutA divalent cation tolerance homolog (E. coli)
- H
- heavy
- L
- light
- NCT
- nicastrin
- TGN
- trans-Golgi network.
REFERENCES
- 1. Glenner G. G., Wong C. W. (1984) Alzheimer disease. Initial report of the purification and characterization of a novel cerebrovascular amyloid protein. Biochem. Biophys. Res. Commun. 120, 885–890 [DOI] [PubMed] [Google Scholar]
- 2. Buée L., Bussière T., Buée-Scherrer V., Delacourte A., Hof P. R. (2000) Tau protein isoforms, phosphorylation, and role in neurodegenerative disorders. Brain Res. Brain Res. Rev. 33, 95–130 [DOI] [PubMed] [Google Scholar]
- 3. Zhang Y. W., Thompson R., Zhang H., Xu H. (2011) APP processing in Alzheimer disease. Mol. Brain 4, 3. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4. Sinha S., Anderson J. P., Barbour R., Basi G. S., Caccavello R., Davis D., Doan M., Dovey H. F., Frigon N., Hong J., Jacobson-Croak K., Jewett N., Keim P., Knops J., Lieberburg I., Power M., Tan H., Tatsuno G., Tung J., Schenk D., Seubert P., Suomensaari S. M., Wang S., Walker D., Zhao J., McConlogue L., John V. (1999) Purification and cloning of amyloid precursor protein β-secretase from human brain. Nature 402, 537–540 [DOI] [PubMed] [Google Scholar]
- 5. Vassar R., Bennett B. D., Babu-Khan S., Kahn S., Mendiaz E. A., Denis P., Teplow D. B., Ross S., Amarante P., Loeloff R., Luo Y., Fisher S., Fuller J., Edenson S., Lile J., Jarosinski M. A., Biere A. L., Curran E., Burgess T., Louis J. C., Collins F., Treanor J., Rogers G., Citron M. (1999) β-Secretase cleavage of Alzheimer amyloid precursor protein by the transmembrane aspartic protease BACE. Science 286, 735–741 [DOI] [PubMed] [Google Scholar]
- 6. Yan R., Bienkowski M. J., Shuck M. E., Miao H., Tory M. C., Pauley A. M., Brashier J. R., Stratman N. C., Mathews W. R., Buhl A. E., Carter D. B., Tomasselli A. G., Parodi L. A., Heinrikson R. L., Gurney M. E. (1999) Membrane-anchored aspartyl protease with Alzheimer disease β-secretase activity. Nature 402, 533–537 [DOI] [PubMed] [Google Scholar]
- 7. Lau K. F., McLoughlin D. M., Standen C., Miller C. C. (2000) X11 α and x11 β interact with presenilin-1 via their PDZ domains. Mol. Cell. Neurosci. 16, 557–565 [DOI] [PubMed] [Google Scholar]
- 8. Luo Y., Bolon B., Kahn S., Bennett B. D., Babu-Khan S., Denis P., Fan W., Kha H., Zhang J., Gong Y., Martin L., Louis J. C., Yan Q., Richards W. G., Citron M., Vassar R. (2001) Mice deficient in BACE1, the Alzheimer β-secretase, have normal phenotype and abolished β-amyloid generation. Nat. Neurosci. 4, 231–232 [DOI] [PubMed] [Google Scholar]
- 9. Ohno M., Sametsky E. A., Younkin L. H., Oakley H., Younkin S. G., Citron M., Vassar R., Disterhoft J. F. (2004) BACE1 deficiency rescues memory deficits and cholinergic dysfunction in a mouse model of Alzheimer disease. Neuron 41, 27–33 [DOI] [PubMed] [Google Scholar]
- 10. Ohno M., Cole S. L., Yasvoina M., Zhao J., Citron M., Berry R., Disterhoft J. F., Vassar R. (2007) BACE1 gene deletion prevents neuron loss and memory deficits in 5XFAD APP/PS1 transgenic mice. Neurobiol. Dis. 26, 134–145 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11. Yang L. B., Lindholm K., Yan R., Citron M., Xia W., Yang X. L., Beach T., Sue L., Wong P., Price D., Li R., Shen Y. (2003) Elevated β-secretase expression and enzymatic activity detected in sporadic Alzheimer disease. Nat. Med. 9, 3–4 [DOI] [PubMed] [Google Scholar]
- 12. Johnston J. A., Liu W. W., Todd S. A., Coulson D. T., Murphy S., Irvine G. B., Passmore A. P. (2005) Expression and activity of β-site amyloid precursor protein-cleaving enzyme in Alzheimer disease. Biochem. Soc. Trans. 33, 1096–1100 [DOI] [PubMed] [Google Scholar]
