Background: RUNX proteins are important for development, but not much is known about the functions of RUNX after development.
Results: RNT-1, the nematode RUNX homolog, was stabilized by oxidative stress through a p38 MAP kinase for proper response to oxidative stress.
Conclusion: RNT-1 plays a role in defensive responses to oxidative stress.
Significance: We identified a new role for a RUNX protein in a postdevelopmental process.
Keywords: Caenorhabditis elegans, p38 MAPK, Phosphorylation, Protein Stability, Stress Response, RUNX
Abstract
RUNX proteins are evolutionarily conserved transcription factors known to be involved in various developmental processes. Here we report a new role for a RUNX protein: a role in stress response. We show that RNT-1, the Caenorhabditis elegans RUNX homolog, is constantly produced and degraded by the ubiquitination-proteasome pathway in the intestine of the nematode. RNT-1 was rapidly stabilized by oxidative stress, and the rnt-1-mutant animals were more sensitive to oxidative stress, indicating that rapid RNT-1 stabilization is a defense response against the oxidative stress. The MAP kinase pathway is required for RNT-1 stabilization, and RNT-1 was phosphorylated by SEK-1/PMK-1 in vitro. ChIP-sequencing analysis revealed a feedback loop mechanism of the MAP kinase pathway by the VHP-1 phosphatase in the RNT-1-mediated oxidative stress response. We propose that rnt-1 is regulated at the protein level for its role in the immediate response to environmental challenges in the intestine.
Introduction
RUNX3 family proteins, evolutionarily well conserved transcription factors, have pivotal roles in various aspects of development such as hematopoiesis, chondrogenesis, neurogenesis, and cancer development (1–4). The effects of RUNX deregulation in various developmental processes have been studied. Deletion or null mutation as well as overexpression of the RUNX family caused aberrant phenotypes such as acute myelogenous leukemia, cleidocranial dysplasia, and gastric adenocarcinoma, suggesting that RUNX exerts its effects in a dose-dependent manner (5). In addition, diverse cellular signaling events that modify both the transcriptional and posttranscriptional status of the RUNX genes affect the amount and the activity of RUNX proteins in response to developmental or environmental cues. Transcriptional regulation, such as chromatin remodeling (6) and DNA methylation (7–10), and posttranscriptional regulation, such as acetylation (11, 12), ubiquitination (13–15), and phosphorylation (16, 17), have been reported as the regulation mechanisms of RUNX activity.
In Caenorhabditis elegans, only one functional RUNX gene, rnt-1, was identified (18). Mutations in rnt-1 caused severe morphological defects induced by improper development of the hypodermis and the intestine (19). In C. elegans, RNT-1 is also involved in cell fate commitment in the seam cell division, which is similar to the functions of mammalian RUNXs (20). The function of RNT-1 in cell cycle regulation is explained by its genetic interaction with cki-1, a cyclin-dependent kinase inhibitor. Transcriptional regulation of rnt-1 has been confirmed in a previous study (21). From nematodes to humans, the biological functions of RUNX proteins have been limited to developmental processes such as normal development or carcinogenesis, and their functions, aside from those in development, have not been fully studied. In this study, we wanted to pursue this issue using C. elegans as a model organism.
Here, we report that RNT-1, a RUNX family protein, has a new role in stress response. We show that the level of RNT-1 proteins was up-regulated by oxidative and osmotic stress via stabilization of the proteins, which are otherwise degraded by proteasome-mediated proteolysis. The stabilization of RNT-1 proteins was mediated by the p38 MAP kinase pathway. We also show that vhp-1, one of the target genes of stabilized RNT-1, was transcriptionally activated by RNT-1 and formed a negative feedback loop by modulating the PMK-1 pathway for appropriate oxidative stress response.
EXPERIMENTAL PROCEDURES
Worm Strains and Culture
C. elegans strains of the genotype of wild-type N2 Bristol and rnt-1(ok351) was obtained from the C. elegans Genome Center (Minneapolis, MN), and the rnt-1(tm388) and rnt-1(tm491) strain was obtained from the National Bioresources Project (Tokyo, Japan). Worms were grown at 20 °C on nematode growth media (NGM) by a standard method (22).
Microinjection and Microscopy
Microinjection of DNA into the gonads of the adult hermaphrodites was carried out according to a standard protocol (23). The pPD-run-FL construct (19), which bears sequences of the full-length rnt-1 promoter and RNT-1 protein-fused GFP, was used for expression analysis. For intestine-specific rescue of the defect in the rnt-1 mutant, 2 kb of act-5 promoter at and cDNA of rnt-1 were cloned to the pPD95.77 vector. The pRF4, which contains the dominant rol-6 (su1006) gene, was used as an injection marker at the concentration of 100 μg/ml (24). All plasmids were injected at the concentration of 100 μg/ml. Plasmid DNAs used for injection were extracted with the Qiagen plasmid midi kit (catalog number 12145). To see the expression pattern of transgenes, the transgenic lines were paralyzed with 5 mm levamisole and mounted on 5% agar pads, and the fluorescence was observed using an Axioplan 2 microscope.
Feeding RNAi Method
For pmk-1 RNAi, we used the HT115 Escherichia coli strain containing the full-length pmk-1 cDNA in the pPD129.36 (L4440) vector. The clone was confirmed by sequencing. For ubq-1, uba-1, math-33, sek-1, nsy-1, and vhp-1 RNAi, we used an RNAi library purchased from MRC (Cambridge, UK). We started RNAi from the L4 stage for pmk-1, sek-1, and nsy-1 RNAi and examined the phenotypes of the L4 progeny. For ubq-1 and uba-1 RNAi, whose RNAi knockdown caused embryonic lethality, we provided L1 larvae with RNAi bacteria and observed the phenotypes when they reached adulthood. For vhp-1, we provided L2 larvae with RNAi bacteria and observed the phenotypes when they reached adulthood. Worms were fed by each RNAi E. coli strain on NGM plates containing 1 mm isopropyl 1-thio-β-d-galactopyranoside.
In Situ Hybridization
Detection of rnt-1 transcripts in embryos, larvae, and adult worms was performed as described in Ref. 25. Digoxigenin-dideoxy UTP-labeled oligonucleotide was synthesized, followed by the protocol (Bionex, Korea). We used worms containing pPD-run-FL and pRF4 to detect rnt-1 RNA.
