Background: Influenza virus NA is transported to the host cell surface.
Results: Cdc42 promotes the transport of NA to the plasma membranes, whereas ARHGAP21 inhibits this process.
Conclusion: Cdc42 positively and ARHGAP21 negatively regulate NA transport to the cell surface and virus replication.
Significance: Identification of host factors involved in regulating NA transport is critical for understanding influenza virus replication.
Keywords: Actin, GTPase, Influenza Virus, Membrane Transport, Viral Protein, ARHGAP21, Cdc42, Protein Transport, Neuraminidase
Abstract
Influenza virus neuraminidase (NA) is transported to the virus assembly site at the plasma membrane and is a major viral envelope component that plays a critical role in the release of progeny virions and in determination of host range restriction. However, little is known about the host factors that are involved in regulating the intracellular and cell surface transport of NA. Here we identified the Cdc42-specific GAP, ARHGAP21 differentially expressed in host cells infected with influenza A virus using cDNA microarray analysis. Furthermore, we have investigated the involvement of Rho family GTPases in NA transport to the cell surface. We found that expression of constitutively active or inactive mutants of RhoA or Rac1 did not significantly affect the amount of NA that reached the cell surface. However, expression of constitutively active Cdc42 or depletion of ARHGAP21 promoted the transport of NA to the plasma membranes. By contrast, cells expressing shRNA targeting Cdc42 or overexpressing ARHGAP21 exhibited a significant decrease in the amount of cell surface-localized NA. Importantly, silencing Cdc42 reduced influenza A virus replication, whereas silencing ARHGAP21 increased the virus replication. Together, our results reveal that ARHGAP21- and Cdc42-based signaling regulates the NA transport and thereby impacts virus replication.
Introduction
Influenza viruses are enveloped viruses that possess segmented negative-strand RNA genomes (vRNA). Influenza A virus, a highly infectious respiratory pathogen, is the major cause of annual epidemics and occasional pandemics. Influenza A virus assembles at the plasma membrane of infected host cells, releases by budding, and possesses a lipid membrane derived from the host cell (1). This envelope contains two important transmembrane glycoproteins: hemagglutinin (HA) and neuraminidase (NA).4 Both HA and NA play critical roles throughout the virus life cycle. HA mediates cell-surface sialic acid receptor binding and virus entry into the cell by fusion of the viral membrane with the endosomal membrane (2). NA possesses an enzyme activity that promotes influenza virus dispersion within mucosal secretions of the respiratory tract (3). Importantly, NA enzymatic activity removes sialic acid from virus and cellular glycoproteins to facilitate the release of progeny virions from the host cells and the spread of infection to new cells (4–6). Because HA and NA are responsible for binding to and release from host cell receptors, they are crucial in determining host specificity (7).
Influenza A virus uses the machineries of host cells to synthesize and transport its own components (8). The viral NA, a type II transmembrane glycoprotein, is translated on the rough endoplasmic reticulum and transported to the virus assembly site at the plasma membrane where it along with the HA, matrix 2 protein, eight distinct influenza virus ribonucleoproteins, and other viral components is packaged into budding virions (9). Recent studies indicate the involvement of NA in virus morphogenesis and budding (10–12). Therefore, efficient cellular transport of the viral protein NA to the surface of the host cell is especially important for virus morphogenesis, budding, and for the release of nascent virions from the infected cells after budding. Although previous studies have identified signals/sequences in NA for translocation, sorting, and raft association (10, 13), little is known about what host cellular factors are involved in the regulation of intracellular and cell surface transport of NA, and the mechanism by which viral NA usurps the components of the host trafficking machineries to undergo the transport remains to be further determined.
Rho family GTPases, which belong to the Ras GTPase superfamily, are intracellular signal transducers known to regulate many features of cell behavior, but the most profound effects of this protein family are on the actin cytoskeletal architecture (14–17). The membrane-associated actin cytoskeleton plays an important role in protein trafficking within the secretory pathway (18–20). Interestingly, increasing evidence implicates actin signaling molecules including Rho family GTPases in intracellular trafficking (14, 15, 21, 22). The three best-characterized Rho family members are Cdc42, RhoA, and Rac1. Similar to other GTPases, they act as molecular switches cycling between an active GTP-bound state and an inactive GDP-bound state. Cdc42, RhoA, and Rac1 have been shown to regulate various aspects of membrane trafficking (21, 23–26). Therefore, these Rho family members are candidates for connecting protein transport to actin cytoskeletal dynamics.
Cdc42, a highly conserved small GTPase, is known to regulate intracellular trafficking and cell polarity (14, 15, 27, 28). As a trafficking regulator, Cdc42 has been shown to function at several different steps of vesicle trafficking including transport into and out of the Golgi apparatus. Previous studies have found that Cdc42 and ADP-ribosylation factor (ARF) regulate dynein recruitment to Golgi membranes in vitro and that these GTP-binding proteins influence several trafficking events in cells (29–31). Previous studies also revealed that Cdc42 mutants had significant effects on the endoplasmic reticulum to Golgi transport of the viral glycoprotein VSV-G (32, 33). Together these data suggest that Cdc42 is an important component of the protein trafficking machinery. However, although progress has been made in our understanding of the role of Cdc42 in the protein trafficking, it is not known whether this small GTPase is employed during influenza virus replication.
Influenza virus assembly requires the completion of viral component sorting and transport to the assembly site at the plasma membrane. This process must involve precise spatial and temporal regulation mediated at least partially by the host cellular trafficking machineries. Constitutively activated or delayed function of the host machineries could lead to increased or inefficient operation of the virus assembly, budding, and virion release. Indeed, in this study we found that overexpression of different forms of Cdc42 or silencing of Cdc42 or Cdc42-specific GTPase-activating protein (GAP), ARHGAP21, had significant effects on influenza A virus NA transport to the host cell surface and thereby affected virus replication.
EXPERIMENTAL PROCEDURES
Antibodies and Reagents
The following antibodies were used in this study: anti-FLAG, anti-GFP, and anti-TGN46 (Sigma), anti-HA tag (Medical & Biological Laboratories Co., Nagoya, Japan), anti-Cdc42 and anti-EEA1 (BD Biosciences). Other antibodies were obtained as described previously (34). Clostridium difficile toxin B was obtained from List Biological Laboratories (Campbell, CA). C3 transferase was obtained from Cytoskeleton Inc (Denver, CA), and cytochalasin D was obtained from Enzo Life Sciences (Farmingdale, NY).
Microarray and Data Analysis
Total RNAs were extracted from three different groups of A549 cells that had been infected with or without WSN for 10 h using TRIzol reagent (Invitrogen). Procedures for cDNA synthesis, labeling, hybridization, and data analysis were carried out as described previously (35). The expression Microarray used was from Arraystar (Rockville, MD). The gene profile data have been submitted to Gene Expression Omnibus (ncbi.nlm.nih.gov; access number GSE32878).
Cdc42 Activation Assay and Immunoblotting
PAK-GST protein beads were purchased from Cytoskeleton lnc. Cdc42 activation assay was performed as described previously (30). Briefly, 293T cells transfected with plasmids expressing GFP-Cdc42 were infected with or without A/WSN/33 influenza virus (H1N1) for 10 h. The cells were harvested and solubilized with the lysis buffer. The lysates were incubated with PAK-GST protein beads at 4 °C for 1 h. The beads were washed, resuspended in SDS sample buffer and processed for SDS-PAGE and Western blotting. The Western blotting was performed as previously described (34). Where indicated, Western blot signals were quantified by densitometry.
