Abstract
Background
Despite the popularity of zebrafish as a research model, its sex determination (SD) mechanism is still unknown. Most cytogenetic studies failed to find dimorphic sex chromosomes and no primary sex determining switch has been identified even though the assembly of zebrafish genome sequence is near to completion and a high resolution genetic map is available. Recent publications suggest that environmental factors within the natural range have minimal impact on sex ratios of zebrafish populations. The primary aim of this study is to find out more about how sex is determined in zebrafish.
Methodology/Principal Findings
Using classical breeding experiments, we found that sex ratios across families were wide ranging (4.8% to 97.3% males). On the other hand, repeated single pair crossings produced broods of very similar sex ratios, indicating that parental genotypes have a role in the sex ratio of the offspring. Variation among family sex ratios was reduced after selection for breeding pairs with predominantly male or female offspring, another indication that zebrafish sex is regulated genetically. Further examinations by a PCR-based “blind assay" and array comparative genomic hybridization both failed to find universal sex-linked differences between the male and female genomes. Together with the ability to increase the sex bias of lines by selective breeding, these data suggest that zebrafish is unlikely to utilize a chromosomal sex determination (CSD) system.
Conclusions/Significance
Taken together, our study suggests that zebrafish sex is genetically determined with limited, secondary influences from the environment. As we have not found any sign for CSD in the species, we propose that the zebrafish has a polygenic sex determination system.
Introduction
Sex determination (SD) establishes the sexual fate of an organism and initiates the gonad differentiation process (reviews: [1], [2], [3]). A variety of signals, including genetic, environmental or even social cues, were found to be sex determinants in vertebrates (see reviews [4], [5], [6]).
The most extensively studied mode of genetic SD is chromosomal sex determination (CSD) as in mammalian and avian species, for example. In this system, sex is determined by a primary switch located on one or both members of a well-differentiated sex chromosomal pair (see e.g. [7], [8], [9], [10]). Since in mammals, including humans, sex is determined by CSD, the vast majority of our knowledge on the molecular regulation of vertebrate sex is based on data collected from such systems.
In the other type of genetic sex determination system, called polygenic (multigenic or multifactorial) sex determination (PGSD), the genes with strong influence on sex determination and/or gonad differentiation are distributed throughout the genome and the combination of their alleles determines the sex of the individual [11], [12]. This form of sex determination has not been studied extensively at the experimental level (for exceptions, see e.g. [13], [14]): European seabass [15] and a handful of cichlid species from Lake Malawi [16] are the only fish species that were shown to utilise this system to date.
Sex can also be determined by signals from the environment and there are several environmental effects known to influence sex of an organism. Temperature is one of the most commonly studied environmental cues for sex determination. In many reptiles, sex is determined by environmental temperature during the thermosensitive periods of embryo development or egg incubation (for reviews see [17], [18]). Examples of other environmental cues that also have an influence on sex include pH in guppy [19] and social interactions in some reef fishes [20].
Over the past decades, zebrafish (Danio rerio) has become an important laboratory model organism for many areas of research (for examples see e.g.: [21], [22], [23], [24], [25]). Despite being a popular model for developmental biology and biomedical research, very little is known about its sexual development (for review see [26]). Moreover, most of the current knowledge on zebrafish sexual development is related to its gonad differentiation (for reviews see [27], [28]) while the mode of its sex determination is still disputed.
Most cytogenetic studies showed that the zebrafish has chromosomes of similar size and morphology. This lack of distinct morphological differences together with poor karyotype banding pattern resulted in difficulties with accurately assigning chromosomal pairs (for review see [29]). Therefore, it is not easy to search for sex chromosomes based on size differences, their distinct trademark in most mammalian and avian species. An alternative approach to cytogenetic approaches would be to search for differences between the two sexes at the level of the whole genome. PCR-based methods such as random amplified polymorphic DNA (RAPD; [30], [31]) and amplified fragment length polymorphism (AFLP; [32]) have been used successfully for identification of sex markers in fishes (see e.g. [33], [34], [35]) and other vertebrates (see e.g. [36], [37], [38]). Earlier, we have developed a new PCR-based mass genotyping technique called fluorescent motif enhanced polymorphism (FluoMEP; [39]) that combines the advantages of RAPD and AFLP. In this study, we have utilized this technology to search for sex-linked DNA markers in the genome of three fish species, including zebrafish.
Another molecular tool used for comparing the male and female zebrafish genome in this study is array comparative genome hybridization (aCGH). This method allows for the detection of differences, called copy number variations (CNV) [40], [41], between two complex DNA samples on a genome-wide scale [42], [43]. The method is based on hybridization of two samples onto a ‘tiling array’ that contains probes scanning through the whole genome at regular intervals. Originally, aCGH was developed for the analysis of chromosomes aberrations in cancer cells [43], [44]. Over the years, this method has also been utilized for various purposes such as studying evolution [45], [46], understanding the impact of CNV on transcriptome [47] and isolation of molecular markers [48].
The aim of this study was to perform a detailed analysis on zebrafish sex determination, by combining the power of traditional and molecular technologies. Through analysis of sex ratios in a large number of families, we show that i) sex ratios vary among different families; ii) parental genotypes have a major effect on the sex ratio; and iii) one of the two sexes can be depleted through systematic selection in a few generations. Moreover, PCR-based screens and aCGH performed by a custom-designed tiling array were both unable to find general differences between the genome of the two sexes in two different zebrafish strains. The above data all point towards a genetic mechanism of sex determination and the lack of a chromosomal sex determination system in the zebrafish. We, therefore, propose that zebrafish sex determination is polygenic.
Materials and Methods
Fish stocks and tail fin samples
Experiments performed at Temasek Life Sciences Laboratory were approved by Temasek Life Sciences Laboratory Institutional Animal Care and Use Committee (approval ID: TLL(F)-10-001) and performed according to its guidelines. Experiments performed at Max-Planck-Institut für Entwicklungsbiologie were registered at Regierungspräsidium Tübingen (approval ID 35/9185.46) and carried out according to the Protection of Animals Act (Tierschutzgesetz) and its guidelines. Zebrafish (Danio rerio) of the AB strain, Tübingen strain and a wild type strain, called Toh, purchased from a local aquarium shop were used in this study. All zebrafish were kept in AHAB (Aquatic Habitats, Apopka, FL, USA) recirculation systems according to standard protocols [49], with the exception of the population density study in which fish were raised in an Aqua Schwartz system. Guppy (Poecilia reticulata) fin clips from visually sexed individuals were kind gifts from Dr. Rob Brooks (UNSW, Sydney, Australia). Visually sexed rosy barb (Puntius conchonius) individuals were purchased from a local fish trading company (Qian Hu Fish Farm, Singapore). Their tail fin samples were collected under anesthesia and stored in absolute ethanol at −20°C until use.
