Background: A central question in translation initiation is how GTPase activity and fMet-tRNA positioning are connected.
Results: NMR shows large structural rearrangements in the IF2-G2 subdomain upon nucleotide binding and considerable flexibility within the fMet-tRNA binding domain.
Conclusion: The GDP-induced rearrangements in G2 are not forwarded toward the fMet-tRNA binding C2 subdomain.
Significance: There appears to be no structural relationship between GTP hydrolysis and fMet-tRNA positioning.
Keywords: GTPase, NMR, Protein Dynamics, Ribosomes, Translation Initiation Factors, IF2
Abstract
Bacterial translation initiation factor IF2 promotes ribosomal subunit association, recruitment, and binding of fMet-tRNA to the ribosomal P-site and initiation dipeptide formation. Here, we present the solution structures of GDP-bound and apo-IF2-G2 of Bacillus stearothermophilus and provide evidence that this isolated domain binds the 50 S ribosomal subunit and hydrolyzes GTP. Differences between the free and GDP-bound structures of IF2-G2 suggest that domain reorganization within the G2-G3-C1 regions underlies the different structural requirements of IF2 during the initiation process. However, these structural signals are unlikely forwarded from IF2-G2 to the C-terminal fMet-tRNA binding domain (IF2-C2) because the connected IF2-C1 and IF2-C2 modules show completely independent mobility, indicating that the bacterial interdomain connector lacks the rigidity that was found in the archaeal IF2 homolog aIF5B.
Introduction
An early step in bacterial protein synthesis is the assembly of the 70 S initiation complex (70 S IC),7 where fMet-tRNA is adjusted in the peptidyltransferase center of the ribosome for the first transpeptidation reaction (1–3). The 70 S IC is assembled in two steps. First, 30 S-bound IF2 recruits fMet-tRNA and promotes its P-site decoding by the mRNA initiation triplet with the help of initiation factors IF1 and IF3 to form a 30 S initiation complex (30 S IC) (4). Subsequently, the 50 S ribosomal subunit joins the 30 S IC to yield 70 S IC through a process accompanied by IF2-dependent GTP hydrolysis and conformational rearrangements of components of the complex. The GTP hydrolysis is not required for the docking of the 50 S subunit to the 30 S IC, which occurs also in the presence of the nonhydrolyzable GTP analog GDPNP (5, 6), but rather to dissociate the acceptor end of fMet-tRNA from the IF2-C2 domain because the dominant-lethal phenotype caused by mutations in IF2-G2, which inactivate the GTPase of IF2 (7), can be suppressed by mutations in the IF2-C2 domain, which drastically reduce the affinity for the initiator tRNA (8). Ultimately, the conformational changes occurring during the 30 S IC to 70 S IC transition bring about the dissociation of IF1 and IF3, the disengagement of inorganic phosphate from IF2·GDP-Pi, the detachment of fMet-tRNA from IF2-C2 (the extreme C-terminal module of IF2), and eventually the release of IF2 from the ribosome (6, 9–11).
Besides their similarity in biological outcome, differences exist between the translation initiation systems of prokaryotes versus those of the highly similar archaea and eukarya. Most importantly, bacterial translation initiation is performed by only three initiation factors, whereas in higher organisms 13 translation initiation factors have already been recognized (12). In terms of amino acid conservation, bacterial IF1 is homologous to eIF1A and IF2 to eIF5B, although IF3 has no apparent eukaryotic homolog. Bacterial IFs and their counterpart eIFs cannot be functionally substituted. The difference in the number of involved initiation factors has two consequences. First, bacterial initiation factors combine the activities of several eukaryotic eIFs. For instance, bacterial IF2 directly interacts with both 30 S and 50 S ribosomal subunits, plays a crucial role in the ribosomal assembly, and also delivers fMet-tRNAfMet to the ribosome. Whereas its eukaryotic homolog eIF5B does interact similarly with the ribosomal subunits (13), it does not interact with initiator amino acid-tRNA in aqueous solution. Alternatively, in eukarya early initiator amino acid-tRNA interactions are mediated by eIF2, which is not present in prokaryotes. Second, although some of the functions of bacterial and eukaryotic IFs may be conserved (for instance, the interaction between eIF5B and eIF1A (14)), eIFs show activities that do not exist in bacterial IFs, for instance to support their mutual interactions.
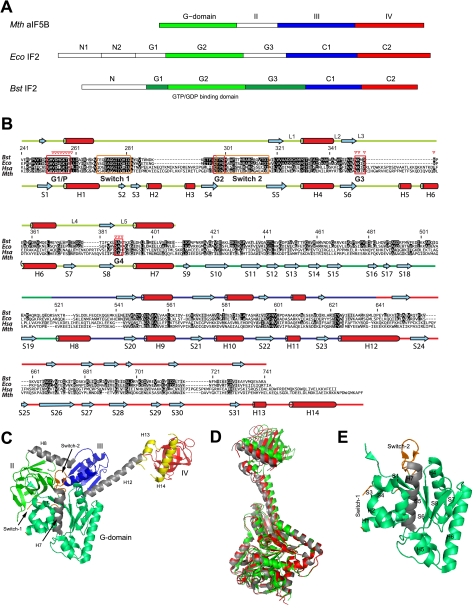
Bacterial IF2 and its homologs from other phylogenetic classes contain multiple modules (schematically represented in Fig. 1A). A mostly unstructured N-terminal domain (N domain), with limited size and sequence conservation (15, 16), anchors IF2 to the 30 S and perhaps also to the 50 S ribosomal subunit. This domain gives IF2 a high affinity for the ribosome, but is dispensable in vitro and in vivo under optimal growth conditions (7, 17–19), and is not present in IF2s of higher organisms. A large (∼42 kDa) G domain, consisting of the three subdomains G1, G2, and G3, follows the N domain. IF2-G1 has unknown function and is likely unstructured. It is not always present in bacterial IF2s and is not present in Methanobacterium thermoautotrophicum aIF5B, an archaeal homolog of bacterial IF2 (15). IF2-G2 and IF2-G3 interact with the 50 S and with the 30 S ribosomal subunit, respectively (19–24). The 19-kDa guanine nucleotide-binding subdomain G2 is the most conserved region of IF2, with amino acid homology to other G-proteins (Fig. 1B) (25, 26). Based on its homology with aIF5B domain II, IF2-G3 is predicted to be a β-barrel module, homologous to domains II of elongation factors EF-G and EF-Tu, and to subdomain C2 of IF2 itself (15). The C-terminal region of IF2 (C domain) includes two modules of similar size, IF2-C1 and IF2-C2 (27), connected by a ∼25-residue long linker. Whereas IF2-C2 recognizes and binds fMet-tRNA (8, 16, 28), no definite function has been assigned to IF2-C1. Structures of both IF2-C1 and IF2-C2 of Bacillus stearothermophilus have been reported (29, 30), and they share extensive homology with domains III and IV of M. thermoautotrophicum aIF5B, respectively.
FIGURE 1.
Domain organization of IF2 and aIF5B. A, domain nomenclature for IF2 homologs from M. thermoautotrophicum (Mth), E. coli (Eco), and B. stearothermophilus (Bst). B, sequence alignment for IF2 homologs from B. stearothermophilus (Bst), E. coli (Eco), Homo sapiens (Hsa), and M. thermoautotrophicum (Mth). Amino acid numbering is for B. stearothermophilus IF2. More and less conserved residues are depicted in black and gray, respectively. Below the sequence alignment the helices (H) and β-strands (S) found in M. thermoautotrophicum aIF5B crystal structures are indicated, connected by colored lines, representing IF2-G2 (light green), IF2-G3 (dark green), IF2-C1 (blue), and IF2-C2 (red). Above the sequence alignment the secondary structure elements as found by NMR on B. stearothermophilus IF2 are shown. Guanine nucleotide binding boxes G1–G4, loops L1-L5, and Switch-1 and Switch-2 regions are indicated; red triangles indicate residues that contact the ligand. C, M. thermoautotrophicum aIF5B domain organization. Domain-connecting helices H7, H8, and H12 are colored gray; Switch-1 and Switch-2 are colored orange. Two C-terminal a/eIF5B helices H13 and H14, not occurring in B. stearothermophilus IF2, are colored yellow. D, crystal structures for the free (gray), the GDP-bound (red), and the GDPNP-bound (green) states of M. thermoautotrophicum aIF5B fit on the G2 domain backbone (excluding the Switch-1 and Switch-2 regions). E, secondary structure elements of the M. thermoautotrophicum aIF5B-G2 domain.