Proteasome Inhibitor Treatment
To test whether the proteasome pathway degrades RNT-1, we treated the worms with the commonly used proteasome inhibitor MG132 (Sigma). The MG132 experiments were performed as described previously, with some modifications (26–28). Briefly, the transgenic animals containing the pPD-run-FL reporter construct were grown on NGM plates. Then, we washed off the plates with M9 (22 mm KH2PO4, 42 mm Na2HPO4, 80 mm NaCl, 1 mm MgSO4) medium and collected into a 1.5-ml microcentrifuge tube. MG132 was added at a final concentration of 50 μm, after which the animals were incubated for 6 h at room temperature on a Nutator mixer and then washed with M9. To see the difference between MG132-treated worms and M9-treated control worms, both lines were mounted on 5% agar pads, and the fluorescence was observed using an Axioplan2 microscope. To perform the Western blot analysis, worms were grown on 100-mm NGM lite media plates and harvested with M9 buffer. The harvested worms were vigorously shaken in 100 ml of M9 with or without 50 μm MG132 for 6 h.
Immunoprecipitation
To show that RNT-1 is ubiquitinated, plasmids harboring a cDNA of RNT-1 under the CMV promoter and an ubiquitin gene, respectively, were transfected to the 293T cell line. After 24-h incubation, we treated the cells with MG132 for 2 h and harvested them. The following immunoprecipitation and Western blotting were performed with a standard protocol.
Stress Assay
To examine whether RNT-1::GFP was stabilized by oxidative stress and osmotic stress, rnt-1::gfp worms were applied to M9 solution containing 0.1 m paraquat, 10 mm t-butyl peroxide, 0.3 m NaCl, and 625 mm sucrose, respectively. After 0, 30, 60, and 240 min, we harvested worms with M9 and then observed them with an Axioplan2 microscope. To give heat shock, we put worms into a 30 °C incubator for 30 min on NGM plates. For cold shock, worms were incubated at 4 °C for 30 min. Hypoxia was induced by soaking the worms for 1 h in M9 solution. Worms were irradiated with UV light (360 nm, 340 W/cm2, XL-1000 UV Crosslinker, Spectronics Co.) for 1 min. After each stress, worms were mounted on 5% agar pads, and the fluorescence was observed using an Axioplan2 microscope (Zeiss). Images were taken by an AxioCam (Zeiss) camera. To analyze the time course expression level of RNT-1 fused to GFP in the intestine, we only measured its fluorescence intestine in the int1 cells for consistency.
To test whether rnt-1 mutant alleles were sensitive to paraquat, we performed a paraquat lethality test. We synchronized N2 and rnt-1 worms and harvested them when they reached the L4 stage. All L4-stage worms were washed off with M9 buffer and put on empty NGM plates. By using 0.4 m paraquat solution, 50 worms were picked up with a mouth pipette and moved into an empty 24-well plate (29). More 0.4 m paraquat solution was dropped on the worms to avoid dehydration. After 5 min of exposure to paraquat, thrashing worms were counted. Each assay was repeated more than three times, and all of them were carried out at 22 °C.
To test the relationship between the MAP kinase pathway and RNT-1::GFP stabilization in oxidative stress, we synchronized control rnt-1::gfp worms and pmk-1, sek-1, nsy-1, and vhp-1 RNAi-treated rnt-1::gfp worms to L4 stage or young adulthood. L4 or young adult worms were harvested and washed off with M9 buffer. We applied 0.1 m paraquat to the animals, and 50–70 worms were transferred to an empty 24-well plate with a mouth pipette.
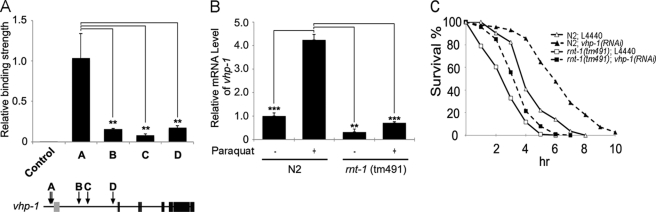
To test whether N2 and rnt-1(tm491) mutants fed with L4440 or vhp-1 RNAi bacteria were sensitive to paraquat, we performed a paraquat lethality test. We synchronized N2 and rnt-1(tm491) strains, fed them with L4440 or vhp-1 RNAi bacteria from the L2 stage, and harvested them when they reached the L4 stage. All L4-stage worms were washed off with M9 buffer, and 50–60 worms were applied to M9 solution containing 0.1 m paraquat at room temperature (22 °C). Dead worms were confirmed by no movement after touching their bodies.
In Vitro Kinase Assay
To test whether pmk-1 kinase activity phosphorylated RNT-1, we performed an in vitro kinase assay. FLAG-tagged PMK-1 and/or MYC-tagged SEK-1 was purified by anti-FLAG and/or anti-MYC-agarose bead from HEK293T cell lysate. Various GST-tagged constructs of RNT-1, PMK-1, and/or SEK-1 immunoprecipitates were incubated with [γ-32P]ATP (10 μCi) in kinase buffer (200 mm HEPES (pH7.3), 200 mm MgCl2, 200 mm MnCl2, 10 mm EDTA, 10 mm NaF, 1 mm Na3VO4, 10 mm DTT, 4 mm PMSF) for 20 min at 30 °C (31). The reaction was stopped by the addition of sample buffer. Phosphorylated RNT-1 was subjected to 9% SDS-polyacrylamide gel electrophoresis and visualized by autoradiography (Fuji, BAS 2000).
In Vitro Site-directed Mutagenesis
To make point mutations in putative phosphorylation sites of RNT-1, each plasmid was mutated by PCR-based mutagenesis with a QuikChange site-directed mutagenesis kit provided by Stratagene.
Chromatin Immunoprecipitation and Further Analyses
We followed the protocol described previously to carry out the ChIP experiments (32). Briefly, mixed-stage RNT-1::GFP transgenic worms were grown on NGM lite plates and harvested. After cross-linking the worm extract, the pellet was washed and resuspended in HEPES lysis buffer. We sonicated the pellet more than eight times. After centrifuging the pellet, supernatant was precleared with salmon sperm DNA/protein A-agarose and incubated with anti-GFP antibody at 4 °C overnight. To identify effector genes downstream of RNT-1 in response to oxidative stress, we performed ChIP-sequencing experiments in the presence or absence of oxidative stress. DNA samples from the ChIP assay were sequenced by next-generation sequencing in the laboratory of one of the authors (Y. J.K.) and Macrogen (Korea), and all data were analyzed by Macrogen (Korea). We compared data sets of the absence and presence of oxidative stress and selected nine genes on the basis of the existence of conserved rnt-1 binding sites and confirmation by real-time quantitative PCR analysis. Real-time quantitative PCR was carried out with TaKaRa SYBR premix Ex Taq in Applied Biosystem Prism 7300 and Bio-Rad iQTM SYBR Green Supermix in iQ5 as described in the manufacturer's manual.