Fish husbandry
Adult zebrafish were kept as mixed sex groups in 2.75 L tanks at a density of <10 individuals per liter. Breeding was carried out in meshed-bottom mouse cages of one liter volume placed into a second cage containing egg water. Breeding pairs were set up at the previous evening in the presence of artificial plants and eggs were collected before noon the next day. Pairs that were reluctant to yield eggs were given a slight cold shock by adding ice-cold egg water (about 20% of the tank volume) 1–2 hours after the start of breeding period. Ripe females that failed to produce eggs with two different males were gently squeezed to aid the removal of eggs, if any, potentially ‘stuck’ in their body and set up for repeated mating one week later.
Fertilized eggs were collected from the bottom of the cage, rinsed on a tea filter and transferred into plastic trays with egg water containing methylene blue. Survivals were recorded at 24 and 48 days post fertilization (dpf). Batches with survival below 50% during this period were discarded and their parents were crossed again later. Embryos were transferred onto the AHAB system before hatching and they were grown there at the following densities (unless indicated otherwise): <100/L for embryos, <80/L for larvae, <20/L for juveniles and <10/L for young adults.
Sexing zebrafish
Zebrafish were sexed visually, based on the following two criteria (unless otherwise noted): i) general body shape; and ii) the presence of ‘genital papilla’ (or cloacal protrusion; [50]) in females (observed on unstressed fish kept in water). Individuals with intermediate body shape and poorly observable papilla were gently squeezed and checked for eggs or sperm. In absence of either, individuals were culled, dissected under a stereo microscope and their gonad was analyzed. Those individuals with unclear sex were not included in the calculation of sex ratio. Only 7 out of the 62 families sexed had such individuals and their ratio was typically less than 5%.
Selection experiment for increased sex bias
We have performed a multi-generation selection experiment in order to increase sex bias. Based on the sex ratio of the offspring we have chosen five lines to be used for selection against males or females (Fig. S1). Pairwise full-sib crosses were performed according to a multifactorial design. The offspring were sexed at about 3 months of age and family sex ratio was recorded. From crosses that produced highly skewed sex ratio, usually three robust males and three robust females were chosen as brooders to produce the next generation.
Population density effects on sex ratio
Rearing density experiments were performed using the Tübingen strain. Embryos and larva were raised at 29°C in petri dishes until 5 dpf at which time they were transferred to 1.5 L of fish water at varying densities. At 10 dpf, 600 ml of fish water was added to each tank to facilitate counting of larvae. At approximately 14 dpf, larval were placed into circulating water, resulting in 2 L of water per tank. From 5 dpf to about 14 dpf, larva were fed powdered fry food two times daily after which time the food source was changed to freshly hatched Artemia nauplia.
Two experiments were performed to test the effect of rearing density on sex ratios. In experiment 1, a total of 44 populations were analyzed spanning a period of about four months. In experiment 2, 30 populations were analyzed spanning about six and a half months. Embryos were collected from pairwise matings on day 0. In experiment 1, embryos from 2 or 3 crosses were often pooled, however more than one pool was often collected per day. Embryos were then sorted into petri dishes containing 50 embryos. In the second experiment, all embryos collected on a given day were pooled before sorting into dishes containing 50 embryos each. On day 5 dpf, larvae were set out at densities of 100, 50 or 25 larvae in 1.5 L of fish water. In experiment 2, the same number of tanks per density were set up out on a given day (e.g. 2 tanks with 100 larvae, 2 tanks with 50 larvae and 2 tanks with 25 larvae) whereas in experiment 1 the number of tanks per density per day was not controlled (Table S1 and Fig. S2). Overall, the difference in the design of the two experiments should have resulted in a lesser degree of genetic diversity between populations in experiment 2 compared to experiment 1. Larva and juvenile fish were counted every 10 days from 10 dpf up to 30 or 40 dpf for most (Table S1 and Fig. S2). In initial experiments, little to no lethality was observed after 30 dpf thus, in experiment 2 counting ceased after 30 dpf for most populations (Table S1 and Fig. S2). All fish were raised to adulthood and then sexed. For the first 30 populations, fish were sexed by dissection and observation of the gonad and subsequent populations were sexed based on coloration.
FluoMEP assay
Genomic DNA (gDNA) samples were extracted from tail fins by digesting them at 55°C overnight in 800 µl of SET buffer (0.5% SDS, 50 mM EDTA, 10 mM Tris/Cl pH 8.0, 200 mM NaCl) and 250 µg/ml Proteinase K (Roche Diagnostics, Indianapolis, IN, USA). Then standard phenol chloroform extraction [51] was performed and the gDNA pellet was dissolved in 100 µL of 1× TE. Pooled male and female samples were generated by combining equal quantity of individual gDNA samples (nine individuals of rosy barb, eight individuals of guppy and four individuals of zebrafish for each sex). FluoMEP screening was carried out as described previously [39]. Bulk segregant analysis [52] was performed using the male and female pooled gDNA samples to screen for potentially sex-linked markers. Potential markers were then subjected to an additional round of analysis on the individuals that formed the pooled samples for confirmation.
Array comparative genomic hybridization
Four families of zebrafish were used for aCGH and each family consisted of the parents, two male offspring and two female offspring individuals. Two of the families were from the AB strain and the other two were from the Toh strain. Genomic DNA samples for aCGH were extracted from tail fins using DNeasy Blood and Tissue kit (Qiagen) according to the manufacturer's instructions with slight modification. Instead of 2 hours incubation, tail fin samples were incubated overnight at 55°C in lysis buffer AL and proteinase K with slow shaking (70 rpm). The quality of extracted gDNA samples was checked on Nanodrop 1000 (Thermo Scientific, Waltham, MA, USA) and by agarose gel electrophoresis.
Individual samples were labelled with NimbleGen Dual-Color DNA Labelling Kit (Roche NimbleGen, Madison, WI, USA) and hybridization was carried out according to the manufacturer's instructions with MAUI hybridization system (BioMicro Systems, Salt Lake City, UT, USA). The oligo array was custom-designed by NimbleGen (Roche NimbleGen) based on zebrafish Zv7 (danre5) genome assembly. During the course of this study Zv8 (danre6) was released, all probes were re-mapped onto the new assembly for data analysis. Each array contained 120 thousand probes (55–70mers) with median spacing of about 10 kb. As preliminary tests have confirmed the accuracy of our procedure, no technical replicates were used for reasons of cost-efficiency.
The array was scanned at 5 µm resolution with Axon GenePix 4000B Microarray scanner (Molecular Devices, Sunnyvale, CA, USA). Raw fluorescent intensity data was retrieved by NimbleScan software (Roche NimbleGen) then imported into Partek Genomic Suite software (Partek Incorporated, St. Louis, MO, USA) for analysis. For copy number detection the genomic segmentation algorithm was used. A minimum of 5 markers were specified, P value threshold set at 0.001 and signal-to-noise ratio set at 0.3. All data was collected according to MIAME guidelines and deposited in NCBI GEO (GSE34338).