The crystal structure of M. thermoautotrophicum aIF5B shows an overall elongated shape (Fig. 1C), with a maximum distance of over 100 Å between its G domain and domain IV. The most striking feature contributing to the extended shape of aIF5B is the long α-helix (H12) connecting domains III and IV. In aIF5B, this helix gives the protein the shape of a pendulum, which upon GTP hydrolysis transmits a conformational change occurring within the “clock” region (domains G, II and III) to the remote C-terminal end of the molecule (Fig. 1D; 15).
Because of the structural conservation of isolated IF2-C1 and IF2-C2 compared with aIF5B domains III and IV, it is tempting to describe the functionality of IF2 extrapolating from the structural similarity between IF2 and aIF5B. In addition, it is conceivable that the IF2 efficiency increases when its activities are partially linked. For instance, an elongated shape in bacterial IF2 would allow the simultaneous placement of the fMet-tRNA anticodon stem loop in the ribosomal 30 S P-site and the acceptor end in the 50 S peptidyltransferase center. However, despite their structural similarity, substantial deviations exist between bacterial IF2 and archaeal/eukaryal a/eIF5B functions. Indeed, although IF2-C2 is crucial for initiator-tRNA binding in bacteria, in archaea this function is not performed by aIF5B but by a/eIF2 (31). More or less subtle structural differences, possibly at the level of the inter-modular contacts, most likely underlie this functional divergence between the different classes of initiation factors. Because no details are yet available concerning the structure of IF2-G2 and the structural adaptability of bacterial IF2 as a result of ligand binding, we isolated the IF2-G2 domain, and after establishing its functionality, we elucidated its structure in the presence and absence of GDP. In addition, subdomain arrangement and overall shape of the C1-C2 region of IF2 were determined. Together, the results demonstrate that the behavior of bacterial IF2 is substantially different from its archaeal homolog aIF5B.
EXPERIMENTAL PROCEDURES
B. stearothermophilus IF2-G2 and IF2 C1-C2 NMR Sample Preparations
B. stearothermophilus IF2-G2 (residues 241–414) was expressed as a GST fusion protein in Escherichia coli strain BL21pLysS (Cmr) grown in M9 minimal medium (32). The fusion protein was purified on a glutathione-Sepharose column (Amersham Biosciences) and subjected to thrombin cleavage to yield IF2-G2, which was further purified by anion-exchange chromatography using a POROS HQ column with a linear 0–0.25 m NaCl gradient in 50 mm Tris-HCl, pH 6.8, followed by Superdex G-75 gel filtration in 20 mm NaPi, pH 6.5, 150 mm KCl, 1 mm DTT, and 0.01% NaN3. B. stearothermophilus IF2 C1-C2 (cloned in pEV1-C) was expressed, and the 26.6-kDa (240 residues) protein product was purified essentially as described (16). Isotopic labeling of the proteins was done with 15NH4Cl and either [12C]- or [13C]glucose. [2H,13C,15N]IF2-G2 was obtained using sodium [13C]acetate and 15NH4Cl in medium containing 97% D2O, 3% H2O.
The following NMR samples were used. (a) 0.4–0.9 mm IF2-G2 ± 0.6–1.2 mm GDP (Sigma) in 20 mm KPi, pH 6.5, 150 mm KCl, 5 mm MgCl2, 1 mm DTT, 0.01% NaN3, 10% D2O, and trace amounts of EDTA-free protease inhibitor mixture (Roche Applied Science). For [15N]IF2-G2·[13C,15N]GDP samples, [13C,15N]GTP (Isotec) was added to [15N]IF2-G2 and left overnight at 37 °C to allow GTP hydrolysis. (b) 0.5 mm IF2 C1-C2 in 20 mm KPi, pH 5.2, 200 mm KCl, 10% D2O, 0.02% NaN3, and EDTA-free protease inhibitor mixture.
NMR Spectroscopy
Unless stated otherwise, NMR experiments were performed at 315 K using Bruker Avance 600 MHz spectrometers. For free and GDP-bound IF2-G2, backbone assignments were performed from triple resonance experiments. For side chain assignments, 15N-FHSQC, H(C)(C)(CO)NH-TOCSY, (H)C(C)(CO)NH-TOCSY, HCCH-TOCSY, 13C-HSQC-CT, and histidine-specific 15N-HSQC spectra were used, either in H2O or D2O. Aromatic side chains were assigned using two-dimensional HB(CB)(CG)(CD)H, (HB)CB(CG)(CD)H, 13C-HSQC-CT, and NOESY spectra.
For distance restraints, two-dimensional NOE and three-dimensional NOESY-15N-HSQC and NOESY-13C-HSQC spectra were recorded on Bruker AVANCE 600 and 900 MHz spectrometers. Also, three-dimensional t1-proton and t1-carbon versions of four-dimensional 13C-HMQC-NOESY-13C-HSQC were recorded for IF2-G2·GDP. In all NOESY experiments, the mixing time was 80 ms.
The experiment to detect through hydrogen bond scalar couplings between GDP phosphate and IF2-G2 amide groups was a variation of a JNP spin-echo difference experiment reported before (33, 34). The 31Pα-15N and 31Pβ-15N spin-echo difference 15N-HSQC-CT spectra for [15N]IF2-G2·GDP were recorded at 298 K on a Bruker AVANCE 500 MHz spectrometer equipped with a z-gradient QNI probe.
The assignment of IF2 C1-C2 was based on the previous assignments of the free C1 and C2 domains (29, 30) and verified and complemented using three-dimensional HNCA, HNCACB, CBCA(CO)NH, and NOESY-15N-HSQC spectra of IF2 C1-C2. 15N relaxation data of IF2 C1-C2 were acquired as described (35). For 15N-R1 rates, relaxation delays of 100, 200, 400, 500, 600, 700, 800, 1000, and 1200 ms were used. 15N-R2 rates were determined from Carr-Purcell-Meiboom-Gill spectra recorded with 8, 16, 24, 36, 48, 76, 96, 120, 148, and 192 ms relaxation delay. 15N{1H}heteronuclear NOEs were determined from normalized signal intensity differences between two experiments as follows: one with proton saturation for 3 s to achieve steady state magnetization and one without proton saturation. The 15N relaxation data for IF2-C1 and IF2-C2 had been recorded previously (29, 30) and were reanalyzed similarly to IF2 C1-C2.
Data Analysis
NMR spectra were processed with NMRPipe (36) and analyzed using NMRView 5.0.3 (37). Chemical shift values were used to determine the presence and position of secondary structure elements employing the programs CSI (38) and TALOS (39). From these analyses, torsion angle restraints were also obtained.
Relaxation rates 15N-R1 and 15N-R2 were determined by two-parameter exponential fitting using Curvefit (A. G. Palmer III). Estimated errors in 15N-R1 and 15N-R2 values were 3% for isolated IF2-C1 and IF2-C2 and 4% for IF2 C1-C2 because of overall line broadening. For 15N{1H} heteronuclear NOE values, errors were fixed at 0.05. For model-free analyses (40), FAST-Modelfree (41) and TENSOR2 (42) were used. Input structures for these programs were the lowest energy structures of IF2-C1 (Protein Data Bank code 1Z9B) or IF2-C2 (Protein Data Bank code 1D1N), reoriented in the frame of inertia using PDBINERTIA (A.G. Palmer III). The output from FAST-Modelfree (using axial symmetric anisotropic rotational diffusion) is shown in the results; TENSOR2, using full tensor asymmetry, gave similar results.