RNA Preparation and Quantitative Real-time PCR Analysis
Total RNA was isolated with TRIzol reagent (Invitrogen) with the freeze-thaw method according to standard protocol of the manufacturer's manual. cDNA was synthesized with RevertAid M-MuLV reverse transcriptase (Fermentas, Canada) using oligo(dT) primer and subjected to PCR amplification.
PCR Primers
Sequences of the primers used in this study are listed in supplemental Table 1.
RESULTS
Proteasome-mediated Degradation of RNT-1 in the Intestine of C. elegans
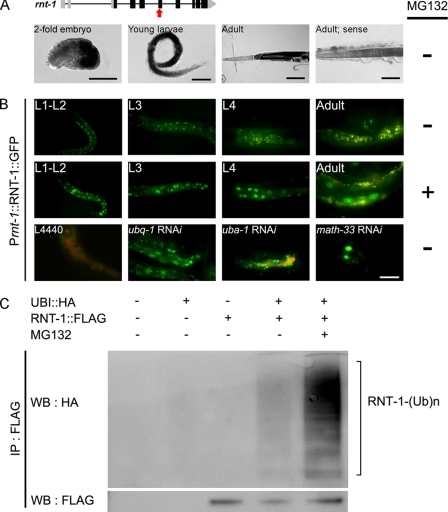
RNT-1 expression in the nematode is tightly regulated in terms of developmental timing and space because GFP reporter assays showed that RNT-1 is expressed in the hypodermis and the intestine from the embryonic stage up to larval stages and that its expression disappears in the adult stage (19, 21). To determine the regulatory mechanism underlying this specific expression, we examined the RNA and protein levels of rnt-1 during development. Interestingly, the in situ hybridization analysis showed that rnt-1 transcription still occurred in the intestine at the adult stage (Fig. 1A), whereas GFP-fused RNT-1 proteins were undetectable (B), indicating that RNT-1 regulation occurs at the posttranscriptional level. Because the rnt-1 transcript was still abundant in the intestine, we examined whether RNT-1 was regulated at the level of protein stability. When the nematodes were treated with MG132, a proteasome inhibitor, the GFP fused RNT-1 protein was stabilized only in the intestine at all developmental stages, including the adult stage (Fig. 1B), indicating that the proteasome continuously degrades RNT-1 in the intestine. We confirmed this result by Western blot analysis (supplemental Fig. 1). Consistent with this, RNAi knockdown of ubq-1, a polyubiquitin gene, and uba-1, the E1 gene, resulted in higher stability of RNT-1 in the intestine (Fig. 1B). In addition, RNAi of a ubiquitin-specific hydrolase, math-33, resulted in the stabilization of RNT-1. However, knockdown of C18E3.2, swsn-2.1, swsn-2.2, T24G10.2, and R07B7.2, which are annotated as homologs of MDM2, an E3 ligase of RUNX3 (15), did not cause stabilization of RNT-1 proteins (data not shown), suggesting that an E3 ligase different from MDM2 may act for RNT-1 in C. elegans. To confirm that RNT-1 is indeed ubiquitinated, we expressed RNT-1 under the CMV promoter in 293T cells together with an ubiquitin construct (Fig. 1C). Ubiquitination of RNT-1 was detected, and the treatment of MG132 increased the level of ubiquitinated RNT-1. These findings indicate that proteasome-mediated degradation is a main process of RNT-1 stability control in the intestine by accelerating its turnover rate.
FIGURE 1.
Continuous degradation of RNT-1 proteins in the intestine. A, the upper panel shows the genomic structure of the rnt-1 gene, and the bottom panels show the in situ hybridization results of rnt-1 at different developmental stages. The white line and the white dotted line in the adult indicate the intestine and gonadal arm, respectively. The red arrow in the upper panel represents the location of the in situ probe of rnt-1. A sense probe was used as the negative control. B, the top two rows show GFP expression in N2 animals containing the RNT-1::GFP transgene in the absence or presence of MG132, respectively. The third row shows the results of knockdown of ubq-1 (polyubiquitin), uba-1 (E1), and math-33 (ubiquitin-specific hydrolase) at the young adult stage. L4440 was used as s negative control. Scale bars = 50 μm. C, ubiquitination of RNT-1 in 293T cells. The ubiquitinated FLAG-tagged RNT-1 was detected in lanes 4 and 5. Treatment of MG132 increased ubiquitinated RNT-1 (lane 5).WB, Western blotting; IP, immunoprecipitation.
Oxidative and Osmotic Stresses Stabilize RNT-1 in the Intestine
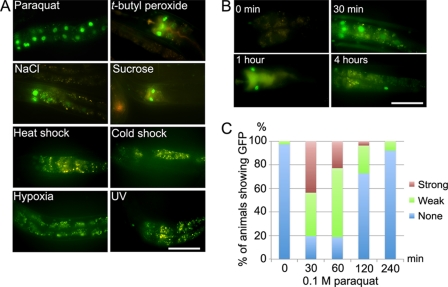
A possible biological implication of RNT-1 regulation at the protein level instead of the transcriptional level was that RNT-1 could be involved in a rapid response to environmental changes, which might require rapid changes in cell physiology. To test this hypothesis, we examined whether RNT-1 in the intestine was stabilized by external stress conditions. Among the various stresses, oxidative stress induced by paraquat and t-butyl peroxide and osmotic stress induced by high concentration of NaCl and sucrose rapidly stabilized RNT-1 in the intestine, which was not affected by heat shock, cold shock, hypoxia, and UV light (Fig. 2A). We could detect the GFP signal from stabilized RNT-1 proteins within 30 min after treating oxidative stress (Fig. 2, B and C). In the oxidative stress condition, stabilized RNT-1 was gradually decreased after 30 min and totally diminished within 4 h. We confirmed that the oxidative stress conditions did not increase the transcript level of rnt-1 (supplemental Fig. 2), again confirming that the protein level, not the transcription level, was affected by the stress conditions. These results raised the possibility that gaining of RNT-1 stability in the intestine might reflect its role in the acute stress response in the intestine.
FIGURE 2.
Oxidative and osmotic stresses stabilize RNT-1::GFP in the intestine. A, oxidative stress (paraquat, t-butyl peroxide) and osmotic stress (NaCl, sucrose) stabilize RNT-1::GFP in wild-type animals expressing a RNT-1::GFP transgene. Heat shock, cold shock, hypoxia, or UV light did not affect the stability of RNT-1. B, time course changes in the RNT-1::GFP abundance after treating paraquat. Late L4 or young adults were used. C, quantification of the animals for their levels of stabilized RNT-1::GFP under oxidative stress conditions. The expressions of GFP in int1 cells were analyzed. Scale bars = 50 μm.