- 13. Walter J., Fluhrer R., Hartung B., Willem M., Kaether C., Capell A., Lammich S., Multhaup G., Haass C. (2001) Phosphorylation regulates intracellular trafficking of β-secretase. J. Biol. Chem. 276, 14634–14641 [DOI] [PubMed] [Google Scholar]
- 14. Huse J. T., Pijak D. S., Leslie G. J., Lee V. M., Doms R. W. (2000) Maturation and endosomal targeting of β-site amyloid precursor protein-cleaving enzyme. The Alzheimer disease β-secretase. J. Biol. Chem. 275, 33729–33737 [DOI] [PubMed] [Google Scholar]
- 15. Huse J. T., Liu K., Pijak D. S., Carlin D., Lee V. M., Doms R. W. (2002) β-Secretase processing in the trans-Golgi network preferentially generates truncated amyloid species that accumulate in Alzheimer disease brain. J. Biol. Chem. 277, 16278–16284 [DOI] [PubMed] [Google Scholar]
- 16. Perrier A. L., Cousin X., Boschetti N., Haas R., Chatel J. M., Bon S., Roberts W. L., Pickett S. R., Massoulié J., Rosenberry T. L., Krejci E. (2000) Two distinct proteins are associated with tetrameric acetylcholinesterase on the cell surface. J. Biol. Chem. 275, 34260–34265 [DOI] [PubMed] [Google Scholar]
- 17. Perrier A. L., Massoulié J., Krejci E. (2002) PRiMA: the membrane anchor of acetylcholinesterase in the brain. Neuron 33, 275–285 [DOI] [PubMed] [Google Scholar]
- 18. Liang D., Nunes-Tavares N., Xie H. Q., Carvalho S., Bon S., Massoulié J. (2009) Protein CutA undergoes an unusual transfer into the secretory pathway and affects the folding, oligomerization, and secretion of acetylcholinesterase. J. Biol. Chem. 284, 5195–5207 [DOI] [PubMed] [Google Scholar]
- 19. Savchenko A., Skarina T., Evdokimova E., Watson J. D., Laskowski R., Arrowsmith C. H., Edwards A. M., Joachimiak A., Zhang R. G. (2004) X-ray crystal structure of CutA from Thermotoga maritima at 1.4 Å resolution. Proteins 54, 162–165 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20. Fong S. T., Camakaris J., Lee B. T. (1995) Molecular genetics of a chromosomal locus involved in copper tolerance in Escherichia coli K-12. Mol. Microbiol. 15, 1127–1137 [DOI] [PubMed] [Google Scholar]
- 21. Arnesano F., Banci L., Benvenuti M., Bertini I., Calderone V., Mangani S., Viezzoli M. S. (2003) The evolutionarily conserved trimeric structure of CutA1 proteins suggests a role in signal transduction. J. Biol. Chem. 278, 45999–46006 [DOI] [PubMed] [Google Scholar]
- 22. Tanaka Y., Tsumoto K., Nakanishi T., Yasutake Y., Sakai N., Yao M., Tanaka I., Kumagai I. (2004) Structural implications for heavy metal-induced reversible assembly and aggregation of a protein. the case of Pyrococcus horikoshii CutA. FEBS Lett. 556, 167–174 [DOI] [PubMed] [Google Scholar]
- 23. Yan R., Han P., Miao H., Greengard P., Xu H. (2001) The transmembrane domain of the Alzheimer β-secretase (BACE1) determines its late Golgi localization and access to β-amyloid precursor protein (APP) substrate. J. Biol. Chem. 276, 36788–36796 [DOI] [PubMed] [Google Scholar]
- 24. Thinakaran G., Borchelt D. R., Lee M. K., Slunt H. H., Spitzer L., Kim G., Ratovitsky T., Davenport F., Nordstedt C., Seeger M., Hardy J., Levey A. I., Gandy S. E., Jenkins N. A., Copeland N. G., Price D. L., Sisodia S. S. (1996) Endoproteolysis of presenilin 1 and accumulation of processed derivatives in vivo. Neuron 17, 181–190 [DOI] [PubMed] [Google Scholar]
- 25. Xu H., Sweeney D., Wang R., Thinakaran G., Lo A. C., Sisodia S. S., Greengard P., Gandy S. (1997) Generation of Alzheimer β-amyloid protein in the trans-Golgi network in the apparent absence of vesicle formation. Proc. Natl. Acad. Sci. U.S.A. 94, 3748–3752 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26. Leem J. Y., Vijayan S., Han P., Cai D., Machura M., Lopes K. O., Veselits M. L., Xu H., Thinakaran G. (2002) Presenilin 1 is required for maturation and cell surface accumulation of nicastrin. J. Biol. Chem. 277, 19236–19240 [DOI] [PubMed] [Google Scholar]