Structure Calculations
IF2-G2 structures were calculated with ARIA1.2 (43) using the Parallhdg5.3 force field with PROLSQ parameters. Structure calculation parameters were default, except for the following: (i) all annealing stages were in Cartesian space, and (ii) the number of MD steps was doubled for every annealing stage. Structure analysis, validation, and visualization were done with Procheck-NMR (44), What-If and What-Check (45, 46), MolMol (47), and PyMOL (W. L. DeLano).
The IF2 C1-C2 homology model was based on amino acid sequence alignments of B. stearothermophilus IF2 and M. thermoautotrophicum aIF5B and built using the M. thermoautotrophicum aIF5B crystal structure (Protein Data Bank code 1G7R) using MODELLER6 version 2 (48). From 10 calculated structures, the lowest energy structure was used as the final model. Predicted values for the overall tumbling properties of particles in aqueous solution were obtained using the program HYDRONMR (49).
RESULTS
In Vitro Activities of Isolated IF2-G2
Prior to structural studies, we tested if the biological properties of the complete IF2 G domain (42 kDa) are preserved in its isolated G2 module (19 kDa). Our results demonstrate that the B. stearothermophilus IF2-G2·GTP complex can weakly bind the B. stearothermophilus 50 S ribosomal subunits (supplemental Fig. S1Aa) but not the 30 S subunits (data not shown). Furthermore, IF2-G2 binds and hydrolyzes GTP in both a ribosome-dependent and ribosome-independent manner (supplemental Fig. S1A, b and c). We conclude that IF2-G2 houses the functional elements responsible for GTP binding, the catalytic center for GTP hydrolysis, and at least part of the region for establishing functional interactions with the 50 S ribosomal subunit, although it does not contain the region responsible for the interaction with the 30 S subunit. Furthermore, these functional properties are similar to those of the complete G domain and even of the native IF2 molecule (18, 19, 50, 51). The structural integrity of IF2-G2 is further underscored by the fact that NMR spectra obtained with this domain can be easily recognized within those of larger IF2 fragments such as the G2-G3·GDP complex (supplemental Fig. S1B).
Structure of IF2-G2·GDP
Unlike IF2-G2·GTP, IF2-G2·GDPNP, and to some extent free IF2-G2, whose stability proved limited in time, IF2-G2·GDP gave a stable complex with invariant spectra during NMR measurements. The HSQC spectra of IF2-G2 and IF2-G2·GDP are quite different, indicating that the structures of free and GDP-bound IF2-G2 diverge substantially (supplemental Fig. S1B). Stepwise titration of IF2-G2 with GDP gave rise to two sets of NMR signals, one from the free protein, and the other from the IF2-G2·GDP complex. This is indicative for a slow exchange regime on the NMR time scale, implying that GDP binds tightly to IF2-G2. Most amino acids in the IF2-G2·GDP complex were assigned, including the non-proline residues of the P-loop (except for Thr258), Switch-1, and boxes G3 and G4. The N-terminal region Gly237–Ile249, preceding β-strand S1, and the region Lys292–Ile317, including the Switch-2 region, could not be assigned. For GDP, we were able to assign the H1, N1, H8, C8, H1′, C1′, H2′, C2′, H4′, C4′, H5′, and C5′ resonances.
The NMR spectra for IF2-G2·GDP displayed several unusual features that were also found before for other GDP-bound G-proteins. Low field resonances for the G1-box/P-loop Asp254 and Lys257 amide protons (10.16 and 10.53 ppm, respectively) suggest their hydrogen bonding to the GDP β-phosphate, like in Ras·GDP (52). Chemical shift differences between the free and protein-bound ligand suggest hydrogen bonding between H1 of GDP and IF2-G2 (53, 54). In preliminary IF2-G2·GDP structures, calculated without explicit intermolecular hydrogen bond restraints, the H1 atom of GDP was always found within 2.5 Å from the carboxyl group of Asp354 of the 351NKMD354 guanine-binding G3-box. Thus, this aspartate probably acts as a hydrogen bond acceptor, exactly like in other G-proteins. The NMR signal of G4-box Ala388 HN, an atom found within 3.0 Å from O6 of the guanine ring in the preliminary IF2-G2·GDP structures, is substantially shifted (0.8 ppm) upon GDP binding (supplemental Fig. S1B), indicating the presence of a contact seen also in other G-proteins (55, 56). Finally, the unusual proton signal of G1-box/P-loop His255 Hϵ2 at 13.1 ppm implies its protection from solvent exchange, possibly because of hydrogen bonding with the carboxyl group of Asp326 that is found within 2.0 Å in the preliminary IF2-G2·GDP structures.
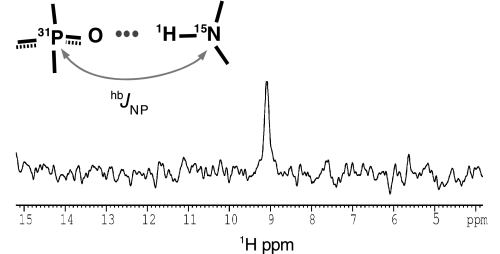
Besides these indirect protein-ligand contacts, hydrogen bonding between G1-box/P-loop Thr259 backbone nitrogen and the GDP α-phosphorus atoms is clearly indicated by a scalar JNP coupling (Fig. 2; supplemental Fig. S2). Although an equivalent hydrogen bond has been repeatedly suggested by crystal structures of GTP/GDP-bound G-proteins (15, 57–59), to our knowledge its existence is experimentally demonstrated here for the first time.
FIGURE 2.
IF2 binds GDP through a direct hydrogen bond. The figure shows the data of the direct detection of intermolecular JNP scalar couplings. One-dimensional 31P–15N spin-echo difference 15N-HSQC spectrum of IF2-G2·GDP and a schematic view of the phosphate-amide proton hydrogen bonds are detectable with the pulse sequence of supplemental Fig. S2.
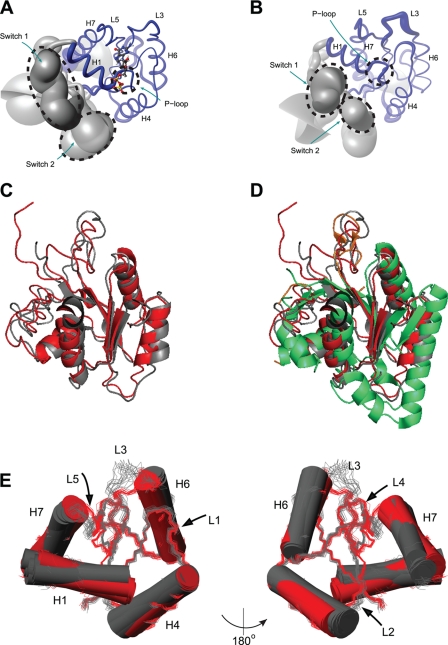
For the final NMR structure calculations, four hydrogen bonds (Asp254-GDP, Lys257-GDP, Thr259-GDP, and Asp354-GDP) and 24 NOE distance restraints were used to position GDP in its complex with IF2-G2. In the structure of the IF2-G2·GDP complex (Fig. 3A and supplemental Table S1), the protein core is folded like GTP/GDP-binding domains of other G-proteins (Figs. 1E and 3D) containing a central four-stranded parallel β-sheet formed by residues 247–251 (S1), 317–322 (S5), 346–351 (S6), and 381–386 (S8), surrounded by the four α-helices H1 (residues 258–266), H4 (330–343), H6 (360–370), and H7 (394–406). Also typical GDP-binding elements (P-loop, G3-box, and G4-box) are well defined, but, as in other free and GDP-bound G-proteins (15, 60–63), Switch-1 and Switch-2 are unstructured, likely because of internal mobility (Fig. 3A).
FIGURE 3.