Validation of aCGH results
For validation, we used the same gDNA samples that were analyzed by aCGH, plus an additional male and female offspring per family were included. All PCR primers (see Table S2 for primer sequences) were designed to target a sub-region of the copy number variable region (CNVR) using Primer3 version 0.4.0 [53]. Quantifast Probe PCR kit (Qiagen) was used for PCR validation of CNVR2 and CNVR5. The amplified products were then analysed by 2% agarose gel. A single-copy exon (DrSC23) was used as reference. For validation of CNVR3, real time quantitative PCR was carried out as described previously [54] using MyIQ real-time PCR detection system (Biorad, Hercules, CA, USA) with IQ Sybr Green Supermix (Biorad). Samples were normalized against two reference loci (DrSC19 and DrSC23), both of which were found to be a single-copy exon [55]. Relative quantification was calculated to estimate gain or loss of copy number with reference to paternal gDNA sample [54].
Results
Wide-ranging sex ratios among zebrafish families
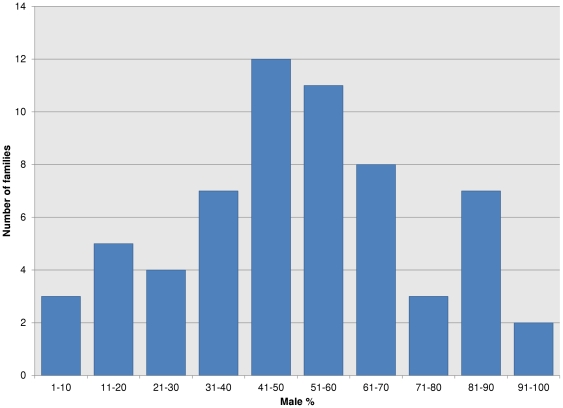
The classical method to determine if a species is using chromosomal sex determination system is to analyze the sex ratio among many families [5]. In the presence of strong CSD, the sex ratio is expected to be close to 50% [5]. In order to elucidate whether CSD is the main sex determination system in zebrafish, the sex ratios of 62 families were analyzed. The percentage of males among the families analyzed ranged from 4.8% to 97.3% with median of 51% (std. dev. ±22.6%; Fig. 1). Such a wide-ranging sex ratio among the families would be highly unusual for a predominantly sex chromosomal system.
Figure 1. Wide-ranging sex ratios were observed among 62 zebrafish families.
We have crossed randomly picked zebrafish individuals, grown their offspring to sexual maturity and determined their sex ratio based on presence/absence of sexual dimorphic phenotypic markers.
Skewed family sex ratios are very likely due to genetic factors
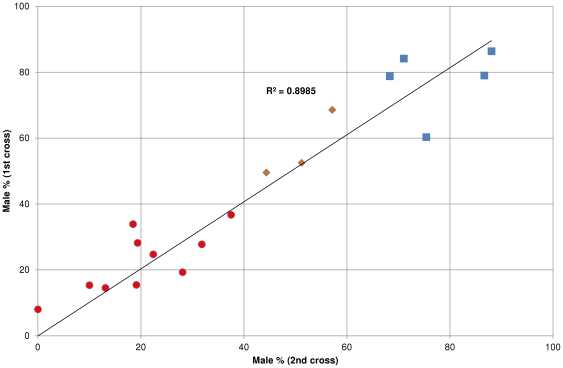
In order to investigate the potential reason for skewed family sex ratios repeated single pair crossing was carried out. Nineteen breeding pairs were crossed twice on different occasions and their offspring were raised at similar, but not identical conditions (i.e. ambient water temperature ranging between 27–29°C, variable densities and amount feed). Based on the sex ratio of the first set of clutches, the breeding pairs were divided into three groups (Table 1): female-biased group (ten pairs; less than 40% males in their offspring), unbiased group (three pairs; 40–60% males in their offspring) and male-biased group (six pairs; more than 60% males in their offspring). In the female-biased group, the difference in mean male percentages for the first and second batches was 1.3%. The biggest difference produced by a female-biased pair was 15.4% (mating pair 16). Three pairs were assigned to the unbiased group and the mean difference between their 1st and 2nd cross was 6%. In the male-biased group, mating pair 6 showed an unusually big, 25.2% drop in the sex ratio (from 82.9% to 57.7% males) that was 1.6 fold higher than the second highest change and 3.7 fold higher than the mean of the rest. We decided to remove this pair from the comparison and used data for the remaining five pairs only, where the mean difference in the male percentages for the first and second batches was 0.2%. The biggest difference produced by a male-biased pair was 15.1% (mating pair 3). Overall, we observed very similar offspring sex ratios between the first and second crosses from the same breeding pair indicated by the high R2 value of 0.8985 (Fig. 2). The fact that sex ratios of different batches of offspring from the same breeding pair were very similar suggests that sex in zebrafish is heritable, whereas wide-ranging sex ratios across the families point towards a complex genetic trait.
Table 1. The percentage of males from repeated single pair mating of 19 randomly selected zebrafish pairs.
| Mating Pair | Cross (male %) | |
| 1st | 2nd | |
| Male-biased offspring | ||
| 1 | 88.1 | 86.4 |
| 2 | 86.7 | 79.0 |
| 3 | 75.4 | 60.3 |
| 4 | 71.0 | 84.2 |
| 5 | 68.3 | 78.8 |
| 6* | 82.9 | 57.7 |
| Mean + | 77.9 | 77.7 |
| Unbiased offspring | ||
| 7 | 57.1 | 68.6 |
| 8 | 51.2 | 52.5 |
| 9 | 44.4 | 49.6 |
| Mean | 50.9 | 56.9 |
| Female-biased offspring | ||
| 10 | 37.5 | 36.7 |
| 11 | 31.8 | 27.8 |
| 12 | 28.1 | 19.3 |
| 13 | 22.4 | 24.7 |
| 14 | 19.4 | 28.2 |
| 15 | 19.1 | 15.5 |
| 16 | 18.5 | 33.9 |
| 17 | 13.1 | 14.5 |
| 18 | 10.0 | 15.4 |
| 19 | 0.0 | 8.0 |
| Mean | 25.2 | 26.6 |
Removed from further analysis.
Does not include mating pair 6.
Figure 2. Sex ratios of offspring groups generated by repeated single pair crossings show close correlation.