RNT-1 Is Involved in the Oxidative Stress Response
To test whether RNT-1 was involved in the defensive mechanism against oxidative stress, we examined the susceptibility of rnt-1 mutant animals to paraquat treatment. After being exposed to 400 mm paraquat for 5 min, approximately 30% of the wild-type worms were still alive, but only 10% of rnt-1 mutant worms survived (Fig. 3A). The defective response to oxidative stress in rnt-1 mutant alleles was rescued with full-length RNT-1 expression under its own promoter (supplemental Fig. 3). We were able to rescue the defective oxidative stress response by expressing RNT-1 under an intestine-specific promoter, act-5 (supplemental Fig. 3), suggesting that RNT-1 in the intestine is necessary for the stress response. Taken together, RNT-1 is required for a defensive response against oxidative stress.
FIGURE 3.
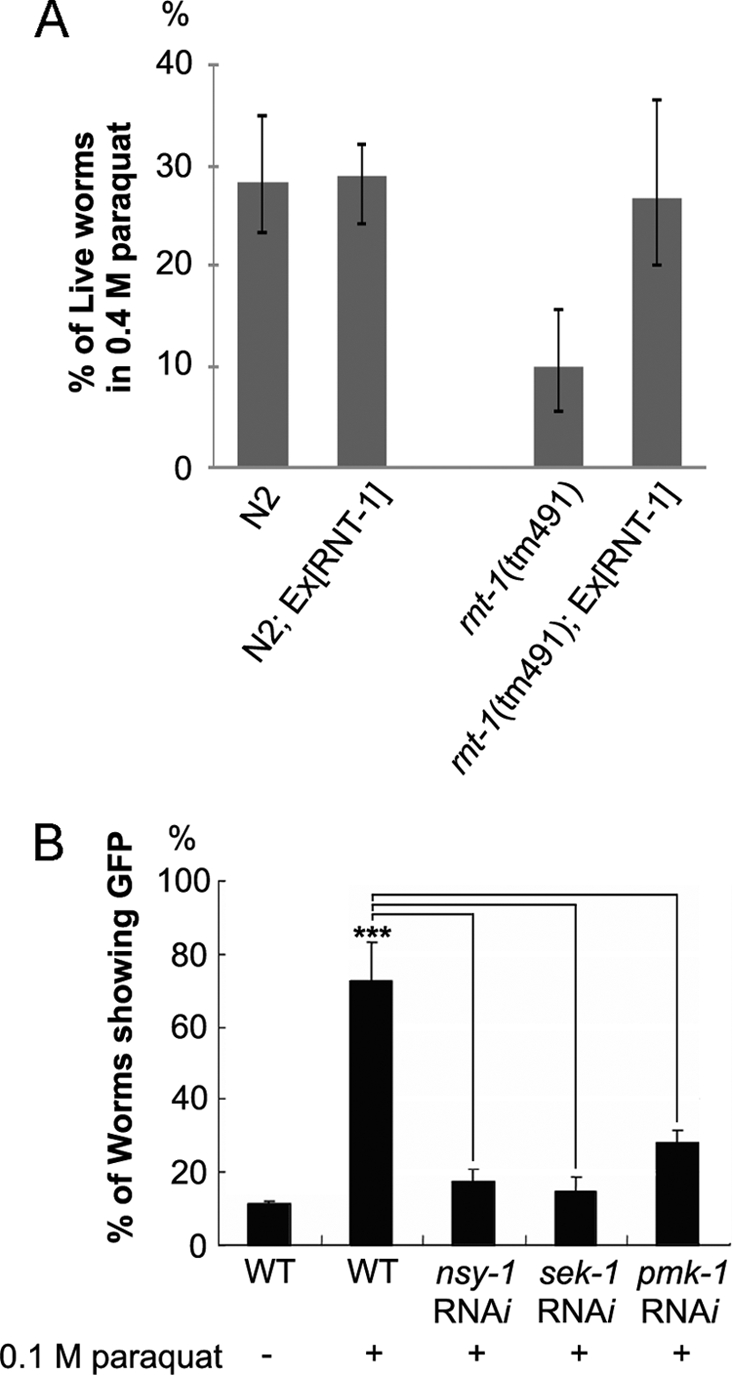
RNT-1 is required for response to oxidative stress. A, survival rates of N2 and rnt-1(tm491) animals after 5-min exposure to 0.4 m paraquat for acute oxidative stress. B, stabilization of RNT-1 in the intestine by oxidative stress is dependent on nsy-1, sek-1, and pmk-1. ***, p <0.001.
The MAPK Pathway Is Responsible for Regulating RNT-1 in the Stress Response
The p38 MAP kinase pathway is responsible for various stress responses in C. elegans (30, 33). To examine whether the p38 MAP kinase pathway is involved in the stabilization of RNT-1 in the presence of oxidative stress, we examined the knockdown effects of nsy-1, sek-1, and pmk-1, which encode MAP3K, MAP2K, and MAPK, respectively. We found that RNAi of any of these genes prevented accumulation of RNT-1 proteins in the intestine after exposure to each stress (Fig. 3B). We excluded the possibility of the involvement of other MAP kinases such as pmk-2, pmk-3 (p38), jnk-1 (JNK), and mpk-1 (ERK) (data not shown). However, RNAi of kgb-1 or mek-1 partially diminished the intensity of the stabilized GFP signal (supplemental Fig. 4), suggesting that these genes may be partially involved. Using an in vitro phosphorylation system, we found that purified SEK-1 and PMK-1, but not SEK-1 or PMK-1 alone, were able to phosphorylate RNT-1 (Fig. 4A). We also confirmed that the C-terminal region of the RNT-1 protein, which bears multiple serine/threonine sites, is responsible for phosphorylation-mediated modification by SEK-1/PMK-1, whereas the Runt domain was dispensable (Fig. 4B). These data suggest that p38 MAPK directly phosphorylates RNT-1 so that RNT-1 can be stabilized. To identify the phosphorylation sites of RNT-1, we first tested whether serine 255 is phosphorylated because p38 MAPK has a conserved phosphorylation site of Pro-X-Ser/Thr-Pro (Fig. 4C and supplemental Fig. 5A). We identified that the polypeptide containing the 251st to the 301st amino acid of RNT-1 was phosphorylated and that the mutation in serine 255 to alanine dramatically decreased the phosphorylation level. We also found that the polypeptide containing the 201st to the 251st amino acid of RNT-1 was phosphorylated (Fig. 4B), but we failed to find the phosphorylation site, although we tried three putative MAPK phosphorylation sites (supplemental Fig. 5B). These data raise the possibility of the existence of other phosphorylation site(s).
FIGURE 4.