- 27. Kwak Y. D., Wang R., Li J. J., Zhang Y. W., Xu H., Liao F. F. (2011) Differential regulation of BACE1 expression by oxidative and nitrosative signals. Mol. Neurodegener. 6, 17. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28. Zhang Y. W., Liu S., Zhang X., Li W. B., Chen Y., Huang X., Sun L., Luo W., Netzer W. J., Threadgill R., Wiegand G., Wang R., Cohen S. N., Greengard P., Liao F. F., Li L., Xu H. (2009) A functional mouse retroposed gene Rps23r1 reduces Alzheimer β-amyloid levels and Tau phosphorylation. Neuron 64, 328–340 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29. Yang J., Yang H., Yan L., Yang L., Yu L. (2009) Characterization of the human CUTA isoform2 present in the stably transfected HeLa cells. Mol. Biol. Rep. 36, 63–69 [DOI] [PubMed] [Google Scholar]
- 30. Tisdale E. J. (2001) Glyceraldehyde-3-phosphate dehydrogenase is required for vesicular transport in the early secretory pathway. J. Biol. Chem. 276, 2480–2486 [DOI] [PubMed] [Google Scholar]
- 31. He W., Lu Y., Qahwash I., Hu X. Y., Chang A., Yan R. (2004) Reticulon family members modulate BACE1 activity and amyloid-β peptide generation. Nat. Med. 10, 959–965 [DOI] [PubMed] [Google Scholar]
- 32. Murayama K. S., Kametani F., Saito S., Kume H., Akiyama H., Araki W. (2006) Reticulons RTN3 and RTN4-B/C interact with BACE1 and inhibit its ability to produce amyloid β-protein. Eur. J. Neurosci. 24, 1237–1244 [DOI] [PubMed] [Google Scholar]
- 33. Shi Q., Prior M., He W., Tang X., Hu X., Yan R. (2009) Reduced amyloid deposition in mice overexpressing RTN3 is adversely affected by preformed dystrophic neurites. J. Neurosci. 29, 9163–9173 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34. He X., Li F., Chang W. P., Tang J. (2005) GGA proteins mediate the recycling pathway of memapsin 2 (BACE). J. Biol. Chem. 280, 11696–11703 [DOI] [PubMed] [Google Scholar]
- 35. Tesco G., Koh Y. H., Kang E. L., Cameron A. N., Das S., Sena-Esteves M., Hiltunen M., Yang S. H., Zhong Z., Shen Y., Simpkins J. W., Tanzi R. E. (2007) Depletion of GGA3 stabilizes BACE and enhances β-secretase activity. Neuron 54, 721–737 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36. Wahle T., Prager K., Raffler N., Haass C., Famulok M., Walter J. (2005) GGA proteins regulate retrograde transport of BACE1 from endosomes to the trans-Golgi network. Mol. Cell. Neurosci. 29, 453–461 [DOI] [PubMed] [Google Scholar]
- 37. Okada H., Zhang W., Peterhoff C., Hwang J. C., Nixon R. A., Ryu S. H., Kim T. W. (2010) Proteomic identification of sorting nexin 6 as a negative regulator of BACE1-mediated APP processing. FASEB J. 24, 2783–2794 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38. Hartmann T., Bieger S. C., Brühl B., Tienari P. J., Ida N., Allsop D., Roberts G. W., Masters C. L., Dotti C. G., Unsicker K., Beyreuther K. (1997) Distinct sites of intracellular production for Alzheimer disease A β40/42 amyloid peptides. Nat. Med. 3, 1016–1020 [DOI] [PubMed] [Google Scholar]
- 39. Greenfield J. P., Tsai J., Gouras G. K., Hai B., Thinakaran G., Checler F., Sisodia S. S., Greengard P., Xu H. (1999) Endoplasmic reticulum and trans-Golgi network generate distinct populations of Alzheimer β-amyloid peptides. Proc. Natl. Acad. Sci. U.S.A. 96, 742–747 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40. Yang J., Li Q., Yang H., Yan L., Yang L., Yu L. (2008) Overexpression of human CUTA isoform2 enhances the cytotoxicity of copper to HeLa cells. Acta Biochim. Pol. 55, 411–415 [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.