Structures of GDP-bound and apo-IF2-G2. A, “sausage” representation of the IF2-G2·GDP NMR ensemble. The thickness of the backbone represents the r.m.s.d. Structural elements are indicated; GDP is shown in “ball-and-stick.” B, sausage representation of apo-IF2-G2. C, backbone overlay of the best structures of the NMR ensembles calculated for apo- (gray) and GDP-bound (red) IF2-G2. D, as C, additionally overlaid with the crystal structure of the G2 domain from M. thermoautotrophicum aIF5B (green). E, different orientations of the reorganization within the B. stearothermophilus IF2-G2 domain between its apo-state (gray) and GDP-bound (red) state.
Free Versus GDP-bound IF2-G2
Despite its reduced sample stability, except for the N terminus and Switch-2 region (because of line broadening), free IF2-G2 could also be studied by NMR. After structure calculation, like the IF2-G2·GDP complex, apo-IF2-G2 also shows a typical G-protein architecture (Fig. 3, B–D and supplemental Table S1). When superimposed on the well defined regions S5–S6 and H6–S8, excluding all G-boxes and Switches (residues 317–350, 361–386), the pairwise backbone r.m.s.d. between the NMR ensembles of IF2-G2·GDP and apo-IF2-G2 was 1.0 Å. The most pronounced differences between the two ensembles are in the orientations of the helices H1 and H6 and in the relative orientations of the loops L3 and L5 (Fig. 3E), as underscored by several large chemical shift changes (supplemental Fig. S1B). These loops, containing the guanosine ring-binding G3- and G4-box elements, move closer to the nucleotide base upon GDP binding. They are more ordered in the IF2G2·GDP structure, which is likely due to their reduced flexibility upon GDP binding. Helix H1 is twisted by ∼15° upon GDP binding, and the N-terminal end of helix H6 is moved by ∼1 Å toward the guanosine base in IF2-G2·GDP. There is a possibility that reorientation of helix H1 in the IF2-G2 structure does not reflect a similar reorganization of this helix in intact IF2, because only a limited number of long range contacts were found for this helix. However, the chemical shift changes at the beginning of helix H6 are large, indicating that the position of this helix is substantially modified. Similar yet much less pronounced structural transitions have been seen in other G-proteins before. However, the differences between free and GDP-bound forms in crystal structures of other G-proteins are much smaller than the 1 Å that we observed in the case of the IF2-G2. In the case of aIF5B (15), the backbone r.m.s.d. difference between free and GDP-bound forms in the regions corresponding to the core of IF2-G2 is only 0.36 Å, whereas for elongation factor EF-Tu (58, 63), this r.m.s.d. is 0.6 Å. It is likely that the pronounced structural changes induced by GDP binding to IF2-G2 also occur in intact IF2, because for several of the residues involved spectral changes were observed in experiments with the G2 domain in the context of a larger IF2 G2-G3 construct (supplemental Fig. S1B). Such rearrangements in the G2 domain are likely propagated to other regions of IF2.
Effect of GDPNP on IF2-G2 Switch-2
When GTP was added to IF2-G2, signals indicative for the GDP-bound form occurred in time due to GTP hydrolysis. This prevented the assignment of the IF2-G2·GTP complex, but at the same time it showed that GTP hydrolysis is accompanied by a conformational change. Titration of IF2-G2 with the GTP analog GDPNP to IF2-G2 did not yield the large spectral changes that GDP did (supplemental Fig. S1B). In this IF2-G2·GDPNP complex, the slow hydrolysis of GDPNP also prevented us from obtaining detailed structural information, and these data clearly suggest that “GTP”-bound and apo-IF2-G2 are structurally highly similar, while in the presence of GDP IF2-G2 adopts a clearly distinct conformation.
Upon GDPNP addition, a number of residues, including Gly251 from the P-loop 251GHVDHGKT258 and Asp354 and Ser387 from the guanine ring-binding G3- and G4-boxes, show behavior that is typical for the “fast-exchange” regime; their resonances shift with increasing concentrations of GDPNP, indicating low affinity for this ligand (Kd > 10 μm). Furthermore, resonances from P-loop His252, His255, Gly256, and Thr259; L1-loop Met329; L3-loop Ile350 and Asn351; and L5 residues Lys385 and Leu386, which are expected to contact the guanine ring, broaden or disappear upon GDPNP addition. A similar situation was observed for the Ras-GDPNP interaction (62), which was explained by intermediate conformational exchange (regional polysterism) in the GDPNP-bound form of Ras. Based on the observed exchange broadening, the rate for such a process in IF2 was estimated to be 102 s−1 < Rex < 103 s−1. Most important, residues near the G2-box/Switch-2 region (Phe295 and Gly300) broaden upon GDPNP binding, suggesting that this region is influenced by the interaction with the GTP analog. Together, these data demonstrate that GDPNP is bound to IF2-G2, but it does not induce large conformational changes beyond the GTP binding region.
In principle, the ligand-dependent conformational changes in IF2-G2 could be communicated to other IF2 domains. Most likely this occurs directly to subdomains G3 and C1, which, based on structural homology between IF2 and aIF5B, are expected to contact Switch-2. However, the information concerning ligand binding or GTP hydrolysis in the guanine nucleotide binding pocket could also be communicated intrinsically all the way to the extreme C-terminal C2 domain of IF2. For this to occur, the rigidity of the interdomain regions, as seen for aIF5B, seems a prerequisite.
Structure and Dynamics of IF2 C1-C2
Signaling by guanine nucleotide-induced conformational transitions, possibly involving contacts of the Switch-2 region with other domains, would permit the IF2 domain orientation to be directly controlled by the G2 subdomain. Besides the Switch-2 region of IF2-G2, the connector between IF2-C1 and IF2-C2, spanning from Arg622 to Lys646 in B. stearothermophilus IF2 and corresponding to α-helix H12 of M. thermoautotrophicum aIF5B, could be a region undergoing functionally relevant motions (Fig. 1, C and D). In aIF5B this long α-helix is in a defined position, folded back over the dorsal face of the central β-sheet of domain III, which keeps domains III and IV at a fixed distance (15). To study the structure and dynamics of this region for bacterial IF2, the connected B. stearothermophilus C1-C2 subdomains were studied.
Because the NMR structures of the isolated domains IF2-C1 and IF2-C2 are in good agreement with the corresponding domains of aIF5B (15, 29, 30), a homology model for the bacterial C1-C2 domain organization was created based on the aIF5B domain organization (Fig. 4A). Minor mismatches between the NMR structures of the separate domains and the homology model are found for the IF2-C1 loop Val566–Arg570, connecting H9 and S21, and the IF2-C1 loop Ala589–Asn593, connecting H10 and S22. In aIF5B both loops make extensive contacts with the region corresponding to helix H12. The fact that in IF2 these loops are poorly conserved (Fig. 1B) suggests that in bacteria they play no role in defining the position of the connector between the C1 and C2 subdomains.
FIGURE 4.
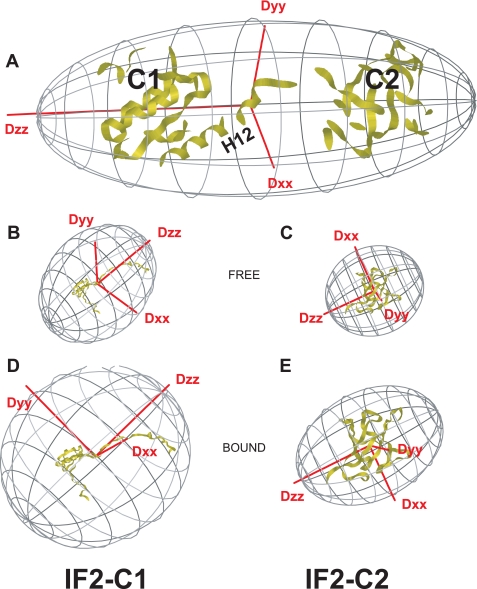
Hydrodynamic analysis of B. stearothermophilus IF2 C1-C2. The figure shows the orientations of the rotational diffusion tensors for free and connected B. stearothermophilus IF2 C1-C2 domains. A, predicted rotational diffusion tensor for the M. thermoautotrophicum aIF5B-based homology model for B. stearothermophilus IF2 C1-C2 assuming a completely rigid linker. B, rotational diffusion tensor for free IF2-C1 using the B. stearothermophilus IF2-C1 NMR structure. C, rotational diffusion tensor for free IF2-C2 using the B. stearothermophilus IF2-C2 NMR structure. D, rotational diffusion tensor for IF2-C1 when bound to IF2-C2, using the B. stearothermophilus IF2-C1 NMR structure. E, rotational diffusion tensor for IF2-C2 when bound to IF2-C1, using the B. stearothermophilus IF2-C2 NMR structure. Note: for direct comparison, the individual domains were manually rotated to the coordinate frame of IF2 C1-C2 in A. Also, the rotational diffusion tensors are scaled with respect to their rotational correlation times.