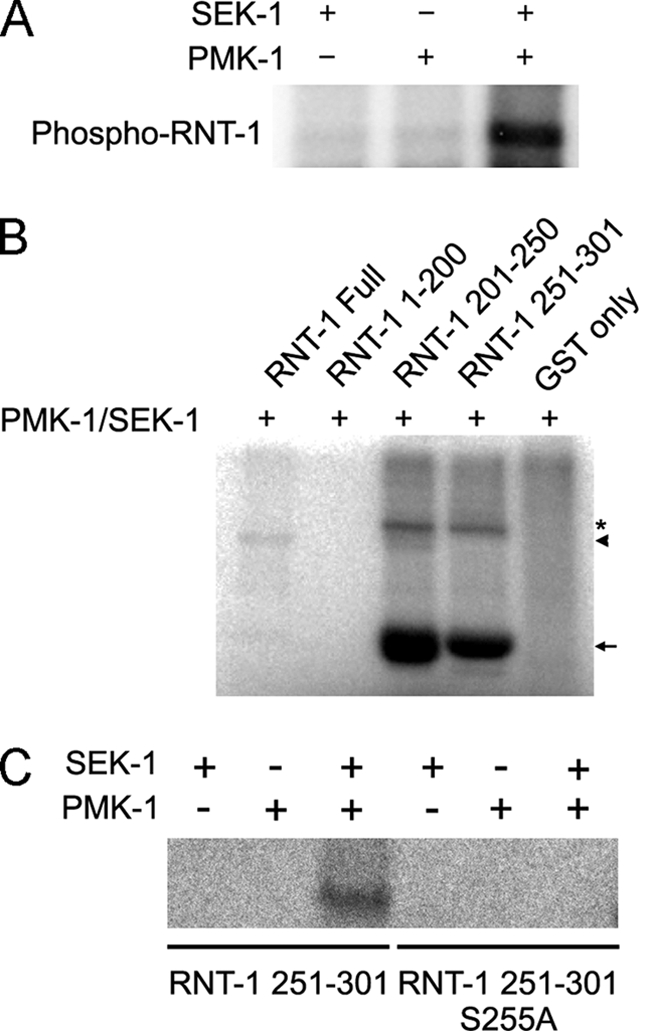
Purified PMK-1 can directly phosphorylate the C-terminal regions of RNT-1 in vitro. A, the result of the in vitro kinase assay of RNT-1 with SEK-1/PMK-1 kinases. B, domain studies of RNT-1 using GST-fused polypeptides containing 1–200, 201–250, or 251–301 amino acids. The asterisk is a nonspecific signal or a dimerized protein signal. The arrowhead is the signal of RNT-1 full sequence. The arrow is the GST-partial RNT-1 fusion protein signal. C, the result of the in vitro kinase assay with wild-type and the mutation of serine 255 to alanine of RNT-1.
vhp-1, a Phosphatase of the MAP Kinase Pathway, Is a Transcriptional Target of RNT-1
As a first attempt to identify downstream target genes of RNT-1 in response to stresses, we performed ChIP-sequencing experiments in the presence or absence of oxidative stress. Among the candidate target genes (listed in supplemental Table 2), we focused on vhp-1 because this gene encodes a phosphatase of MAP kinase. vhp-1 has been reported to function in various stress responses that are related to the MAP kinase pathway (34–37). All five conserved rnt-1 binding sites are at the promoter of vhp-1. We tested where RNT-1 actually binds by ChIP-quantitative PCR analysis. We found that the binding site between 9683 and 9731 nucleotides upstream of the translational start site of vhp-1, which bears two conserved RNT-1 binding sequences (marked A in Fig. 5A), was responsible for RNT-1 binding in vivo. Next, we checked whether transcription of vhp-1 was regulated by RNT-1 in the oxidative stress condition (Fig. 5B). In wild-type animals, the transcription of vhp-1 was increased by paraquat treatment. In rnt-1-mutant animals, however, the transcription level of vhp-1 was dramatically decreased, both in the presence and absence of paraquat treatment. We also tested whether the disruption of vhp-1 gene activity could affect the sensitivity to oxidative stress (Fig. 5C and supplemental Fig. 6). Knockdown of vhp-1 by RNAi increased the resistance to 100 mm paraquat in the wild-type background but only partially in the rnt-1 mutant background. The observation that RNAi of vhp-1 still caused a partially increased resistance in the rnt-1 mutant background suggests that vhp-1 is involved in rnt-1-independent response pathways. Transcription of vhp-1 was not activated by osmotic stress, and knockdown of vhp-1 did not show significant resistance to high osmolality (data not shown), suggesting that vhp-1 functions as a negative regulator of the oxidative stress response but not the osmotic stress response.
FIGURE 5.
vhp-1 is a target of RNT-1 in oxidative stress response. A, chromatin immunoprecipitation quantitative PCR analysis on the promoter of vhp-1. The arrows indicate the locations of the putative RNT-1 binding sequences. In A, two putative RNT-1 binding sequences are closely located. B, relative mRNA levels of vhp-1 in the absence or presence of 0.1 m paraquat in wild-type N2 and rnt-1 mutant animals. C, effects of vhp-1 RNAi on the survival rates of N2 and rnt-1(tm491) in M9 buffer containing 0.1 m paraquat. Additional experiments are presented in supplemental Fig. 4. **, p <0.01; ***, p <0.001.
A Negative Feedback Loop in the Oxidative Stress Response
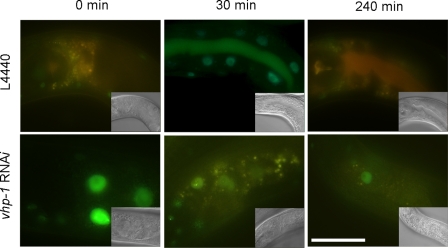
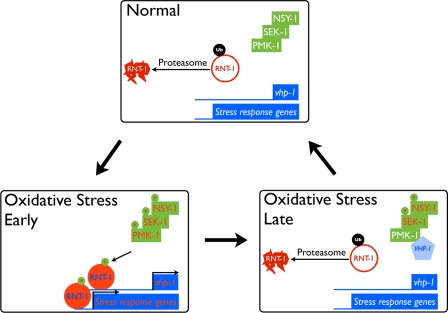
Even with continuous exposure to oxidative stress, the stabilization of RNT-1 proteins was abolished after 4 h (Fig. 2B). One possibility is that the transcriptional activation of vhp-1 by RNT-1 could in turn diminish the stabilization of RNT-1 by dephosphorylating PMK-1, resulting in a decrease in the amount of phosphorylated RNT-1 proteins. To test this, we measured the extent of the stabilization of RNT-1 GFP proteins after 100 mm paraquat treatment in the vhp-1 RNAi background (Fig. 6). In wild-type control animals, stabilized RNT-1 was increased by 30 min but abolished after 4-h treatment of 100 mm paraquat. In vhp-1 RNAi animals, however, RNT-1 proteins were detected in the intestine even before paraquat treatment. Moreover, RNT-1 remained stabilized after 4-h exposure. These results suggest that stabilized RNT-1 is diminished by the decrease in active PMK-1, which is mediated by vhp-1 transcription activated by RNT-1.
FIGURE 6.
The effect of the vhp-1 RNAi on the stability of RNT-1 in the oxidative stress condition. The photos show time course changes in the intestinal GFP of wild-type animals containing an RNT-1::GFP transgene in the control (L4440, upper panels) and vhp-1 RNAi background (bottom panels). Scale bar = 25 μm.
DISCUSSION
In this study, we revealed a novel function of RNT-1 in oxidative stress response in C. elegans. The data presented here show that ubiquitin-mediated degradation of RNT-1 is blocked by acute oxidative stress and that RNT-1 regulates stress response genes to increase viability. Stabilized RNT-1 activated the transcription of vhp-1, which, in turn, turns off the signal cascade of the p38 MAP kinase pathway. Overall, we propose a new role for rnt-1 in the stress response with a regulatory feedback loop involving vhp-1 (Fig. 7). Stabilized RNT-1 also activated stress response genes to cope with the stresses. The precise mechanism of the defensive role of RNT-1 target genes should be pursued further in the future. Further analysis of target genes in oxidative stress (supplemental Table 2) will extend the understanding of the role of RNT-1 in stress responses.
FIGURE 7.
A model of RNT-1 action in oxidative stress. Oxidative stress induces the transcription of vhp-1, a phosphatase, which, in turn, negatively regulates the stabilization of RNT-1 in time under the oxidative stress condition.
So far, studies of RNT-1 in the nematode have been focused on its role in development, such as proliferation and differentiation, and its transcriptional regulation. In this report, we, for the first time, showed that RNT-1 is controlled at the posttranslational level by environmental stress signals in the intestine. In C. elegans, one of the first organs that confront the changes of the environment is the intestine. Therefore, it is plausible that continuous expression and degradation of RNT-1 proteins in the intestine, not the regulation of rnt-1 expression at the level of transcription, may be beneficial to the worms so that they can more rapidly respond to the environmental changes to protect them from damage. Degradation of RNT-1 proteins, seemingly in vain, may not be a waste of energy in nature because it is probable that the wild environment is not always as favorable to the worms as the laboratory culture condition and that RNT-1 may not be degraded all the time in the wild.
The MAP kinase pathways have been identified as the signaling pathways that transduce the environmental changes from outside of the cell or of an organism to the inside (38). In particular, the p38 MAP kinase has been known to have pivotal roles in stress response, immunological regulation, apoptosis, senescence, and cell cycle checkpoint. The p38 MAP kinase pathway in C. elegans also has well conserved functions in development, various stress responses, and innate immunity (39, 40). Phosphorylation of PMK-1, an ortholog of p38 MAP kinase, is increased by oxidative stress, pathogens, and osmotic shock (30, 33, 41). Among the downstream effector genes of the PMK-1 pathway there are several key transcription factors. One such protein is SKN-1, which is translocated from the cytosol to the nucleus upon oxidative stress (42). Recently, another transcription factor, ATF-7, was reported as a regulator of innate immunity controlled by the PMK-1 pathway (43). Our finding that oxidative stress induced the stabilization of RNT-1 protein in the nucleus through the PMK-1 pathway adds to the repertoires of the MAP kinase-regulated stress response mechanism.
It is not clear at this point how phosphorylation of RNT-1 proteins can protect them from ubiquitination-mediated proteasome action. However, protein stability augmentation by phosphorylation is not unprecedented. For example, c-Jun, a component of the AP-1 transcription factor complex, is an oncoprotein and is tightly regulated by its degradation mediated by three E3 ligases, COP1, Itch, and Fbw7 (44–46). c-Jun N-terminal Kinases (JNKs) are one of the kinases of c-Jun and protect c-Jun from degradation (47). The specific characteristics of E3 ligase for RNT-1 and its dynamic regulatory mechanisms upon oxidative stress still remain to be established. Identifying the E3 ligase of RNT-1 may contribute to the understanding of a main concept regarding the immediate turnover mechanism of RUNX at the protein level. Excess responses and defects in the turning off of signaling events may cause diseases such as autoimmune disease and cancer development. The fact that RUNX family is involved in immune cell development and carcinoma in mammals encourages further study to discover a conserved feedback loop by posttranslational modification.
The MAP kinase phosphatase vhp-1, which detaches phosphorylation from MAP kinases, has been identified in other studies to regulate heavy metal stress and innate immunity in C. elegans (34, 35). We demonstrated the existence of the posttranslational modification mechanism for RNT-1 and a distinct type of regulatory feedback loop in the intestine of C. elegans.
Recently, there was a report that RUNX2 induces vesicular calcification in response to oxidative stress in mice (48), which supports the possibility of functional conservation in the oxidative stress response from nematodes to mammals. Others suggested that genetic alteration caused by impairment of the defense system against external chemicals or pathogenic bacteria, for example Helicobacter pylori, is the main cause of neoplastic transformation (49). In this context, RUNX3 might be required for various stress responses to protect the gastric epithelial cells from cellular transformation. At least in C. elegans, RNT-1 has a critical role in the stress response in the intestine, which is a primitive organ of mammalian gastrointestinal track systems. It would be interesting to examine the control of RUNX genes at the level of protein stability and their role in the stress response in addition to their developmental roles.
Supplementary Material
Acknowledgments
We thank H. Lee (Seoul National University) for a vector, A. Fire (Stanford University, CA) for vectors, J. Ahringer (MRC, London) for RNAi clones, and the National Bioresources Project (Tokyo, Japan) and the Caenorhabditis Genetics Center (Minneapolis, MN) for C. elegans strains.
This work was supported by the World Class University program and Research Center for Functional Cellulomics.

This article contains supplemental Tables 1 and 2, Figs. 1–6, and references.
- RUNX
- runt-related transcription factor
- MAPK
- Mitogen-activated Protein Kinase
- NGM
- nematode growth medium.