Nineteen randomly selected breeding pairs were crossed twice; eighteen of them are shown here. The high R2 value indicates that sex ratios between 1st and 2nd crosses from the same breeding pair are very similar. Red circles indicate pairs producing offspring with female-biased sex ratio, orange diamond labels the pairs with unbiased sex ratio, whereas blue squares indicate pairs producing offspring with male-biased sex ratio.
Enhancement and maintenance of sex-biased lines through multiple generations by full-sib selective breeding
This experiment was carried out with the aim to determine if sex-biased ratios in lines can be maintained or increased by selecting for breeding pairs that produced brood with highly skewed sex ratios through several generations.
A total of 5 lines were established and followed through two to four generations. Several lines were split into sub-lines that were later split further depending on the sex ratios resulting from the multifactorial crosses. Altogether, offspring from 26 fourth generation families were grown to maturity and sexed (Fig. S1). In two families, we managed to generate an all-male offspring in the F3 generation (Fig. S1), whereas our efforts to generate all-female offspring were unsuccessful.
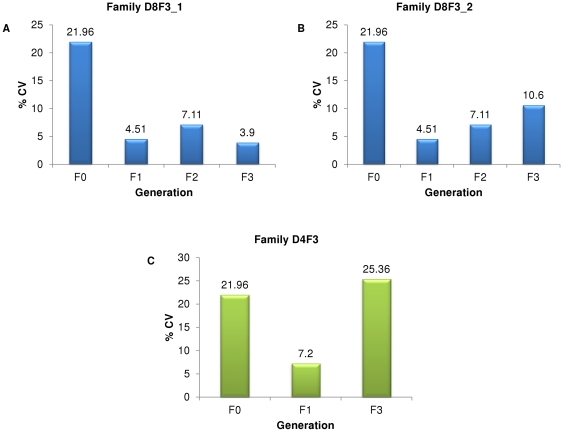
Here, we describe two male-biased families that we managed to maintain for two generations through selection from a single line and split in the third generation (Fig. S1). The F3 mean sex ratios of the two families were 96.8% (family D8F3_1) and 93.3% (family D8F3_2) males. We then analyzed the family sex ratio variation for each generation by calculating coefficients of variation (CV). It was observed that after selection the family sex ratio CV decreased at least two-folds when compared to the F0 generation (Fig. 3A & B). To verify that the decrease was due to selection pressure, we performed a control experiment with the D4F3 family by doing a mass cross for the F2 generation without any selection (Fig. 3C). The control family sex ratio CV for the F0 generation was 21.96% and after selection the F1 generation family sex ratio CV was 7.2%, about three-fold lower. However, after F2 mass cross the family sex ratio CV of F3 increased to 25.36%. The “bouncing back" of F3 family sex ratio CV to a level similar to F0 indicates that selection pressure was indeed maintaining the highly skewed family sex ratio. This indicates that zebrafish sex is a genetic trait and the fact that we were able to keep highly skewed sex ratios - and even eliminate one of the two sexes in some cases - suggests the absence of a strong effect by sex chromosomes on sex determination.
Figure 3. Coefficients of variation for each generation family sex ratios show selection effect on sex ratio.
Panels A & B: For both families, CV for the F0 generation (unselected) was more than two-folds higher than those for the subsequent generations, which underwent selection. Panel C: In the control experiment, after selecting for pairs that produced high proportion of males at F0 generation, CV for F1 generation family sex ratio decreased by about three-folds. However, when selection pressure was removed at F2 generation by performing a random mass cross, CV for F3 generation family sex ratio returned to a level similar to that of unselected F0 generation.
FluoMEP assay identified sex-linked DNA markers from guppy and rosy barb, but not from zebrafish
FluoMEP assay was used to search for DNA markers tightly associated with sex from three different fish species' genomes. The first species was the guppy (Poecilia reticulata) which has XX/XY sex chromosomes [56]. Altogether, 144 different primer combinations utilizing the same common motif primer were tested. They yielded three male-specific sex markers (Fig. S3A), that showed 100% agreement with phenotypic sex in eight individuals tested (data not shown). Next, we screened the genome of rosy barb (Puntius conchonius) that is also known to have XX/XY sex chromosomes [57], [58], with 386 primer combinations (based on two common primers) and obtained two male-specific sex markers (Fig. S3B). When tested on eight individuals, the sexing efficiency of the two markers was also 100% (data not shown). These results demonstrated that FluoMEP is able to isolate sex-linked DNA markers from fish genomes with substantial differences between the male and female genomes.
In order to search for sex-linked DNA markers in zebrafish, we used a total of 258 FluoMEP primer combinations (based on 29 common primers) to screen pooled male and female zebrafish genomic DNA samples. However, no sex-linked DNA marker was found suggesting that there are no substantial differences between zebrafish male and female genomes.
No universal sex-linked CNV was detected in four zebrafish families by aCGH
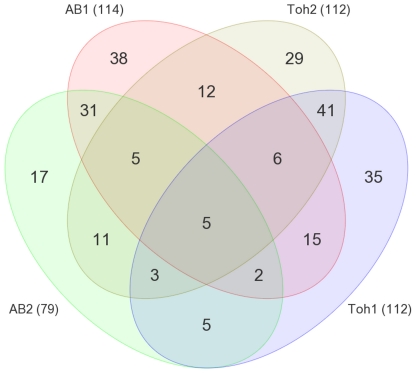
We continued our investigation for sex-linked differences at the genome level by aCGH. We used a custom-designed oligonucleotide microarray containing 120,000 probes covering the assembled zebrafish genome (Zv7). By testing samples from two families each of the AB and Toh strains, a total of 255 CNV regions (CNVRs) were detected (Fig. 4). Among them, 64 CNVRs were present in both strains, 105 were unique to the Toh strain and 86 were present only in the AB strain. Five CNVRs were common to all the four families screened (Fig. 4). As we expected that a sex-determining chromosomal region would be present in all strains, we analyzed the five common CNVRs on individuals, but none of them turned out to be inherited in a sex-linked pattern (Fig. S4).
Figure 4. Comparative analysis of CNVRs in four zebrafish families.
Out of 255 CNVRs detected, only five were present in all four families tested, however, those common CNVRs have not shown any association with sex. The number of CNVRs detected for each family is indicated in the bracket.
