Background: The Paf1 complex associates with RNA polymerase II during elongation.
Results: The Cdc73 C-domain adopts a Ras-like fold that contributes to histone methylation and Paf1C recruitment to active genes.
Conclusion: Cdc73 C-domain is important for Paf1 complex recruitment to genes.
Significance: Rtf1 and Cdc73 C-domain combine to couple Paf1 complex to elongating polymerase.
Keywords: Histone Methylation, RNA Polymerase II, Structural Biology, Transcription Elongation Factors, X-ray Crystallography, Cdc73, Paf1 Complex, Parafibromin, Ras, Transcription Elongation
Abstract
The conserved Paf1 complex localizes to the coding regions of genes and facilitates multiple processes during transcription elongation, including the regulation of histone modifications. However, the mechanisms that govern Paf1 complex recruitment to active genes are undefined. Here we describe a previously unrecognized domain within the Cdc73 subunit of the Paf1 complex, the Cdc73 C-domain, and demonstrate its importance for Paf1 complex occupancy on transcribed chromatin. Deletion of the C-domain causes phenotypes associated with elongation defects without an apparent loss of complex integrity. Simultaneous mutation of the C-domain and another subunit of the Paf1 complex, Rtf1, causes enhanced mutant phenotypes and loss of histone H3 lysine 36 trimethylation. The crystal structure of the C-domain reveals unexpected similarity to the Ras family of small GTPases. Instead of a deep nucleotide-binding pocket, the C-domain contains a large but comparatively flat surface of highly conserved residues, devoid of ligand. Deletion of the C-domain results in reduced chromatin association for multiple Paf1 complex subunits. We conclude that the Cdc73 C-domain probably constitutes a protein interaction surface that functions with Rtf1 in coupling the Paf1 complex to the RNA polymerase II elongation machinery.
Introduction
Eukaryotic gene expression is facilitated by a large number of accessory factors that function at various steps in RNA synthesis and maturation, including the recruitment of RNA polymerase (pol)4 II to gene promoters, the modification or remodeling of chromatin, and the processing of nascent transcripts. Transcription can be loosely divided into three stages, initiation, elongation, and termination, with each stage utilizing different accessory factors. Many of these proteins bind chromatin directly, whereas others associate with the polymerase or recognize the RNA transcript (reviewed in Ref. 1). Although the recruitment and assembly of transcription factors within the initiation stage have been intensely studied, less is known about how proteins involved in transcription elongation associate with RNA pol II.
The Paf1 complex (Paf1C), which in Saccharomyces cerevisiae is composed of the five subunits Paf1, Ctr9, Cdc73, Rtf1, and Leo1, was originally identified as a protein complex that co-purifies with RNA pol II (2, 3). Although initially implicated in transcription initiation (2–4), Paf1C now has established roles in post initiation events, including the transcription-coupled modification of histones (5–9), the recruitment of RNA 3′-end processing factors (10–12), the modulation of RNA pol II C-terminal domain phosphorylation (10, 13), and the recruitment of the chromatin remodeling factor Chd1 to open reading frames (ORFs) (14).
Paf1C is required for the establishment of several histone modifications during transcription elongation. No intrinsic enzymatic activity has been attributed to either Paf1C as a whole or any of its subunits. Rather, it is thought that Paf1C may regulate the activity or localization of enzyme complexes responsible for histone modifications. Mutational studies indicate that the Rtf1 subunit plays a prominent role in promoting monoubiquitylation of histone H2B lysine 123 by the Rad6-Bre1 ubiquitin conjugase-ligase complex (6, 8, 15, 16), a modification that is required for subsequent di- and trimethylation of histone H3 Lys-4 and Lys-79 by the Set1/COMPASS and Dot1 methyltransferases, respectively (7, 9, 17–19). In addition, the Paf1 and Ctr9 subunits are required for proper levels of H3 Lys-36 trimethylation on the coding regions of active genes and downstream effects on H3 and H4 acetylation (5). The importance of Paf1C-mediated histone modifications is highlighted by their broad impact on gene expression patterns, both in yeast and human cells, and their connections to human cancers and stem cell pluripotency (20–25).
Paf1C associates with RNA pol II throughout the coding regions of genes (26, 27) and engages in physical interactions with the elongation factors Spt16-Pob3/FACT and Spt4-Spt5/DSIF as well as RNA pol II (10, 13, 28–30). Consistent with its important functions during elongation, mutations in genes encoding Paf1C subunits cause sensitivity to 6-azauracil (6-AU) and mycophenolic acid (MPA), phenotypes associated with defects in transcription elongation (28). Last, transcription elongation efficiency is reduced in paf1Δ or cdc73Δ cell extracts in vitro and in Paf1C mutant strains in vivo (31, 32).
Despite the importance of its roles in directing elongation-coupled processes, the mechanisms underlying Paf1C recruitment to the transcriptional machinery remain unclear. Current data implicate several different elongation factors in recruiting the Paf1C to RNA pol II. These include Spt4-Spt5/DSIF, Spt16-Pob3/FACT, Spt6, the Ccr4-NOT complex, and the Bur1-Bur2 kinase complex (30, 33–39). In addition, Paf1C recruitment is facilitated by phosphorylation of serine-5 residues on the C-terminal domain of RNA pol II (30). Of these factors, the involvement of Spt4-Spt5 in Paf1C recruitment is the best characterized. Studies in yeast have shown that the Bur1-Bur2 kinase phosphorylates the C-terminal repeat domain of Spt5, and this domain is important for the association of Paf1C with ORFs (35, 39).
Little is known about regions within Paf1C that govern its interactions with RNA pol II and/or elongation factors and thereby restrict its localization to active genes. However, chromatin immunoprecipitation (ChIP) and co-immunoprecipitation assays have revealed prominent roles for both the Rtf1 and Cdc73 subunits in mediating Paf1C-RNA pol II interactions. Deletion of either of these subunits reduces the levels of Paf1C that immunoprecipitate with RNA pol II or localize to transcribed genes (10, 13). Mutational studies have identified the Plus-3 domain of Rtf1 as being required for Paf1C occupancy on active genes (16), and interestingly, biochemical experiments have revealed binding of the Rtf1 Plus-3 domain to DNA substrates that mimic the transcription bubble (40).
The domains of Cdc73 important for promoting association of Paf1C with chromatin have not been investigated. Cdc73 (parafibromin in humans) is the smallest subunit within Paf1C, and like other Paf1C subunits, it is conserved throughout eukaryotes. The gene encoding parafibromin/hCdc73 has long been recognized as a tumor suppressor gene in humans. Inactivating germ line mutations account for ∼50% of patients with hyperparathyroidism-jaw tumor syndrome (HPT-JT) (41) and are associated with a variety of related cancers, including parathyroid carcinomas and ossifying fibromas (42, 43). Interestingly, parafibromin can function as a tumor suppressor by recruiting histone methyltransferases to inhibit cyclin D1 expression (44, 45) and as an oncogenic factor by activating Wnt signaling (46). These apparently antagonistic functions of parafibromin utilize sequences not found within the yeast protein (47). Based on the high degree of overall sequence identity between parafibromin and Cdc73 from yeast, we reasoned that there might be a previously unrecognized region or domain within Cdc73 that is playing a conserved function in transcription.
Here we describe the identification of a domain within the highly conserved C terminus of S. cerevisiae Cdc73, which we call the C-domain. We show that deletion of the C-domain causes phenotypes associated with defects in transcription elongation while having little impact on the ability of Paf1C to assemble. We also demonstrate that deletion of the C-domain or a single amino acid substitution within the C-domain enhances the mutant phenotypes and histone modification defects of an rtf1Δ strain, revealing functional redundancy between these components of the complex. We have determined the structure of the C-domain and unexpectedly found that it adopts a fold that is highly similar to GTPases of the Ras superfamily. The canonical nucleotide binding pocket is altered in Cdc73, and we do not observe a nucleotide ligand. Finally, we show that the C-domain is important for the association of Paf1C with chromatin. Together, our data indicate that this newly identified Ras-like domain within Cdc73 functions together with the Plus-3 domain of Rtf1 to facilitate association of Paf1C with active genes.
EXPERIMENTAL PROCEDURES
Protein Expression and Purification
The coding sequence for the S. cerevisiae Cdc73 C-domain (residues 230–393) was amplified by PCR and cloned into the bacterial expression vector pET151-D/TOPO (Invitrogen). Upon induction, the resulting protein contains an N-terminal His6-tag as well as a TEV cleavage site. BL21-DE3 Codon-Plus Escherichia coli cells were grown in ZY Autoinduction medium (48) at room temperature for 24–30 h, harvested by centrifugation, and lysed via homogenization in 25 mm Tris, pH 8.0, 500 mm NaCl, 10% glycerol, 5 mm imidazole, 1 mm β-mercaptoethanol. Lysate was cleared by centrifugation at 100,000 × g, and Cdc73 C-domain was purified by nickel affinity chromatography. Following overnight digestion with TEV protease, a second round of nickel affinity purification was performed to remove the cleaved His-tag, TEV protease, and nonspecific contaminants. Cation exchange chromatography (HiTrap-SP) was used to remove contaminating nucleic acids, and any aggregates were removed by gel filtration using a Sephacryl S-200 column (GE Healthcare). Peak fractions were concentrated to 20 mg/ml in 15 mm Tris, pH 7.0, 25 mm NaCl, 0.0% glycerol, and 1 mm β-mercaptoethanol using a Vivaspin concentrator (Millipore) prior to crystallization. The purity was >99% as verified by SDS-PAGE. Selenomethionine substituted Cdc73 C-domain was expressed using PASM medium (48). Purification was the same as the native protein.
Crystallization of S. cerevisiae Cdc73 C-domain
Initial poorly formed crystals were obtained by the sitting drop vapor diffusion method at 20 °C against a reservoir solution containing 0.1 m Bicine, pH 8.5, and 20% PEG 6000. Extensive optimization of the crystallization conditions combined with microseeding eventually resulted in the formation of small single crystals grown at 4 °C overnight against a reservoir solution containing 0.1 m HEPES at pH 8.0 and 15% PEG 8000. The crystals were cryoprotected by transition of the crystal into reservoir solution supplemented with increasing concentrations of PEG 8000 and glycerol to final concentrations of 30% PEG 8000 and 15% glycerol. The cryoprotected crystals were flash frozen under liquid nitrogen prior to data collection. Crystals of selenomethionine-containing Cdc73 C-domain were grown in the same initial conditions as the native crystals, and were microseeded and cryoprotected using the same technique.
Structure Determination
The quality of Cdc73 crystals was high, with most crystals having low mosaicity and diffracting to high resolution (1.7–1.5 Å). The crystals belong to space group P41212 with a = b = 53.3 Å, c = 130.2 Å. Diffraction data from all Cdc73 C-domain crystals were collected at beamline X25 at the National Synchrotron Light Source. Integration, scaling, and merging of diffraction data were performed using HKL2000 (49). Experimental phasing was performed by the single wavelength anomalous dispersion technique using a selenomethionine-containing C-domain crystal collected at peak wavelength. Phases were calculated using AutoSol within PHENIX (50), and an initial model was built using the Autobuild routine. This initial model was then refined against native data at 1.55 Å using positional refinement and anisotropic B-factor refinement. The final model has no Ramachandran outliers and good geometry. Analysis of model quality was monitored using MolProbity (51).
Yeast Strains, Media, and Growth Assays
S. cerevisiae strains (Table 1) are isogenic to FY2 (52) and were constructed by standard methods of transformation and tetrad dissection (53, 54). Rich (YPD), synthetic complete (SC), and minimal (SD) media were as described (54). 6-AU and MPA were added to SC−Ura medium at final concentrations of 50 μg/ml or 20 μg/ml, respectively. For dilution growth assays, strains were grown to saturation in appropriate media and washed, suspended, and serially diluted in sterile water, prior to being pipetted onto solid media.
TABLE 1.
S. cerevisiae strains used in this study
| Strain | Genotype | |
|---|---|---|
| GHY1185a | MATa | his4-912δ lys2-128δ leu2Δ (0 or 1) trp1Δ63 CTR9-6×MYC::LEU2 LEO1-3×HA::HIS3 SPT5-FLAG |
| KY703 | MATα | his4-912δ lys2-128δ leu2Δ1 (0 or 1) rtf1Δ101::LEU2 cdc73Δ::KanMX |
| KY1021 | MATa | his4-912δ lys2-128δ leu2Δ1 trp1Δ63 |
| KY1789 | MATa | his4-912δ lys2-128δ leu2Δ1 trp1Δ63 rtf1Δ101::LEU2 cdc73ΔC-TAP::TRP1 |
| KY1791 | MATa | his4-912δ lys2-128δ leu2Δ1 trp1Δ63 cdc73ΔC-TAP::TRP1 |
| KY1797 | MATα | his4-912δ lys2-128δ leu2Δ1 trp1Δ63 rtf1Δ101::LEU2 CDC73-TAP::TRP1 |
| KY1799 | MATα | his4-912δ lys2-128δ leu2Δ1 trp1Δ63 CDC73-TAP::TRP1 |
| KY1802 | MATa | his4-912δ lys2-128δ leu2Δ1 trp1Δ63 rtf1Δ101::LEU2 |
| KY1857 | MATα | his4-912δ cdc73Δ::KanMX |
| KY1858 | MATa | his4-912δ trp1Δ63 cdc73Δ::KanMX |
| KY1978 | MATα | his3Δ200 his4-912δ lys2-128δ leu2Δ (0 or 1) trp1Δ63 CDC73-TAP::TRP1 CTR9-6×MYC::LEU2 LEO1–3×HA::HIS3 SPT5-FLAG |
| KY1979 | MATα | his3Δ200 his4-912δ lys2-128δ leu2Δ (0 or 1) trp1Δ63 cdc73ΔC-TAP::TRP1 CTR9–6×MYC::LEU2 LEO1-3×HA::HIS3 SPT5-FLAG |
| KY2191 | MATa | his3Δ200 lys2-128δ leu2Δ (0 or 1) trp1Δ63 cdc73Δ::KanMX CTR9–6×MYC::LEU2 |
| KY2195 | MATα | rtf1Δ101::LEU2 cdc73Δ::KanMX his4-912δ leu2Δ1 trp1Δ63 |
a From G. Hartzog.
Plasmids
Plasmids were constructed by standard techniques (53). To generate a plasmid expressing Cdc73, a PciI-EcoRV fragment of PCR-amplified genomic DNA (550 bp 5′ to the ATG to 538 bp 3′ of the stop codon) was subcloned into pRS314 (55), yielding pBTB4. A plasmid expressing Cdc73 with an N-terminal triple HA tag (3×HA-Cdc73) was created by first introducing an NdeI site in pBTB4 at the ATG of CDC73 by site-directed mutagenesis (Stratagene) to create pCD3. A fragment encoding the 3×HA tag was amplified from pMR2307 (56) with primers incorporating NdeI sites and ligated to pCD3 cut with NdeI to create pWR4. To generate pCD8, a plasmid expressing a C-terminally truncated form of 3×HA-Cdc73, amino acids 230–393 were deleted from pBTB4 using the QuikChange protocol (Stratagene) to create pWR5. Then a SalI-MscI fragment from pWR4 containing the N-terminal 3×HA tag was inserted into pWR5 cut with the same enzymes to create pCD8. To create amino acid substitutions within Cdc73, pWR4 was subjected to site-directed mutagenesis using the QuikChange protocol. All plasmids created through PCR or site-directed mutagenesis were verified by DNA sequencing.
Western Analysis
Strains were grown to a density of ∼3 × 107 cells/ml, and extracts were prepared by glass bead lysis in radioimmune precipitation buffer or by a rapid boiling method (15, 16). Samples were resolved by SDS-PAGE, transferred to nitrocellulose or PVDF, exposed to primary and secondary antibodies, and visualized by chemiluminescent detection. Primary antibodies detected H3 Lys-36 trimethyl (me3) (Abcam), H3 Lys-36 dimethyl (me2) (Millipore), H3 Lys-4 me3 (Abcam), H3 Lys-4 me2 (Millipore), H3 Lys-79 me2 and me3 (Abcam), and total H3 (a gift of LeAnn Howe). Membranes were also probed with antibody specific for glucose-6-phosphate dehydrogenase (Sigma). Levels of HA-Rtf1 or HA-Cdc73, both tagged at their N termini with a triple HA sequence, were measured using antibody specific for the HA tag (Roche Applied Science). For some experiments, polyclonal antiserum against Rtf1 was used (28).
ChIP Assays
Plasmid transformants of strains KY1858 and KY2191 were grown in SC−Trp medium to a density of ∼1 × 107 cells/ml. Cross-linking and chromatin preparation were performed essentially as described (57). Sonicated chromatin was incubated with antibodies specific for Rtf1 (28), Rpb3 (Neoclone), HA-agarose (Santa Cruz Biotechnology, Inc.), or Myc-agarose (Santa Cruz Biotechnology, Inc.). Samples were incubated with protein A- or protein G-conjugated agarose (GE Healthcare) to immunoprecipitate Rtf1 or Rpb3, respectively. Dilutions of purified input DNA and immunoprecipitated DNA were analyzed by quantitative real-time PCR with SYBR Green detection (Fermentas) using primers to indicated loci. Values represent the mean and S.D. of at least three biological replicates.
One-step TAP and Mass Spectrometry
Affinity purifications were performed using a protocol adapted from Ref. 58. Whole-cell extracts were incubated with rabbit IgG-coated Dynabeads (Invitrogen). Bound proteins, isolated by a magnet, were incubated with TEV protease for 3 h at 16 °C. Proteins released by TEV cleavage were analyzed by Western blotting or mass spectrometry. For the latter, proteins were run 1 cm into an 8% SDS-polyacrylamide gel, and excised gel slices were treated with trypsin. Peptides were identified by tandem MS using an Orbitrap mass spectrometer (Fred Hutchinson Cancer Research Center Proteomics Facility). Peptide Prophet (59) was used to evaluate the validity of peptide identifications, eliminating peptides with a score of <0.85 (<2.5% false discovery rate). Values represent the sum of identified peptides for bound fractions prepared from three biological replicates.
RESULTS
C-terminal Domain within Cdc73 Plays Role in Transcription Elongation
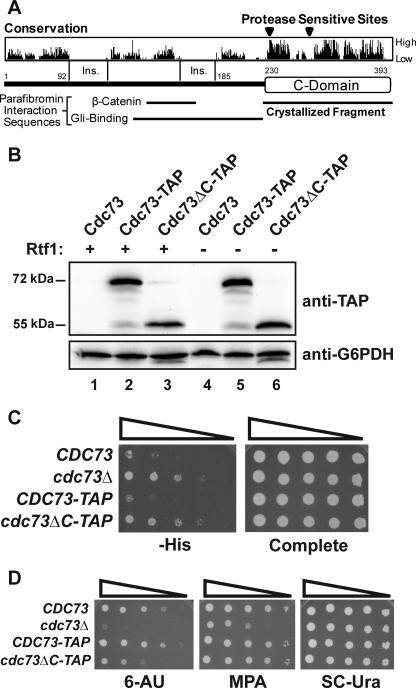
Genetic and biochemical data point to an important role for Cdc73 in transcription elongation. However, there are no currently recognized domains that could account for the roles of Cdc73 in transcription, despite apparent sequence conversation among Cdc73 proteins. Therefore, we sought to determine whether there was a stably folded domain within Cdc73 that could account for its activities by performing limited proteolysis on full-length, 393-amino acid S. cerevisiae Cdc73 and analyzing the resulting fragments by N-terminal sequencing. Using this approach, we noted the presence of two stable fragments beginning at residues 236 and 286 and likely extending to the end of the protein (data not shown). Interestingly, the fragment beginning at amino acid 236 contains the entirety of a region of high sequence conservation (Fig. 1A) that is predicted to contain numerous elements of secondary structure. Integration of a TAP tag to replace residues 231–393 of chromosomally expressed Cdc73 did not affect the stability of the remaining Cdc73 protein in vivo (Fig. 1B), indicating that residues 1–230 may fold and function independently of the rest of Cdc73. Based on these data, we hypothesized that Cdc73 is composed of two regions, an N-terminal protease-sensitive region containing residues 1–235 and a C-terminal protease-resistant domain composed of residues 236–393, which we refer to as the C-domain.
FIGURE 1.
Deletion of the Cdc73 C-domain causes phenotypes associated with defects in transcription elongation. A, domain architecture of Cdc73. Sequence conservation is indicated, as are insertions conserved within higher eukaryotes and the locations of known binding interactions for parafibromin. B, Western analysis of tagged Cdc73 proteins. Extracts from strains with the indicated genotypes (KY1021, KY1799, KY1791, KY1802, KY1797, and KY1789) were probed with peroxidase-antiperoxidase antibody to detect the TAP tag. C, dilution analysis assessing the Spt− phenotype of strains with the indicated genotypes (KY1021, KY1857, KY1799, and KY1791). 5-Fold dilutions starting from 1.0 × 108 cells/ml were spotted to SD medium lacking histidine (−His) and incubated at 30 °C for 4 days. D, dilution analyses assessing 6-AU and MPA sensitivity of the strains used in C. 10-Fold dilutions starting from 1.0 × 108 cells/ml were spotted to medium containing 6-AU or MPA and incubated at 30 °C for 4 days.
To investigate the functional importance of the Cdc73 C-domain, we tested whether deletion of amino acids 231–393 from chromosomally expressed Cdc73 caused phenotypes previously described for a cdc73 null mutation. Compared with a control strain expressing C-terminally TAP-tagged full-length Cdc73 (CDC73-TAP), the C-domain deletion strain (cdc73ΔC-TAP) exhibited two mutant phenotypes commonly ascribed to defects in transcription and/or chromatin structure. As indicated by growth on plates lacking histidine, the C-domain deletion suppressed the effects of a Ty δ-element insertion mutation within the promoter of the HIS4 gene, the his4-912δ allele, nearly as well as a complete deletion of CDC73 (Fig. 1C). This level of growth indicates that the C-domain deletion confers a moderate Spt− (suppressor of Ty) phenotype (60).
We also asked if deletion of the C-domain affected growth on medium containing 6-AU or MPA. 6-AU is a base analog, which inhibits enzymes in the ribonucleotide synthesis pathway (61), and MPA alters expression of IMD2, a gene encoding IMP dehydrogenase, thereby reducing synthesis of GMP (62, 63). The reduction in ribonucleotide levels caused by these compounds has been proposed to impair RNA pol II transcription elongation and elevate the requirement for elongation factors, although more recent studies have revealed that MPA sensitivity correlates with altered transcription start site selection within the IMD2 promoter (63). Our analysis indicates that deletion of the C-domain caused increased sensitivity to 6-AU, although not to the level observed for the cdc73 null mutation (Fig. 1D). In contrast, deletion of the C-domain did not significantly impair growth on MPA-containing media at the concentrations tested (Fig. 1D). Taken together, these results identify a domain at the C terminus of Cdc73 whose deletion causes phenotypes associated with defects in transcription.
Cdc73 C-domain Is Not Required for Paf1C Assembly
One possible interpretation of the genetic data is that the Cdc73 C-domain is required for either Cdc73 incorporation into Paf1C or for assembly of a complete Paf1C. To address this possibility, we performed one-step purifications (58) of TAP-tagged Cdc73ΔC truncation and TAP-tagged full-length Cdc73 proteins from yeast lysates and analyzed the co-purifying material for the presence of other Paf1C subunits by Western blotting (Fig. 2A). The results show that both full-length Cdc73-TAP and Cdc73ΔC-TAP could effectively pull-down Paf1C subunits, whereas an untagged Cdc73 control protein could not (Fig. 2A). To confirm these results by a complementary approach and avoid the use of strains in which multiple Paf1C subunits are tagged, a situation that could influence complex stability, we purified Cdc73-TAP and Cdc73ΔC-TAP from yeast lysates in which the only tagged Paf1C subunit was Cdc73. Mass spectrometry analysis of Cdc73-associated proteins from these lysates revealed similar numbers of peptides for the various Paf1C subunits, independent of the presence of the C-domain (Fig. 2B). In addition, Western analysis of these lysates confirmed that the levels of Rtf1 associated with Cdc73 are unaffected by deletion of the C-domain (Fig. 2C). Therefore, although we cannot rule out the possibility that the C-domain truncation may cause subtle defects in Paf1C stability, these results indicate that the C-domain of Cdc73 is not required for Paf1C assembly.
FIGURE 2.
A C-domain truncation does not prevent Paf1C assembly. A, Western analysis of crude extract and bound fractions from one-step affinity purifications of untagged Cdc73, Cdc73-TAP, and Cdc73ΔC-TAP from strains GHY1185, KY1978, and KY1979. Extracts were probed with indicated antibodies to detect the presence of Paf1C subunits. *, nonspecific band. B, summary of mass spectrometry data. The numbers represent the sums of peptides detected in three independent affinity purifications of untagged Cdc73, Cdc73-TAP, or Cdc73ΔC-TAP from strains KY1021, KY1799, and KY1791. C, Western analysis of Rtf1 levels in bound fractions from one-step affinity purifications using the same strains as in the mass spectrometry analysis.
Cdc73 C-domain Adopts a Ras-like Fold
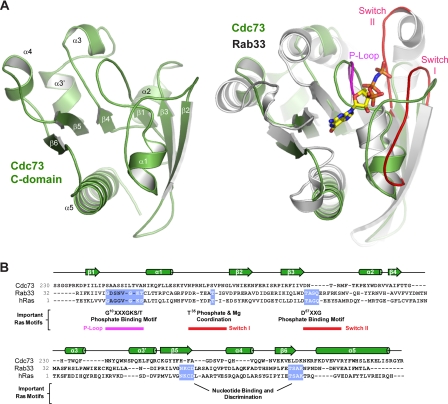
To investigate the molecular basis behind the role of the Cdc73 C-domain in transcription, we crystallized and determined the structure of the C-domain from S. cerevisiae Cdc73 using x-ray crystallography. Phases were determined using the single wavelength anomalous dispersion method, and the final model was refined against 1.55 Å native data to Rwork and Rfree values of 17.3 and 21.0%, respectively (see Table 2 and “Experimental Procedures” for a complete description of the crystallization and structure determination process). Cdc73 crystals contained one molecule in the asymmetric unit. As shown in Fig. 3A, the C-domain is a tightly packed mixed α-β fold. Central to the fold of the protein is a six-stranded β sheet with short stretches of α helix contained within the loops that connect the strands. Finally, there is a long helical segment at the very C terminus of the protein.
TABLE 2.
Data collection and refinement statistics for the yeast Cdc73 C-domain
| Selenomethionine | Native | |
|---|---|---|
| Data collection | ||
| Space group | P41212 | P41212 |
| Cell dimensions | ||
| a (Å) | 53.3 | 53.3 |
| b (Å) | 53.3 | 53.3 |
| c (Å) | 130.0 | 130.2 |
| Resolution (Å) | 50-1.96 | 37.7-1.55 |
| (1.99-1.96)a | (1.58-1.55) | |
| Unique reflections | 26,002 | 52,612 |
| Rmergeb | 5.7 (38.0.5) | 5.4 (38.2) |
| I/σI | 31.1 (8.3) | 58.0 (5.9) |
| Completeness (%) | 100 (100) | 99.3 (97.8) |
| Redundancy | 8.3 (8.4) | 7.1 (6.9) |
| Refinement | ||
| Resolution (Å) | 37.7-1.55 | |
| (1.60-1.55) | ||
| Rworkc/Rfreed (%) | 17.3/21.0 | |
| (15.4/23.6) | ||
| No. of atoms | ||
| Protein | 2879 | |
| Water | 145 | |
| Root mean square deviations | ||
| Bond lengths (Å) | 0.007 | |
| Bond angles (degrees) | 1.068 | |
| Average isotropic B values (Å2) | ||
| Protein | 29.3 | |
| Solvent | 39.5 | |
| Ramachandran (%) | ||
| Outliers | 0.00 | |
| Allowed | 0.00 | |
| Favored | 100 | |
a Values in parentheses correspond to those in the outer resolution shell.
b Rmerge = (|(ΣI − 〈I〉)|)/(ΣI), where 〈I〉 is the average intensity of multiple measurements.
c Rwork = Σhkl‖Fo(hkl)‖ − Fc (hkl)‖/Σhkl|Fo(hkl)|.
d Rfree represents the cross-validation R factor for 5% of the reflections against which the model was not refined.
FIGURE 3.
Structure of the yeast Cdc73 C-domain. A, schematic representation of the yeast Cdc73 C-domain. Elements of secondary structure are indicated and named using the Ras nomenclature. Structural overlap of Cdc73 C-domain (green) and Rab33 (gray) is shown. Bound GTP analog from the Rab33 structure is shown as sticks. The positions of the P-loop (magenta) and the Switch I and II loops (red) important for Ras function are shown. B, primary sequence alignment of S. cerevisiae Cdc73, H-Ras, and Rab33. Above are the secondary structure elements, as determined from the Cdc73 structure. Sequence motifs that are important for Ras function are indicated.
Remarkably, alignments of the Cdc73 C-domain against the structural data base using DALI (64) revealed significant structural similarity with small GTPases of the Ras superfamily. Hundreds of Ras-related structural homologs were identified in this search; however, sequence identity was typically less than 12%, explaining why primary sequence searches failed to recognize this domain. For clarity, we will focus our analysis on a comparison with H-Ras (65), the founding member of the family, as well as Rab33 (66), which displays the highest structural identity to the Cdc73 C-domain. The structural overlap is quite strong, with a root mean square deviation of 3.2 Å over 120 Cα atoms for Rab33 (Fig. 3A, right) and 3.3 Å over 112 Cα atoms for H-Ras.
The Ras superfamily can be subdivided into eight subfamilies based on sequence, structure, and function. These include the Ras, Rab, Ran, Rho, Rap, and Arf families with functions ranging from cell proliferation, membrane trafficking, and cytoskeletal dynamics to nuclear import. Despite the diversity in biological function, there are common features beyond their structural fold that unite this group (reviewed in Ref. 67). They are small nucleotide-binding proteins, most using a guanine nucleotide. GTP hydrolysis is typically stimulated by activating proteins, whereas nucleotide exchange factors remove the GDP and facilitate GTP binding. These additional activating and exchange factors allow Ras proteins to toggle between GDP- and GTP-bound states. These two states are correlated with binding to additional effector proteins, thus allowing nucleotide binding to serve as a molecular switch that modulates protein-protein interactions.
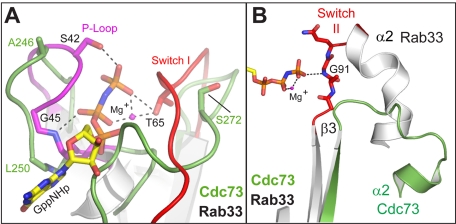
A number of interesting features distinguish the C-domain structure from the structures of canonical small GTPases, such as Ras. Ras family proteins utilize a series of loops to recognize nucleotide, aid in hydrolysis, and communicate information to the effector-binding region via a loaded spring mechanism (68). The most important of these regions are the P-loop, which makes extensive contacts with the γ-phosphate; Switch I, which interacts with both phosphate and the coordinated Mg2+ ion; and Switch II, which also interacts with the γ-phosphate. Structural superposition of the Cdc73 C-domain with Rab33 highlights these differences (Fig. 3A, right). First, the C-domain loop corresponding to the P-loop projects away from the position of the Rab33 bound nucleotide. The Switch I loop is different as well; in this case, the equivalent C-domain loop initially extends in toward the binding pocket and then projects away near the connection with β2 (Fig. 4A). Switch II is dramatically different in Cdc73 and bears little resemblance to Rab33 (Fig. 4B). The cumulative effect of these differences in loop conformation is to alter the surface where nucleotide would bind in canonical Ras proteins such that there is no discernible binding pocket in the C-domain. In addition to the differences in conformation, important sequence motifs within the P-loop, Switch I, and Switch II as well as important guanine recognition motifs are not conserved within the Cdc73 C-domain (Fig. 3B). We observe no evidence of a nucleotide or other ligand within our C-domain crystals. Attempts to co-crystallize the C-domain with a variety of nucleotides or to affinity-purify the C-domain with GTP-agarose both failed to demonstrate an interaction. Together, these observations suggest that the C-domain is probably functioning as a modulator of a protein-protein interaction and is unlikely to be sensing a guanine nucleotide in a manner analogous to the Ras family GTPases.
FIGURE 4.
Loops important for nucleotide binding in Ras adopt noncanonical conformations in the Cdc73 C-domain. Structural superposition of the Cdc73 C-domain (green) with Rab33 bound to a GTP analog (Protein Data Bank entry 1Z06). Within the Rab33 structure, the P-loop (magenta), and Switch I and II (red) are indicated, and the position of the GTP analog is shown in sticks. A, loops corresponding to P-loop and Switch I are altered in Cdc73. Residues making important hydrogen bonding interactions with nucleotide and/or Mg ion in Rab33 are indicated as are equivalent residues in Cdc73. B, the Switch II loop region is shifted within Cdc73. Switch II from Rab33 (red) and its interaction with the terminal phosphate is indicated, whereas the position of the equivalent loop in the C-domain is shown in green.
Conserved Surfaces within Cdc73 C-domain
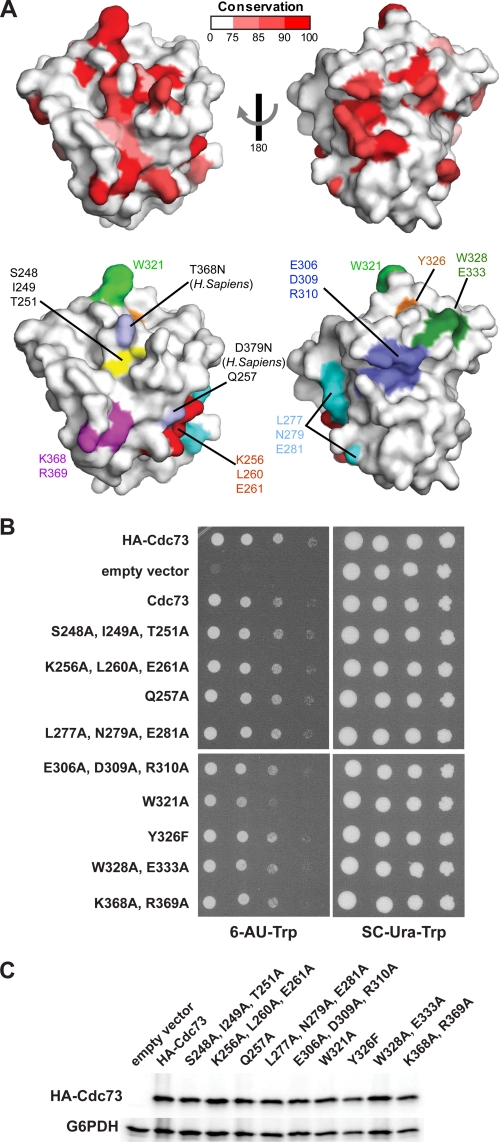
We next mapped evolutionary conservation onto the surface of the Cdc73 C-domain structure. To do this, we performed a multiple-sequence alignment on a collection of 14 Cdc73 sequences from model organisms using ClustalW (69). The conservation was scored at each amino acid position using the RISLER matrix within ESPRIPT (70, 71) and mapped onto the surface of the C-domain structure (Fig. 5A). Unlike similar maps for small GTPases, which are highly conserved within the nucleotide binding pocket, we find that the Cdc73 surface has several dispersed patches with significant sequence conservation. The most prominent of these is an extended surface adjacent to the site where nucleotide would bind in Ras proteins (Fig. 5A, left). There is also a second prominent patch of conserved residues on the opposite face of the protein (Fig. 5A, right).
FIGURE 5.
A conserved surface within the Cdc73 C-domain. A, sequence conservation from a multiple sequence alignment of Cdc73 sequences was scored and displayed on the C-domain surface with low conservation in white and invariant residues in red. The residues indicated were mutated and subjected to phenotypic analysis. Residues mutated in HPT-JT patients are indicated by the substitution in Homo sapiens. B, analysis of the 6-AU sensitivity of strains expressing HA-tagged Cdc73 mutant proteins with the indicated amino acid substitutions in the C-domain. Media contained 6-AU and lacked tryptophan for the selection of CEN/ARS plasmid-encoded CDC73 derivatives in strain KY1858. Strains were incubated at 30 °C for 3 days. C, Western analysis of HA-tagged wild-type and mutant Cdc73 proteins. Extracts from the strains analyzed in B were prepared, and Western blots were probed with antibodies specific to the HA tag and glucose-6-phosphate dehydrogenase (G6PDH), which served as a loading control.
Despite being physically separated on the surface, the conserved regions may all contribute toward a common function of Cdc73, and therefore mutations that alter these regions might lead to common mutant phenotypes. To address this possibility, we introduced a series of single and multisite amino acid substitutions within the C-domain to disturb patches of sequence conservation found on the surface. Several additional mutants were designed to disrupt regions of sequence important within Ras family members (Figs. 3B and 5A, bottom). These substitutions were generated within the context of N-terminally HA-tagged, full-length Cdc73 and expressed from yeast CEN/ARS plasmids. The ability of the mutations to complement the 6-AU sensitivity of a cdc73Δ strain was then examined (Fig. 5B). Unexpectedly, of the nine site-directed mutants tested, we found that only a single substitution (W321A) on the C-domain surface caused modest sensitivity to 6-AU (Fig. 5, A and B). Trp-321 is surprisingly exposed for a tryptophan residue and is located at the periphery of the large conserved region (Fig. 5A, left). The position of the Trp-321 side chain is stabilized by interactions with the neighboring residue Asn-324 but is otherwise exposed and accessible for potential binding interactions. Importantly, all of our mutant proteins are similarly expressed and are stable, implying that our results are only due to the substitution in question (Fig. 5C). Although our assay may not be sensitive enough to reveal more subtle effects of the other substitutions, the fact that the W321A substitution does not fully complement the cdc73Δ mutation suggests that this residue may be involved in a molecular interaction, the loss of which results in 6-AU sensitivity.
Role of the Cdc73 C-domain in Histone Modifications
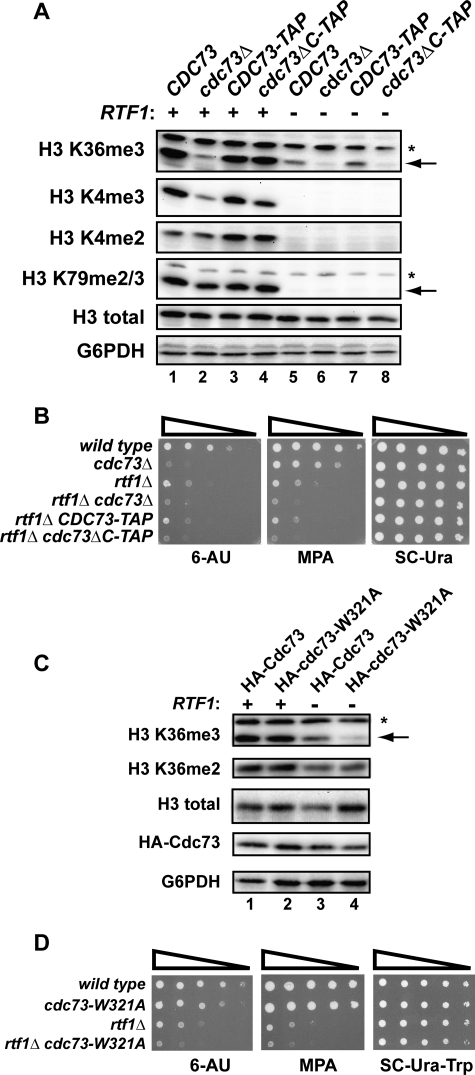
Several subunits within Paf1C, including Cdc73, are required for the proper modification of histones during transcription. Among deletions of Paf1C subunits, paf1Δ and ctr9Δ mutations cause the most severe defects in H3 Lys-36 trimethylation; however, deletion of CDC73 also leads to a significant reduction in this modification (5). In addition, Cdc73 is required for normal levels of H3 Lys-4 trimethylation in yeast cells (34). We therefore asked whether the Cdc73 C-domain is important for H3 Lys-36 trimethylation, H3 Lys-4 di- or trimethylation, or H3 Lys-79 di- or trimethylation, using Western analysis of strains that expressed untagged Cdc73, Cdc73-TAP, or Cdc73ΔC-TAP from the endogenous CDC73 locus (Fig. 6A). Antibody specificities were confirmed by probing levels of histone modifications in set1Δ, set2Δ, and dot1Δ control extracts (data not shown). Our results show that H3 Lys-4 dimethylation and H3 Lys-79 di-/trimethylation levels were essentially unaffected by deletion of the full CDC73 gene or by deletion of the C-domain. In addition, deletion of the C-domain did not reduce H3 Lys-4 trimethylation or H3 Lys-36 trimethylation levels significantly, although both of these modifications were decreased by the complete deletion of CDC73. These results suggest that the Cdc73 C-domain either lacks an important role in promoting Paf1C-dependent histone modifications or functions redundantly with other members of Paf1C. To address the latter hypothesis, we tested the effect of the cdc73ΔC mutation on histone modifications in strains deleted for RTF1. As expected from previous studies (7, 8, 16), the rtf1Δ mutation caused a complete loss of the H3 Lys-4 and Lys-79 methyl marks irrespective of the CDC73 genotype, demonstrating that these modifications are primarily controlled by Rtf1. In contrast, H3 Lys-36 trimethylation levels were only partially reduced in rtf1Δ strains that express full-length CDC73. Interestingly, deletion of either the full CDC73 gene or the region encoding the C-domain strongly enhanced the H3 Lys-36 trimethylation defect of the rtf1Δ strain. The synthetic effect of the C-domain deletion was not due to reduced Cdc73 protein levels in the rtf1Δ genetic background (Fig. 1B). Therefore, our results suggest that the Cdc73 C-domain functions redundantly with Rtf1 in promoting H3 Lys-36 trimethylation.
FIGURE 6.
Deletion of both the Cdc73 C-domain and Rtf1 leads to enhanced mutant phenotypes and defects in histone modifications. A and C, Western analysis on extracts from strains with the indicated genotypes (KY1021, KY1857, KY1799, KY1791, KY1802, KY703, KY1797, and KY1789) to detect levels of Paf1C-dependent H3 modifications. The arrows indicate the band of interest, and nonspecific bands are marked by an asterisk. B and D, dilution analyses assessing 6-AU and MPA sensitivity of the indicated strains (KY1021, KY1857, KY1802, KY703, KY1797, and KY1789). 10-Fold dilutions starting from 1.0 × 108 cells/ml were spotted to medium containing 6-AU or MPA and incubated at 30 °C for 2 days (D, SC−Ura−Trp), 4 days (B and D, 6-AU), or 8 days (D, MPA). For C and D, cdc73Δ (KY1858) or cdc73Δ rtf1Δ (KY2195) strains were transformed with TRP1-marked CEN/ARS plasmids that express either HA-tagged Cdc73 or HA-tagged Cdc73-W321A. Transformants were grown on media lacking tryptophan.
To further address the possibility that Rtf1 and the Cdc73 C-domain have overlapping functions, we asked if deletion of the C-domain could enhance the 6-AU and MPA sensitivity phenotypes of an rtf1Δ strain. Deletion of RTF1 leads to a partial sensitivity to both compounds, and this sensitivity was noticeably enhanced by deletion of the C-domain or the full CDC73 gene (Fig. 6B).
To investigate further the importance of amino acid Trp-321 in the C-domain, we tested whether the W321A substitution conferred defects in H3 Lys-36 methylation either alone or in combination with rtf1Δ. Like the C-domain deletion, the W321A substitution resulted in wild type levels of H3 Lys-36 trimethylation in an RTF1 background but an enhanced reduction in H3 Lys-36 trimethylation levels in an rtf1Δ background. Cdc73 protein levels were unaffected in this genetic background (Fig. 6C). Additionally, the W321A substitution did not affect H3 Lys-36 dimethylation levels in the presence or absence of RTF1 (Fig. 6C). In agreement with the histone modification data, cdc73-W321A rtf1Δ double mutant strains were more sensitive to 6-AU and MPA than either single mutant strain (Fig. 6D). These data are consistent with the idea that Trp-321 is a functionally important residue within the C-domain. Taken together, our findings suggest that the C-domain has important functions in transcription elongation and that the deletion of RTF1 exacerbates the effects of losing these functions.
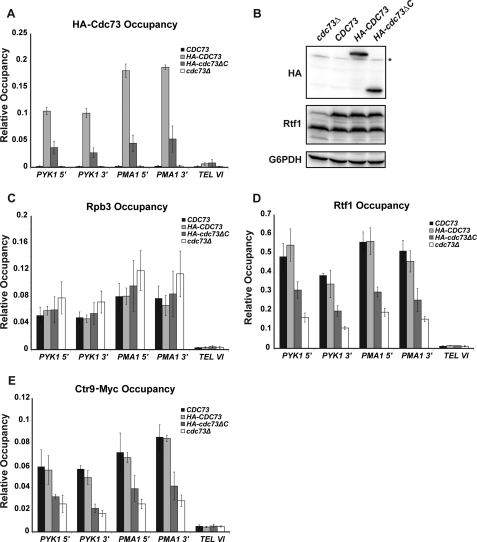
C-domain Is Important for Paf1C Localization to Active Genes
Previous studies have shown that deletion of either CDC73 or RTF1 greatly reduces the occupancy of other Paf1C subunits on active genes (13). The functional region within Rtf1 that is important for recruiting Paf1C to RNA pol II-transcribed genes has been mapped to the central Plus3 domain (16). However, the role of Cdc73 in directing the localization of Paf1C to active genes remains largely undefined. Given the genetic redundancy between RTF1 and CDC73, we asked whether the Cdc73 C-domain played a role in promoting the association of Paf1C with chromatin by performing ChIP assays. Using HA-tagged Cdc73 or an HA-tagged Cdc73 derivative lacking the C-domain, we found that the levels of Cdc73 associated with the active genes PYK1 and PMA1 were significantly reduced in the absence of the C-domain (Fig. 7A). The reduction in Cdc73 occupancy was not due to decreased levels of the HA-Cdc73ΔC protein compared with the HA-tagged full-length protein (Fig. 7B) or to reduced levels of RNA pol II on the genes (Fig. 7C). To ask if the C-domain is required for full recruitment of other Paf1C subunits, we measured the occupancy of Rtf1 and Ctr9-Myc at the PYK1 and PMA1 genes in strains lacking the full Cdc73 protein or just the C-domain. Consistent with previous studies (13), the occupancies of Rtf1 and Ctr9-Myc were reduced in the cdc73Δ strain (Fig. 7, D and E). Importantly, a similar reduction in occupancy for these proteins was observed in the cdc73ΔC strain. Our TAP results indicate that the decreased chromatin association of Rtf1 and Ctr9 in the cdc73ΔC strain is not due to reduced protein levels or severe defects in Paf1C assembly (Fig. 2). Collectively, our results suggest that the Ras-like fold in Cdc73 plays a role in recruiting Paf1C to RNA pol II and/or stabilizing this association.
FIGURE 7.
Deletion of the Cdc73 C-domain reduces Paf1C occupancy on actively transcribed genes. ChIP analysis of HA-tagged Cdc73 (A), Rpb3 (C), Rtf1 (D), and Myc-tagged Ctr9 (E) was measured at the 5′ end of PYK1 (+253 to +346, relative to ATG), 3′ end of PYK1 (+1127 to +1270), 5′ end of PMA1 (+214 to +319), 3′ end of PMA1 (+2107 to +2194) and at an untranscribed region proximal to the telomere of chromosome VI (TEL VI; chromosome coordinates 269,495–269,598). Values represent the average of at least three independent samples with S.D. (error bars). B, Western analysis of HA-tagged Cdc73 levels in strains utilized for ChIP. Extracts from a cdc73Δ strain (KY1858) transformed with empty vector or plasmids expressing full-length untagged Cdc73, full-length HA-tagged Cdc73, or truncated HA-tagged Cdc73ΔC were probed with antibodies specific for the HA tag (Cdc73), Rtf1, or glucose-6-phosphate dehydrogenase (G6PDH) as a loading control. *, nonspecific band. The two bands detected with the Rtf1 antiserum represent full-length Rtf1 and a consistently observed breakdown product.
DISCUSSION
Proper gene expression requires a large number of accessory factors, which must be appropriately recruited and localized to the transcription complex. Paf1C is a transcription regulatory complex with multiple important functions; however, our understanding of how it is recruited to RNA pol II is limited. Previous work has demonstrated that two subunits of yeast Paf1C, Cdc73 and Rtf1, play prominent roles in directing the complex to active genes (10, 13). Deletion analysis indicates that the Plus-3 domain of Rtf1 is important for localizing Paf1C to chromatin; however, the role of Cdc73 in Paf1C recruitment is unclear. Here we describe the identification of a previously unrecognized domain at the C terminus of Cdc73, which we call the C-domain. We have crystallized and determined the structure of the Cdc73 C-domain and, despite a lack of sequence conservation, found that the C-domain adopts a fold that is highly similar to the small GTPase Ras. We found that deletion of the C-domain confers an Spt− phenotype and sensitivity to 6-AU, consistent with a role for this domain in transcription elongation. However, we found only moderate effects of a C-domain deletion on histone modifications, suggesting that the main role of the C-domain lies outside of modifications to chromatin.
Given the propensity for Ras family members to mediate protein-protein interactions, it seemed reasonable to consider that the C-domain could be functioning as a scaffold for interactions with other Paf1C subunits. TAP purifications of Paf1C performed from extracts lacking various Paf1C subunits indicate that Rtf1 levels within the purified complexes are reduced in the absence of Cdc73 (10). Because rtf1Δ strains are 6-AU sensitive (16), one could explain the 6-AU sensitivity of a Cdc73 C-domain deletion mutant if the C-domain was required for incorporation of Rtf1 into Paf1C. Our data, however, show that all of the subunits of Paf1C can still assemble in the absence of the C-domain, suggesting a role for the C-domain outside of maintaining overall Paf1C architecture. This is in general agreement with data on parafibromin in which residues within the central portion of the protein have been implicated in the assembly of human Paf1C (23). The central region of parafibromin has also been shown to mediate binding to Gli and β-catenin, further supporting a role for this region of the protein as a recognition module in higher eukaryotes (46, 72). Our data demonstrate that yeast Paf1C is capable of assembling in the absence of the Cdc73 C-domain; however, given the proximity of the C-domain to sequences required for Paf1C assembly, we cannot discount the possibility that the C-domain has a more subtle effect on Paf1C stability or the conformation of the complex.
Importantly, we find that deletion of the Cdc73 C-domain significantly reduces Paf1C occupancy on active genes. We interpret this result to mean that the Cdc73 C-domain is playing a role in promoting association of Paf1C with chromatin just like the Rtf1 Plus-3 domain. This is in agreement with coimmunoprecipitation experiments (10) and suggests that Paf1C uses two independent domains for chromatin association and possibly recognizes two independent protein or ligand binding partners. The discovery that the Cdc73 C-domain has a Ras-like fold is an intriguing result in light of this finding; this domain has long been recognized as a protein-protein and protein-ligand interaction partner. In the case of the yeast C-domain, however, the canonical ligand binding pocket has been altered, and it seems unlikely that it would bind a nucleotide ligand. However, sequence conservation of residues near the canonical ligand binding surface is high, indicating that this surface is probably important for an evolutionarily conserved interaction. Two possible explanations seem most probable. First, that Paf1C uses a different small molecule ligand as a regulatory signal to modulate a protein-protein interaction, just as Ras does with GTP. As mentioned before, GTP binding seems unlikely, given differences in important nucleotide recognition sequences within the C-domain; however, this does not preclude binding to another yet to be determined ligand. It would seem, however, that a significant structural rearrangement would be needed to facilitate binding. The second possibility is that in Cdc73, the Ras fold has been repurposed to bind a protein. The surface of the Cdc73 C-domain that is equivalent to the canonical GTP-binding pocket is relatively flat. Using this surface to interact with another protein would bypass the need for a deep binding pocket.
Any of the proteins involved in the transcription complex could be potential binding partners for the Cdc73 C-domain; however, the FACT, RNA pol II, and Spt4-Spt5 complexes seem most likely because they are already known to interact with Paf1C. RNA pol II and the Spt4-Spt5 complex are particularly interesting candidates for this interaction because both contain C-terminal repeating sequences. Such sequences are likely to be highly flexible and could adapt to fit the extended conserved surface found on the yeast Cdc73 C-domain. Our finding that, with the exception of W321A, substitutions of individual, exposed amino acids or small clusters of amino acids failed to yield an observable mutant phenotype is consistent with the idea that the C-domain comprises an extensive, multivalent interaction surface. Interestingly, Rtf1 uses a different and unrelated domain to direct Paf1C to chromatin. One possibility is that the Cdc73 C-domain and the Rtf1 Plus-3 domain bind the same protein ligand. The extended repeat domains within Spt5 or RNA pol II could facilitate simultaneous binding of the C-domain and the Plus-3 domain to either protein. Another possibility is that the Cdc73 C-domain functions in conjunction with the Plus-3 domain of Rtf1 to form a bivalent recruitment module with two distinct attachment points on the RNA pol II elongation machinery.
Mutations within the gene for human Cdc73, parafibromin, are found in patients with HPT-JT as well as other cancers. Most are frameshift mutations near the 5′ end of the parafibromin gene, which would be predicted to produce severely truncated, unstable, or nonfunctional proteins. Although these mutations are most likely functioning as null alleles, the discovery of point mutations from HPT-JT patients that alter specific amino acids within the C-domain emphasizes the importance of this domain in human disease (20). These mutations would substitute asparagines at residue Thr-369 or Ser-379 within parafibromin. Interestingly, the equivalent residues within the yeast protein are located on the large conserved surface near the Ras nucleotide binding site and would not be predicted to affect protein stability. The Cdc73 proteins of humans and other higher eukaryotes contain conserved sequences not found within the S. cerevisiae protein. These sequences interact with transcription regulatory proteins, including Gli and β-catenin. The physical proximity between the binding sites for regulatory factors and the C-domain could mean that, in higher organisms, binding of those factors influences the function of the C-domain. Additionally, the C-domain could direct the recruitment of regulatory factors to the core transcription elongation machinery. An informative area of future study will be the identification of potential binding partners for the yeast Cdc73 protein.
Acknowledgments
We are grateful to Erica McGreevy, Kristianna Ricchio, and John Hempel for technical support. We thank Brett Tomson and Alan Hinnebusch for critical reading of the manuscript and Alan Hinnebusch for communication of unpublished results. Operations at the National Synchrotron Light Source are supported by the Department of Energy, Office of Basic Energy Sciences, and the National Institutes of Health. Data collection at the National Synchrotron Light Source was funded by the National Center for Research Resources.
This work was supported, in whole or in part, by National Institutes of Health Grants R01GM52593 (to K. M. A.) and R21RR025787 (to R. G. G.). This work was also supported by fellowships from the Howard Hughes Medical Institute (to C. P. D. and W. P. R.) and a Beckman Scholars Award (to C. P. D.).
The atomic coordinates and structure factors (code 3V46) have been deposited in the Protein Data Bank, Research Collaboratory for Structural Bioinformatics, Rutgers University, New Brunswick, NJ (http://www.rcsb.org/).
- pol
- polymerase
- 6-AU
- 6-azauracil
- MPA
- mycophenolic acid
- HPT-JT
- hyperparathyroidism-jaw tumor syndrome
- TEV
- tobacco etch virus
- Bicine
- N,N-bis(2-hydroxyethyl)glycine
- me2 and me3
- di- and trimethyl, respectively
- TAP
- tandem affinity purification.
REFERENCES
- 1. Sikorski T. W., Buratowski S. (2009) The basal initiation machinery. Beyond the general transcription factors. Curr. Opin. Cell Biol. 21, 344–351 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2. Shi X., Finkelstein A., Wolf A. J., Wade P. A., Burton Z. F., Jaehning J. A. (1996) Paf1p, an RNA polymerase II-associated factor in Saccharomyces cerevisiae, may have both positive and negative roles in transcription. Mol. Cell. Biol. 16, 669–676 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3. Wade P. A., Werel W., Fentzke R. C., Thompson N. E., Leykam J. F., Burgess R. R., Jaehning J. A., Burton Z. F. (1996) A novel collection of accessory factors associated with yeast RNA polymerase II. Protein Expr. Purif. 8, 85–90 [DOI] [PubMed] [Google Scholar]
- 4. Stolinski L. A., Eisenmann D. M., Arndt K. M. (1997) Identification of RTF1, a novel gene important for TATA site selection by TATA box-binding protein in Saccharomyces cerevisiae. Mol. Cell. Biol. 17, 4490–4500 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5. Chu Y., Simic R., Warner M. H., Arndt K. M., Prelich G. (2007) Regulation of histone modification and cryptic transcription by the Bur1 and Paf1 complexes. EMBO J. 26, 4646–4656 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6. Wood A., Schneider J., Dover J., Johnston M., Shilatifard A. (2003) The Paf1 complex is essential for histone monoubiquitination by the Rad6-Bre1 complex, which signals for histone methylation by COMPASS and Dot1p. J. Biol. Chem. 278, 34739–34742 [DOI] [PubMed] [Google Scholar]
- 7. Krogan N. J., Dover J., Wood A., Schneider J., Heidt J., Boateng M. A., Dean K., Ryan O. W., Golshani A., Johnston M., Greenblatt J. F., Shilatifard A. (2003) The Paf1 complex is required for histone H3 methylation by COMPASS and Dot1p. Linking transcriptional elongation to histone methylation. Mol. Cell 11, 721–729 [DOI] [PubMed] [Google Scholar]
- 8. Ng H. H., Dole S., Struhl K. (2003) The Rtf1 component of the Paf1 transcriptional elongation complex is required for ubiquitination of histone H2B. J. Biol. Chem. 278, 33625–33628 [DOI] [PubMed] [Google Scholar]
- 9. Ng H. H., Robert F., Young R. A., Struhl K. (2003) Targeted recruitment of Set1 histone methylase by elongating Pol II provides a localized mark and memory of recent transcriptional activity. Mol. Cell 11, 709–719 [DOI] [PubMed] [Google Scholar]
- 10. Nordick K., Hoffman M. G., Betz J. L., Jaehning J. A. (2008) Direct interactions between the Paf1 complex and a cleavage and polyadenylation factor are revealed by dissociation of Paf1 from RNA polymerase II. Eukaryot. Cell 7, 1158–1167 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11. Rozenblatt-Rosen O., Nagaike T., Francis J. M., Kaneko S., Glatt K. A., Hughes C. M., LaFramboise T., Manley J. L., Meyerson M. (2009) The tumor suppressor Cdc73 functionally associates with CPSF and CstF 3′ mRNA processing factors. Proc. Natl. Acad. Sci. U.S.A. 106, 755–760 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12. Sheldon K. E., Mauger D. M., Arndt K. M. (2005) A Requirement for the Saccharomyces cerevisiae Paf1 complex in snoRNA 3′ end formation. Mol. Cell 20, 225–236 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13. Mueller C. L., Porter S. E., Hoffman M. G., Jaehning J. A. (2004) The Paf1 complex has functions independent of actively transcribing RNA polymerase II. Mol. Cell 14, 447–456 [DOI] [PubMed] [Google Scholar]
- 14. Simic R., Lindstrom D. L., Tran H. G., Roinick K. L., Costa P. J., Johnson A. D., Hartzog G. A., Arndt K. M. (2003) Chromatin remodeling protein Chd1 interacts with transcription elongation factors and localizes to transcribed genes. EMBO J. 22, 1846–1856 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15. Tomson B. N., Davis C. P., Warner M. H., Arndt K. M. (2011) Identification of a role for histone H2B ubiquitylation in noncoding RNA 3′-end formation through mutational analysis of Rtf1 in Saccharomyces cerevisiae. Genetics 188, 273–289 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16. Warner M. H., Roinick K. L., Arndt K. M. (2007) Rtf1 is a multifunctional component of the Paf1 complex that regulates gene expression by directing cotranscriptional histone modification. Mol. Cell. Biol. 27, 6103–6115 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17. Briggs S. D., Xiao T., Sun Z. W., Caldwell J. A., Shabanowitz J., Hunt D. F., Allis C. D., Strahl B. D. (2002) Gene silencing. Trans-histone regulatory pathway in chromatin. Nature 418, 498. [DOI] [PubMed] [Google Scholar]
- 18. Dover J., Schneider J., Tawiah-Boateng M. A., Wood A., Dean K., Johnston M., Shilatifard A. (2002) Methylation of histone H3 by COMPASS requires ubiquitination of histone H2B by Rad6. J. Biol. Chem. 277, 28368–28371 [DOI] [PubMed] [Google Scholar]
- 19. Sun Z. W., Allis C. D. (2002) Ubiquitination of histone H2B regulates H3 methylation and gene silencing in yeast. Nature 418, 104–108 [DOI] [PubMed] [Google Scholar]
- 20. Bradley K. J., Cavaco B. M., Bowl M. R., Harding B., Cranston T., Fratter C., Besser G. M., Conceicao Pereira M., Davie M. W., Dudley N., Leite V., Sadler G. P., Seller A., Thakker R. V. (2006) Parafibromin mutations in hereditary hyperparathyroidism syndromes and parathyroid tumors. Clin. Endocrinol. 64, 299–306 [DOI] [PubMed] [Google Scholar]
- 21. Carpten J. D., Robbins C. M., Villablanca A., Forsberg L., Presciuttini S., Bailey-Wilson J., Simonds W. F., Gillanders E. M., Kennedy A. M., Chen J. D., Agarwal S. K., Sood R., Jones M. P., Moses T. Y., Haven C., Petillo D., Leotlela P. D., Harding B., Cameron D., Pannett A. A., Höög A., Heath H., 3rd, James-Newton L. A., Robinson B., Zarbo R. J., Cavaco B. M., Wassif W., Perrier N. D., Rosen I. B., Kristoffersson U., Turnpenny P. D., Farnebo L. O., Besser G. M., Jackson C. E., Morreau H., Trent J. M., Thakker R. V., Marx S. J., Teh B. T., Larsson C., Hobbs M. R. (2002) HRPT2, encoding parafibromin, is mutated in hyperparathyroidism-jaw tumor syndrome. Nat. Genet. 32, 676–680 [DOI] [PubMed] [Google Scholar]
- 22. Ding L., Paszkowski-Rogacz M., Nitzsche A., Slabicki M. M., Heninger A. K., de Vries I., Kittler R., Junqueira M., Shevchenko A., Schulz H., Hubner N., Doss M. X., Sachinidis A., Hescheler J., Iacone R., Anastassiadis K., Stewart A. F., Pisabarro M. T., Caldarelli A., Poser I., Theis M., Buchholz F. (2009) A genome-scale RNAi screen for Oct4 modulators defines a role of the Paf1 complex for embryonic stem cell identity. Cell Stem Cell 4, 403–415 [DOI] [PubMed] [Google Scholar]
- 23. Rozenblatt-Rosen O., Hughes C. M., Nannepaga S. J., Shanmugam K. S., Copeland T. D., Guszczynski T., Resau J. H., Meyerson M. (2005) The parafibromin tumor suppressor protein is part of a human Paf1 complex. Mol. Cell. Biol. 25, 612–620 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24. Shema E., Tirosh I., Aylon Y., Huang J., Ye C., Moskovits N., Raver-Shapira N., Minsky N., Pirngruber J., Tarcic G., Hublarova P., Moyal L., Gana-Weisz M., Shiloh Y., Yarden Y., Johnsen S. A., Vojtesek B., Berger S. L., Oren M. (2008) The histone H2B-specific ubiquitin ligase RNF20/hBRE1 acts as a putative tumor suppressor through selective regulation of gene expression. Genes Dev. 22, 2664–2676 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25. Mutiu A. I., Hoke S. M., Genereaux J., Liang G., Brandl C. J. (2007) The role of histone ubiquitylation and deubiquitylation in gene expression as determined by the analysis of an HTB1(K123R) Saccharomyces cerevisiae strain. Mol. Genet. Genomics 277, 491–506 [DOI] [PubMed] [Google Scholar]
- 26. Kim M., Ahn S. H., Krogan N. J., Greenblatt J. F., Buratowski S. (2004) Transitions in RNA polymerase II elongation complexes at the 3′ ends of genes. EMBO J. 23, 354–364 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27. Mayer A., Lidschreiber M., Siebert M., Leike K., Söding J., Cramer P. (2010) Uniform transitions of the general RNA polymerase II transcription complex. Nat. Struct. Mol. Biol. 17, 1272–1278 [DOI] [PubMed] [Google Scholar]
- 28. Squazzo S. L., Costa P. J., Lindstrom D. L., Kumer K. E., Simic R., Jennings J. L., Link A. J., Arndt K. M., Hartzog G. A. (2002) The Paf1 complex physically and functionally associates with transcription elongation factors in vivo. EMBO J. 21, 1764–1774 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29. Krogan N. J., Kim M., Ahn S. H., Zhong G., Kobor M. S., Cagney G., Emili A., Shilatifard A., Buratowski S., Greenblatt J. F. (2002) RNA polymerase II elongation factors of Saccharomyces cerevisiae. A targeted proteomics approach. Mol. Cell. Biol. 22, 6979–6992 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30. Qiu H., Hu C., Wong C. M., Hinnebusch A. G. (2006) The Spt4p subunit of yeast DSIF stimulates association of the Paf1 complex with elongating RNA polymerase II. Mol. Cell. Biol. 26, 3135–3148 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31. Rondón A. G., Gallardo M., García-Rubio M., Aguilera A. (2004) Molecular evidence indicating that the yeast PAF complex is required for transcription elongation. EMBO Rep. 5, 47–53 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32. Tous C., Rondón A. G., García-Rubio M., González-Aguilera C., Luna R., Aguilera A. (2011) A novel assay identifies transcript elongation roles for the Nup84 complex and RNA processing factors. EMBO J. 30, 1953–1964 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33. Kaplan C. D., Holland M. J., Winston F. (2005) Interaction between transcription elongation factors and mRNA 3′-end formation at the Saccharomyces cerevisiae GAL10-GAL7 locus. J. Biol. Chem. 280, 913–922 [DOI] [PubMed] [Google Scholar]
- 34. Laribee R. N., Krogan N. J., Xiao T., Shibata Y., Hughes T. R., Greenblatt J. F., Strahl B. D. (2005) BUR kinase selectively regulates H3 K4 trimethylation and H2B ubiquitylation through recruitment of the PAF elongation complex. Curr. Biol. 15, 1487–1493 [DOI] [PubMed] [Google Scholar]
- 35. Liu Y., Warfield L., Zhang C., Luo J., Allen J., Lang W. H., Ranish J., Shokat K. M., Hahn S. (2009) Phosphorylation of the transcription elongation factor Spt5 by yeast Bur1 kinase stimulates recruitment of the PAF complex. Mol. Cell. Biol. 29, 4852–4863 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36. Mulder K. W., Brenkman A. B., Inagaki A., van den Broek N. J., Timmers H. T. (2007) Regulation of histone H3K4 trimethylation and PAF complex recruitment by the Ccr4-Not complex. Nucleic Acids Res. 35, 2428–2439 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37. Pavri R., Zhu B., Li G., Trojer P., Mandal S., Shilatifard A., Reinberg D. (2006) Histone H2B monoubiquitination functions cooperatively with FACT to regulate elongation by RNA polymerase II. Cell 125, 703–717 [DOI] [PubMed] [Google Scholar]
- 38. Pruneski J. A., Hainer S. J., Petrov K. O., Martens J. A. (2011) The Paf1 complex represses SER3 transcription in Saccharomyces cerevisiae by facilitating intergenic transcription-dependent nucleosome occupancy of the SER3 promoter. Eukaryot. Cell 10, 1283–1294 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39. Zhou K., Kuo W. H., Fillingham J., Greenblatt J. F. (2009) Control of transcriptional elongation and cotranscriptional histone modification by the yeast BUR kinase substrate Spt5. Proc. Natl. Acad. Sci. U.S.A. 106, 6956–6961 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40. de Jong R. N., Truffault V., Diercks T., Ab E., Daniels M. A., Kaptein R., Folkers G. E. (2008) Structure and DNA binding of the human Rtf1 Plus3 domain. Structure 16, 149–159 [DOI] [PubMed] [Google Scholar]
- 41. Cascón A., Huarte-Mendicoa C. V., Javier Leandro-García L., Letón R., Suela J., Santana A., Costa M. B., Comino-Méndez I., Landa I., Sánchez L., Rodríguez-Antona C., Cigudosa J. C., Robledo M. (2011) Detection of the first gross CDC73 germ line deletion in an HPT-JT syndrome family. Genes Chromosomes Cancer 50, 922–929 [DOI] [PubMed] [Google Scholar]
- 42. Pimenta F. J., Gontijo Silveira L. F., Tavares G. C., Silva A. C., Perdigão P. F., Castro W. H., Gomez M. V., Teh B. T., De Marco L., Gomez R. S. (2006) HRPT2 gene alterations in ossifying fibroma of the jaws. Oral Oncol. 42, 735–739 [DOI] [PubMed] [Google Scholar]
- 43. Tan M. H., Morrison C., Wang P., Yang X., Haven C. J., Zhang C., Zhao P., Tretiakova M. S., Korpi-Hyovalti E., Burgess J. R., Soo K. C., Cheah W. K., Cao B., Resau J., Morreau H., Teh B. T. (2004) Loss of parafibromin immunoreactivity is a distinguishing feature of parathyroid carcinoma. Clin. Cancer Res. 10, 6629–6637 [DOI] [PubMed] [Google Scholar]
- 44. Woodard G. E., Lin L., Zhang J. H., Agarwal S. K., Marx S. J., Simonds W. F. (2005) Parafibromin, product of the hyperparathyroidism-jaw tumor syndrome gene HRPT2, regulates cyclin D1/PRAD1 expression. Oncogene 24, 1272–1276 [DOI] [PubMed] [Google Scholar]
- 45. Yang Y. J., Han J. W., Youn H. D., Cho E. J. (2010) The tumor suppressor, parafibromin, mediates histone H3 K9 methylation for cyclin D1 repression. Nucleic Acids Res. 38, 382–390 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46. Mosimann C., Hausmann G., Basler K. (2006) Parafibromin/Hyrax activates Wnt/Wg target gene transcription by direct association with β-catenin/Armadillo. Cell 125, 327–341 [DOI] [PubMed] [Google Scholar]
- 47. Newey P. J., Bowl M. R., Thakker R. V. (2009) Parafibromin. Functional insights. J. Intern. Med. 266, 84–98 [DOI] [PubMed] [Google Scholar]
- 48. Studier F. W. (2005) Protein production by autoinduction in high density shaking cultures. Protein Expr. Purif. 41, 207–234 [DOI] [PubMed] [Google Scholar]
- 49. Otwinowski Z., Minor W. (1997) Processing of X-ray diffraction data collected in oscillation mode. Methods Enzymol. 276, 307–326 [DOI] [PubMed] [Google Scholar]
- 50. Adams P. D., Afonine P. V., Bunkóczi G., Chen V. B., Davis I. W., Echols N., Headd J. J., Hung L. W., Kapral G. J., Grosse-Kunstleve R. W., McCoy A. J., Moriarty N. W., Oeffner R., Read R. J., Richardson D. C., Richardson J. S., Terwilliger T. C., Zwart P. H. (2010) PHENIX. A comprehensive Python-based system for macromolecular structure solution. Acta Crystallogr. D Biol. Crystallogr. 66, 213–221 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51. Davis I. W., Leaver-Fay A., Chen V. B., Block J. N., Kapral G. J., Wang X., Murray L. W., Arendall W. B., 3rd, Snoeyink J., Richardson J. S., Richardson D. C. (2007) MolProbity. All-atom contacts and structure validation for proteins and nucleic acids. Nucleic Acids Res. 35, W375–W383 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52. Winston F., Dollard C., Ricupero-Hovasse S. L. (1995) Construction of a set of convenient Saccharomyces cerevisiae strains that are isogenic to S288C. Yeast 11, 53–55 [DOI] [PubMed] [Google Scholar]
- 53. Ausubel F., Brent R., Kingston R., Moore D., Smith J. A., Seidman J., Struhl K. (1988) Short Protocols in Molecular Biology : A Compendium of Methods from Current Protocols in Molecular Biology. Greene Publishing Associates, New York [Google Scholar]
- 54. Rose M. D., Winston F., Hieter P. (1990) Methods in Yeast Genetics, pp. 179–180, Cold Spring Harbor Laboratory, Cold Spring Harbor, NY [Google Scholar]
- 55. Sikorski R. S., Hieter P. (1989) A system of shuttle vectors and yeast host strains designed for efficient manipulation of DNA in Saccharomyces cerevisiae. Genetics 122, 19–27 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56. Tyers M., Tokiwa G., Nash R., Futcher B. (1992) The Cln3-Cdc28 kinase complex of S. cerevisiae is regulated by proteolysis and phosphorylation. EMBO J. 11, 1773–1784 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57. Shirra M. K., Rogers S. E., Alexander D. E., Arndt K. M. (2005) The Snf1 protein kinase and Sit4 protein phosphatase have opposing functions in regulating TATA-binding protein association with the Saccharomyces cerevisiae INO1 promoter. Genetics 169, 1957–1972 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58. Oeffinger M., Wei K. E., Rogers R., DeGrasse J. A., Chait B. T., Aitchison J. D., Rout M. P. (2007) Comprehensive analysis of diverse ribonucleoprotein complexes. Nat. Methods 4, 951–956 [DOI] [PubMed] [Google Scholar]
- 59. Keller A., Nesvizhskii A. I., Kolker E., Aebersold R. (2002) Empirical statistical model to estimate the accuracy of peptide identifications made by MS/MS and database search. Anal. Chem. 74, 5383–5392 [DOI] [PubMed] [Google Scholar]
- 60. Winston F., Sudarsanam P. (1998) The SAGA of Spt proteins and transcriptional analysis in yeast. Past, present, and future. Cold Spring Harb. Symp. Quant. Biol. 63, 553–561 [DOI] [PubMed] [Google Scholar]
- 61. Exinger F., Lacroute F. (1992) 6-Azauracil inhibition of GTP biosynthesis in Saccharomyces cerevisiae. Curr. Genet. 22, 9–11 [DOI] [PubMed] [Google Scholar]
- 62. Jenks M. H., Reines D. (2005) Dissection of the molecular basis of mycophenolate resistance in Saccharomyces cerevisiae. Yeast 22, 1181–1190 [DOI] [PubMed] [Google Scholar]
- 63. Kuehner J. N., Brow D. A. (2008) Regulation of a eukaryotic gene by GTP-dependent start site selection and transcription attenuation. Mol. Cell 31, 201–211 [DOI] [PubMed] [Google Scholar]
- 64. Holm L., Kääriäinen S., Rosenström P., Schenkel A. (2008) Searching protein structure databases with DaliLite v.3. Bioinformatics 24, 2780–2781 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 65. Pai E. F., Krengel U., Petsko G. A., Goody R. S., Kabsch W., Wittinghofer A. (1990) Refined crystal structure of the triphosphate conformation of H-Ras p21 at 1.35 Å resolution. Implications for the mechanism of GTP hydrolysis. EMBO J. 9, 2351–2359 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 66. Eathiraj S., Pan X., Ritacco C., Lambright D. G. (2005) Structural basis of family-wide Rab GTPase recognition by rabenosyn-5. Nature 436, 415–419 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 67. Wennerberg K., Rossman K. L., Der C. J. (2005) The Ras superfamily at a glance. J. Cell Sci. 118, 843–846 [DOI] [PubMed] [Google Scholar]
- 68. Vetter I. R., Wittinghofer A. (2001) The guanine nucleotide-binding switch in three dimensions. Science 294, 1299–1304 [DOI] [PubMed] [Google Scholar]
- 69. Thompson J. D., Higgins D. G., Gibson T. J. (1994) ClustalW. Improving the sensitivity of progressive multiple sequence alignment through sequence weighting, position-specific gap penalties, and weight matrix choice. Nucleic Acids Res. 22, 4673–4680 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 70. Gouet P., Courcelle E., Stuart D. I., Métoz F. (1999) ESPript. Analysis of multiple sequence alignments in PostScript. Bioinformatics 15, 305–308 [DOI] [PubMed] [Google Scholar]
- 71. Risler J. L., Delorme M. O., Delacroix H., Henaut A. (1988) Amino acid substitutions in structurally related proteins. A pattern recognition approach. Determination of a new and efficient scoring matrix. J. Mol. Biol. 204, 1019–1029 [DOI] [PubMed] [Google Scholar]
- 72. Mosimann C., Hausmann G., Basler K. (2009) The role of Parafibromin/Hyrax as a nuclear Gli/Ci-interacting protein in Hedgehog target gene control. Mech. Dev. 126, 394–405 [DOI] [PubMed] [Google Scholar]