Abstract
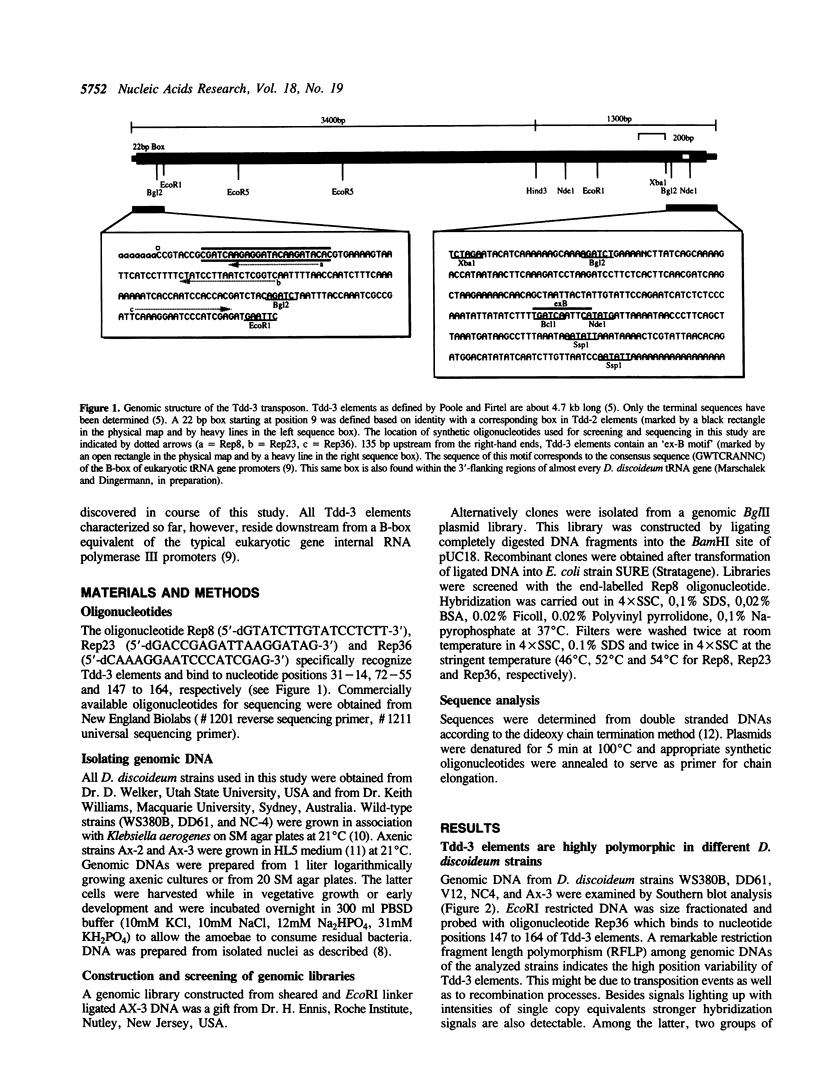
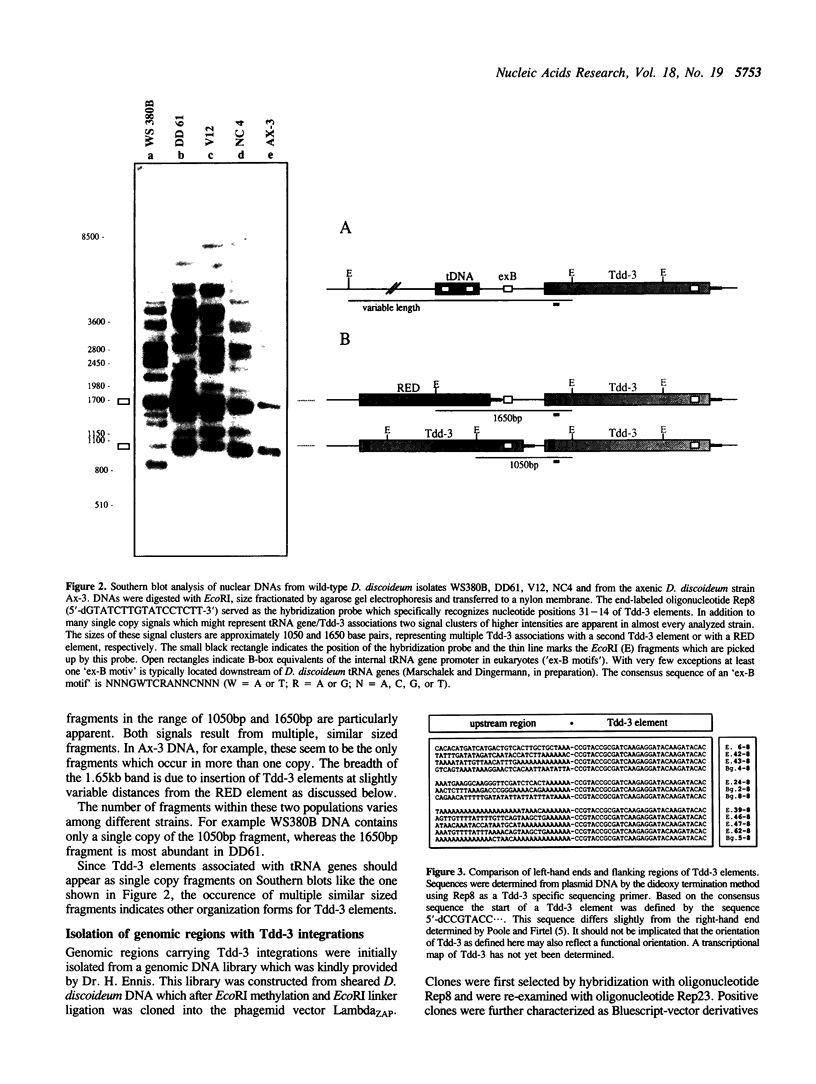
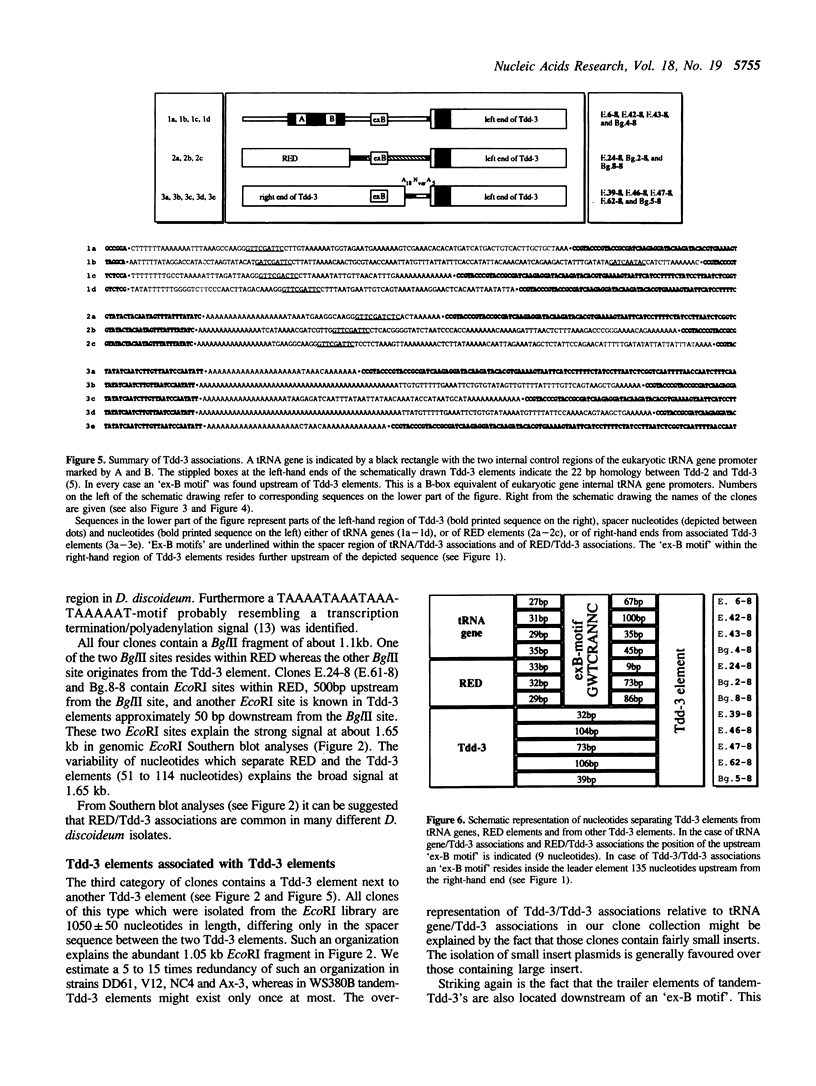
The transposable element Tdd-3 from D. discoideum has been described originally in 1984 (Poole and Firtel, 1984). Additional copies of this element were discovered in the course of a recent study on tRNA gene organization in D. discoideum. Five out of 24 independently isolated tRNA genes proved to be associated with Tdd-3 elements. The surprising observation that all the elements always occurred within the 3'-flanking sequences of the Dictyostelium tRNA genes suggested the possibility of a general position specific integration of Tdd-3 elements upon transposition. Therefore we isolated additional Tdd-3 elements from various genomic D. discoideum libraries in order to test this hypothesis. Several new Tdd-3 elements were found associated with various tRNA genes. Additionally we identified Tdd-3 elements organized in tandem array or in association with RED (Repetitive Element of Dictyostelium), another repetitive element recently identified by our laboratory. In all cases a B-box equivalent of the eukaryotic gene-internal RNA polymerase III promoter was identified upstream of all Tdd-3 elements.
Full text
PDF



Images in this article
Selected References
These references are in PubMed. This may not be the complete list of references from this article.
- Aksoy S., Williams S., Chang S., Richards F. F. SLACS retrotransposon from Trypanosoma brucei gambiense is similar to mammalian LINEs. Nucleic Acids Res. 1990 Feb 25;18(4):785–792. doi: 10.1093/nar/18.4.785. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dingermann T., Amon-Böhm E., Bertling W., Marschalek R., Nerke K. A family of non-allelic tRNA(ValGUU) genes from the cellular slime mold Dictyostelium discoideum. Gene. 1988 Dec 20;73(2):373–384. doi: 10.1016/0378-1119(88)90502-1. [DOI] [PubMed] [Google Scholar]
- Dingermann T., Amon E., Williams K. L., Welker D. L. Chromosomal mapping of tRNA genes from Dictyostelium discoideum. Mol Gen Genet. 1987 Apr;207(1):176–187. doi: 10.1007/BF00331507. [DOI] [PubMed] [Google Scholar]
- Dingermann T., Brechner T., Marschalek R., Amon-Böhm E., Welker D. L. tRNAGlu(GAA) genes from the cellular slime mold Dictyostelium discoideum. DNA. 1989 Apr;8(3):193–204. doi: 10.1089/dna.1.1989.8.193. [DOI] [PubMed] [Google Scholar]
- Geiduschek E. P., Tocchini-Valentini G. P. Transcription by RNA polymerase III. Annu Rev Biochem. 1988;57:873–914. doi: 10.1146/annurev.bi.57.070188.004301. [DOI] [PubMed] [Google Scholar]
- Ikenaga H., Saigo K. Insertion of a movable genetic element, 297, into the T-A-T-A box for the H3 histone gene in Drosophila melanogaster. Proc Natl Acad Sci U S A. 1982 Jul;79(13):4143–4147. doi: 10.1073/pnas.79.13.4143. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Inouye S., Yuki S., Saigo K. Sequence-specific insertion of the Drosophila transposable genetic element 17.6. 1984 Jul 26-Aug 1Nature. 310(5975):332–333. doi: 10.1038/310332a0. [DOI] [PubMed] [Google Scholar]
- Kassavetis G. A., Braun B. R., Nguyen L. H., Geiduschek E. P. S. cerevisiae TFIIIB is the transcription initiation factor proper of RNA polymerase III, while TFIIIA and TFIIIC are assembly factors. Cell. 1990 Jan 26;60(2):235–245. doi: 10.1016/0092-8674(90)90739-2. [DOI] [PubMed] [Google Scholar]
- Marschalek R., Brechner T., Amon-Böhm E., Dingermann T. Transfer RNA genes: landmarks for integration of mobile genetic elements in Dictyostelium discoideum. Science. 1989 Jun 23;244(4911):1493–1496. doi: 10.1126/science.2567533. [DOI] [PubMed] [Google Scholar]
- Poole S. J., Firtel R. A. Genomic instability and mobile genetic elements in regions surrounding two discoidin I genes of Dictyostelium discoideum. Mol Cell Biol. 1984 Apr;4(4):671–680. doi: 10.1128/mcb.4.4.671. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sandmeyer S. B., Bilanchone V. W., Clark D. J., Morcos P., Carle G. F., Brodeur G. M. Sigma elements are position-specific for many different yeast tRNA genes. Nucleic Acids Res. 1988 Feb 25;16(4):1499–1515. doi: 10.1093/nar/16.4.1499. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sanger F., Nicklen S., Coulson A. R. DNA sequencing with chain-terminating inhibitors. Proc Natl Acad Sci U S A. 1977 Dec;74(12):5463–5467. doi: 10.1073/pnas.74.12.5463. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sharp S. J., Schaack J., Cooley L., Burke D. J., Söll D. Structure and transcription of eukaryotic tRNA genes. CRC Crit Rev Biochem. 1985;19(2):107–144. doi: 10.3109/10409238509082541. [DOI] [PubMed] [Google Scholar]
- Watts D. J., Ashworth J. M. Growth of myxameobae of the cellular slime mould Dictyostelium discoideum in axenic culture. Biochem J. 1970 Sep;119(2):171–174. doi: 10.1042/bj1190171. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Weiner A. M., Deininger P. L., Efstratiadis A. Nonviral retroposons: genes, pseudogenes, and transposable elements generated by the reverse flow of genetic information. Annu Rev Biochem. 1986;55:631–661. doi: 10.1146/annurev.bi.55.070186.003215. [DOI] [PubMed] [Google Scholar]
- Welker D. L., Williams K. L. A genetic map of Dictyostelium discoideum based on mitotic recombination. Genetics. 1982 Dec;102(4):691–710. doi: 10.1093/genetics/102.4.691. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Xiong Y. E., Eickbush T. H. Functional expression of a sequence-specific endonuclease encoded by the retrotransposon R2Bm. Cell. 1988 Oct 21;55(2):235–246. doi: 10.1016/0092-8674(88)90046-3. [DOI] [PubMed] [Google Scholar]
- Xiong Y., Eickbush T. H. The site-specific ribosomal DNA insertion element R1Bm belongs to a class of non-long-terminal-repeat retrotransposons. Mol Cell Biol. 1988 Jan;8(1):114–123. doi: 10.1128/mcb.8.1.114. [DOI] [PMC free article] [PubMed] [Google Scholar]