DNA Construction
cDNA encoding NA of the A/Anhui/1/2005 (H5N1) virus strain (accession number EU128239) was subcloned into the pcDNA3.1(−) vector with an HA tag in the COOH terminus. NA(H274Y) mutation was generated using a QuikChange site-directed mutagenesis kit (Stratagene, La Jolla, CA). Wild-type RhoA, Rac1, Cdc42, and GAP domains (ARF-BD/Rho-GAP) of human ARHGAP21 were subcloned into the pEGFP-C1 vector, and their mutants (RhoA(G14V), RhoA(T19N), Rac1(G12V), Rac1(T17N), Cdc42(Q61L), and Cdc42(T17N)) were generated by site-directed mutagenesis or obtained as described previously (29).
Immunofluorescence
Immunofluorescence was performed as described previously (29, 34). Images were acquired using a confocal microscope (model LSCMFV500) and a 60× objective (both from Olympus Optical, Japan) with an NA of 1.40.
RT-PCR and Quantitative Real-time PCR
Total RNA was extracted from cells using TRIzol reagent (Invitrogen). The isolated RNA was used to synthesize cDNA with M-MLV reverse transcriptase (Promega, Madison, WI), and followed by PCR using rTaq DNA Polymerase and quantitative PCR using SYBR® Premix Ex TaqTM II (Takara, Tokyo, Japan) with the following primers: human Cdc42 forward, (5′-TCTCCTGATATCCTACACAAC-3′ and reverse, 5′-CTCAATAGTAGAGGGGTCAT-3′); human ARHGAP21 forward (5′-CACGTCGAGAGGTCCACATAAA-3′) and reverse (5′-TGATCAATGCTAGGGCTAGTTGGT-3′). Expression level of β-actin was used as a control.
NA Activity Assay
NA enzymatic activity assay was performed as described previously (36, 37), and samples were analyzed using a neuraminidase assay kit (Beyotime Institute of Biotechnology, Haimen, Jiangsu, China). Briefly, 293T cells and HeLa cells that express NA were harvested by centrifugation and then resuspended in the assay buffer (15 mm MOPS, 145 mm sodium chloride, 2.7 mm potassium chloride, and 4.0 mm calcium chloride, pH 7.4) containing 2% fetal bovine serum. After the addition of NA fluorogenic substrate, NA activity on the cell surface was measured by the multifunctional microplate reader (supplemental Methods). Cleavage of the substrate by NA activity produces a fluorescence that emits an emission wavelength of 440 nm with an excitation wavelength of 360 nm. The intensity of fluorescence reflects the activity of NA, and the activities were quantified as the fluorescence above the background from untransfected cells.
Cell Surface Biotinylation
Cell surface biotinylation was performed as described previously (38). Briefly, HeLa cells transfected with NA were serum-starved for 4 h. All subsequent manipulations were performed at 4 °C. Cells were rinsed with borate buffer (154 mm NaCl, 1.0 mm boric acid, 7.2 mm KCl, and 1.8 mm CaCl2, pH 9.0) and exposed to 1 mg/ml sulfo-NHS-SS-biotin (Thermo Scientific, Waltham, MA) in borate buffer for 40 min. Cells were then lysed with N+ buffer (60 mm HEPES-NaOH, pH 7.4, 150 mm NaCl, 3 mm KCl, 5 mm EDTA, 3 mm EGTA, and 1% Triton X-100) and incubated with streptavidin-agarose for 12 h at 4 °C. The proteins bound to the beads were analyzed by immunoblotting.
Endocytosis and Anterograde Transport of NA to Plasma Membrane
Endocytosis and anterograde transport of NA were measured as previously described (38). For analysis of endocytosis, HeLa cells expressing NA were surface-labeled with sulfo-NHS-SS-biotin. Cells were then incubated at 37 °C for 30 min to allow endocytosis in the presence or absence of toxin B. Surface biotin was cleaved by washing with 50 mm Tris-HCl, 150 mm reduced glutathione (GSH). The biotin bound to freshly endocytosed proteins was protected from cleavage with GSH. Cells were lysed, and biotinylated proteins were retrieved with streptavidin-agarose beads. The internalized NA was examined by Western blotting.
To measure anterograde transport of NA, HeLa cells were first treated with sulfo-NHS-acetate to block the NHS reactive sites on the cell surface. Cells were rinsed with phosphate-buffered saline (PBS)-Ca2+-Mg2+ (0.1 mm CaCl2 and 1 mm MgCl2 in PBS) at 4 °C and then exposed to 1.5 mg/ml sulfo-NHS-acetate for 2 h. After quenching for 20 min, cells were treated with or without toxin B for 15 or 30 min at 37 °C. Cells were incubated with 1 mg/ml sulfo-NHS-SS-biotin and solubilized in N+ buffer. The biotinylated fraction, which represents newly inserted surface proteins, was precipitated by streptavidin-agarose beads and analyzed by Western blotting.
shRNA-based Knockdown of Rho Family Members or ARHGAP21 and Generation of Cell lines
The pSIH-H1-Puro shRNA lentivector system was purchased from System Biosciences (SBI, Mountain View, CA). To directly monitor the lentiviral infection efficiency, we replaced the sequence expressing puromycin in the vector with that of GFP and named it as pSIH-H1-GFP. The sequences used in the shRNAs targeting Cdc42, ARHGAP21, RhoA, Rac1, and luciferase (the control shRNA) were as follows: 5′-GCCTATCACTCCAGAGACT-3′, 5′-GGATCTGTGTCGCAGTTTA-3′, 5′-AAGGCAGAGATATGGCAAACA-3′, 5′-AGTGGTATCCTGAGGTGCG, and 5′-CTTACGCTGAGTACTTCGA-3′. Stable cell lines expressing the shRNAs targeting Cdc42, ARHGAP21, RhoA, Rac1, or luciferase were generated using a lentiviral spin infection. The cells were sorted by GFP with flow cytometry. Western blotting and real-time PCR were performed to determine the interference efficiency.
Plaque Assay and Hemagglutinin (HA) Assay
A549 cells were infected with A/WSN/33 virus at an multiplicity of infection of 0.05. After adsorption for 1 h at 37 °C, the cells were washed with PBS and maintained in α-minimal essential medium containing 2 μg/ml trypsin. The supernatants of cell cultures were harvested at the indicated times and examined for the viral titers by plaque assay and hemagglutinin assay. For plaque assay, Madin-Darby canine kidney cells were infected with serial dilutions of the viruses. Cells were then washed with PBS and overlaid with α-minimal essential medium containing 1.5% low melting point agarose and 2 μg/ml TPCK (l-1-tosylamido-2-phenylethyl chloromethyl ketone)-treated trypsin. After 72 h of incubation at 37 °C, plaques were stained with 0.165 mg/ml neutral red and counted. For hemagglutinin assay, the supernatants were diluted with PBS and mixed with an equal volume of 0.5% chicken erythrocytes. The viral titers were counted from the highest dilution factors that produced a positive reading.
RESULTS
Infection of Influenza A Virus Causes Decreased Expression of ARHGAP21 Revealed by Microarray Analyses and Increase in GTP-bound Cdc42
In an attempt to provide insights into the mechanisms by which the host cell interacts with influenza A virus, cDNA microarray was used to determine the differentially expressed genes in A549 human alveolar epithelial cells in response to infection with A/WSN/33 influenza virus (H1N1). As shown in Fig. 1A, all the genes whose expressions were altered by at least 2-fold were clustered and displayed. The expression microarray analysis revealed 337 up-regulated genes and 1024 down-regulated genes after the viral infection. Many genes are involved in innate immunity, inflammatory response, and intracellular transport. Interestingly, expression of ARHGAP21, a Cdc42-specific GAP, was found to be markedly down-regulated by influenza A virus infection. This finding was further confirmed by the RT-PCR and quantitative real-time PCR (Fig. 1, B and C). On the other hand, we observed that expression of Cdc42 was not significantly changed after the viral infection using microarray analysis and quantitative real-time PCR (Fig. 1C).
FIGURE 1.
Infection of influenza A virus causes a decreased expression of ARHGAP21 revealed by microarray analyses and a increase in GTP-bound Cdc42. A, a gene expression microarray analysis was used to determine the differentially expressed genes in A549 cells infected with or without A/WSN/33 influenza virus (H1N1). All the genes whose expressions were changed by at least 2-fold were clustered and displayed. Shown are results from three independent experiments. B, RT-PCR was performed to examine the expression of ARHGAP21 in A549 cells infected with or without WSN viruses. C, quantitative real-time PCR was performed to examine the expression of Cdc42 and ARHGAP21 in A549 cells infected with or without WSN viruses. Plotted are the average expression levels from three independent experiments. The error bars represent the S.E. *, p < 0.01. D, Cdc42-transfected cells were infected with or without WSN viruses, and the cell lysates were used for pulldown Cdc42 activation assays with PAK-GST protein beads. The lysates and the bound material were fractionated by SDS-PAGE and probed with antibodies as indicated. These results are representative of three identical experiments. E, the relative levels of active Cdc42 in D were quantified by densitometric analyses of the immunoblots. Shown are the average values for control cells and cells infected with WSN (n = 3; *, p < 0.01). The error bars represent the S.E.
Because expression of ARHGAP21 is greatly reduced in influenza virus-infected cell, next we wondered whether the viral infection activates Cdc42 signaling. To this end, 293T cells were infected with the influenza virus followed by a GST pulldown assay by which the levels of Cdc42-GTP bound to the p21 binding domain (PBD) of PAK1 fused to GST (GST-PAK-PBD) were measured. We found that infection of the influenza virus caused a significant increase in Cdc42-GTP levels as compared with the control (Fig. 1, D and E). Based on these findings, a tentative conclusion is that influenza virus infection results in reduction in ARHGAP21 level and thereby activates Cdc42 by decreasing GTP hydrolysis on Cdc42.
Rho Family GTPases Are Required for Efficient Transport of Influenza A Virus NA to Cell Surface
Given that Rho GTPases are known regulators of actin dynamics and that both Rho GTPases and actin cytoskeleton have been implicated in regulating protein trafficking (14, 18, 29), we investigated whether Rho GTPases are employed during the transport of influenza A virus NA to the cell surface. For this, we wished to establish a system for examination of the cell surface NA. The NA is a type II membrane glycoprotein that contains a COOH-terminal globular head carrying the enzyme active site. Because NA enzyme active site locates outside the cell, we used an NA enzyme activity assay to determine the amount of NA transported to the cell surface by measuring the NA activity expressed at the cell surface. Additionally, we determined the amount of NA transported to the cell surface by immunofluorescence analysis.
Cellular distribution of NA protein was examined in HeLa cells transfected with cDNA encoding the wild-type or mutant NA (H274Y) of A/Anhui/1/2005 (H5N1) influenza virus. As shown in Fig. 2A, although wild-type NA was found throughout the cytoplasm, a significant amount of NA was detected in the rim of plasma membranes, indicating that NA was transported to the plasma membrane and accumulated on the cell surface. To confirm such subcellular localization of NA, we used a well characterized NA H274Y mutant as a control (36). As expected, H274Y mutation decreased the amount of NA protein that reached the cell surface (Fig. 2A). To further determine whether cell surface NA enzyme activity corresponds to its cell surface expression, we quantified the NA activity on the cell surface by using a fluorogenic substrate. Indeed, cells expressing NA H274Y showed ∼50% reduction in total surface activity compared with wild-type NA (Fig. 2B and supplemental Fig. S1), whereas the total amount of cellular NA protein was not affected by this mutation (Fig. 2C). These findings are well consistent with the previous observation that H274Y hampered the efficiency with which NA was transported to the plasma membranes (36). Previous studies demonstrated that H274Y mutation had no effect on the catalytic activity per unit enzyme (36, 39), suggesting that the differences are due to less NA protein at the cell surface.
FIGURE 2.
Rho-family GTPases are required for efficient transport of NA to the cell surface. A, B, and C, a studying system for examination of the cell surface NA was established and verified. A, immunofluorescence staining was performed using an anti-HA antibody to detect HA-tagged NA protein. Shown are confocal micrographs of HeLa cells transfected with NA wild-type (NA(WT)) or NA(H274Y) mutant. The nuclei were stained with DAPI. Bar, 10 μm. B, 293T cells transfected with NA(WT) or NA(H274Y) were examined for the cell surface NA enzymatic activity. The NA(WT) activity is 100, and the result from NA(H274Y) is shown as the percentage difference in the NA activity with respect to NA(WT). Plotted is the average NA activity from three independent experiments. The error bars represent the S.E. (n = 3; *, p < 0.01). C, lysates derived from cells in B were analyzed by Western blotting using indicated antibodies. D, shown are confocal micrographs of HeLa cells transfected with NA(WT). The cells were mock-treated or treated with toxin B (100 ng/ml) for 15 min. Bar, 10 μm. E, 293 T cells expressing NA were treated as described in D, and cell surface NA activity was analyzed. The average NA activity from three experiments is plotted such that the NA activity in mock treatment is 100. The error bars represent the S.E. (n = 3; *, p < 0.01). F, HeLa cells expressing NA were treated as described in D. For surface labeling of NA, cells were biotinylated with sulfo-NHS-SS-biotin. The labeled surface proteins were then retrieved from the whole cell lysate (WCL) by streptavidin precipitation and examined by immunoblotting with indicated antibodies. G, the relative levels of NA in F were quantified by densitometry and normalized to input (WCL). Plotted are the results from three experiments. The error bars represent the S.E. (n = 3; *, p < 0.01).
Next, we used this established system to investigate the contribution of Rho GTPases to influenza virus NA transport to the cell surface. Rho-GTPase activity was acutely inhibited in cells treated with C. difficile toxin B, as toxin B glucosylates all members of the Rho family and inhibits their effector interactions (30, 40–42). Studies using confocal microscopy showed that there was an accumulation of NA at the juxtanuclear Golgi region, and the distribution of NA in the cell surface was decreased in cells treated with toxin B for 15 min (Fig. 2D). Using co-localization assay by confocal microscopy, we observed that an increased amount of NA was present at the trans-Golgi network in treated cells (supplemental Fig. S2), suggesting that transport of NA from the Golgi to the plasma membranes was inhibited by toxin B treatment. By contrast, there was clearly more NA at the cell surface in untreated cells. To further verify the effect of Rho family inhibitor on NA transport, we examined the NA activity on the cell surface after treatment with toxin B. As expected, toxin B-treated cells showed more than 40% reduction in NA activity as compared with untreated cells (Fig. 2E).
To further confirm these observations, we used an independent approach to test the effects of toxin B on NA transport to the plasma membranes. The amounts of NA on the plasma membranes were measured by biotinylation of cell surface proteins as described (38). Consistent with the results from NA activity assay, toxin B-treated cells exhibited 45% reduction in total surface NA compared with the control (Fig. 2, F and G). Accordingly, we concluded that Rho family GTPases are required for efficient transport of influenza virus NA to the cell surface.
Alteration of RhoA or Rac1 Function Has No Significant Effect on Transport of NA to Cell Surface
In an attempt to gain a better understanding of the molecular basis of the altered NA transport in toxin B-treated cells, we investigated which Rho GTPase family member(s) is involved in the NA transport to the cell surface. The Rho GTPase family is defined by several subfamilies represented by RhoA, Rac1, and Cdc42. To test whether RhoA functions in the NA cell surface transport, we used another pharmacological reagent C3 transferase, an ADP-ribosyltransferase that selectively ribosylates Rho proteins in the effector binding domain and inactivates them (43). Importantly, it has extremely low affinity for other members of the Rho family such as Cdc42 and Rac1. Therefore, C3 transferase was used to specifically block RhoA activity to identify if it contributes to NA transport.
HeLa cells transfected with NA encoding cDNA were processed for immunofluorescence study. The addition of C3 transferase to the cell culture did not significantly alter the amount of cell surface immunofluorescence (Fig. 3A). To confirm this finding, we performed the cell surface NA activity assay. Again, no significant changes in total NA enzymatic activity on the plasma membranes were observed in C3 transferase-treated cells when compared with untreated cells (Fig. 3B).
FIGURE 3.
Alteration of RhoA function has no significant effect on the transport of NA to the cell surface. A, HeLa cells transfected with NA cDNA were incubated with or without C3 transferase (1 μg/ml) for 15–30 min and followed by immunofluorescence staining. Shown are confocal micrographs. Bar, 10 μm. B, 293T cells expressing NA were treated with C3 transferase as described in A. The cell surface NA activity was measured as described in Fig. 1. Plotted is the average NA activity from three independent experiments. The error bars represent the S.E. C, HeLa cells were co-transfected with plasmid encoding NA and either pEGFP empty vector, GFP-RhoA(WT), GFP-RhoA(G14V), or GFP-RhoA(T19N). Shown are confocal micrographs. Bar, 10 μm. D, cell transfection was performed as described in C. The cell surface NA activity was quantified, and the average NA activity from three independent experiments is plotted. The error bars represent the S.E. E, lysates from cells in D were analyzed by Western blotting using indicated antibodies.
To further determine whether RhoA plays a role in NA cellular trafficking, we co-expressed NA with dominant-negative or constitutively active mutants of RhoA and determined their influences on NA transport. Because Rho GTPases cycle between an active GTP-bound state and an inactive GDP-bound state, these mutants can disrupt cellular processes by altering the GTP-binding/hydrolysis cycle. However, expression of either constitutively active GFP-RhoA(G14V) or dominant-negative GFP-RhoA(T19N) did not significantly affect the amount of NA protein localized at the cell surface as compared with the expression of RhoA wild-type (GFP-RhoA(WT)) or GFP alone (Fig. 3C). Similar results were obtained from the analysis of cell surface NA activity (Fig. 3D). Western blotting analysis demonstrated that the total amount of cellular NA protein was not significantly affected by ectopic expression of RhoA proteins (Fig. 3E). Together, these results suggest that RhoA is not required for NA intracellular and cell surface transport.
Rac1 has been implicated previously in vesicle trafficking (44). Hence, we next examined whether it is involved in NA cell surface transport. To this end, we used constitutively active and dominant-negative mutants of Rac1 and examined their effects on NA transport to the plasma membrane. Like cells expressing mutant RhoA, cells expressing constitutively active or inactive mutants of Rac1 protein maintained a similar cellular distribution of NA as compared with the cells expressing wild-type Rac1 or GFP alone (Fig. 4A). To determine whether there were more subtle defects in NA trafficking, we quantified the amount of NA that had arrived at the cell surface by measuring the NA activity. As shown in Fig. 4B, expression of wild-type Rac1(WT) or dominant-negative Rac1(T17N) slightly but not significantly reduced the levels of NA activity on the cell surface. Additionally, the total amount of NA protein appeared to be normal in cells ectopically expressing Rac1 or its mutants (Fig. 4C). These observations suggest that there does not exist an absolute requirement for Rac1 function during NA transport to the cell surface.
FIGURE 4.
Rac1 function is not absolutely required for the transport of NA to the cell surface. A, HeLa cells co-transfected with NA cDNA, and either pEGFP vector, GFP-Rac1(WT), GFP-Rac1(G12V), or GFP-Rac1(T17N) were stained with an anti-HA-tagged NA antibody. Shown are confocal micrographs. Bar, 10 μm. B, cell transfection was performed as described in A. The cell surface NA activity was measured. Plotted are the results from three independent experiments. The error bars represent the S.E. C, lysates from cells in B were analyzed by Western blotting using the indicated antibodies. D, shRNA-based knockdown of RhoA was analyzed by immunoblot probed with antibodies as indicated (upper panel). The relative levels of RhoA protein in the upper panel were quantified by densitometry and normalized to the actin bands (lower panel). Plotted are the results from three experiments. The error bars represent the S.E. (n = 3; *, p < 0.001). E, shRNA-based knockdown of Rac1 was analyzed as described in D. Plotted are the average Rac1 levels from three independent experiments. The error bars represent the S.E. (n = 3; *, p < 0.001). F, cells expressing NA and shRNAs targeting either RhoA, Rac1, or luciferase were measured for the cell surface NA activity. Plotted are the results from three independent experiments. The error bars represent the S.E.
To further confirm these observations, we used the shRNA to disrupt the expression of RhoA or Rac1 in HeLa cells. Western blotting showed that expression of RhoA or Rac1 was strongly diminished in cells expressing related shRNA (Fig. 4, D and E). However, depletion of RhoA or Rac1 had no significant effect on the transport of NA to the cell surface (Fig. 4F and supplemental Fig. S3). Taken together, these results demonstrated that neither RhoA nor Rac1 disruption closely mimicked the consequences of toxin B treatment, implying that another Rho family member(s) may be involved in regulating NA transport.
Cdc42 Regulates Transport of NA to Cell Surface
It has been documented that the small GTP-binding protein, Cdc42, regulates actin dynamics and is linked to several protein transport steps in cells (14, 29–31, 33). However, despite extensive studies of the Cdc42 function in the early secretory pathway, little is known about its role in viral protein intracellular transport. Thus, we explored whether Cdc42 is involved in NA trafficking to the cell surface. For this, we carried out several experiments using dominant-negative and constitutively active mutant forms of Cdc42 and an shRNA targeting Cdc42. Surprisingly, we found that unlike cells expressing mutant RhoA or Rac1, cells expressing wild-type Cdc42 or constitutively active Cdc42(Q61L) showed an increased amount of NA on the cell surface as compared with GFP-expressing control cells (Fig. 5A). Interestingly, partial co-localization of Cdc42 with NA was observed on the surface of these cells (Fig. 5A). By contrast, cells expressing dominant-negative Cdc42(T17N) displayed a moderately decreased amount of cell surface-localized NA and some NA in trans-Golgi network (Fig. 5A and supplemental Fig. S4). To confirm these observations, we measured the cell surface NA activity. Consistent with the immunocytochemical analysis, quantification of NA activity revealed that levels of total enzyme activity on the plasma membranes were significantly increased in cells expressing Cdc42(Q61L) and increased to a lesser extent in cells expressing wild-type Cdc42, whereas expression of Cdc42(T17N) caused somewhat reduced NA activity (Fig. 5B). Of note, expression of these forms of Cdc42 had no overt effects on the total amount of NA protein (Fig. 5C). Experiments using biotinylation of cell surface proteins further demonstrated that the active form of Cdc42 promoted NA transport to the cell surface (supplemental Fig. S5).
FIGURE 5.
Cdc42 regulates the transport of NA to the cell surface. A, shown are confocal micrographs of HeLa cells co-transfected with NA and either pEGFP vector, GFP-Cdc42(WT), GFP-Cdc42(Q61L), or GFP-Cdc42(T17N). Bar, 10 μm. B, cell transfection was performed as described in A. The cell surface NA activity was measured, and the results from three independent experiments are plotted. The error bars represent the S.E. (n = 3; **, p < 0.01; ***, p < 0.05). C, lysates from cells in B were analyzed by immunoblotting using indicated antibodies. D, the interference efficiency of shRNAs targeting Cdc42 was analyzed by real-time PCR. Plotted are the average expression levels from three identical experiments. The error bars represent the S.E. *, p < 0.001. E, shRNA-based knockdown of Cdc42 was analyzed by Western blotting using the antibodies as indicated (left panel). The levels of Cdc42 protein in the left panel were quantified by densitometry and normalized to the actin bands (right panel). Plotted are the average Cdc42 levels from three experiments. The error bars represent the S.E. (n = 3; *, p < 0.001). F, shown are HeLa cells expressing NA and GFP + shRNA vectors targeting Cdc42, ARHGAP21, or luciferase control. Bar, 10 μm. G, cells expressing NA and shRNAs targeting Cdc42, ARHGAP21, or luciferase were measured for the cell surface NA activity. Plotted are the results from three independent experiments. The error bars represent the S.E. (n = 3; **, p < 0.01).
Because the expression of mutant RhoA and mutant Rac1 had little effects on NA trafficking, the consequence of the overexpression of Cdc42 proteins appeared to be specific. To directly demonstrate the involvement of endogenous Cdc42 in NA transport, we employed an shRNA targeting Cdc42. Quantitative RT-PCR and Western blotting revealed that the shRNA strongly down-regulated the expression of Cdc42 compared with the control shRNA to luciferase (Fig. 5, D and E). Interestingly, silencing Cdc42 increased accumulation of NA at the juxtanuclear region, and decreased NA at the cell surface (Fig. 5F). NA activity assay provided additional evidence that transport of NA to the cell surface was significantly retarded when Cdc42 was silenced (Fig. 5G). The fact that disrupting Cdc42 expression caused defects in NA trafficking suggests that Cdc42 likely is the target of toxin B that affects NA trafficking.
Change in Cell Surface NA Amount Is Because of Altered Anterograde Transport
The changes in cell surface NA amount could be caused by changes in anterograde transport and/or in retrograde transport of NA. To clarify which mechanism was involved in the regulation of surface NA amount, endocytosis and anterograde transport of NA were measured in HeLa cells treated with toxin B or expressing the constitutively active form of Cdc42. We found that endocytosis of NA was not detectable (Fig. 6, A and D). In addition, NA was not seen in EEA1-positive endosomes (supplemental Fig. S6). Interestingly, the transport of NA to the plasma membrane was blocked by treatment with toxin B (Fig. 6, B and C). In contrast, expression of the active form of Cdc42 increased the anterograde transport of NA to the cell surface (Fig. 6, E and F). These results are consistent with those from NA activity assay and immunofluorescence study presented above. Collectively, our experiments demonstrated that Cdc42 is required for efficient transport of influenza virus NA toward the plasma membranes.
FIGURE 6.
The change in cell surface NA amount is because of altered anterograde transport. A, to determine the endocytosis of NA, HeLa cells transfected with NA cDNA were biotinylated with sulfo-NHS-SS-biotin at 4 °C. Cells were then warmed to 37 °C and mock-treated or treated with toxin B (100 ng/ml) followed by treatment with reduced GSH to cleave surface biotin. Control cells were biotinylated and maintained at 4 °C. The biotinylated proteins that were freshly endocytosed were precipitated with streptavidin-bound agarose and examined by immunoblotting using the indicated antibodies. Shown are representative data from three independent experiments with similar results. B, to examine the anterograde transport of NA, HeLa cells transfected with NA cDNA were first blocked by sulfo-NHS-acetate and then treated with toxin B at 37 °C. Cell surface biotinylation was then performed, and biotinylated proteins were precipitated with streptavidin-bound beads and examined by immunoblotting as described in A. C, the levels of NA protein transported to the plasma membrane in B were quantified by densitometry and normalized to input (whole cell lysate (WCL)). Plotted are the results from three independent experiments. The error bars represent the S.E. (n = 3; *, p < 0.01). D, HeLa cells were co-transfected with NA and either pEGFP vector or Cdc42(Q61L). The endocytosis of NA was examined as described in A. E, HeLa cells were co-transfected with NA and either pEGFP vector or Cdc42(Q61L). The transport of NA to the cell surface was examined as described in B. F, the levels of NA protein transported to the plasma membrane in E were quantified as described in C. Plotted are the average values from three independent experiments. The error bars represent the S.E. (n = 3; *, p < 0.01).
ARHGAP21 Is Involved in Regulating Transport of NA to Cell Surface
Recent studies have shown that ARHGAP21 functions as a Cdc42-specific GAP and acts downstream of the small GTP binding ADP-ribosylation factor 1 (ARF1) to play an important role in regulating Golgi structure and function and in protein trafficking through the control of Cdc42 activity (30, 31, 45–47). Because our experiments revealed that Cdc42(Q61L) promotes, but Cdc42(T17N) inhibits, NA trafficking to the plasma membranes, we sought to explore whether altering GTPase cycle had any effect on NA trafficking. To this end, ARHGAP21 was overexpressed or silenced, and the influences of these treatments on NA transport were examined in HeLa cells.
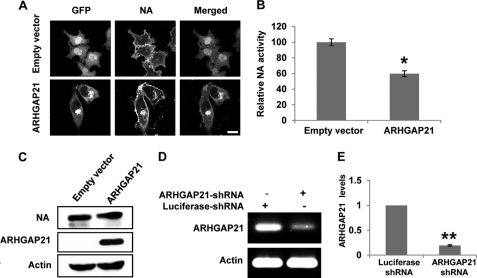
To start to elucidate the function of the ARHGAP21 in NA trafficking, HeLa cells were transiently co-transfected with NA-coding cDNA and a plasmid encoding a fragment of ARHGAP21 containing the ARF binding and GAP domains as described previously (30). We observed that overexpression of ARHGAP21 caused pleiotropic defects in cell morphology but had no effect on cell viability (supplemental Fig. S7). Interestingly, we found that although NA protein was seen on the plasma membranes, a significant amount of NA was enriched in the juxtanuclear punctuate structures where it co-localized with ARHGAP21 (Fig. 7A). Furthermore, quantification of NA activity on the cell surface confirmed that NA trafficking to the plasma membranes was significantly reduced in cells overexpressing ARHGAP21 compared with the control cells (Fig. 7B). The total amount of NA protein was not affected by ARHGAP21 (Fig. 7C).
FIGURE 7.
ARHGAP21 is involved in regulating transport of NA to the cell surface. A, shown are HeLa cells co-transfected with NA-coding cDNA and either pEGFP vector or plasmid encoding a GFP-fused ARHGAP21 containing the ARF-binding and GAP domains. Bar, 10 μm. B, 293T cells were co-transfected with plasmids as described in A, and the cell surface NA activity was quantified. Plotted is the average NA activity from three independent experiments. The error bars represent the S.E. (n = 3; *, p < 0.01). C, lysates from cells in B were analyzed by immunoblotting using the indicated antibodies. D, the interference efficiency of shRNAs targeting ARHGAP21 was analyzed by RT-PCR. E, the interference efficiency of shRNAs targeting ARHGAP21 was analyzed by quantitative real-time PCR. Plotted are the average expression levels from three independent experiments. The error bars represent the S.E. (n = 3; *, p < 0.001).
To further verify that ARHGAP21 indeed plays a role in NA transport to the cell surface, we used shRNA to silence ARHGAP21 in HeLa cells and monitored the NA transport compared with luciferase shRNA control. The shRNA targeting sequence of ARHGAP21 was previously characterized (45). RT-PCR and quantitative real-time PCR analysis demonstrated that ARHGAP21 transcript levels were reduced ∼80% in these cells (Fig. 7, D and E). Interestingly, we found that silencing ARHGAP21 resulted in an increased amount of NA localized at the plasma membranes, indicating that disrupting ARHGAP21 expression facilitates NA trafficking toward the plasma membranes (Fig. 5F). To confirm that knockdown of ARHGAP21 expression increases the amount of NA that reaches the cell surface, we performed the NA activity assay. The results revealed that reduced expression of ARHGAP21 caused an increase in total NA enzyme activity on the cell surface (Fig. 5G). Together, these findings suggest that ARHGAP21 negatively regulates NA transport to the cell surface.
Silencing Cdc42 or ARHGAP21 Has Significant Effects on Influenza A Virus Replication
In influenza A virus-infected cells, various intracellular factors can affect the efficiency of viral replication. However, there is little information available about factors related to the regulation of viral replication efficiency by control of viral protein traffic. Our results established a role of Cdc42 and ARHGAP21 in the trafficking of influenza A virus NA to the cell surface. These observations prompted us to evaluate the function of these host factors in influenza A virus replication.
We generated several A549 human alveolar epithelial cell lines stably expressing the shRNAs targeting either Cdc42, ARHGAP21, or control luciferase using a lentiviral expression system (supplemental Fig. S8). The interference efficiency of these shRNAs in cells was analyzed by real-time PCR and Western blotting analysis. Expression of Cdc42 or ARHGAP21 was greatly diminished in cells expressing related shRNAs (Figs. 5, D and E, and 7, D and E). The cells were then infected with A/WSN/33 influenza virus at multiplicity of infection of 0.05. The supernatants from infected cell culture were harvested to examine the viral titers by hemagglutinin assay at different time points post-infection. We observed that the viral titers in the supernatants were clearly reduced when the endogenous Cdc42 in A549 cell was knocked down (Fig. 8A). In contrast, silencing ARHGAP21 resulted in an increase in viral titers (Fig. 8A). To further substantiate this finding, we performed the plaque assay of A/WSN/33 influenza viruses in Madin-Darby canine kidney cells using the supernatants from infected A549 cell culture (post-infection = 24 h). Consistent with the results obtained from hemagglutinin assay, we found that depletion of Cdc42 decreased the amount of progeny virions, whereas disrupting ARHGAP21 expression caused an increase in the viral titers (Fig. 8, B and C). These results indicate that Cdc42 is required for efficient replication of influenza A virus, whereas ARHGAP21 inhibits this process.
FIGURE 8.

Silencing of Cdc42 or ARHGAP21 has significant effects on influenza A virus replication. A, A549 cell lines stably expressing the shRNAs targeting either Cdc42, ARHGAP21, or control luciferase were infected with A/WSN/33 influenza virus at multiplicity of infection of 0.05. The supernatants of cell culture were harvested and examined for the viral titers by hemagglutinin assay at different time points post-infection as indicated. B and C, Cdc42 knockdown A549 cells, ARHGAP21 knockdown A549 cells, and control cells were infected with influenza A virus as described in A. Viral titers in the supernatants of these cells were examined by plaque assay (post-infection = 24 h) in Madin-Darby canine kidney cells. B, shown are representative of plaques formed by influenza A virus and stained with 0.165 mg/ml neutral red. C, the average viral titers from three independent experiments are plotted. The error bars represent the S.E. (n = 3; *, p < 0.01; **, p < 0.005). PFU, plaque-forming units.
DISCUSSION
The NA protein of influenza A virus cleaves sialic acid moieties from sialyloligosaccharides and facilitates the release of nascent virions. However, the biological importance of NA is not only its property of removing the virus receptor in the budding stage but also the role it plays in promoting influenza virus dispersion within mucosal secretions of the respiratory tract through its enzymatic activity. Importantly, NA inhibitors, a class of drugs designed to block the enzyme active site, have been widely used to control influenza in humans. Despite these findings, the exact mechanism that underlies the intracellular and cell surface transport of NA is not fully understood. In particular, the host components that facilitate the completion of NA transport remain unclear.
In this study we have analyzed the involvement of Rho family GTPases in NA transport, especially in NA transport to the cell surface where influenza virus assembly occurs. We found that Cdc42 and its specific GAP ARHGAP21 are both involved in NA transport to the plasma membranes. Our results represent the first report of the role of Rho family member in regulating influenza viral protein intracellular trafficking. To further confirm the functional relevance of Cdc42 and ARHGAP21 in influenza virus life cycle, we generated the Cdc42 knockdown and ARHGAP21 knockdown cell lines. We observed that depletion of Cdc42 inhibited the NA trafficking to the plasma membranes and decreased the replication efficiency of influenza A virus. These experiments provide evidence that Cdc42 is required for efficient transport of NA to the host cell surface and for efficient replication of influenza A virus. On the other hand, silencing ARHGAP21 caused an increase in the amount of NA that reached the cell surface and an increase in the amount of progeny virions. Notably, overexpression of a constitutively active GTPase-defective mutant form of Cdc42 (Cdc42Q61L) in HeLa cells similarly increased the amount of NA expressed at the cell surface. Therefore, a very likely possibility is that the increase in the amount of NA transported to the cell surface and progeny virions upon knocking down ARHGAP21 may be an indirect consequence of Cdc42 activation. Together, these data suggest that influenza viruses may cause activation of Cdc42 signaling to complete their life cycle in the host cell.
Recently, there is increasing evidence implicating Rho family GTPases in intracellular trafficking. A previous study found that RhoA, Rac1, and Cdc42 each regulate distinct aspects of Shiga toxin trafficking from the cell surface to the Golgi apparatus (30). RhoA regulates the internalization of Shiga toxin, whereas Cdc42 regulates the retrograde motility of Shiga toxin to the juxtanuclear Golgi apparatus (30). Surprisingly, here we found that expression of constitutively active or inactive mutants of RhoA or Rac1 proteins in HeLa cells did not significantly affect the amount of NA that reached the cell surface. By contrast, cells that express the constitutively active or inactive mutants of Cdc42 or its GAP ARHGAP21 or that express the shRNAs targeting Cdc42 or ARHGAP21 exhibited a significant change in the amount of cell surface-localized NA. These results suggest that among three members of the Rho family examined, Cdc42 is the only small GTPase to have a specific function to regulate the anterograde transport of influenza virus NA protein. Each of the trafficking steps during intracellular and cell surface transport of NA remains to be further investigated to determine the precise role of Cdc42 in this process.
The contribution of the actin cytoskeleton during the cell surface transport of influenza virus proteins is unclear. However, actin cytoskeleton is thought to contribute to protein transport in several ways, including facilitating vesicle formation and providing microfilament tracks and force for vesicle movement through actin dynamics (15, 18, 48). Cdc42 and its specific GAP ARHGAP21 have been shown to regulate Arp2/3-dependent actin dynamics and motor proteins at the Golgi and the cell surface (15, 20, 29, 45) and are implicated in several protein transport steps (14, 29–31, 33). For example, previous experiments demonstrated that Cdc42 signaling led to the assembly of a specific pool of actin on Golgi membranes that is involved in protein transport through Golgi (19, 20). In addition, we have found previously that microtubule/dynein-dependent transport is sensitive to disrupting Cdc42-mediated actin signaling (29). Interestingly, our current data presented above suggest that Cdc42 and ARHGAP21 are both involved in NA transport in cells. Therefore, we reasoned that actin dynamics is important for NA transport to the cell surface.
To address this possibility, we performed experiments using actin toxin cytochalasin D. An apparent reduction in the levels of cell surface-localized NA protein was observed when actin dynamics were disrupted with cytochalasin D (supplemental Fig. S9). After the cytochalasin D treatment, NA protein remained dispersed in the cells, and an accumulation of NA at the juxtanuclear Golgi region was seen, whereas no such distribution of NA was seen in Mock-treated cells, indicating that cytochalasin D inhibits the transport of NA from the Golgi to the plasma membranes (supplemental Fig. S9A). Similar results were obtained from the NA activity assay (supplemental Fig. S9B). These data indicate a functional relevance of actin in NA transport to the cell surface. Therefore, there exists the possibility that overexpression or depletion of Cdc42 or its GAP ARHGAP21 results in actin cytoskeleton remodeling, which affects the efficiency of NA transport in host cells and thereby impacts influenza A virus replication. Activation of Cdc42 has emerged as one of the crucial signaling mechanisms in intracellular transport. Further analyzing the regulation of cytoskeleton and molecular motor proteins such as dynein by ARHGAP21- and Cdc42-based signaling during the transport of viral components will, therefore, be valuable in understanding the mechanism of Cdc42-regulated transport and virus replication. On the other hand, we observed that Cdc42 knockdown cells showed only a 40% reduction in NA transport to the cell surface and in virus replication as compared with wild-type cells. There could be several explanations for this finding. One possibility is that Cdc42 was not completely depleted in the cells and the residual protein still functions to regulate NA trafficking. Alternatively, there exist other cellular factors that are involved in regulating NA trafficking. Further investigation will likely identify additional host factors that regulate multiple specific aspects of the transport of influenza A viral components and thereby affect virus life cycle.
Supplementary Material
Acknowledgments
We thank members of the Chen laboratory for helpful discussions.
This work was supported by National Basic Research Program (973) of China (2010CB534004), National Key Technology R&D Program (2009BA183B01-8), Intramural grant of the Chinese Academy of Sciences (2010-Biols-CAS-0204), and Hundreds of Talents Program of Chinese Academy of Sciences 2009-2014.

This article contains supplemental Methods and Figs. S1–S9.
The gene profile data have been submitted to Gene Expression Omnibus (GSE32878).
- NA
- neuraminidase
- ARF
- ADP-ribosylation factor
- GAP
- GTPase-activating protein.
REFERENCES
- 1. Wiley D. C., Skehel J. J. (1987) The structure and function of the hemagglutinin membrane glycoprotein of influenza virus. Ann. Rev. Biochem. 56, 365–394 [DOI] [PubMed] [Google Scholar]
- 2. Weis W., Brown J. H., Cusack S., Paulson J. C., Skehel J. J., Wiley D. C. (1988) Structure of the influenza virus hemagglutinin complexed with its receptor, sialic acid. Nature 333, 426–431 [DOI] [PubMed] [Google Scholar]
- 3. Gubareva L. V. (2004) Molecular mechanisms of influenza virus resistance to neuraminidase inhibitors. Virus Res. 103, 199–203 [DOI] [PubMed] [Google Scholar]
- 4. Liu C., Eichelberger M. C., Compans R. W., Air G. M. (1995) Influenza type A virus neuraminidase does not play a role in viral entry, replication, assembly, or budding. J. Virol. 69, 1099–1106 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5. Ohuchi M., Asaoka N., Sakai T., Ohuchi R. (2006) Roles of neuraminidase in the initial stage of influenza virus infection. Microbes Infect. 8, 1287–1293 [DOI] [PubMed] [Google Scholar]
- 6. Li Q., Qi J., Zhang W., Vavricka C. J., Shi Y., Wei J., Feng E., Shen J., Chen J., Liu D., He J., Yan J., Liu H., Jiang H., Teng M., Li X., Gao G. F. (2010) The 2009 pandemic H1N1 neuraminidase N1 lacks the 150-cavity in its active site. Nat. Struct. Mol. Biol. 17, 1266–1268 [DOI] [PubMed] [Google Scholar]
- 7. Liu D., Liu X., Yan J., Liu W. J., Gao G. F. (2009) Interspecies transmission and host restriction of avian H5N1 influenza virus. Sci. China C Life Sci. 52, 428–438 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8. Whittaker G. R. (2001) Intracellular trafficking of influenza virus: clinical implications for molecular medicine. Expert Rev. Mol. Med. 3, 1–13 [DOI] [PubMed] [Google Scholar]
- 9. Brass A. L., Huang I. C., Benita Y., John S. P., Krishnan M. N., Feeley E. M., Ryan B. J., Weyer J. L., van der Weyden L., Fikrig E. (2009) The IFITM proteins mediate cellular resistance to influenza A H1N1 virus, West Nile virus, and dengue virus. Cell 139, 1243–1254 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10. Barman S., Adhikary L., Chakrabarti A. K., Bernas C., Kawaoka Y., Nayak D. P. (2004) Role of transmembrane domain and cytoplasmic tail amino acid sequences of influenza a virus neuraminidase in raft association and virus budding. J. Virol. 78, 5258–5269 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11. Chen B. J., Leser G. P., Morita E., Lamb R. A. (2007) Influenza virus hemagglutinin and neuraminidase, but not the matrix protein, are required for assembly and budding of plasmid-derived virus-like particles. J. Virol. 81, 7111–7123 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12. Nayak D. P., Balogun R. A., Yamada H., Zhou Z. H., Barman S. (2009) Influenza virus morphogenesis and budding. Virus Res. 143, 147–161 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13. Barman S., Nayak D. P. (2000) Analysis of the transmembrane domain of influenza virus neuraminidase, a type II transmembrane glycoprotein, for apical sorting and raft association. J. Virol. 74, 6538–6545 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14. Cerione R. A. (2004) Cdc42. New roads to travel. Trends Cell Biol 14, 127–132 [DOI] [PubMed] [Google Scholar]
- 15. Harris K. P., Tepass U. (2010) Cdc42 and vesicle trafficking in polarized cells. Traffic 11, 1272–1279 [DOI] [PubMed] [Google Scholar]
- 16. Melendez J., Grogg M., Zheng Y. (2011) Signaling role of Cdc42 in regulating mammalian physiology. J. Biol. Chem. 286, 2375–2381 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17. Calvo F., Sanz-Moreno V., Agudo-Ibáñez L., Wallberg F., Sahai E., Marshall C. J., Crespo P. (2011) RasGRF suppresses Cdc42-mediated tumorcell movement, cytoskeletal dynamics and transformation. Nat. Cell Biol. 13, 819–826 [DOI] [PubMed] [Google Scholar]
- 18. Stamnes M. (2002) Regulating the actin cytoskeleton during vesicular transport. Curr. Opin. Cell Biol. 14, 428–433 [DOI] [PubMed] [Google Scholar]
- 19. Fucini R. V., Chen J. L., Sharma C., Kessels M. M., Stamnes M. (2002) Golgi vesicle proteins are linked to the assembly of an actin complex defined by mAbp1. Mol. Biol. Cell 13, 621–631 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20. Chen J. L., Lacomis L., Erdjument-Bromage H., Tempst P., Stamnes M. (2004) Cytosol-derived proteins are sufficient for Arp2/3 recruitment and ARF/coatomer-dependent actin polymerization on Golgi membranes. FEBS Lett. 566, 281–286 [DOI] [PubMed] [Google Scholar]
- 21. Balklava Z., Pant S., Fares H., Grant B. D. (2007) Genome-wide analysis identifies a general requirement for polarity proteins in endocytic traffic. Nat. Cell Biol. 9, 1066–1073 [DOI] [PubMed] [Google Scholar]
- 22. Luna A., Matas O. B., Martínez-Menárguez J. A., Mato E., Durán J. M., Ballesta J., Way M., Egea G. (2002) Regulation of protein transport from the Golgi complex to the endoplasmic reticulum by CDC42 and N-WASP. Mol. Biol. Cell 13, 866–879 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23. Etienne-Manneville S., Hall A. (2002) Rho GTPases in cell biology. Nature 420, 629–635 [DOI] [PubMed] [Google Scholar]
- 24. Sabharanjak S., Sharma P., Parton R. G., Mayor S. (2002) GPI-anchored proteins are delivered to recycling endosomes via a distinct cdc42-regulated, clathrin-independent pinocytic pathway. Dev. Cell 2, 411–423 [DOI] [PubMed] [Google Scholar]
- 25. Malyukova I., Murray K. F., Zhu C., Boedeker E., Kane A., Patterson K., Peterson J. R., Donowitz M., Kovbasnjuk O. (2009) Macropinocytosis in Shiga toxin 1 uptake by human intestinal epithelial cells and transcellular transcytosis. Am. J. Physiol. Gastrointest. Liver Physiol. 296, G78–G92 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26. Hall A. (2005) Rho GTPases and the control of cell behavior. Biochem. Soc. Trans. 33, 891–895 [DOI] [PubMed] [Google Scholar]
- 27. Osmani N., Peglion F., Chavrier P., Etienne-Manneville S. (2010) Cdc42 localization and cell polarity depend on membrane traffic. J. Cell Biol. 191, 1261–1269 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28. Slaughter B. D., Das A., Schwartz J. W., Rubinstein B., Li R. (2009) Dual modes of cdc42 recycling fine-tune polarized morphogenesis. Dev. Cell 17, 823–835 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29. Chen J. L., Fucini R. V., Lacomis L., Erdjument-Bromage H., Tempst P., Stamnes M. (2005) Coatomer-bound Cdc42 regulates dynein recruitment to COPI vesicles. J. Cell Biol. 169, 383–389 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30. Hehnly H., Longhini K. M., Chen J. L., Stamnes M. (2009) Retrograde Shiga toxin trafficking is regulated by ARHGAP21 and Cdc42. Mol. Biol. Cell 20, 4303–4312 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31. Hehnly H., Xu W., Chen J. L., Stamnes M. (2010) Cdc42 regulates microtubule-dependent Golgi positioning. Traffic 11, 1067–1078 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32. Wu W. J., Erickson J. W., Lin R., Cerione R. A. (2000) The γ-subunit of the coatomer complex binds Cdc42 to mediate transformation. Nature 405, 800–804 [DOI] [PubMed] [Google Scholar]
- 33. Matas O. B., Fritz S., Luna A., Egea G. (2005) Membrane trafficking at the ER/Golgi interface. Functional implications of RhoA and Rac1. Eur. J. Cell Biol. 84, 699–707 [DOI] [PubMed] [Google Scholar]
- 34. Guo G., Qiu X., Wang S., Chen Y., Rothman P. B., Wang Z., Chen Y., Wang G., Chen J. L. (2010) Oncogenic E17K mutation in the pleckstrin homology domain of AKT1 promotes v-Abl-mediated pre-B-cell transformation and survival of Pim-deficient cells. Oncogene 29, 3845–3853 [DOI] [PubMed] [Google Scholar]
- 35. Li Z., Nie F., Wang S., Li L. (2011) Histone H4 Lys-20 monomethylation by histone methylase SET8 mediates Wnt target gene activation. Proc. Natl. Acad. Sci. U.S.A. 108, 3116–3123 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36. Bloom J. D., Gong L. I., Baltimore D. (2010) Permissive secondary mutations enable the evolution of influenza oseltamivir resistance. Science 328, 1272–1275 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37. Liu S., Li R., Zhang R., Chan C. C., Xi B., Zhu Z., Yang J., Poon V. K., Zhou J., Chen M. (2011) CL-385319 inhibits H5N1 avian influenza A virus infection by blocking viral entry. Eur. J. Pharmacol. 660, 460–467 [DOI] [PubMed] [Google Scholar]
- 38. Lee-Kwon W., Kawano K., Choi J. W., Kim J. H., Donowitz M. (2003) Lysophosphatidic acid stimulates brush border Na+/H+ exchanger 3 (NHE3) activity by increasing its exocytosis by an NHE3 kinase A regulatory protein-dependent mechanism. J. Biol. Chem. 278, 16494–16501 [DOI] [PubMed] [Google Scholar]
- 39. Collins P. J., Haire L. F., Lin Y. P., Liu J., Russell R. J., Walker P. A., Skehel J. J., Martin S. R., Hay A. J., Gamblin S. J. (2008) Crystal structures of oseltamivir-resistant influenza virus neuraminidase mutants. Nature 453, 1258–1261 [DOI] [PubMed] [Google Scholar]
- 40. Aktories K., Just I. (1995) Monoglucosylation of low molecular mass GTP-binding Rho proteins by clostridial cytotoxins. Trends Cell Biol. 5, 441–443 [DOI] [PubMed] [Google Scholar]
- 41. Wilkins T. D., Lyerly D. M. (1996) Clostridium difficile toxins attack Rho. Trends Microbiol. 4, 49–51 [DOI] [PubMed] [Google Scholar]
- 42. Aktories K., Schmidt G., Just I. (2000) Rho GTPases as targets of bacterial protein toxins. Biol. Chem. 381, 421–426 [DOI] [PubMed] [Google Scholar]
- 43. Aktories K., Just I. (1995) In vitro ADP-ribosylation of Rho by bacterial ADP-ribosyltransferases. Methods Enzymol. 256, 184–195 [DOI] [PubMed] [Google Scholar]
- 44. Ridley A. J. (2006) Rho GTPases and actin dynamics in membrane protrusions and vesicle trafficking. Trends Cell Biol 16, 522–529 [DOI] [PubMed] [Google Scholar]
- 45. Dubois T., Paléotti O., Mironov A. A., Fraisier V., Stradal T. E., De Matteis M. A., Franco M., Chavrier P. (2005) Golgi-localized GAP for Cdc42 functions downstream of ARF1 to control Arp2/3 complex and F-actin dynamics. Nat. Cell Biol. 7, 353–364 [DOI] [PubMed] [Google Scholar]
- 46. Ménétrey J., Perderiset M., Cicolari J., Dubois T., Elkhatib N., El Khadali F., Franco M., Chavrier P., Houdusse A. (2007) Structural basis for ARF1-mediated recruitment of ARHGAP21 to Golgi membranes. EMBO J. 26, 1953–1962 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47. Kumari S., Mayor S. (2008) ARF1 is directly involved in dynamin-independent endocytosis. Nat. Cell Biol. 10, 30–41 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48. Valderrama F., Luna A., Babía T., Martinez-Menárguez J. A., Ballesta J., Barth H., Chaponnier C., Renau-Piqueras J., Egea G. (2000) The golgi-associated COPI-coated buds and vesicles contain β/γ-actin. Proc. Natl. Acad. Sci. U.S.A. 97, 1560–1565 [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.