REFERENCES
- 1. Collins A., Littman D. R., Taniuchi I. (2009) RUNX proteins in transcription factor networks that regulate T-cell lineage choice. Nat. Rev. Immunol. 9, 106–115 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2. Stifani S., Ma Q. (2009) “Runxs and regulations” of sensory and motor neuron subtype differentiation. Implications for hematopoietic development. Blood Cells Mol. Dis. 43, 20–26 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3. Yoshida C. A., Komori T. (2005) Role of Runx proteins in chondrogenesis. Crit. Rev. Eukaryot. Gene Expr. 15, 243–254 [DOI] [PubMed] [Google Scholar]
- 4. Ito Y. (2008) RUNX genes in development and cancer. Regulation of viral gene expression and the discovery of RUNX family genes. Adv. Cancer Res. 99, 33–76 [DOI] [PubMed] [Google Scholar]
- 5. Cohen M. M., Jr. (2009) Perspectives on RUNX genes: an update. Am. J. Med. Genet. Part A 149A, 2629–2646 [DOI] [PubMed] [Google Scholar]
- 6. Lee Y. M. (2011) Control of RUNX3 by histone methyltransferases. J. Cell. Biochem. 112, 394–400 [DOI] [PubMed] [Google Scholar]
- 7. Villagra A., Gutiérrez J., Paredes R., Sierra J., Puchi M., Imschenetzky M., Wijnen Av A., Lian J., Stein G., Stein J., Montecino M. (2002) Reduced CpG methylation is associated with transcriptional activation of the bone-specific rat osteocalcin gene in osteoblasts. J. Cell. Biochem. 85, 112–122 [PubMed] [Google Scholar]
- 8. Kim W. J., Kim E. J., Jeong P., Quan C., Kim J., Li Q. L., Yang J. O., Ito Y., Bae S. C. (2005) RUNX3 inactivation by point mutations and aberrant DNA methylation in bladder tumors. Cancer Res. 65, 9347–9354 [DOI] [PubMed] [Google Scholar]
- 9. Lau Q. C., Raja E., Salto-Tellez M., Liu Q., Ito K., Inoue M., Putti T. C., Loh M., Ko T. K., Huang C., Bhalla K. N., Zhu T., Ito Y., Sukumar S. (2006) RUNX3 is frequently inactivated by dual mechanisms of protein mislocalization and promoter hypermethylation in breast cancer. Cancer Res. 66, 6512–6520 [DOI] [PubMed] [Google Scholar]
- 10. Kim E. J., Kim Y. J., Jeong P., Ha Y. S., Bae S. C., Kim W. J. (2008) Methylation of the RUNX3 promoter as a potential prognostic marker for bladder tumor. J. Urol. 180, 1141–1145 [DOI] [PubMed] [Google Scholar]
- 11. Jin Y. H., Jeon E. J., Li Q. L., Lee Y. H., Choi J. K., Kim W. J., Lee K. Y., Bae S. C. (2004) Transforming growth factor β stimulates p300-dependent RUNX3 acetylation, which inhibits ubiquitination-mediated degradation. J. Biol. Chem. 279, 29409–29417 [DOI] [PubMed] [Google Scholar]
- 12. Sierra J., Villagra A., Paredes R., Cruzat F., Gutierrez S., Javed A., Arriagada G., Olate J., Imschenetzky M., Van Wijnen A. J., Lian J. B., Stein G. S., Stein J. L., Montecino M. (2003) Regulation of the bone-specific osteocalcin gene by p300 requires Runx2/Cbfa1 and the vitamin D3 receptor but not p300 intrinsic histone acetyltransferase activity. Mol. Cell. Biol. 23, 3339–3351 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13. Huang G., Shigesada K., Ito K., Wee H. J., Yokomizo T., Ito Y. (2001) Dimerization with PEBP2β protects RUNX1/AML1 from ubiquitin-proteasome-mediated degradation. EMBO J. 20, 723–733 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14. Shen R., Wang X., Drissi H., Liu F., O'Keefe R. J., Chen D. (2006) Cyclin D1-cdk4 induce runx2 ubiquitination and degradation. J. Biol. Chem. 281, 16347–16353 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15. Chi X. Z., Kim J., Lee Y. H., Lee J. W., Lee K. S., Wee H., Kim W. J., Park W. Y., Oh B. C., Stein G. S., Ito Y., van Wijnen A. J., Bae S. C. (2009) Runt-related transcription factor RUNX3 is a target of MDM2-mediated ubiquitination. Cancer Res. 69, 8111–8119 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16. Wee H. J., Huang G., Shigesada K., Ito Y. (2002) Serine phosphorylation of RUNX2 with novel potential functions as negative regulatory mechanisms. EMBO Rep. 3, 967–974 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17. Qiao M., Shapiro P., Fosbrink M., Rus H., Kumar R., Passaniti A. (2006) Cell cycle-dependent phosphorylation of the RUNX2 transcription factor by cdc2 regulates endothelial cell proliferation. J. Biol. Chem. 281, 7118–7128 [DOI] [PubMed] [Google Scholar]
- 18. Bae S. C., Lee J. (2000) cDNA cloning of run, a Caenorhabditis elegans Runt domain-encoding gene. Gene 241, 255–258 [DOI] [PubMed] [Google Scholar]
- 19. Nam S., Jin Y. H., Li Q. L., Lee K. Y., Jeong G. B., Ito Y., Lee J., Bae S. C. (2002) Expression pattern, regulation, and biological role of runt domain transcription factor, run, in Caenorhabditis elegans. Mol. Cell. Biol. 22, 547–554 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20. Nimmo R., Antebi A., Woollard A. (2005) Mab-2 encodes RNT-1, a C. elegans Runx homologue essential for controlling cell proliferation in a stem cell-like developmental lineage. Development 132, 5043–5054 [DOI] [PubMed] [Google Scholar]
- 21. Shim J., Lee J. (2008) Regulation of rnt-1 expression mediated by the opposing effects of BRO-1 and DBL-1 in the nematode Caenorhabditis elegans. Biochem. Biophys. Res. Commun. 367, 130–136 [DOI] [PubMed] [Google Scholar]
- 22. Brenner S. (1974) The genetics of Caenorhabditis elegans. Genetics 77, 71–94 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23. Mello C. C., Kramer J. M., Stinchcomb D., Ambros V. (1991) Efficient gene transfer in C. elegans. Extrachromosomal maintenance and integration of transforming sequences. EMBO J. 10, 3959–3970 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24. Kramer J. M., French R. P., Park E. C., Johnson J. J. (1990) The Caenorhabditis elegans rol-6 gene, which interacts with the sqt-1 collagen gene to determine organismal morphology, encodes a collagen. Mol. Cell. Biol. 10, 2081–2089 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25. Seydoux G., Fire A. (1995) Whole-mount in situ hybridization for the detection of RNA in Caenorhabditis elegans embryos. Methods Cell Biol. 48, 323–337 [DOI] [PubMed] [Google Scholar]