In IF2 C1-C2 only four residues of the connector could be unambiguously assigned, i.e. Ile624, Tyr625, Asn626, and Val627. These are the most conserved residues of the connector helix (Fig. 1B) and directly contact the GTPase domain in the homology model based on the crystal structure of aIF5B (15). The chemical shift values of these residues and the strong sequential HN–HN contacts for these residues indicate the presence of helicity (supplemental Fig. S3A), which supports the homology model for IF2 C1-C2 with at least one helical turn at the N terminus of the connector. Because, apart from the connector, the NMR spectra of full IF2 C1-C2 and the separate C1 and C2 domains are very similar, the physical connection of the two domains introduces neither large differences in their environments nor extensive interdomain contacts. However, some residues in IF2-C1, clustered on the surface where it contacts the connector in the model, show small signal shifts or intensity increases, indicating that their environment is slightly changed. In addition, the enhanced signal intensity observed for the core residues Ile597 and Gly598 suggests that the connector causes increased rigidity within IF2-C1 (data not shown).
In aIF5B, the rigid helix H12 supposedly acts as a lever, transmitting conformational changes caused by GTP hydrolysis from the “G-II-III” region all the way to domain IV (Fig. 1D; 15). Although we could not verify the dynamics within the connector of B. stearothermophilus IF2 directly because of the lack of assignments for this region, we could establish its mobility indirectly after comparing the tumbling properties of IF2 C1-C2 with those of free IF2-C1 and IF2-C2. Table 1 summarizes the results obtained from Model-free analyses of the 15N-T1, -T2, and 15N{1H}heteronuclear NOE relaxation rates for IF2-C1, IF2-C2, and IF2-C1-C2 (see supplemental Fig. S3, B and C). The experimental rotational correlation times (τc) and rotational diffusion tensor axis ratios were compared with the ones predicted for the NMR structures of the free C1 and C2 modules and for the rigid homology model of IF2 C1-C2 based on the x-ray structure of aIF5B. For the homology model of IF2 C1-C2, a τc value is predicted that is more than twice as large as for the free modules, while the elongated molecule would also show very different diffusion tensor axis lengths. The experimental data, however, clearly show that this is not the case. The τc values of both C1 and C2 within IF2 C1-C2 are similar to those of the free modules and much smaller than the values predicted for the rigid homology model. Also the diffusion tensor shapes obtained by the Model-free analysis do not change drastically upon domain connection (Table 1), and the tensors do not align with those for the homology model, but they also do not change for IF2-C1 and IF2-C2 upon their mutual attachment (Fig. 4). These findings can only be explained by an independent modular tumbling of the C1 and C2 domains. The 15N relaxation data and the Model-free analyses do show an increase in τc values for full IF2 C1-C2 compared with free IF2-C1 and IF2-C2, so the tumbling of C1 and C2 within IF2 C1-C2 is somewhat affected by their partners. This can be explained by the presence of motions slower than the rotational correlation times of the two modules. Therefore, the presence of a dynamic equilibrium between a helical stalk and a predominantly unfolded H12 region cannot be excluded. The increase in τc, however, remains much less than predicted for a completely rigid homology model.
TABLE 1.
Tumbling properties of free and connected B. stearothermophilus IF2-C1 and IF2-C2 domains
For isolated and connected IF2-C1 and -C2 domains, NMR relaxation data were analyzed by Modelfree analysis. Extracted rotational correlation times and diffusion tensors were compared with predictions based on structure models.
| Domain | C1 | C1-C2a | C1-C2b | C2 |
|---|---|---|---|---|
| Rotational correlation times τc (ns) | ||||
| Predictedc | 5.19 | 18.4 | 18.4 | 6.95 |
| Experimental | 9.02 ± 0.04 | 11.59 ± 0.07 | 11.44 ± 0.05 | 7.34 ± 0.03 |
| Rotational diffusion tensor axis length ratios | ||||
| Predictedc,d | ||||
| Dxx/Dzz | 0.94 | 0.40 | 0.40 | 0.75 |
| Dyy/Dzz | 0.98 | 0.40 | 0.40 | 0.83 |
| Experimentale | ||||
| Dxx/Dzz | 0.73 ± 0.02 | 0.85 ± 0.03 | 0.84 ± 0.02 | 0.80 ± 0.02 |
a Domain C1 is considered when connected to domain C2.
b Domain C2 is considered when connected to domain C1.
c Predictions were done with HYDRONMR on lowest energy NMR structures of isolated IF2-C1 and IF2-C2 and on the homology model of connected IF2 C1-C2.
d Assumed is: Dzz > Dyy ≥ Dxx.
e Model-free analyses were performed with FAST-Modelfree assuming axial symmetry (Dxx = Dyy, hence η = 0).
The positional mobility of the C2 domain is likely also present in intact IF2, because NMR signals of the free C2 domain and of C2 as part of C1-C2 are easily recognized in spectra recorded for IF2 “G2-G3-C1-C2” (data not shown). In addition, preliminary analysis of signal intensities indicates that also in complete IF2-GC the C2 domain tumbles independently from the rest of the protein. Taken together, the relaxation data indicate that in IF2 the physical connection between the C1 and C2 modules does not lead to the slower and asymmetric tumbling that is expected if the interdomain connector were completely rigid. We conclude that from this aspect bacterial IF2 behaves very differently from archaeal aIF5B.
DISCUSSION
The allosteric communication between different IF2 domains emerged clearly from earlier studies. For instance, IF2·GTP displays higher affinity for the 30 S ribosomal subunit than IF2·GDP (64), a difference that is magnified when the N-terminal domain that anchors IF2 to the 30 S ribosomal subunit is missing (18, 19). Because IF2-G3 is the module responsible for functional IF2–30 S interactions (19, 21–24), this different affinity likely results from a conformational transition between GTP-bound and GDP-bound IF2-G2 being transferred to IF2-G3. The nature of its nucleotide ligand influences also other IF2 activities. In fact, 30 S-bound IF2·GTP allows the rapid docking of the 50 S subunit to the 30 S IC to yield 70 S IC (5), whereas the alarmone ppGpp, bound to IF2 instead of GTP under nutritional stress, selectively inhibits IF2 functioning (27). Furthermore, only nucleotide-free and GDP-bound IF2 are able to dissociate from the 70 S IC, whereas IF2·GDPNP cannot, suggesting that conformational changes caused by GTP hydrolysis are a prerequisite for IF2 recycling (65).