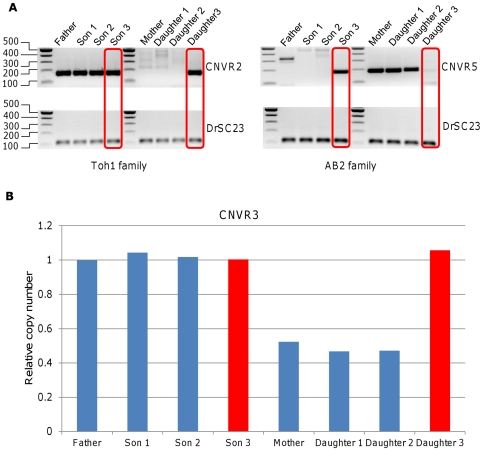
Additional five CNVRs showed apparent family-specific sex linkage (Table 2) and were further analyzed by PCR-based methods with additional two offspring individuals (one male and one female) from the same families. Multiple primers were designed for the first two CNVRs, but failed to yield a PCR product, presumably due to differences between the Zv7 genome assembly used for the probe design and Zv8 used for the analysis of results. Two of the remaining three CNVRs were found not to be sex-linked by PCR (Fig. 5A), while the last one was found not to be sex-specific by real time quantitative PCR (qPCR; Fig. 5B). In fact, none of the additional offspring individuals analyzed did show sex-linked inheritance pattern for any of these three markers. Therefore, we concluded that no family-specific, sex-linked CNVR was identified from the four zebrafish families analyzed.
Table 2. CNV regions selected for further validation due to their apparent association to sex based on preliminary aCGH.
| Chromosome | Start | End | Length (Kb) | Family | Copy number | |
| CNVR1 | 1 | 56,170,986 | 57,093,117 | 922 | AB2 | Gain |
| CNVR2 | 7 | 664 | 308,167 | 308 | Toh1 | Loss |
| CNVR3 | 8 | 30,107,209 | 30,331,905 | 225 | Toh2 | Loss |
| CNVR4 | 8 | 43,154,384 | 43,205,453 | 51 | AB2 | Gain |
| CNVR5 | 8 | 47,481,469 | 47,612,299 | 131 | AB2 | Gain |
Figure 5. PCR-based validation of aCGH results that showed apparent family specific sex-linked inheritance pattern confirms that none of the three CNVRs analyzed are sex-linked.
A) The lack of sex-linkage for CNV regions 2 and 5 as confirmed by PCR. Size of the amplified fragments for CNVR2 and CNVR5 are 157 bp and 183 bp, respectively. CNVR2 was present only in males from the Toh1 family (Father and Son 1 and 2), while CNVR5 was only seen in female samples from the AB2 family (Mother and Daughter 1 and 2). As they showed a family-specific, sex-linked pattern, additional offspring (one son and one daughter; red boxes) were used for the validation. Upon further validation, CNVR2 and CNVR5 were found not to be sex-linked. B) CNV region 3 could only be validated by real time qPCR. As the three female samples from Toh2 family used for aCGH showed a loss with reference to the father's genome, additional offspring (one son and one daughter; red bar) were used for validation. Further validation also showed that this is not a sex-linked CNVR.
Rearing density has a limited effect on sex ratios in zebrafish
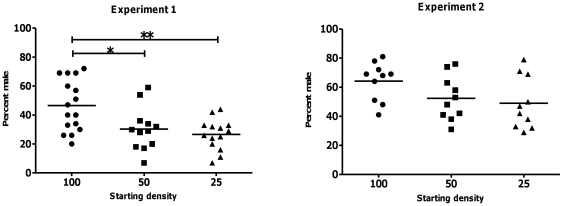
Density has been shown to affect sex ratios in some fish species (see [59] for review). To ask whether rearing density influences sex determination in zebrafish, we raised groups of zebrafish at three different densities from 5 dpf to adulthood and assayed the sex ratios of the resulting adults. The three groups had starting densities of 100, 50, and 25 larvae per 1.5 liters of water, respectively. Two independent experiments were performed that varied slightly in their design (see Materials and Methods) yet resulted in similar outcomes with respect to relative sex ratios across different rearing densities. However, in each experiment, we observed wide-ranging sex ratios at all the tested starting densities (Fig. 6). This profile was similar to that observed in the breeding experiment (see Fig. 1). We found that zebrafish reared at high density had approximately twenty per cent more males on average than those raised at middle or low densities indicating a modest effect of large differences in rearing density on sex determination in zebrafish (Fig. 6).
Figure 6. High rearing densities yield higher male to female sex ratios compared to lower ones.
Two individual experiments were performed consisting of forty-four populations for experiment 1 and thirty populations for experiment 2. The final percentage of males was assayed for each population and the averages for each population group, denoted by the starting density, were calculated. For each experiment the overall sex ratios varied, but both showed about a twenty percent increase in male percentage in populations with starting densities of 100 fish per 1.5 liters compared to populations with starting densities of 50 or 25 fish per 1.5 liters. Each datapoint represents the percentage of males for a given parental pair, whereas the horizontal line indicates the mean male ratio.
As the time window in which sex determination occurs is not well defined, we wanted to account for potential changes in population densities due to larval or juvenile death in the above experiments. The number of fish was counted every 10 days beginning on day 10 post fertilization. The highest degree of lethality was typically between 10 and 20 dpf, which corresponded to the period at which both food and water regimes were altered (see Materials and Methods). After 20 dpf, limited loss was observed in most populations and after 30 dpf most individuals survived (Table S1 and Fig. S2). Despite some larval lethality, the average percentage of dead fish in each density group from experiment 2 did not differ significantly indicating that larval death did not contribute to the observed higher percentage of males in the high density group (Table S1 and Fig. S2).
Discussion
Molecular and breeding data suggest a genetic sex determination system without a predominant sex chromosome in zebrafish
Although zebrafish has become one of the prime vertebrate models for developmental biology, its sex determination mechanism is still unknown. Therefore, the primary aim of this project was to find out more about the sex determination of this species. The first question we asked was: does zebrafish use a chromosomal sex determination system?
So far, several cytogenetic analyses were performed on zebrafish karyotypes to search for a size-heteromorphic chromosomal pair, which is a hallmark of CSD with highly differentiated sex chromosomes. However, the accurate assignment of chromosomal pairs is hampered by lack of substantial size differences among the zebrafish chromosomes and their poor staining. Ten teams reported the lack of a heteromorphic chromosomal pair in the zebrafish karyotype [60], [61], [62], [63], [64], [65], [66], [67], [68], [69], while only two publications described the presence of such a pair [70], [71]. Researchers have also tried to look for sex chromosomes in the zebrafish genome by searching for sex bivalent synaptonemal complexes [72] and performing comparative genomic hybridization (CGH) between male and female gDNAs [73]. Negative results from both latter studies – together with the vast majority of cytogenetic data - suggest that zebrafish does not possess heteromorphic sex chromosomes.
Our breeding data also indicate the absence of chromosomal sex determination in zebrafish, whereby the inheritance of a particular chromosome would be the predominant determiner of sex. We observed variable family sex ratios from 62 clutches of offspring from different breeding pairs (Fig. 1). Broods from species that have strong chromosomal sex determination system typically exhibit a narrow range of family sex ratios that do not divert substantially from 1∶1 (male to female; e.g. Nile tilapia [4] and rainbow trout [74]). Moreover, we were able to obtain several strongly male-biased zebrafish families by selective crossing of brooders that produced higher proportion of male offspring over a few generations. In the chromosomal sex determination system, the chance for the occurrence of such sex-biased families would be very low, because the ratio of male and female would tend to ‘bounce back’ close to 1∶1 in the next generation. These breeding data also indicate that zebrafish sex determination is unlikely to be based primarily on sex chromosomes.
To further prove the absence of chromosomal sex determination system, we screened the zebrafish genome for sex-linked differences with molecular tools. The first experiments we performed were a series of comparative FluoMEP assays [39]. After screening through 258 FluoMEP primer combinations, no confirmed sex marker was obtained from zebrafish. On the other hand, sex markers for guppy and rosy barb were detected by using the same method. The latter data prove that the FluoMEP assay is suitable for isolating sex-linked markers from genomes known to contain heteromorphic sex chromosomes. The fact that we were unable to obtain sex-linked markers from zebrafish with the same method provides an additional indication that no substantial differences exist between the male and female genomes. Even if there are sex chromosomes in zebrafish, they will be likely showing limited differences at sequence level and therefore undergo recombination with each other along the majority of their length. In this case, the identification of such sex chromosomes through the analysis of pools generated based on phenotypes would be extremely difficult.
Next, we performed array comparative genomic hybridization (aCGH) on four families of zebrafish. Through the analysis of 120 thousand genomic locations, a total of 255 CNVRs were observed and most showed a pattern of Mendelian inheritance. However, no universal sex-linked CNVR was found among the four families of zebrafish tested. The aCGH results suggest that the possibility for highly differentiated, heteromorphic sex chromosomes in the zebrafish genome is quite low. The caveat of our current aCGH approach is that the assembly of zebrafish genome (Zv8) on the basis of which the probes were analyzed managed to assemble only about 89% of the total sequences obtained [75]. Therefore, there is still a possibility that there are sex-linked CNVRs “hiding" in the remaining 11% of the genome. Furthermore, the probes present on the custom-made oligo array have a median spacing of 10 kb intervals and by setting the window of detection to 5 probes per window allows for a resolution of around 50 kb. This means that any genomic difference with less than 50 kb in length will not be picked up by our aCGH approach. However, we argue that the size difference for most active sex chromosomal pairs will likely exceed 50 kb in length, as in case of the medaka, the only known SD region described from teleosts so far [76]. Recently a high resolution zebrafish CNV map was published by analyzing 80 genomes with 1.4 kb probe spacing CNV array [77]. Although it has higher resolution than our CNV array the study did not performed comparative analysis of the male and female genome.
Considering the combined data of the FluoMEP and aCGH approaches, the majority of the (assembled) zebrafish genome was probed for sex-linked sequences in this study. Although we still cannot completely rule out the presence of a sex chromosomal pair, it is unlikely that a single predominant sex-determining region exists. As our breeding data also do not support the presence of a sex chromosome, we provide a strong case against CSD in zebrafish.
Very recently, a genome-wide association study was performed for the identification of sex determining regions with a SNP array containing over 5,300 features [78]. The authors reported two regions on two separate chromosomes (Chr5 and Chr16) accounting for 16% variance of the trait, providing a direct experimental evidence for a polygenic sex determination system in the zebrafish [78]. These data further strengthen the notion that zebrafish sex is not determined by a sex chromosomal pair.
Our data and results from vast majority of the above studies contradict a recent suggestion that zebrafish has a female dominant (ZZ/ZW) sex determination system [79]. Although the results described in that publication seem to support the possibility of a ZZ/ZW sex chromosome system, their data do not conclusively demonstrate that this mode of sex determination is actually in place. Attempts to identify the genetic factor(s) regulating sex or the proposed sex chromosomes were not made in their study.
Zebrafish sex is determined genetically
Since molecular and breeding studies failed to identify heteromorphic sex chromosomes or their effect, we next sought to find out if genetic factors are involved in zebrafish sex determination. We performed repeated single pair mating in which 19 randomly selected breeding pairs were bred twice. The environmental factors such as ambient temperature, amount of food given and rearing density were not tightly controlled. Even so, broods derived from the same breeding pair did not exhibit major sex ratio differences between repeated crossings of 18 out of 19 breeding pairs tested (Fig. 2). This indicates that the wide-ranging sex ratios normally observed are most likely due to the parental genotypes. In addition, we showed that sex ratio variation decreases substantially under selective pressure, a strong indication that sex is a genetic trait. Another interesting phenomenon we observed was that after three generations of selection we were able to obtain two all-male families while attempts to produce all-female families were unsuccessful. We do not have an explanation for this difference and we propose that further investigations are needed to elucidate the underlying reasons. Nevertheless, our data show that zebrafish uses primarily genetic sex determination system. As we have also demonstrated that CSD is not likely the mode of sex determination in zebrafish, we propose that a PGSD is in place. Based on our data and the recent aforementioned association study [78], we propose that the number of genes contributing to the sex determination process might be far more than just a handful.
Polygenic sex determination might be more common among vertebrates than expected
The vast majority of our knowledge about vertebrate sex determination was obtained from species using sex chromosomal systems. On the other hand, over 90% of the fish species analyzed through karyotyping does not show the presence of differentiated sex chromosomes (see [5] for review).
Recently, it was proposed that multiple parallel sex determining pathways are likely to operate in species with CSD and this mode of SD could be extended to species with PGSD as well [80]. In such scenario, both systems might have a more similar regulation than expected, differing only in the location of the factors: all would map onto the sex chromosomes in CSD, whereas in PGSD some (or all) of them would be located on the autosomes. Therefore, analysis of zebrafish and other fish species utilizing the PGSD system could be important for basic research and potentially useful for aquaculture projects as well.
Environmental factors have limited influence on zebrafish sex ratio
Temperature is the most commonly studied environmental cue for sex determination. It is utilized by many reptile species [18], [81], [82], [83] and some fish species [74], [84]. In animals with temperature-based sex determination (TSD), substantial fluctuations in the environmental temperature will likely cause significant changes in the offspring sex ratio [85]. Two papers reported that the temperature at natural habitat of zebrafish ranges from 26 to 38°C [86], [87]. However, it is believed that 26 to 29°C is the temperature range for normal zebrafish development and rearing them within this range did not result in significant sex ratio changes [85]. It was also observed that exposure to increased temperature (35–37°C) either during early development (5–48 hpf) [88] or between 17–27 dpf [89] resulted in male-biased sex ratio. On the other hand, at our laboratories we observed high mortality if zebrafish larvae were grown at 37°C from the beginning. Therefore, temperature is unlikely to be the primary signal for zebrafish sex determination, but might exert secondary effects on its sexual development.
Rearing density is another environmental cue known to influence sex ratio of some fish species such as the American eel [90]. The exact underlying mechanisms of how rearing density directs sexual development are still unknown. We have tested the effect of rearing density on zebrafish sex, and found a substantial increase of males at high density (100 individuals per 1.5 liters of water). In another study, slow growth rate as a result of limited food supply - usually experienced at high rearing density - had been suggested to influence zebrafish sex differentiation leading to higher percentage of males [91]. Nonetheless, we think that rearing density is unlikely to be the primary determinant for zebrafish sex, as we observed wide ranging sex ratios at all three densities tested. A strong determinant should produce broods of very similar sex ratio. In addition, the response to these environmental factors seems to differ between families indicating that influence of rearing density on sex ratio is most likely conferred by the genotype of the fish.
Another environmental factor that is known to have an effect on zebrafish sex ratio is oxygen level [92]. It was found that under hypoxic conditions there was a reduction of estrogen synthesis leading to an increase of androgen to estrogen ratio which favors male development [93]. However, the decreased oxygen level had only limited effect on the sex ratio of zebrafish leading to higher percentage of males (12.5% differences) [92]. This is unlikely the cause of wide ranging sex ratio observed in the zebrafish.
Published data and our results both seem to suggest that non-extreme environmental factors do not have a major effect on zebrafish sex ratio. No drastic change in sex ratio upon environmental effects experienced at the natural surroundings of the species was observed in any of the studies. Furthermore, response to environmental factors varies among the treatment groups. This indicates that the underlying genotype of each individual is directing sexual development in response to environmental stimulus.
Conclusions
For this study, we performed classical breeding experiments together with large-scale genomic analyses to show that zebrafish sex is determined genetically with no sign of a chromosomal sex determination system. The characteristics of sex ratios observed in zebrafish were as follows: i) wide variation among different families; ii) strong influence from parental genotypes; and iii) the ability to eliminate one of the sexes by selection. All these features point toward a species without a predominant chromosomal sex determination system [11]. Our in-depth investigation by molecular tools (i.e. FluoMEP and aCGH) also failed to identify any difference between the male and female genomes. Several studies, which investigated environmental impacts on zebrafish sex ratio, found them either to result in limited change or show strong effects outside of the physiological range of the species. This cannot account for the wide-ranging sex ratios among families; hence we reckon that zebrafish does not use a primary environmental sex determination system.
Taken together, the above data indicate either the lack of sex chromosomes in zebrafish or the presence of very weak ones that are frequently over-ridden by strong modifier genes. In our opinion, these two situations are principally the same, as there are several genes distributed throughout the genome with major effects on sexual development in both; therefore we propose that zebrafish sex determination should be considered polygenic.
Earlier, others have indicated the possibility of a polygenic sex determination system for zebrafish based on a single set of experiment each (see e.g. [94] & [78]). Our study adds data obtained by four different methods that all point to a polygenic sex determination system, creating a tipping point in this argument.
Supporting Information
Multifactorial selective breeding was carried out over a few generations for five lines of zebrafish to select for pairs that produced a sex-biased family. All multifactorial crosses were set up using full siblings. The selective breeding process involved selecting for pairs that produce highly biased sex ratio (highlighted by different colour box) then offspring from the selected pairs were used to set up multifactorial crosses in the next generation. This was repeated for a few generations. The two all-male families were from the D8 line (D8F3_1 and D8F3_2). The control family D4F3 had a mass cross performed in the F2 generation without selection.
(XLSX)
Plots of the number of fish present in each population over time. Each line represents one population, housed in a single tank. Data points of fish counts are represented by diamonds. Populations with higher than the overall average percentage of males are colored orange while those populations with a lower than average male percentage are colored blue. These data are from experiment 1 and 2 shown in Table S1.
(PPTX)
Sex-linked FluoMEP markers obtained by bulk segregant analysis performed on pooled male and female samples of guppy ( Poecilia reticulata). A) and rosy barb (Puntius conchonius; B). Primer combinations are indicated on the top right corner of the peak profiles. Red boxes indicate the sex-linked markers that were confirmed through individual testing. The remaining differences are false positives that have occurred with similar frequency in both species depicted here, as well as in the zebrafish (not shown).
(PPTX)
Inheritance pattern of the five CNVRs universal to all four families. None of them showed sex-linked inheritance pattern. Gain in copy number is indicated by blue box while loss in copy number is indicated by red box.
(PPTX)
Data tables for rearing density experiment 1 and 2.
(XLSX)
Sequences of all primers used.
(XLSX)
Acknowledgments
We thank Prof Christiane Nüsslein-Volhard for her support of the project, Dr Rob Brooks for guppy samples, Dr Alex Chang and Qian Hu Fish Farm for helping to obtain the rosy barb samples, Miss Rashmi Sukumaran for developing the FluoMEP marker screening program, TLL fish facility for taking care of the zebrafish and TLL DNA sequencing facility for performing the FluoMEP electrophoretic analysis.
Footnotes
Competing Interests: The corresponding author has served as an Academic Editor of PLoS ONE since 2006. This does not alter the authors' adherence to all the PLoS ONE policies on sharing data and materials. All other authors have declared that no competing interests exist.
Funding: This study was supported by the Ministry of National Development, Agri-Food and Veterinary Authority of Singapore, and internal research funds from Temasek Life Sciences Laboratory. KRS was supported by a Human Frontiers in Science Program post doctoral fellowship and the Max Planck Society during the course of this work. The funders had no role in study design, data collection and analysis, decision to publish, or preparation of the manuscript.
References
- 1.DeFalco T, Capel B. Gonad morphogenesis in vertebrates: Divergent means to a convergent end. Annu Rev Cell Dev Biol. 2009;25:457–482. doi: 10.1146/annurev.cellbio.042308.13350. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Morrish BC, Sinclair AH. Vertebrate sex determination: many means to an end. Reproduction. 2002;124:447–457. doi: 10.1530/rep.0.1240447. [DOI] [PubMed] [Google Scholar]
- 3.Wilhelm D, Palmer S, Koopman P. Sex determination and gonadal development in mammals. Physiol Rev. 2007;87:1–28. doi: 10.1152/physrev.00009.2006. [DOI] [PubMed] [Google Scholar]
- 4.Baroiller JF, D'Cotta H, Bezault E, Wessels S, Hoerstgen-Schwark G. Tilapia sex determination: Where temperature and genetics meet. Comp Biochem Physiol A Mol Integr Physiol. 2009;153:30–38. doi: 10.1016/j.cbpa.2008.11.018. [DOI] [PubMed] [Google Scholar]
- 5.Devlin RH, Nagahama Y. Sex determination and sex differentiation in fish: an overview of genetic, physiological, and environmental influences. Aquaculture. 2002;208:191–364. [Google Scholar]
- 6.Manolakou P, Lavranos G, Angelopoulou R. Molecular patterns of sex determination in the animal kingdom: a comparative study of the biology of reproduction. Reprod Biol Endocrinol. 2006;4:59. doi: 10.1186/1477-7827-4-59. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Charlesworth B. The evolution of chromosomal sex determination. Hoboken, NJ: John Wiley and Sons Ltd; 2002. pp. 207–224. [PubMed] [Google Scholar]
- 8.Graves J. The evolution of mammalian sex chromosomes and the origin of sex determining genes. Philos T Roy Soc B. 1995;350:305–311. doi: 10.1098/rstb.1995.0166. [DOI] [PubMed] [Google Scholar]
- 9.Graves JAM. Sex chromosome specialization and degeneration in mammals. Cell. 2006;124:901–914. doi: 10.1016/j.cell.2006.02.024. [DOI] [PubMed] [Google Scholar]
- 10.Wilson M, Makova K. Genomic analyses of sex chromosome evolution. Annu Rev Genom Hum G. 2009;10:333–354. doi: 10.1146/annurev-genom-082908-150105. [DOI] [PubMed] [Google Scholar]
- 11.Bulmer MG, Bull JJ. Models for polygenic sex determination and sex ratio control. Evolution. 1982;36:13–26. doi: 10.1111/j.1558-5646.1982.tb05005.x. [DOI] [PubMed] [Google Scholar]
- 12.Kosswig C. Polygenic sex determination. Experientia. 1964;20:190–199. doi: 10.1007/BF02135395. [DOI] [PubMed] [Google Scholar]
- 13.Franco M, Rubini P, Vecchi M. Sex-determinants and their distribution in various populations of Musca domestica L. of Western Europe. Genet Res. 1982;40:279–293. doi: 10.1017/s0016672300019157. [DOI] [PubMed] [Google Scholar]
- 14.Voordouw MJ, Anholt BR. Heritability of sex tendency in a harpacticoid copepod, Tigriopus californicus. Evolution. 2002;56:1754–1763. doi: 10.1111/j.0014-3820.2002.tb00189.x. [DOI] [PubMed] [Google Scholar]
- 15.Vandeputte M, Dupont-Nivet M, Chavanne H, Chatain B. A polygenic hypothesis for sex determination in the European sea bass Dicentrarchus labrax. Genetics. 2007;176:1049–1057. doi: 10.1534/genetics.107.072140. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Ser JR, Roberts RB, Kocher TD. Multiple interacting loci control sex determination in lake Malawi cichlid fish. Evolution. 2010;64:486–501. doi: 10.1111/j.1558-5646.2009.00871.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Modi W, Crews D. Sex chromosomes and sex determination in reptiles: Commentary. Curr Opin Genet Dev. 2005;15:660–665. doi: 10.1016/j.gde.2005.09.009. [DOI] [PubMed] [Google Scholar]
- 18.Pieau C, Dorizzi M, Richard-Mercier N. Temperature-dependent sex determination and gonadal differentiation in reptiles. Cell Mol Life Sci. 1999;55:887–900. doi: 10.1007/s000180050342. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Barón B, Bückle F, Espina S. Environmental factors and sexual differentiation in Poecilia sphenops Valenciennes (Pisces: Poeciliidae). Aquac Res. 2002;33:615–619. [Google Scholar]
- 20.Godwin J. Social determination of sex in reef fishes. Semin Cell Dev Biol. 2009;20:264–270. doi: 10.1016/j.semcdb.2008.12.003. [DOI] [PubMed] [Google Scholar]
- 21.Eimon P, Ashkenazi A. The zebrafish as a model organism for the study of apoptosis. Apoptosis. 2010;15:331–349. doi: 10.1007/s10495-009-0432-9. [DOI] [PubMed] [Google Scholar]
- 22.Grunwald D, Eisen J. Headwaters of the zebrafish emergence of a new model vertebrate. Nat Rev Genet. 2002;3:717–724. doi: 10.1038/nrg892. [DOI] [PubMed] [Google Scholar]
- 23.Hortopan G, Dinday M, Baraban S. Zebrafish as a model for studying genetic aspects of epilepsy. Disease Model Mech. 2010;3:144–148. doi: 10.1242/dmm.002139. [DOI] [PubMed] [Google Scholar]
- 24.Mione M, Trede N. The zebrafish as a model for cancer. Disease Model Mech. 2010;3:517–523. doi: 10.1242/dmm.004747. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Norton W, Bally-Cuif L. Adult zebrafish as a model organism for behavioural genetics. BMC Neurosci. 2010;11:90. doi: 10.1186/1471-2202-11-90. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Siegfried KR. In search of determinants: gene expression during gonadal sex differentiation. J Fish Biol. 2010;76:1879–1902. doi: 10.1111/j.1095-8649.2010.02594.x. [DOI] [PubMed] [Google Scholar]
- 27.Orban L, Sreenivasan R, Olsson PE. Long and winding roads: testis differentiation in zebrafish. Mol Cell Endocrinol. 2009;312:35–41. doi: 10.1016/j.mce.2009.04.014. [DOI] [PubMed] [Google Scholar]
- 28.von Hofsten J, Olsson P. Zebrafish sex determination and differentiation: involvement of FTZ-F1 genes. Reprod Biol Endocrinol. 2005;3:63. doi: 10.1186/1477-7827-3-63. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Sola L, Gornung E. Classical and molecular cytogenetics of the zebrafish, Danio rerio (Cyprinidae, Cypriniformes): an overview. Genetica. 2001;111:397–412. doi: 10.1023/a:1013776323077. [DOI] [PubMed] [Google Scholar]
- 30.Welsh J, McClelland M. Fingerprinting genomes using PCR with arbitrary primers. Nucleic Acids Res. 1990;18:7213–7218. doi: 10.1093/nar/18.24.7213. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Williams JG, Kubelik AR, Livak KJ, Rafalski JA, Tingey SV. DNA polymorphisms amplified by arbitrary primers are useful as genetic markers. Nucleic Acids Res. 1990;18:6531–6535. doi: 10.1093/nar/18.22.6531. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Vos P, Hogers R, Bleeker M, Reijans M, van de Lee T, et al. AFLP: a new technique for DNA fingerprinting. Nucleic Acids Res. 1995;23:4407–4414. doi: 10.1093/nar/23.21.4407. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Ezaz MT, Harvey SC, Boonphakdee C, Teale AJ, McAndrew BJ, et al. Isolation and physical mapping of sex-linked AFLP markers in Nile tilapia (Oreochromis niloticus L.). Mar Biotechnol (NY) 2004;6:435–445. doi: 10.1007/s10126-004-3004-6. [DOI] [PubMed] [Google Scholar]
- 34.Iturra P, Medrano JF, Bagley M, Lam N, Vergara N, et al. