Background: PAI-2 (SERPINB2) is a serine protease inhibitor that is highly up-regulated in response to cellular stress.
Results: The upstream transactivator region of the SERPINB2 promoter was localized to an AP-1 site that overcomes PAUSE-1 silencer activity.
Conclusion: The AP-1 site binds factors that promote the derepression of the SERPINB2 promoter.
Significance: These data provide molecular insight into cell-specific derepression of the PAI-2 gene during cell differentiation and cancer.
Keywords: AP1 transcription factor, Fos, Gene regulation, Phorbol esters, Promoters, Serpin, PAI-2, Derepression, Transactivator
Abstract
Transcriptional up-regulation of the plasminogen activator inhibitor type-2 (PAI-2) gene is a major response to cellular stress. The expression of PAI-2 is induced by a variety of cytokines and growth factors that act in a cell type- and differentiation stage-dependent manner. We previously reported that the human SERPINB2 gene promoter is controlled by three major transcription regulatory domains: an inducible proximal promoter, an upstream silencer (PAUSE-1), and a distal transactivator region between −5100 and −3300, which appears to overcome inhibition mediated by the silencer. The distal transactivator region is inducible by the phorbol ester PMA, a potent activator of the protein kinase C (PKC) pathway that is a powerful inducer of PAI-2 gene expression in monocytes, macrophages, and myelomonocytic cells as well as in epidermal keratinocytes. Here we show that a 21-bp region (−4952/−4932), containing an AP-1 element, is both necessary and sufficient for PMA-induced transactivator activity in PAI-2-expressing U937 cells. This site specifically binds FosB in PAI-2-expressing U937 cells but not in HeLa cells that do not express PAI-2, and overexpression of FosB, c-Fos, or c-Jun in HeLa cells is sufficient to cause derepression of transcription from the SERPINB2 promoter. Although FosB is likely to be involved in transactivator-mediated derepression of PAI-2 transcription in macrophage-like cells, as exemplified by the U937 cell line, c-Jun may be functional in other cell types. These data suggest a model for the transcriptional control of the human PAI-2 gene and further our understanding of the molecular basis for its tissue-specific expression.
Introduction
The plasminogen activator inhibitor type 2 (PAI-2, SERPINB2)2 gene is transcriptionally repressed in a range of cell types but may be rapidly and robustly induced in response to certain inflammatory or cell stress stimuli. PAI-2 is a member of a large family of structurally related proteins called serine protease inhibitors (serpins), which are recognized to be key regulators of a range of biological processes such as complement activation, fibrinolysis, coagulation, cellular differentiation, tumor suppression, apoptosis, and cell motility (1–3). PAI-2 was originally characterized as an inhibitor of the urokinase type plasminogen activator (4), and intracellular PAI-2 has been associated with macrophage survival (5), monocyte (6) and keratinocyte (7) differentiation, cytoprotection (8, 9), apoptosis (10), signal transduction, and modulation of immune responses (11–14). Accurate control of PAI-2 gene expression is crucial, because its dysregulation is associated with inflammatory diseases such as asthma, periodontal disease, pre-eclampsia, and cancer (14–19).
PAI-2 gene expression is tightly regulated, and constitutive expression is highly restricted to certain cell and tissue types, specifically keratinocytes, trophoblasts, macrophages, neurons, and pre-adipocytes (20, 21). PMA (phorbol 12-myristate 13-acetate) is a strong inducer of PAI-2 gene expression in many cells, including cells of the monocytic lineages. PMA, a potent tumor promoter, is a naturally derived organic compound whose endogenous analog is diacylglycerol. It is well established that a major cellular receptor for PMA and other phorbol esters is protein kinase C (PKC), a family of at least 11 serine/threonine protein kinase isoenzymes with selective tissue distributions, activators, and substrates (22, 23). PMA-induced changes in gene transcription are primarily mediated through the specific binding of AP-1 complexes to the DNA sequence 5′-TGA(G/C)TCA-3′, which has been termed the AP-1 binding site or PMA responsive element (24, 25). AP-1 is a dimeric transcription factor complex composed primarily of members of the Jun and Fos protein families (26–28). Jun family members (c-Jun, JunB, and JunD) can homo- or heterodimerize with other Jun or Fos family proteins. Fos family proteins (c-Fos, FosB, Fra-1, and Fra-2) are unable to homo- or heterodimerize with other Fos family proteins and are therefore only found as dimers with Jun partners.
To define regulatory mechanisms responsible for the limited and cell-type-specific expression of PAI-2, we have characterized transcriptional regulatory promoter elements important for expression of the SERPINB2 gene (29, 30). Our previous studies revealed that the SERPINB2 gene promoter was transcriptionally controlled by an inducible proximal promoter, an upstream silencer (PAUSE-1), and a more distal transactivator region. While the proximal promoter was PMA-inducible in U937 cells and constitutively active in HeLa cells, the silencer region repressed transcription in both cell types. The transactivator region was functional only in U937 cells, and hypothesized to derepress transcription in PAI-2-expressing U937 cells, but not in HeLa cells that do not express PAI-2. This transactivator region was located between 3.3 and 5.0 kb upstream of the PAI-2 transcription initiation site. In the present study, we have defined the minimal DNA region responsible for PAI-2 transactivator activity, and we have identified specific members of the activator protein-1 (AP-1) superfamily of proteins that bind to the PAI-2 transactivator region and functionally derepress the SERPINB2 promoter in both PAI-2-expressing and -non-expressing cells. This mechanism may offer valuable insight into differential PAI-2 gene expression during cell differentiation and in certain cancers.
EXPERIMENTAL PROCEDURES
Cell Culture
Human histiocytic lymphoma U937 cells (European Collection of Cell Cultures no. 85011440) and human cervical carcinoma HeLa cells (ATCC no. CCL-2.2) were maintained in RPMI 1640 media (Invitrogen), supplemented with 2 mm l-glutamine, 10% serum supreme (BioWhittaker), 200 μg/ml penicillin, 100 μg/ml streptomycin, 25 mm HEPES, and 25 mm sodium bicarbonate, in 5% CO2 and 95% humidified air atmosphere at 37 °C. Bacterial lipopolysaccharide, an activator of U937 cells, was undetectable in cell cultures. Cell viability was determined by trypan blue dye exclusion. All cultures were routinely checked to exclude mycoplasma infection. Cells were stimulated with 40 ng/ml PMA for 18 h, unless otherwise indicated, prior to harvesting. U937 cells expressed a low level of PAI-2 mRNA, which was increased as early as 2 h following treatment with PMA, rising to a maximum at 10 h of more than 30-fold that of constitutive levels (supplemental Fig. S1).
Construction of hPAI-2 Reporter Gene Plasmids
An 8.8-kb genomic fragment containing 5 kb of the SERPINB2 promoter as well as the first exon and first intron of the human SERPINB2 gene has been described (31). The nucleotide sequence of the first 2 kb of the human SERPINB2 promoter (32) (GenBankTM accession number M22469) and the sequence of the most distal 1.6 kb of this genomic fragment (31) (GenBankTM accession number L19065) have been reported. We report the sequence of the interim 1.4 kb, which has been deposited in GenBankTM with accession number AF071400. With the available complete sequence of the human PAI-2 promoter region, chloramphenicol acetyltransferase (CAT) reporter plasmids described previously (29, 31) were renamed to reflect the position of the deletions in the deletion constructs described previously. These included pCAT-4952 (pNatCAT), pCAT-3306 (pCAT5′-3.3), and pCAT-1977 (pCAT5′-1.9), which contain the human SERPINB2 gene 5′-flanking sequence from −4952, −3306, and −1977 nucleotides, respectively, to the 3′-end of the first intron (+3729).
The deletion constructs pCAT-4622, pCAT-4276, pCAT-3993, and pCAT-3628 were created by PCR amplification from pCAT-4952 using the sense primers, BShPt2, 5′-CTCCAAGCTTCCTAAGCCTATGCTTATTCA-3′; BShPt3, 5′-TAATAAGCTTGATCTTTGCTATAAATTAAC-3′; BShPt4, 5′-TACAAAGCTTCAAACATGAGTAAGTCATTC-3′; and BShPt5, 5′-TTAAAAGCTTTGCATGCCTATTATGGAAAA-3′, and the antisense primer BShPt1, 5′-GATTAAGCTTTCTCAAATAACCTGAAATAG-3′. Each primer contained a HindIII restriction site (underlined). The 5′-nucleotide of each new construct is indicated in bold type. PCR products were digested with HindIII and cloned into the HindIII site (−3306) of pCAT-3306.
pCAT-1977+330 and pCAT-1977+330R contain the 330-bp transactivator region −4952/−4623 cloned into the HindIII site immediately upstream of nucleotide −1977 of pCAT-1977 in the wild-type and reverse orientation, respectively. The −4952/−4623 region was cloned from pCAT-4952 by a PCR approach using the sense primer BS5′CAT (5′-ACCGGGAAGCTTGAATTCATGACTCACAGTGTT-3′) and the antisense primer BShPt6 (5′-AAGCATAAGCTTAGGCCACCGGGAGG-3′). Both primers contain a HindIII restriction site (underlined) that enable digestion of the PCR product with HindIII and subsequent cloning into the HindIII site of pCAT-1977.
pCAT-1977+AP-1 contains the transactivator region −4952/−4932 cloned into the HindIII site immediately upstream of nucleotide −1977 of pCAT-1977 in the wild-type orientation. This construct was prepared by annealing the synthesized oligonucleotides BSAP1c (5′-AGCTTGAATTCATGACTCACAGTGTTA-3′) and BSAP1n (5′-AGCTTAACACTGTGAGTCATGAATTCA-3′), phosphorylating their 5′-termini and cloning the resultant product into the HindIII site of pCAT-1977.
CAT reporter constructs containing mutations of the PAI-2 transactivator region AP-1, AP-2, and NF-κB-like sites (pCAT-4952mAP-1, pCAT-4952mAP-2, and pCAT-4952mNF-κB, respectively) were produced by one of two methods. pCAT-4952mAP-1 was generated from pCAT-4952 using the PCR primers BSmAP1c (5′-ACCGGGAAGCTTGAATTCACATATTCCAGTGTTCTGAGGCTGCTCT-3′) (mutated bases in bold and the HindIII restriction site underlined) and BShPt6, and cloning the HindIII-digested PCR product into pCAT-3306. pCAT-4952mAP-2 and pCAT-4952NFkB were created by the splicing by overlap extension recombinant PCR approach of Ho et al. (33). The internal, partially overlapping, complementary oligonucleotide primer pairs used to introduce mutations were as follows: BSmAP2c, 5′-TAGCTGTACTATTTCCACTTCTTAAAATAGGTGAGG-3′; BSmAP2n, 5′-GAAGTGGAAATAGTACAGCTATGAGTGTGAGAAAGT-3′; BSmNFkBc, 5′-TCTCTCTATTAAGTAGCTCCCGGTGGCCTAAGCCTA-3′; and BSmNFkBn, 5′-ACCGGGAGCTACTTAATAGAGAGACAGGGCAAAGAAAA-3′ (mutated bases in bold). The flanking oligonucleotide primers used were BS5′CAT and BShPt1. PCR products were digested with HindIII and cloned into the HindIII restriction site of pCAT-3306. All constructs were verified by DNA (Applied Biosystems) sequence analysis. All plasmid DNA preparations were twice purified by equilibrium centrifugation in cesium chloride-ethidium bromide gradients prior to transfection.
CAT Reporter Gene Assays
Cells (1 × 107 HeLa and 3 × 107 U937) were transfected with 20 μg of plasmid reporter DNA in a total volume of 250 μl by electroporation (0.25 kV and 960 microfarads). For co-transfection experiments, 20 μg of reporter plasmid and 5 μg of expression plasmid were used. Transfected cells were transferred to 10 ml of media and incubated 16–18 h prior to harvesting. CAT assays were performed using 50 μg of whole cell lysate essentially as described previously (29). Acetylation of [14C]chloramphenicol was detected using a PhosphorImager (Molecular Dynamics), and signals were quantitated using ImageQuant version 4.2a (Molecular Dynamics). CAT activity was expressed as the percentage conversion of [14C]chloramphenicol per microgram of whole cell lysate and normalized to the value of the positive control plasmid, pCAT-Control (Promega), to allow comparison of the results of independent experiments. Protein concentration was determined by using the Bradford assay (Bio-Rad).
Electrophoretic Mobility Shift Assays
Radiolabeled, double-stranded oligonucleotide probes for electrophoretic mobility shift assays (EMSA) were prepared using T4 polynucleotide kinase in the presence of [γ-32P]ATP and purified using Sephadex G-25 (NAPTM 5) columns (Amersham Biosciences). HeLa and U937 nuclear extracts were prepared essentially as described (34). DNA-binding reactions (20 μl) contained 20 mm HEPES (pH 7.9), 50 mm KCl, 15 mm MgCl2, 1 mm EDTA, 50 μg/ml BSA, 1 mm DTT, 7.5% glycerol, 2 μg of poly(dI-dC) (Roche Applied Science), 5 μg of nuclear extract, and 10,000–20,000 cpm (0.05–0.2 ng) of radiolabeled probe. Binding reactions were incubated 20 min at room temperature. EMSAs were performed on 5% polyacrylamide gels (29:1 acrylamide:bisacrylamide (Bio-Rad)) cast in 1× TBE (50 mm Tris borate, 1 mm EDTA, pH 8.0). Dried gels were exposed to Kodak XK-1 film between intensifying screens at −70 °C.
For supershift assays, nuclear extracts were preincubated 1 h on ice with 2 μg of antibody prior to the DNA-protein-binding reaction. Antibodies used were: anti-c-Jun (sc-45), anti-JunB (sc-46), anti-JunD (sc-74), anti-c-Fos (sc-52), anti-FosB (sc-7203), anti-Fra-1 (sc-183), and anti-Fra-2 (sc-604) (Santa Cruz Biotechnology). Jun and Fos family pCMV expression plasmids (35) were kind gifts from Dr. X. Wang.
Northern Blot Analysis
Total RNA for Northern analysis and RT-PCR was prepared using TRIzol reagent (Invitrogen). Gel electrophoresis was performed using denaturing 1.2% agarose gels, and RNA transfer was to Hybond N membranes (Amersham Biosciences). cDNA probes were radiolabeled with [γ-32P]dCTP (Amersham Biosciences) using a Megaprime DNA labeling system (Amersham Biosciences). Membranes were hybridized in the presence of probe at 65 °C and washed to a final stringency of 0.5× SSC/0.1% SDS at 65 °C before exposure to Kodak XK-1 film.
RT-PCR
RT-PCR was performed using SuperscriptTM II reverse transcriptase (Invitrogen) for first strand cDNA synthesis followed by PCR amplification with AmpliTaqTM DNA polymerase. For first strand cDNA synthesis, 5 μg of total RNA and 500 ng of oligo(dT) (Invitrogen), in a volume of 12 μl, were first incubated together at 70 °C for 10 min then chilled on ice. The reaction was then made up to a final volume of 20 μl, containing 0.5 mm each of dATP, dCTP, dGTP, and dTTP, and 200 units of SuperscriptTM II reverse transcriptase in its supplied buffer (50 mm Tris-HCl (pH 8.3), 75 mm KCl, 3 mm MgCl2, 10 mm DTT). The reaction was incubated at 42 °C for 90 min followed by heat inactivation at 70 °C for 15 min. PCR was performed using 3 μl of the first strand cDNA synthesis reaction, 25 ng (3–4 pmol) of each oligonucleotide primer, 0.4 mm each of dATP, dCTP, dGTP, and dTTP, 1.5 mm MgCl2, and 0.5 unit of AmpliTaqTM DNA polymerase in its supplied buffer (10 mm Tris-HCl (pH 8.3), 50 mm KCl), in a total reaction volume of 25 μl. Cycle conditions were 94 °C for 5 min, followed by 40 cycles of 94 °C for 30 s, 56 °C for 30 s, and 72 °C for 90 s.
Quantitative PCR
RNeasy Mini Kit (Qiagen) was used to isolate total RNA from PMA-treated HeLa cells that were transiently transfected with Jun and Fos family pCMV expression plasmids (Lipofectamine, Invitrogen). For first-strand cDNA synthesis, 1 μg of total RNA was used as a template with random primers and TaqMan® reverse transcription reagents (Applied Biosystems). For quantitative PCR, inventoried TaqMan® Gene Expression 20× primers for SERPINB2 and β-actin were used, and the assay was performed according to the manufacturer's instructions (Applied Biosystems).
RESULTS
PAI-2 Transactivator Activity Localizes to the −4952/−4623 (330 bp) Region of the Human PAI-2 Promoter
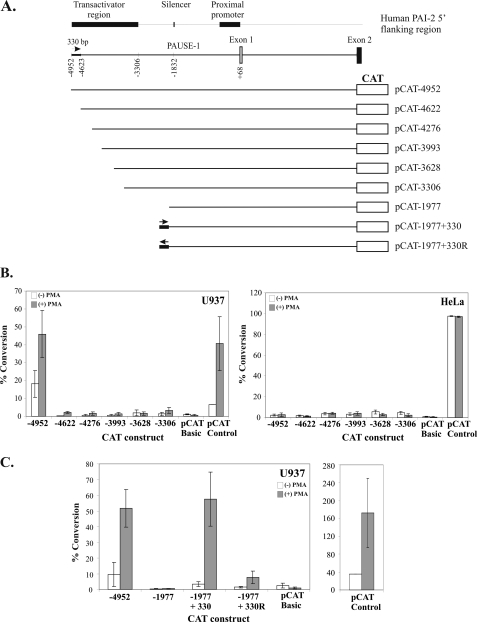
The constitutive and PMA-inducible transactivator activity of the SERPINB2 promoter is present within a 1.6-kb region (−3306 to −4952) upstream of the PAI-2 transcription initiation site (29). To localize this transactivator activity, a series of CAT reporter plasmids was constructed from pCAT-4952 with progressively greater 5′-deletions to −3306 (Fig. 1A). U937 cells were transiently transfected with these reporter plasmids, and CAT activity was measured in the presence or absence of 40 ng/ml PMA. As shown in Fig. 1B, constitutive and PMA-inducible CAT activity was abolished upon deletion of the PAI-2 promoter to position −4622 in U937 cells. Furthermore, deletion beyond nucleotide −4622 resulted in neither additional loss nor significant restoration of PAI-2 promoter-directed CAT activity. In contrast, no significant constitutive nor PMA-inducible PAI-2 promoter-directed CAT activity was seen with any of the CAT reporter constructs in HeLa cells (Fig. 1B). Together these results demonstrate that the cell-type-specific activity of the human PAI-2 transactivator is located within a 330-bp region between nucleotides −4952 and −4623 of the human PAI-2 promoter.
FIGURE 1.
Transactivator activity maps to the −4952/−4623 region of the human SERPINB2 promoter and is position-independent, but orientation-dependent. A, schematic diagram of CAT reporter gene constructs. Deletions are derived from the 8.8-kb SERPINB2 gene fragment containing 5.1 kb of the 5′-flanking sequence and including the first intron. Deletions were made in the 1.6-kb region between the EcoRI and HindIII restriction enzyme sites; the exact position of the deletion is reflected in the construct name. The location of the PAUSE-1 silencer element is indicated. pCAT-1977+330 and pCAT-1977+330R consist of the 330-bp transactivator element inserted into a minimum construct containing the PAUSE-1 element in the wild-type and reverse orientations, respectively. B, CAT reporter gene analysis of deletion constructs in U937 and HeLa cells in the presence (+) or absence (−) of PMA. C, CAT reporter gene analysis of constructs in U937 cells with or without PMA showing position-independent, but orientation-dependent transactivator activity. CAT activities are reported as the percentage conversion of chloramphenicol normalized to the average value for untreated pCAT control. The results represent the mean ± S.E. from at least three independent transfections performed in triplicate.
The 330-bp Region (−4952/−4623) of the Human PAI-2 Promoter Is Sufficient for PAI-2 Transactivator Activity
To investigate whether the 330-bp region (−4952 to −4623) of the human SERPINB2 transactivator contained the one or more necessary cis-elements for mediating PAI-2 transactivator activity, this region was inserted immediately upstream of nucleotide −1977 in pCAT-1977 in both the wild-type and reverse orientations, denoted as pCAT-1977+330 and pCAT-1977+330R, respectively (Fig. 1A). pCAT-1977 contains the PAUSE-1 silencer element, which effectively silences transcription from the SERPINB2 promoter in both U937 and HeLa cells (29, 30). Transient transfection experiments in U937 cells demonstrated that the 330-bp transactivator region, when positioned immediately upstream of nucleotide −1977 in the wild-type orientation, effectively reproduced the transactivator activity detected with pCAT-4952 (Fig. 1C), thereby derepressing silencer activity in pCAT-1977. In contrast, inversion of the 330-bp transactivator was found to greatly reduce transactivator activity (Fig. 1C). Taken together, these results indicated that the 330-bp region of the human SERPINB2 promoter (−4952/−4623) contains the necessary cis-element(s) required for transactivator function and that this activity is orientation-dependent. Moreover, the data further indicate that the activity of the 330-bp region can occur independently of nucleotides −4622 to −1978.
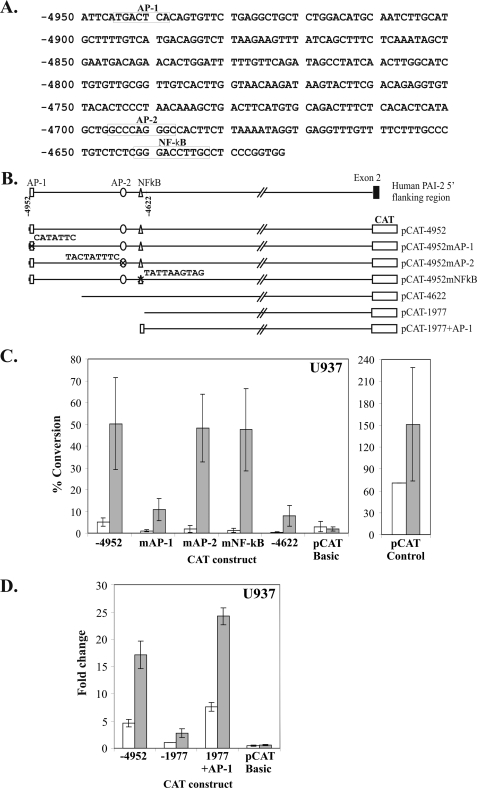
PAI-2 Transactivator Function Is Dependent on an AP-1 Site at −4945/−4939
Analysis of the nucleotide sequence of the 330-bp transactivator region revealed several potential candidates for mediating transactivator function (Fig. 2A). These include an AP-1 site, an AP-2 site, and an NF-κB motif, each of which are recognized binding sites for transcription factors transducing PMA-induced cell signaling (24, 36, 37). To investigate whether these cis-elements may play a role in transactivator function, each element was mutated in pCAT-4952 by single nucleotide substitution to disrupt transcription factor-binding activity, generating pCAT-4952mAP-1, pCAT-4952mAP-2, and pCAT-4952mNF-κB (Fig. 2B). Transient transfection of U937 cells shows that, although mutation of either the AP-2 or NF-κB site had no effect on PMA-inducible PAI-2 transactivator function, mutation of the AP-1 site at −4945/−4939 reduced pCAT-4952-directed CAT activity to the level observed for pCAT-4622 (Fig. 2C). This result demonstrates that PAI-2 transactivator function is dependent on an intact AP-1 site at −4945/−4939 and is possibly mediated by one or more members of the AP-1 transcription factor family.
FIGURE 2.
A distal AP-1 site at −4952/−4932 is necessary and sufficient for human PAI-2 transactivator activity in U937 cells. A, nucleotide sequence of the 330-bp transactivator region between −4952 and −4623 showing the positions of consensus sequences for AP-1, AP-2, and NF-κB elements. B, schematic diagram of the mutant transactivator CAT reporter gene constructs showing mutations at positions −4945/−4939 (AP-1), −4696/4688 (AP-2), and −4642/−4633 (NF-κB). In addition, pCAT-1977+AP-1 consists of a minimum (21 bp) region containing the AP-1 element inserted into pCAT-1977 in the wild-type orientation. C, CAT reporter gene analysis of constructs in U937 with or without PMA showing that the AP-1 site is required for transactivator activity. D, U937 cells transiently transfected with the indicated expression plasmids with or without PMA, showing that the 21-bp AP-1 site is able to derepress promoter activity. CAT activities are reported as the percentage conversion of chloramphenicol normalized to the average value for untreated pCAT control. The results represent the mean ± S.E. from at least three independent transfections performed in triplicate.
A 21-bp Region (−4952/−4932) Containing the Consensus AP-1 Site, Is Sufficient for Transactivator Function
To determine whether the AP-1 site was sufficient for transactivator function, a 21-bp region (−4952/−4932), containing the consensus AP-1 site, was inserted immediately upstream of pCAT-1977 in the wild-type orientation (pCAT-1977+AP-1) (Fig. 2D). Reporter gene analysis shows that the AP-1 site effectively derepressed silencing of pCAT-1977 in U937 cells, comparable with the transactivator activity detected using pCAT-4952 (Fig. 2E). These data demonstrate that the AP-1 site in the 21-bp region between −4952 and −4932 is sufficient to reproduce PAI-2 transactivator activity.
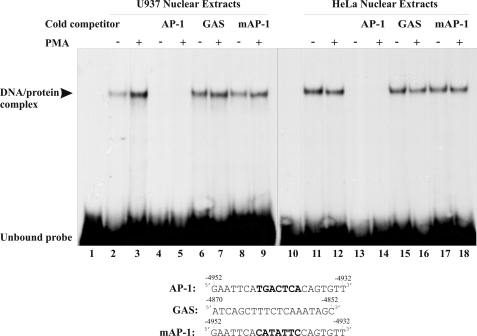
The 21-bp Transactivator Region (−4952/−4932) Is Bound by Nuclear Factors in Both U937 and HeLa Cells
To investigate nuclear protein binding to the 21-bp AP-1 site (−4952/−4932), EMSAs were performed with nuclear extracts from both untreated and PMA-treated U937 and HeLa cells (Fig. 3). A single DNA-protein complex was detected with the 21-bp (−4952/−4932) transactivator probe, the mobility of which was shifted relative to that of the unbound or free probe. Although the intensity of this complex was approximately the same with both untreated and PMA-treated HeLa nuclear extracts, it was consistently observed to increase with PMA treatment with U937 nuclear extracts. The specificity of this interaction was investigated using a 100-fold molar excess of unlabeled competitor oligonucleotides (Fig. 3). Although the unlabeled wild-type 21-bp double-stranded oligonucleotide (−4952/−4932) successfully competed for nuclear extract binding to the radiolabeled 21-bp probe, neither a mutated AP-1 oligonucleotide nor a control oligonucleotide containing a GAS element (nonspecific oligonucleotide) competed for binding. These data confirmed that nuclear proteins from both U937 and HeLa cells specifically bound to the human SERPINB2 promoter −4952/−4932 region in vitro and, furthermore, that a consensus AP-1 cis-element is required for binding of these nuclear proteins. These data indicate that human PAI-2 transactivator function might involve one or more members of the AP-1 transcription factor family.
FIGURE 3.
The −4952/−4932 AP-1 region is a site for nuclear binding factors from both U937 and HeLa cells. EMSA analysis of nuclear extracts prepared from U937 or HeLa cells cultured in the presence or absence of PMA for 4 h. A 21-bp 32P-labeled double-stranded −4952/−4932 oligonucleotide was used as a probe. Cold competition reactions were performed using a 100-fold molar excess of the following unlabeled specific double-stranded oligonucleotides: AP-1, the −4952/−4932 region containing the distal AP-1 consensus sequence; mAP-1, the −4952/−4932 region containing a mutant AP-1 site; and GAS, a GAS region containing an unrelated oligonucleotide from the −4870/−4852 region of the human PAI-2 promoter.
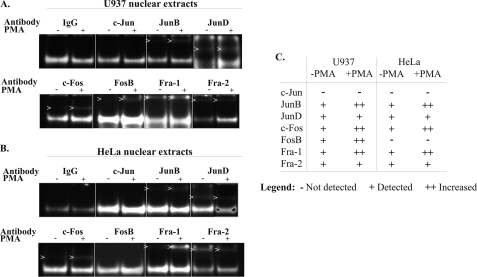
The Human PAI-2 Transactivator Binds Multiple Jun and Fos Family Proteins
To identify the specific transcription factors involved in the 21-bp (−4952/−4932) transactivator binding complex, EMSA supershift assays were performed using antibodies specific for each Jun and Fos family member. As shown in Fig. 4, both U937 and HeLa nuclear extracts contained multiple Jun and Fos family proteins, which bound the 21-bp (−4952/−4932) double-stranded oligonucleotide in vitro. JunB, JunD, c-Fos, Fra-1, and Fra-2, present in nuclear extracts from both U937 and HeLa cells, each bound the 21-bp transactivator probe prior to treatment of the cells with PMA, with increased binding observed for JunB, c-Fos, and Fra-1 following PMA treatment. In contrast, neither U937 nor HeLa nuclear extracts showed detectable c-Jun-binding activity before or after treatment with PMA. Significantly, PMA-inducible binding of the 21-bp transactivator probe by FosB was detected in U937 nuclear extracts but was absent in HeLa nuclear extracts. Immunoblot analysis showed that U937 and HeLa cells both expressed constitutive c-Jun and FosB (data not shown). Fig. 4C summarizes the findings from three sets of independent supershift experiments showing the binding of individual Jun and Fos family members to the 21-bp transactivator probe using both U937 and HeLa nuclear extracts, in the presence and absence of PMA.
FIGURE 4.
The human PAI-2 transactivator AP-1 site binds multiple Jun and Fos family proteins. Supershift assays were performed with nuclear extracts prepared from U937 cells (A) and HeLa cells (B) cultured in the presence (+) or absence (−) of PMA for 4 h and using the 32P-labeled double stranded −4952/−4932 oligonucleotide as a probe. Nuclear extracts were preincubated with 4 μg of specific antibody at 4 °C for 2 h. Images have been inverted for clarity. C, table summarizing the findings from three sets of independent supershift experiments.
The absence of c-Jun in the EMSA supershift assays was somewhat surprising, given that c-Jun previously has been shown to be induced in both U937 and HeLa cells by PMA treatment (38, 39) and has been shown to bind a double-stranded oligonucleotide containing an AP-1 site by EMSA supershift assay (40). Because induction of c-Jun may be a transient event (41), EMSA supershift assays were performed using nuclear extracts prepared at various times following PMA treatment of both U937 and HeLa cells. Although recombinant c-Jun was demonstrated to be capable of binding to the 21-bp transactivator probe, no c-Jun bound with nuclear extracts prepared from either U937 or HeLa cells treated for up to 4 h with PMA (supplemental Fig. S2).
These data demonstrate that the 21-bp region of the SERPINB2 transactivator binds multiple Jun and Fos family members present in both U937 and HeLa cells. Although most of the bound Jun and Fos family-binding proteins are common to both PAI-2-expressing U937 cells and PAI-2-non-expressing HeLa cells, FosB bound the transactivator only in U937 cells and not HeLa cells.
Transient Transfection of HeLa Cells with FosB or c-Jun Derepresses Transcriptional Silencing of the Human PAI-2 Promoter
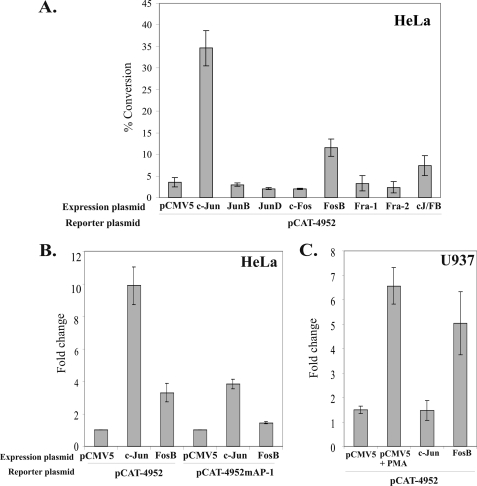
To investigate whether the absence of detectable nuclear FosB or c-Jun binding to the 21-bp transactivator region reflected a functional consequence for human PAI-2 promoter activity in HeLa cells, HeLa cells were transiently co-transfected with expression vectors for FosB and c-Jun, as well as each of the other Jun and Fos family members, together with the CAT reporter vector pCAT-4952. As shown in Fig. 5A, transient overexpression of JunB, JunD, c-Fos, Fra-1, or Fra-2 had no effect on PAI-2 promoter-directed CAT activity. However, transient overexpression of FosB or c-Jun increased PAI-2 promoter-directed CAT activity ∼10-fold and 35-fold, respectively, in HeLa cells. Mutation of the transactivator AP-1 site resulted in a 6- and 2-fold decrease in CAT activity following transient overexpression of c-Jun and FosB, respectively (Fig. 5B), indicating that the increase in CAT activity is dependent on a functional AP-1 site. Transient overexpression of both FosB and c-Jun together, however, had an antagonistic effect on promoter activity when compared with overexpression of either factor alone, with PAI-2 promoter-directed CAT activity increasing only slightly more than 2-fold with this combination (Fig. 1A). Thus overexpression of either FosB or c-Jun, two AP-1 constituent factors that did not bind to the 21-bp SERPINB2 transactivator region in either untreated or PMA-treated HeLa cells by EMSA supershift assay, resulted in a functional derepression of the normally transcriptionally silenced SERPINB2 promoter in HeLa cells.
FIGURE 5.
Overexpression of c-Jun or FosB overcomes transcriptional silencing in HeLa cells, but overexpression of FosB, not c-Jun, overcomes transcriptional silencing in U937 cells. A, HeLa cells were transiently transfected with the indicated expression plasmids together with pCAT-4952 or (B) pCAT-4952mAP-1 in the presence of PMA. C, U937 cells were transiently transfected with the indicated expression plasmids together with pCAT-4952 in the presence of PMA. CAT activities are reported as the percentage conversion of chloramphenicol normalized to the average value for untreated pCAT control. The results represent the mean ± S.E. from at least three independent transfections performed in triplicate.
Transient Transfection of U937 Cells with FosB, but Not c-Jun, Derepresses Transcriptional Silencing of the Human PAI-2 Promoter
Since transient overexpression of FosB or c-Jun was found to derepress transcriptional silencing of the SERPINB2 promoter in HeLa cells, an effect mediated, at least in part, by the transactivator AP-1 site, the effect of FosB and c-Jun overexpression on PAI-2 promoter activity was also investigated in U937 cells. As shown in Fig. 5C, transient overexpression of FosB, but not c-Jun, resulted in transactivation of the SERPINB2 promoter in U937 cells. Thus, whereas transient overexpression of either FosB or c-Jun will derepress PAI-2 promoter silencing in HeLa cells, only transient overexpression of FosB and not c-Jun was found to derepress silencing of the SERPINB2 promoter in U937 cells.
Transient Transfection of HeLa Cells with c-Jun, c-Fos, or FosB Expression Plasmids Induces Low Level Expression of the Endogenous PAI-2 Gene
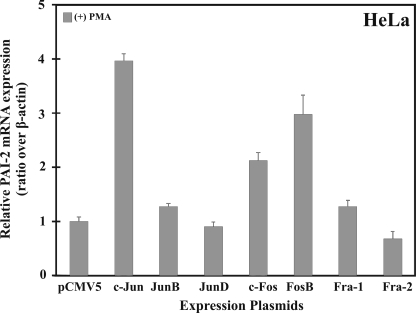
The experimental data demonstrated that overexpression of both FosB and c-Jun in HeLa cells could lead to derepression of silencing of a PAI-2 promoter-CAT reporter gene. To investigate whether FosB or c-Jun overexpression could modulate endogenous PAI-2 mRNA expression in HeLa cells, HeLa cells were transiently transfected with vectors encoding AP-1 family members and analyzed by quantitative PCR after PMA treatment. Endogenous PAI-2 mRNA could be induced at least 2-fold in HeLa cells after transient transfection with c-Jun, c-Fos, or FosB compared with the empty vector alone and was not detected following transfection with the other Jun and Fos family member plasmids (Fig. 6). These data provide evidence that overexpression of FosB or c-Jun may be sufficient to cause derepression and transactivation of the endogenously silenced SERPINB2 gene, although clearly additional factors are required for full induction of PAI-2 gene expression.
FIGURE 6.
Transient expression of FosB, c-Jun, and c-Fos induces low level expression of the endogenous PAI-2 gene. HeLa cells were transiently transfected with the indicated expression plasmids in the presence of PMA. RNA was isolated, and PAI-2 mRNA was detected by quantitative PCR. PAI-2 Ct values were normalized to that of β-actin.
DISCUSSION
Transcriptional derepression is an important mechanism for the accurate control of gene expression. Here we have shown that a distal AP-1 site is both necessary and sufficient for transactivator function in both PAI-2-expressing and -non-expressing cells. Our studies indicated that the AP-1 site mediated PAI-2 promoter activity in a location-independent and orientation-dependent manner. Supershift assays revealed that the PAI-2 transactivator bound specific Jun and Fos family transcription factors in both U937 and HeLa cells. Functional promoter activity studies revealed that overexpression of FosB in HeLa and U937 cells resulted in transactivation of the SERPINB2 promoter, but only when the transactivator AP-1 site was intact. Similarly, overexpression of c-Jun was also found to transactivate the human SERPINB2 promoter in HeLa cells, but not U937 cells, an effect also mediated at least in part by the transactivator AP-1 site. Remarkably, endogenous PAI-2 mRNA transcripts could be induced in HeLa cells, a cell line in which PAI-2 is transcriptionally silent, following transient overexpression of c-Jun, c-Fos, or FosB.
The SERPINB2 promoter contains several AP-1 or AP-1-like consensus sequences, however, their presence does not automatically imply functionality in a given gene promoter. The SERPINB2 promoter contains a consensus AP-1 site between nucleotides −1568 and −1562, yet this site has not been implicated in either constitutive or PMA-stimulated PAI-2 gene expression (29). In contrast, two AP-1-like sites in the proximal SERPINB2 promoter have been found to contribute to both constitutive and PMA-stimulated transcription of PAI-2 (42). Because various Jun and Fos family members differ in their transactivation potential (26), differences in promoter-bound AP-1 complexes may be involved in cell-type-specific differences in gene transcription. The data presented here suggest that this is the case for the PAI-2 transactivator AP-1 site and its differential action in derepressing transcription from the SERPINB2 promoter in U937 and HeLa cells. FosB, presumably exerting its effect on the PAI-2 transactivator AP-1 site as a JunB-FosB or JunD-FosB dimer, appears to be the mechanism by which U937 cells derepress transcriptional silencing of the SERPINB2 promoter. Alternatively, c-Jun may possibly be the most important Jun family member mediating derepression of the SERPINB2 gene in HeLa cells.
The identification of FosB as the mediator of PAI-2 transactivator function explains the orientation dependence observed for PAI-2 transactivator activity (Fig. 1A). Because FosB must form a heterodimer with a Jun partner to bind DNA, and Jun-Fos heterodimers bind DNA in a preferred orientation (43, 44), Jun-FosB binding to the transactivator AP-1 site is likely to be unidirectional. Reversing the transactivator sequence will reverse the orientation of the bound Jun-FosB AP-1 factor, an action that is likely to impair its ability to interact with other DNA-bound factors in the SERPINB2 promoter and thus its activity.
The observation that c-Jun was a strong transactivator of PAI-2 promoter reporter gene expression in HeLa cells, but not in U937 cells, was a surprising finding. However its explanation may lie with the AP-1 dimer preference of the transactivator AP-1 site, the presence of detectable FosB binding in U937 cells but not HeLa cells, and the probability of successful competition for binding to the transactivator AP-1 site by c-Jun in HeLa cells versus U937 cells. Thus, although recombinant c-Jun bound to the transactivator AP-1 site in the absence of nuclear extracts, the absence of c-Jun in transactivator AP-1 complexes formed with both HeLa and U937 nuclear extracts may indicate that this site has greater affinity for AP-1 complexes that do not contain c-Jun. Furthermore, the presence of detectable FosB in transactivator AP-1 complexes formed with U937 nuclear extracts suggests that this factor binds readily as a JunB-FosB or JunD-FosB heterodimer rather than as a c-Jun-FosB heterodimer, an observation made previously with the Bal-17 B cell line (45). Thus given the possible preference of the PAI-2 transactivator AP-1 site for AP-1 complexes not containing c-Jun and the added competition for binding to this site by FosB in U937 cells and not HeLa cells, transient transfection with c-Jun may produce sufficient c-Jun to successfully compete for transactivator AP-1 site binding in HeLa cells. This hypothesis is supported by our evidence showing that transient overexpression of c-Jun in HeLa cells induces the highest endogenous PAI-2 mRNA expression, when compared with overexpression of any other Jun or Fos family member.
The observation that both FosB and c-Jun can derepress transcription from the SERPINB2 promoter may have general significance for PAI-2-expressing cells. Although the expression pattern of Jun and Fos family proteins in the placenta is yet to be suitably examined, an immunostaining analysis of human skin (46) suggests that it might. In this study, FosB was demonstrated to be expressed in both the basal and spinous layers of the epidermis and disappeared in the granular layer. Although staining was localized to both the nuclei and cytoplasm in the basal layer, the intensity of nuclear staining increased in cells of the spinous layers, suggesting nuclear translocation of FosB in these cells. In contrast immunostaining for c-Jun showed that it occurred in a sharply defined band confined to the granular layer of the epidermis in both a nuclear and cytoplasmic distribution. In comparison, PAI-2 expression in the epidermis is low in the basal keratinocyte layer, increases through the spinous layers, and is highest in the granular layers before the transition to cornified dead squames (47). With this non-overlapping and possibly complementary pattern of FosB and c-Jun expression, it is tempting to speculate that the coordinate expression of these two factors in turn may be responsible for the observed expression of PAI-2 in the epidermis. Although expression of FosB by keratinocytes of the basal and spinous layers of the epidermis may result in low to moderate levels of PAI-2 expression in these cell layers, the switch to c-Jun expression by keratinocytes of the granular layers may lead to increased PAI-2 expression in this layer.
With the large body of evidence implicating alterations in PAI-2 expression to a host of inflammatory diseases, it is important that the precise mechanisms for its expression, repression, and derepression be elucidated. With respect to cancer in particular, PAI-2 has been implicated as both a positive and negative prognostic indicator of tumor progression by microarray analysis as well as several in vivo studies (19, 21). Clinically, tumor-associated PAI-2 expression is associated with favorable patient prognosis in several cancers, namely breast, ovarian, squamous cell carcinoma, and pancreatic cancer (19, 48, 49). On the other hand, PAI-2 expression is associated with poor prognosis in endometrial and colorectal cancers (50, 51). Our present findings make an important contribution to an evolving model for the transcriptional control of the human PAI-2 gene and toward understanding the molecular basis for its tissue specific expression.
Supplementary Material
This work was supported, in whole or in part, by National Institutes of Health Grant R01CA098369. This work was also supported by the National Health and Medical Research Council, Australia.

This article contains supplemental Figs. S1 and S2.
- PAI-2
- plasminogen activator inhibitor type 2
- CAT
- chloramphenicol acetyl transferase
- EMSA
- electrophoretic mobility shift assay
- PMA
- phorbol 12-myristate 13-acetate
- PAUSE-1
- PAI-2 upstream silencer element-1
- NF-κB
- nuclear factor-κB
- AP-1
- activator protein-1.
REFERENCES
- 1. Potempa J., Korzus E., Travis J. (1994) The serpin superfamily of proteinase inhibitors. Structure, function, and regulation. J. Biol. Chem. 269, 15957–15960 [PubMed] [Google Scholar]
- 2. Silverman G. A., Bird P. I., Carrell R. W., Church F. C., Coughlin P. B., Gettins P. G., Irving J. A., Lomas D. A., Luke C. J., Moyer R. W., Pemberton P. A., Remold-O'Donnell E., Salvesen G. S., Travis J., Whisstock J. C. (2001) The serpins are an expanding superfamily of structurally similar but functionally diverse proteins. Evolution, mechanism of inhibition, novel functions, and a revised nomenclature. J. Biol. Chem. 276, 33293–33296 [DOI] [PubMed] [Google Scholar]
- 3. Law R. H., Zhang Q., McGowan S., Buckle A. M., Silverman G. A., Wong W., Rosado C. J., Langendorf C. G., Pike R. N., Bird P. I., Whisstock J. C. (2006) An overview of the serpin superfamily. Genome Biol. 7, 216. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4. Kawano T., Morimoto K., Uemura Y. (1968) Urokinase inhibitor in human placenta. Nature 217, 253–254 [DOI] [PubMed] [Google Scholar]
- 5. Park J. M., Greten F. R., Wong A., Westrick R. J., Arthur J. S., Otsu K., Hoffmann A., Montminy M., Karin M. (2005) Signaling pathways and genes that inhibit pathogen-induced macrophage apoptosis. CREB and NF-κB as key regulators. Immunity 23, 319–329 [DOI] [PubMed] [Google Scholar]
- 6. Kruithof E. K., Baker M. S., Bunn C. L. (1995) Biological and clinical aspects of plasminogen activator inhibitor type 2. Blood 86, 4007–4024 [PubMed] [Google Scholar]
- 7. Risse B. C., Brown H., Lavker R. M., Pearson J. M., Baker M. S., Ginsburg D., Jensen P. J. (1998) Differentiating cells of murine stratified squamous epithelia constitutively express plasminogen activator inhibitor type 2 (PAI-2). Histochem. Cell Biol. 110, 559–569 [DOI] [PubMed] [Google Scholar]
- 8. Andela V. B., Schwarz E. M., Puzas J. E., O'Keefe R. J., Rosier R. N. (2000) Tumor metastasis and the reciprocal regulation of prometastatic and antimetastatic factors by nuclear factor kB. Cancer Res. 60, 6557–6562 [PubMed] [Google Scholar]
- 9. Shafren D. R., Gardner J., Mann V. H., Antalis T. M., Suhrbier A. (1999) Picornavirus receptor down-regulation by plasminogen activator inhibitor type 2. J. Virol. 73, 7193–7198 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10. Dickinson J. L., Bates E. J., Ferrante A., Antalis T. M. (1995) Plasminogen activator inhibitor type 2 inhibits tumor necrosis factor α-induced apoptosis. Evidence for an alternate biological function. J. Biol. Chem. 270, 27894–27904 [DOI] [PubMed] [Google Scholar]
- 11. Kumar S., Baglioni C. (1991) Protection from tumor necrosis factor-mediated cytolysis by overexpression of plasminogen activator inhibitor type-2. J. Biol. Chem. 266, 20960–20964 [PubMed] [Google Scholar]
- 12. Gan H., Newman G. W., Remold H. G. (1995) Plasminogen activator inhibitor type 2 prevents programmed cell death of human macrophages infected with Mycobacterium avium, serovar 4. J. Immunol. 155, 1304–1315 [PubMed] [Google Scholar]
- 13. Zhou H. M., Bolon I., Nichols A., Wohlwend A., Vassalli J. D. (2001) Overexpression of plasminogen activator inhibitor type 2 in basal keratinocytes enhances papilloma formation in transgenic mice. Cancer Res. 61, 970–976 [PubMed] [Google Scholar]
- 14. Schroder W. A., Gardner J., Le T. T., Duke M., Burke M. L., Jones M. K., McManus D. P., Suhrbier A. (2010) SerpinB2 deficiency modulates Th1Th2 responses after schistosome infection. Parasite Immunol. 32, 764–768 [DOI] [PubMed] [Google Scholar]
- 15. Ohkuchi A., Minakami H., Aoya T., Haga T., Kimura H., Suzuki M., Sato I. (2001) Expansion of the fraction of Th1 cells in women with preeclampsia. Inverse correlation between the percentage of Th1 cells and the plasma level of PAI-2. Am. J. Reprod. Immunol. 46, 252–259 [DOI] [PubMed] [Google Scholar]
- 16. Roes E. M., Sweep C. G., Thomas C. M., Zusterzeel P. L., Geurts-Moespot A., Peters W. H., Steegers E. A. (2002) Levels of plasminogen activators and their inhibitors in maternal and umbilical cord plasma in severe preeclampsia. Am. J. Obstet. Gynecol. 187, 1019–1025 [DOI] [PubMed] [Google Scholar]
- 17. Woodruff P. G., Boushey H. A., Dolganov G. M., Barker C. S., Yang Y. H., Donnelly S., Ellwanger A., Sidhu S. S., Dao-Pick T. P., Pantoja C., Erle D. J., Yamamoto K. R., Fahy J. V. (2007) Genome-wide profiling identifies epithelial cell genes associated with asthma and with treatment response to corticosteroids. Proc. Natl. Acad. Sci. U.S.A. 104, 15858–15863 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18. Beikler T., Peters U., Prior K., Eisenacher M., Flemmig T. F. (2008) Gene expression in periodontal tissues following treatment. BMC Med. Genomics 1, 30. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19. Croucher D. R., Saunders D. N., Lobov S., Ranson M. (2008) Revisiting the biological roles of PAI2 (SERPINB2) in cancer. Nat. Rev. Cancer 8, 535–545 [DOI] [PubMed] [Google Scholar]
- 20. Medcalf R. L., Stasinopoulos S. J. (2005) The undecided serpin. The ins and outs of plasminogen activator inhibitor type 2. FEBS J. 272, 4858–4867 [DOI] [PubMed] [Google Scholar]
- 21. Medcalf R. L. (2011) Plasminogen activator inhibitor type 2. Still an enigmatic serpin but a model for gene regulation. Methods Enzymol. 499, 105–134 [DOI] [PubMed] [Google Scholar]
- 22. Musashi M., Ota S., Shiroshita N. (2000) The role of protein kinase C isoforms in cell proliferation and apoptosis. Int. J. Hematol. 72, 12–19 [PubMed] [Google Scholar]
- 23. Ron D., Kazanietz M. G. (1999) New insights into the regulation of protein kinase C and novel phorbol ester receptors. FASEB J. 13, 1658–1676 [PubMed] [Google Scholar]
- 24. Angel P., Imagawa M., Chiu R., Stein B., Imbra R. J., Rahmsdorf H. J., Jonat C., Herrlich P., Karin M. (1987) Phorbol ester-inducible genes contain a common cis-element recognized by a TPA-modulated trans-acting factor. Cell 49, 729–739 [DOI] [PubMed] [Google Scholar]
- 25. Chiu R., Imagawa M., Imbra R. J., Bockoven J. R., Karin M. (1987) Multiple cis- and trans-acting elements mediate the transcriptional response to phorbol esters. Nature 329, 648–651 [DOI] [PubMed] [Google Scholar]
- 26. Angel P., Karin M. (1991) The role of Jun, Fos and the AP-1 complex in cell-proliferation and transformation. Biochim. Biophys. Acta 1072, 129–157 [DOI] [PubMed] [Google Scholar]
- 27. Shaulian E. (2010) AP-1–The Jun proteins. Oncogenes or tumor suppressors in disguise? Cell Signal. 22, 894–899 [DOI] [PubMed] [Google Scholar]
- 28. Milde-Langosch K. (2005) The Fos family of transcription factors and their role in tumourigenesis. Eur. J. Cancer 41, 2449–2461 [DOI] [PubMed] [Google Scholar]
- 29. Antalis T. M., Costelloe E., Muddiman J., Ogbourne S., Donnan K. (1996) Regulation of the plasminogen activator inhibitor type-2 gene in monocytes. Localization of an upstream transcriptional silencer. Blood 88, 3686–3697 [PubMed] [Google Scholar]
- 30. Ogbourne S. M., Antalis T. M. (2001) Characterisation of PAUSE-1, a powerful silencer in the human plasminogen activator inhibitor type 2 gene promoter. Nucleic Acids Res. 29, 3919–3927 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31. Antalis T. M., Godbolt D., Donnan K. D., Stringer B. W. (1993) Southwestern blot mapping of potential regulatory proteins binding to the DNA encoding plasminogen activator inhibitor type 2. Gene 134, 201–208 [DOI] [PubMed] [Google Scholar]
- 32. Kruithof E. K., Cousin E. (1988) Plasminogen activator inhibitor 2. Isolation and characterization of the promoter region of the gene. Biochem. Biophys. Res. Commun. 156, 383–388 [DOI] [PubMed] [Google Scholar]
- 33. Ho S. N., Hunt H. D., Horton R. M., Pullen J. K., Pease L. R. (1989) Site-directed mutagenesis by overlap extension using the polymerase chain reaction. Gene 77, 51–59 [DOI] [PubMed] [Google Scholar]
- 34. Antalis T. M., Godbolt D. (1991) Isolation of intact nuclei from hematopoietic cell types. Nucleic Acids Res. 19, 4301. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35. Liberati N. T., Datto M. B., Frederick J. P., Shen X., Wong C., Rougier-Chapman E. M., Wang X. F. (1999) Smads bind directly to the Jun family of AP-1 transcription factors. Proc. Natl. Acad. Sci. U.S.A. 96, 4844–4849 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36. Imagawa M., Chiu R., Karin M. (1987) Transcription factor AP-2 mediates induction by two different signal-transduction pathways. Protein kinase C and cAMP. Cell 51, 251–260 [DOI] [PubMed] [Google Scholar]
- 37. Edbrooke M. R., Burt D. W., Cheshire J. K., Woo P. (1989) Identification of cis-acting sequences responsible for phorbol ester induction of human serum amyloid A gene expression via a nuclear factor kB-like transcription factor. Mol. Cell Biol. 9, 1908–1916 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38. Pulverer B. J., Hughes K., Franklin C. C., Kraft A. S., Leevers S. J., Woodgett J. R. (1993) Co-purification of mitogen-activated protein kinases with phorbol ester-induced c-Jun kinase activity in U937 leukaemic cells. Oncogene 8, 407–415 [PubMed] [Google Scholar]
- 39. Yoshioka K., Deng T., Cavigelli M., Karin M. (1995) Antitumor promotion by phenolic antioxidants. Inhibition of AP-1 activity through induction of Fra expression. Proc. Natl. Acad. Sci. U.S.A. 92, 4972–4976 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40. Doyle G. A., Pierce R. A., Parks W. C. (1997) Transcriptional induction of collagenase-1 in differentiated monocyte-like (U937) cells is regulated by AP-1 and an upstream C/EBP-β site. J. Biol. Chem. 272, 11840–11849 [DOI] [PubMed] [Google Scholar]
- 41. Lamph W. W., Wamsley P., Sassone-Corsi P., Verma I. M. (1988) Induction of proto-oncogene JUN/AP-1 by serum and TPA. Nature 334, 629–631 [DOI] [PubMed] [Google Scholar]
- 42. Cousin E., Medcalf R. L., Bergonzelli G. E., Kruithof E. K. (1991) Regulatory elements involved in constitutive and phorbol ester-inducible expression of the plasminogen activator inhibitor type 2 gene promoter. Nucleic Acids Res. 19, 3881–3886 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43. Rajaram N., Kerppola T. K. (1997) DNA bending by Fos-Jun and the orientation of heterodimer binding depend on the sequence of the AP-1 site. EMBO J. 16, 2917–2925 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44. Ramirez-Carrozzi V. R., Kerppola T. K. (2001) Long-range electrostatic interactions influence the orientation of Fos-Jun binding at AP-1 sites. J. Mol. Biol. 305, 411–427 [DOI] [PubMed] [Google Scholar]
- 45. Dickinson J. A., Amato S. F., McManus B. J., Chiles T. C. (1995) Membrane immunoglobulin receptor cross-linking induces fosB mRNA expression in mature B lymphocytes. Assembly of distinct FosB-containing nucleoprotein complexes during B cell stimulation. Cell Immunol. 165, 92–100 [DOI] [PubMed] [Google Scholar]
- 46. Welter J. F., Eckert R. L. (1995) Differential expression of the fos and jun family members c-fos, fosB, Fra-1, Fra-2, c-jun, junB and junD during human epidermal keratinocyte differentiation. Oncogene 11, 2681–2687 [PubMed] [Google Scholar]
- 47. Lyons-Giordano B., Loskutoff D., Chen C. S., Lazarus G., Keeton M., Jensen P. J. (1994) Expression of plasminogen activator inhibitor type 2 in normal and psoriatic epidermis. Histochemistry 101, 105–112 [DOI] [PubMed] [Google Scholar]
- 48. Smith R., Xue A., Gill A., Scarlett C., Saxby A., Clarkson A., Hugh T. (2007) High expression of plasminogen activator inhibitor-2 (PAI-2) is a predictor of improved survival in patients with pancreatic adenocarcinoma. World J. Surg. 31, 493–502 [DOI] [PubMed] [Google Scholar]
- 49. Chambers S. K., Ivins C. M., Carcangiu M. L. (1997) Expression of plasminogen activator inhibitor-2 in epithelial ovarian cancer. A favorable prognostic factor related to the actions of CSF-1. Int. J Cancer 74, 571–575 [DOI] [PubMed] [Google Scholar]
- 50. Nordengren J., Fredstorp Lidebring M., Bendahl P. O., Brünner N., Fernö M., Högberg T., Stephens R. W., Willén R., Casslén B. (2002) High tumor tissue concentration of plasminogen activator inhibitor 2 (PAI-2) is an independent marker for shorter progression-free survival in patients with early stage endometrial cancer. Int. J Cancer 97, 379–385 [DOI] [PubMed] [Google Scholar]
- 51. Ganesh S., Sier C. F., Heerding M. M., van Krieken J. H., Griffioen G., Welvaart K., van de Velde C. J., Verheijen J. H., Lamers C. B., Verspaget H. W. (1997) Contribution of plasminogen activators and their inhibitors to the survival prognosis of patients with Dukes' stage B and C colorectal cancer. Br. J Cancer 75, 1793–1801 [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.