In this study we determined NMR structures for the G2 subdomain of IF2 in free and GDP-bound forms. Because we have ascertained that ribosome binding and GTP hydrolysis properties of IF2-G2 are essentially identical to those of the complete G domain of IF2 and correspond to at least some of those of the native factor, the structures presented here are likely relevant for the functioning of this domain in the context of the whole protein. The structural organization of IF2-G2 is typical of guanine nucleotide-binding proteins. For instance, both unbound and GDP-bound forms of this domain clearly display nucleotide-binding elements such as the P-loop/G1-box and the G3- and G4-boxes, and interactions between these boxes and the guanine ring were observed in the IF2-G2·GDP complex. In free and GDP-bound IF2-G2, the guanine- and phosphate-binding elements possess almost identical structures, indicating that the former is already amenable for guanine nucleotide binding. However, in other parts of the subdomain, the two forms of IF2-G2 display significant structural differences. In particular, loops L3 and L5 differ in rigidity, whereas helix H6 undergoes a significant reorientation. Furthermore, because the NMR spectra indicate that the structures of apo-IF2-G2 and GDPNP-bound IF2-G2 are overall very similar, the changes in reorientation of loops L3 and L5 and helix H6 upon GDP binding to IF2-G2 could reflect structure changes that occur when IF2-bound GTP is hydrolyzed and the phosphate dissociates to yield IF2·GDP. These conformational changes could be forwarded to other regions of IF2 during different stages of translation initiation and could also regulate, for instance, interactions between IF2 and ribosomal subunits and/or be part of the mechanism by which IF2 promotes the transition from 30 S IC to 70 S IC. Indeed, in the crystal structure of aIF5B, helix H6 contacts the N-terminal part of helix H12, and guanine nucleotide binding or hydrolysis could be communicated from the G2 subdomain to this region of intact IF2. In addition, it is possible that in intact IF2 the reorientation of loops L3 and L5 and helix H6 modulates the affinity of the factor for the 50 S subunit.
Because loop L3, loop L5, and helix H6 do not contact other domains of IF2, at least based on the crystal structures of M. thermoautotrophicum aIF5B, it is unlikely that these elements are involved in the intra-molecular signaling initiated by GTP hydrolysis. Therefore, like in other GTPases Switch-1 and Switch-2 are probably involved in intramolecular signaling. These regions could be flexible in apo- or GTP-bound IF2-G2 and rigidify upon GTP hydrolysis. Unfortunately, we could not confirm this hypothesis directly because these Switches remained undefined in our structures, and the NMR investigation of IF2-G2·GTP and IF2-G2·GDPNP was hampered by the instability of the complexes. Nevertheless, some resonances of residues near the G2-box/Switch-2 region get broadened in the spectrum for IF2-G2·GDPNP indicating the existence of contacts between the GTP analog and its anticipated binding site and the occurrence of changes in Switch-2 dynamics. This situation could be similar to that seen in aIF5B, where Switch-2 is less well ordered in the presence of GDP, but becomes stabilized (15) upon contacting the GTP γ-phosphate and Mg2+.
Events occurring during the transition from 30 S IC to 70 S IC could be more efficient if IF2 domains were allowed to rearrange after the initial interactions with the 30 S subunit. In fact, a rigid connection between the GTP-binding/GTPase region and the C-terminal fMet-tRNAfMet-binding domain seems incompatible with the positioning of the various IF2 domains on different 30 S and 50 S regions within the 70 S IC. The paradox regarding beneficial molecular elongation and levering versus advantageous internal conformational freedom is resolved by our NMR relaxation analysis of free and connected IF2-C1 and IF2-C2. It was shown that, despite their physical connection, the C1 and C2 domains do not interact with each other and that their overall tumbling is uncorrelated. This implies that the connecting region in IF2, although partly helical, is not a continuous, rigid α-helix. Taken together, unless the flexible connector rigidifies in the presence of cofactors, our data do not support the levering model described for the C-terminal domain of M. thermoautotrophicum aIF5B but indicate a conformational arrangement that allows rotation and repositioning of the IF2-C2 module (Fig. 5), which could be required to facilitate its dissociation from fMet-tRNA. Thus, although many structural features are conserved between aIF5B and bacterial IF2, a functionally important difference is seen for the motional freedom of their extreme C-terminal domain.
FIGURE 5.
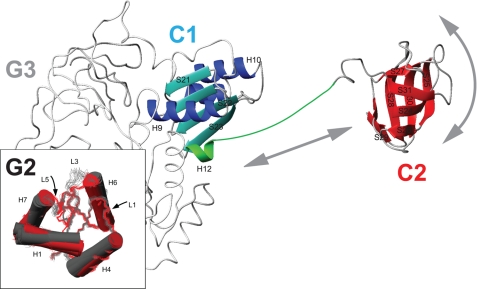
Model for the positional mobility of B. stearothermophilus IF2-C2. The figure shows a backbone representation of B. stearothermophilus IF2 C1-C2. Domain connector “H12” (Arg622–Lys646) is shown in green, and residues belonging to IF2-C1 are blue, and residues forming IF2-C2 are in red. The G23 domains are shown (in gray) based on sequence homology and crystal structures of M. thermoautotrophicum aIF5B. Arrows indicate the motional freedom of the C2 domain compared with the rest of the protein. The inset shows the structural changes detected for IF2-G2 upon GDP binding.
Acknowledgments
We thank Marc Guenneugues, Sylvie Meunier, and Snjezana Rothkrantz-Kos for their contributions at an early stage of the project.
This work was supported in part by European Union EU-NMR Contract RII3-026145, Bio-NMR Contract 261863, the Netherlands Foundation for Scientific Research, and MIUR-PRIN 2003 Grant (to C. O. G.).

This article contains supplemental Figs. S1–S3 and Table S1.
The atomic coordinates and structure factors (codes 2LKD and 2LKC) have been deposited in the Protein Data Bank, Research Collaboratory for Structural Bioinformatics, Rutgers University, New Brunswick, NJ (http://www.rcsb.org/).
1H, 13C, and 15N chemical shifts for free (6946) and GDP-bound (6995) IF2-G2 have been deposited in the Biological Magnetic Resonance Bank.
- IC
- initiation complex
- IF
- initiation factor
- r.m.s.d.
- root mean square deviation
- GDPNP
- guanosine 5′-(β,γ-imido)-triphosphate.
REFERENCES
- 1. Boelens R., Gualerzi C. O. (2002) Structure and function of bacterial initiation factors. Curr. Protein Pept. Sci. 3, 107–119 [DOI] [PubMed] [Google Scholar]
- 2. Laursen B. S., Sørensen H. P., Mortensen K. K., Sperling-Petersen H. U. (2005) Initiation of protein synthesis in bacteria. Microbiol. Mol. Biol. Rev. 69, 101–123 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3. Gualerzi C. O., Fabbretti A., Brandi L., Milon P., Pon C. L. (2010) Role of the initiation factors in mRNA start site selection and fMet-tRNA recruitment by bacterial ribosomes. Isr. J. Chem. 50, 80–94 [Google Scholar]
- 4. Milon P., Carotti M., Konevega A. L., Wintermeyer W., Rodnina M. V., Gualerzi C. O. (2010) The ribosome-bound initiation factor 2 recruits initiator tRNA to the 30 S initiation complex. EMBO Rep. 11, 313–316 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5. Antoun A., Pavlov M. Y., Tenson T., Ehrenberg M. (2004) Ribosome formation from subunits studied by stopped-flow and Rayleigh light scattering. Biol. Proced. Online 6, 35–54 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6. Grigoriadou C., Marzi S., Kirillov S., Gualerzi C. O., Cooperman B. S. (2007) A quantitative kinetic scheme for 70 S translation initiation complex formation. J. Mol. Biol. 373, 562–572 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7. Laalami S., Putzer H., Plumbridge J. A., Grunberg-Manago M. (1991) A severely truncated form of translational initiation factor 2 supports growth of Escherichia coli. J. Mol. Biol. 220, 335–349 [DOI] [PubMed] [Google Scholar]
- 8. Guenneugues M., Caserta E., Brandi L., Spurio R., Meunier S., Pon C. L., Boelens R., Gualerzi C. O. (2000) Mapping the fMet-tRNA(f)(Met)-binding site of initiation factor IF2. EMBO J. 19, 5233–5240 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9. Tomsic J., Vitali L. A., Daviter T., Savelsbergh A., Spurio R., Striebeck P., Wintermeyer W., Rodnina M. V., Gualerzi C. O. (2000) Late events of translation initiation in bacteria. A kinetic analysis. EMBO J. 19, 2127–2136 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10. Milon P., Konevega A. L., Peske F., Fabbretti A., Gualerzi C. O., Rodnina M. V. (2007) Transient kinetics, fluorescence, and FRET in studies of initiation of translation in bacteria. Methods Enzymol. 430, 1–30 [DOI] [PubMed] [Google Scholar]
- 11. Milon P., Konevega A. L., Gualerzi C. O., Rodnina M. V. (2008) Kinetic checkpoint at a late step in translation initiation. Mol. Cell 30, 712–720 [DOI] [PubMed] [Google Scholar]
- 12. Marintchev A., Wagner G. (2004) Translation initiation: structures, mechanisms, and evolution. Q. Rev. Biophys. 37, 197–284 [DOI] [PubMed] [Google Scholar]
- 13. Unbehaun A., Marintchev A., Lomakin I. B., Didenko T., Wagner G., Hellen C. U., Pestova T. V. (2007) Position of eukaryotic initiation factor eIF5B on the 80 S ribosome mapped by directed hydroxyl radical probing. EMBO J. 26, 3109–3123 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14. Marintchev A., Kolupaeva V. G., Pestova T. V., Wagner G. (2003) Mapping the binding interface between human eukaryotic initiation factors 1A and 5B. A new interaction between old partners. Proc. Natl. Acad. Sci. U.S.A. 100, 1535–1540 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15. Roll-Mecak A., Cao C., Dever T. E., Burley S. K. (2000) X-ray structures of the universal translation initiation factor IF2/eIF5B: conformational changes on GDP and GTP binding. Cell 103, 781–792 [DOI] [PubMed] [Google Scholar]
- 16. Spurio R., Brandi L., Caserta E., Pon C. L., Gualerzi C. O., Misselwitz R., Krafft C., Welfle K., Welfle H. (2000) The C-terminal subdomain (IF2 C-2) contains the entire fMet-tRNA-binding site of initiation factor IF2. J. Biol. Chem. 275, 2447–2454 [DOI] [PubMed] [Google Scholar]
- 17. Moreno J. M., Drskjøtersen L., Kristensen J. E., Mortensen K. K., Sperling-Petersen H. U. (1999) Characterization of the domains of E. coli initiation factor IF2 responsible for recognition of the ribosome. FEBS Lett. 455, 130–134 [DOI] [PubMed] [Google Scholar]
- 18. Caserta E., Tomsic J., Spurio R., La Teana A., Pon C. L., Gualerzi C. O. (2006) Translation initiation factor IF2 interacts with the 30 S ribosomal subunit via two separate binding sites. J. Mol. Biol. 362, 787–799 [DOI] [PubMed] [Google Scholar]
- 19. Caserta E., Ferrara C., Milon P., Fabbretti A., Rocchetti A., Tomsic J., Pon C. L., Gualerzi C. O., La Teana A. (2010) Ribosomal interaction of Bacillus stearothermophilus translation initiation factor IF2: characterization of the active sites. J. Mol. Biol. 396, 118–129 [DOI] [PubMed] [Google Scholar]
- 20. Marzi S., Knight W., Brandi L., Caserta E., Soboleva N., Hill W. E., Gualerzi C. O., Lodmell J. S. (2003) Ribosomal localization of translation initiation factor IF2. RNA 9, 958–969 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21. Allen G. S., Zavialov A., Gursky R., Ehrenberg M., Frank J. (2005) The cryo-EM structure of a translation initiation complex from Escherichia coli. Cell 121, 703–712 [DOI] [PubMed] [Google Scholar]
- 22. Myasnikov A. G., Marzi S., Simonetti A., Giuliodori A. M., Gualerzi C. O., Yusupova G., Yusupov M., Klaholz B. P. (2005) Conformational transition of initiation factor 2 from the GTP- to GDP-bound state visualized on the ribosome. Nat. Struct. Mol. Biol. 12, 1145–1149 [DOI] [PubMed] [Google Scholar]
- 23. Simonetti A., Marzi S., Myasnikov A. G., Fabbretti A., Yusupov M., Gualerzi C. O., Klaholz B. P. (2008) Structure of the 30 S translation initiation complex. Nature 455, 416–420 [DOI] [PubMed] [Google Scholar]
- 24. Julián P., Milon P., Agirrezabala X., Lasso G., Gil D., Rodnina M. V., Valle M. (2011) The cryo-EM structure of a complete 30 S translation initiation complex from Escherichia coli. PLoS Biol. 9, 1–11 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25. Cenatiempo Y., Deville F., Dondon J., Grunberg-Manago M., Sacerdot C., Hershey J. W. (1987) The protein synthesis initiation factor 2 G domain. Study of a functionally active C-terminal 65-kilodalton fragment of IF2 from Escherichia coli. Biochemistry 26, 5070–5076 [DOI] [PubMed] [Google Scholar]
- 26. Vachon G., Laalami S., Grunberg-Manago M., Julien R., Cenatiempo Y. (1990) Purified internal G domain of translational initiation factor IF-2 displays guanine nucleotide binding properties. Biochemistry 29, 9728–9733 [DOI] [PubMed] [Google Scholar]
- 27. Misselwitz R., Welfe K., Krafft C., Gualerzi C. O., Welfle H. (1997) Translational initiation factor IF2 from Bacillus stearothermophilus. A spectroscopic and microcalorimetric study of the C domain. Biochemistry 36, 3170–3178 [DOI] [PubMed] [Google Scholar]
- 28. Szkaradkiewicz K., Zuleeg T., Limmer S., Sprinzl M. (2000) Interaction of fMet-tRNAfMet and fMet-AMP with the C-terminal domain of Thermus thermophilus translation initiation factor 2. Eur. J. Biochem. 267, 4290–4299 [DOI] [PubMed] [Google Scholar]
- 29. Wienk H., Tomaselli S., Bernard C., Spurio R., Picone D., Gualerzi C. O., Boelens R. (2005) Solution structure of the C1 subdomain of Bacillus stearothermophilus translation initiation factor IF2. Protein Sci. 14, 2461–2468 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30. Meunier S., Spurio R., Czisch M., Wechselberger R., Guenneugues M., Gualerzi C. O., Boelens R. (2000) Structure of the fMet-tRNA(fMet)-binding domain of B. stearothermophilus initiation factor IF2. EMBO J. 19, 1918–1926 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31. Kyrpides N. C., Woese C. R. (1998) Archaeal translation initiation revisited. The initiation factor 2 and eukaryotic initiation factor 2B α-β-δ subunit families. Proc. Natl. Acad. Sci. U.S.A. 95, 3726–3730 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32. Milon P., Tischenko E., Tomsic J., Caserta E., Folkers G., La Teana A., Rodnina M. V., Pon C. L., Boelens R., Gualerzi C. O. (2006) The nucleotide-binding site of bacterial translation initiation factor 2 (IF2) as a metabolic sensor. Proc. Natl. Acad. Sci. U.S.A. 103, 13962–13967 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33. Mishima M., Hatanaka M., Yokoyama S., Ikegami T., Wälchli M., Ito Y., Shirakawa M. (2000) Intermolecular 31P-15N and 31P-1H scalar couplings across hydrogen bonds formed between a protein and a nucleotide. J. Am. Chem. Soc. 122, 5883–5884 [Google Scholar]
- 34. Löhr F., Mayhew S. G., Rüterjans H. (2000) Detection of scalar couplings across NH…OP and OH…OP hydrogen bonds in a flavoprotein. J. Am. Chem. Soc. 122, 9289–9295 [Google Scholar]
- 35. Houben K., Dominguez C., van Schaik F. M., Timmers H. T., Bonvin A. M., Boelens R. (2004) Solution structure of the ubiquitin-conjugating enzyme UbcH5B. J. Mol. Biol. 344, 513–526 [DOI] [PubMed] [Google Scholar]
- 36. Delaglio F., Grzesiek S., Vuister G. W., Zhu G., Pfeifer J., Bax A. (1995) NMRPipe. A multidimensional spectral processing system based on UNIX pipes. J. Biomol. NMR 6, 277–293 [DOI] [PubMed] [Google Scholar]
- 37. Johnson B. A., Blevins R. A. (1994) NMR View: a computer program for the visualization and analysis of NMR data. J. Biomol. NMR 4, 603–614 [DOI] [PubMed] [Google Scholar]
- 38. Wishart D. S., Sykes B. D. (1994) The 13C chemical-shift index. A simple method for the identification of protein secondary structure using 13C chemical-shift data. J. Biomol. NMR 4, 171–180 [DOI] [PubMed] [Google Scholar]
- 39. Cornilescu G., Delaglio F., Bax A. (1999) Protein backbone angle restraints from searching a database for chemical shift and sequence homology. J. Biomol. NMR 13, 289–302 [DOI] [PubMed] [Google Scholar]
- 40. Lipari G., Szabo A. (1982) Model-free approach to the interpretation of nuclear magnetic resonance relaxation in macromolecules. 1. Theory and range of validity. J. Am. Chem. Soc. 104, 4546–4559 [Google Scholar]
- 41. Cole R., Loria J. P. (2003) FAST-Modelfree. A program for rapid automated analysis of solution NMR spin-relaxation data. J. Biomol. NMR 26, 203–213 [DOI] [PubMed] [Google Scholar]
- 42. Dosset P., Hus J. C., Blackledge M., Marion D. (2000) Efficient analysis of macromolecular rotational diffusion from heteronuclear relaxation data. J. Biomol. NMR 16, 23–28 [DOI] [PubMed] [Google Scholar]
- 43. Linge J. P., O'Donoghue S. I., Nilges M. (2001) Automated assignment of ambiguous nuclear overhauser effects with ARIA. Methods Enzymol. 339, 71–90 [DOI] [PubMed] [Google Scholar]
- 44. Laskowski R. A., Rullmannn J. A., MacArthur M. W., Kaptein R., Thornton J. M. (1996) AQUA and PROCHECK-NMR. Programs for checking the quality of protein structures solved by NMR. J. Biomol. NMR 8, 477–486 [DOI] [PubMed] [Google Scholar]
- 45. Vriend G. (1990) WHAT IF. A molecular modeling and drug design program. J. Mol. Graph. 8, 52–56 [DOI] [PubMed] [Google Scholar]
- 46. Hooft R. W., Vriend G., Sander C., Abola E. E. (1996) Errors in protein structures. Nature 381, 272. [DOI] [PubMed] [Google Scholar]
- 47. Koradi R., Billeter M., Wüthrich K. (1996) MOLMOL. A program for display and analysis of macromolecular structures. J. Mol. Graph. 14, 51–55 [DOI] [PubMed] [Google Scholar]
- 48. Martí-Renom M. A., Stuart A. C., Fiser A., Sánchez R., Melo F., Sali A. (2000) Comparative protein structure modeling of genes and genomes. Annu. Rev. Biophys. Biomol. Struct. 29, 291–325 [DOI] [PubMed] [Google Scholar]
- 49. García de la Torre J., Huertas M. L., Carrasco B. (2000) HYDRONMR. Prediction of NMR relaxation of globular proteins from atomic level structures and hydrodynamic calculations. J. Magn. Reson. 147, 138–146 [DOI] [PubMed] [Google Scholar]
- 50. Gualerzi C. O., Severini M., Spurio R., La Teana A., Pon C. L. (1991) Molecular dissection of translation initiation factor IF2. Evidence for two structural and functional domains. J. Biol. Chem. 266, 16356–16362 [PubMed] [Google Scholar]
- 51. Severini M., Spurio R., La Teana A., Pon C. L., Gualerzi C. O. (1991) Ribosome-independent GTPase activity of translation initiation factor IF2 and of its G domain. J. Biol. Chem. 266, 22800–22802 [PubMed] [Google Scholar]
- 52. Redfield A. G., Papastavros M. Z. (1990) NMR study of the phosphoryl binding loop in purine nucleotide proteins: evidence for strong hydrogen bonding in human N-ras p21. Biochemistry 29, 3509–3514 [DOI] [PubMed] [Google Scholar]
- 53. Giessner-Prettre C., Pullman B., Caillet J. (1977) Theoretical study on the proton chemical shifts of hydrogen bonded nucleic acid bases. Nucleic Acids Res. 4, 99–116 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54. Wijmenga S. S., Van Buuren B. N. (1998) The use of NMR methods for conformational studies of nucleic acids. Prog. NMR Spectrosc. 32, 287–387 [Google Scholar]
- 55. Sprang S. R. (1997) G protein mechanisms. Insights from structural analysis. Annu. Rev. Biochem. 66, 639–678 [DOI] [PubMed] [Google Scholar]
- 56. Vetter I. R., Wittinghofer A. (2001) The guanine nucleotide-binding switch in three dimensions. Science 294, 1299–1304 [DOI] [PubMed] [Google Scholar]
- 57. Milburn M. V., Tong L., deVos A. M., Brünger A., Yamaizumi Z., Nishimura S., Kim S. H. (1990) Molecular switch for signal transduction. Structural differences between active and inactive forms of protooncogenic ras proteins. Science 247, 939–945 [DOI] [PubMed] [Google Scholar]
- 58. al-Karadaghi S., Aevarsson A., Garber M., Zheltonosova J., Liljas A. (1996) The structure of elongation factor G in complex with GDP. Conformational flexibility and nucleotide exchange. Structure 4, 555–565 [DOI] [PubMed] [Google Scholar]
- 59. Song H., Parsons M. R., Rowsell S., Leonard G., Phillips S. E. (1999) Crystal structure of intact elongation factor EF-Tu from Escherichia coli in GDP conformation at 2.05 Å resolution. J. Mol. Biol. 285, 1245–1256 [DOI] [PubMed] [Google Scholar]
- 60. Kraulis P. J., Domaille P. J., Campbell-Burk S. L., Van Aken T., Laue E. D. (1994) Solution structure and dynamics of ras p21.GDP determined by heteronuclear three- and four-dimensional NMR spectroscopy. Biochemistry 33, 3515–3531 [DOI] [PubMed] [Google Scholar]
- 61. Farrar C. T., Halkides C. J., Singel D. J. (1997) The frozen solution structure of p21 ras determined by ESEEM spectroscopy reveals weak coordination of Thr-35 to the active site metal ion. Structure 5, 1055–1066 [DOI] [PubMed] [Google Scholar]
- 62. Ito Y., Yamasaki K., Iwahara J., Terada T., Kamiya A., Shirouzu M., Muto Y., Kawai G., Yokoyama S., Laue E. D., Wälchli M., Shibata T., Nishimura S., Miyazawa T. (1997) Regional polysterism in the GTP-bound form of the human c-Ha-Ras protein. Biochemistry 36, 9109–9119 [DOI] [PubMed] [Google Scholar]
- 63. AEvarsson A., Brazhnikov E., Garber M., Zheltonosova J., Chirgadze Y., al-Karadaghi S., Svensson L. A., Liljas A. (1994) Three-dimensional structure of the ribosomal translocase. Elongation factor G from Thermus thermophilus. EMBO J. 13, 3669–3677 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 64. Pon C. L., Paci M., Pawlik R. T., Gualerzi C. O. (1985) Structure-function relationship in Escherichia coli initiation factors. Biochemical and biophysical characterization of the interaction between IF-2 and guanosine nucleotides. J. Biol. Chem. 260, 8918–8924 [PubMed] [Google Scholar]
- 65. Luchin S., Putzer H., Hershey J. W., Cenatiempo Y., Grunberg-Manago M., Laalami S. (1999) In vitro study of two dominant inhibitory GTPase mutants of Escherichia coli translation initiation factor IF2. Direct evidence that GTP hydrolysis is necessary for factor recycling. J. Biol. Chem. 274, 6074–6079 [DOI] [PubMed] [Google Scholar]