- 26. Bush K. T., Goldberg A. L., Nigam S. K. (1997) Proteasome inhibition leads to a heat shock response, induction of endoplasmic reticulum chaperones, and thermotolerance. J. Biol. Chem. 272, 9086–9092 [DOI] [PubMed] [Google Scholar]
- 27. Courbard J. R., Fiore F., Adélaïde J., Borg J. P., Birnbaum D., Ollendorff V. (2002) Interaction between two ubiquitin-protein isopeptide ligases of different classes, CBLC and AIP4/ITCH. J. Biol. Chem. 277, 45267–45275 [DOI] [PubMed] [Google Scholar]
- 28. Rocca A., Lamaze C., Subtil A., Dautry-Varsat A. (2001) Involvement of the ubiquitin/proteasome system in sorting of the interleukin 2 receptor β chain to late endocytic compartments. Mol. Biol. Cell 12, 1293–1301 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29. Dillin A., Crawford D. K., Kenyon C. (2002) Timing requirements for insulin/IGF-1 signaling in C. elegans. Science 298, 830–834 [DOI] [PubMed] [Google Scholar]
- 30. Solomon A., Bandhakavi S., Jabbar S., Shah R., Beitel G. J., Morimoto R. I. (2004) Caenorhabditis elegans OSR-1 regulates behavioral and physiological responses to hyperosmotic environments. Genetics 167, 161–170 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31. Akiba H., Nakano H., Nishinaka S., Shindo M., Kobata T., Atsuta M., Morimoto C., Ware C. F., Malinin N. L., Wallach D., Yagita H., Okumura K. (1998) CD27, a member of the tumor necrosis factor receptor superfamily, activates NF-κB and stress-activated protein kinase/c-Jun N-terminal kinase via TRAF2, TRAF5, and NF-κB-inducing kinase. J. Biol. Chem. 273, 13353–13358 [DOI] [PubMed] [Google Scholar]
- 32. Oh S. W., Mukhopadhyay A., Dixit B. L., Raha T., Green M. R., Tissenbaum H. A. (2006) Identification of direct DAF-16 targets controlling longevity, metabolism and diapause by chromatin immunoprecipitation. Nat. Genet. 38, 251–257 [DOI] [PubMed] [Google Scholar]
- 33. Inoue H., Hisamoto N., An J. H., Oliveira R. P., Nishida E., Blackwell T. K., Matsumoto K. (2005) The C. elegans p38 MAPK pathway regulates nuclear localization of the transcription factor SKN-1 in oxidative stress response. Genes Dev. 19, 2278–2283 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34. Mizuno T., Hisamoto N., Terada T., Kondo T., Adachi M., Nishida E., Kim D. H., Ausubel F. M., Matsumoto K. (2004) The Caenorhabditis elegans MAPK phosphatase VHP-1 mediates a novel JNK-like signaling pathway in stress response. EMBO J. 23, 2226–2234 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35. Kim D. H., Liberati N. T., Mizuno T., Inoue H., Hisamoto N., Matsumoto K., Ausubel F. M. (2004) Integration of Caenorhabditis elegans MAPK pathways mediating immunity and stress resistance by MEK-1 MAPK kinase and VHP-1 MAPK phosphatase. Proc. Natl. Acad. Sci. U.S.A. 101, 10990–10994 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36. Richardson C. E., Kooistra T., Kim D. H. (2010) An essential role for XBP-1 in host protection against immune activation in C. elegans. Nature 463, 1092–1095 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37. Rohlfing A. K., Miteva Y., Hannenhalli S., Lamitina T. (2010) Genetic and physiological activation of osmosensitive gene expression mimics transcriptional signatures of pathogen infection in C. elegans. PLoS ONE 5, e9010. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38. Plotnikov A., Zehorai E., Procaccia S., Seger R. (2011) The MAPK cascades. Signaling components, nuclear roles and mechanisms of nuclear translocation. Biochim. Biophys. Acta 1813, 1619–1633 [DOI] [PubMed] [Google Scholar]
- 39. Sundaram M. V. (2006) RTK/Ras/MAPK signaling. WormBook, 1–1918050474 [Google Scholar]
- 40. Ewbank J. J. (2006) Signaling in the immune response. WormBook, 1–12 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41. Kim D. H., Feinbaum R., Alloing G., Emerson F. E., Garsin D. A., Inoue H., Tanaka-Hino M., Hisamoto N., Matsumoto K., Tan M. W., Ausubel F. M. (2002) A conserved p38 MAP kinase pathway in Caenorhabditis elegans innate immunity. Science 297, 623–626 [DOI] [PubMed] [Google Scholar]
- 42. An J. H., Blackwell T. K. (2003) SKN-1 links C. elegans mesendodermal specification to a conserved oxidative stress response. Genes Dev. 17, 1882–1893 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43. Shivers R. P., Pagano D. J., Kooistra T., Richardson C. E., Reddy K. C., Whitney J. K., Kamanzi O., Matsumoto K., Hisamoto N., Kim D. H. (2010) Phosphorylation of the conserved transcription factor ATF-7 by PMK-1 p38 MAPK regulates innate immunity in Caenorhabditis elegans. PLoS Genet. 6, e1000892. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44. Dornan D., Wertz I., Shimizu H., Arnott D., Frantz G. D., Dowd P., O'Rourke K., Koeppen H., Dixit V. M. (2004) The ubiquitin ligase COP1 is a critical negative regulator of p53. Nature 429, 86–92 [DOI] [PubMed] [Google Scholar]
- 45. Gao M., Labuda T., Xia Y., Gallagher E., Fang D., Liu Y. C., Karin M. (2004) Jun turnover is controlled through JNK-dependent phosphorylation of the E3 ligase Itch. Science 306, 271–275 [DOI] [PubMed] [Google Scholar]
- 46. Nateri A. S., Riera-Sans L., Da Costa C., Behrens A. (2004) The ubiquitin ligase SCFFbw7 antagonizes apoptotic JNK signaling. Science 303, 1374–1378 [DOI] [PubMed] [Google Scholar]
- 47. Musti A. M., Treier M., Bohmann D. (1997) Reduced ubiquitin-dependent degradation of c-Jun after phosphorylation by MAP kinases. Science 275, 400–402 [DOI] [PubMed] [Google Scholar]
- 48. Byon C. H., Javed A., Dai Q., Kappes J. C., Clemens T. L., Darley-Usmar V. M., McDonald J. M., Chen Y. (2008) Oxidative stress induces vascular calcification through modulation of the osteogenic transcription factor Runx2 by AKT signaling. J. Biol. Chem. 283, 15319–15327 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49. Friedrich M. J., Rad R., Langer R., Voland P., Hoefler H., Schmid R. M., Prinz C., Gerhard M. (2006) Lack of RUNX3 regulation in human gastric cancer. J. Pathol. 210, 141–146 [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.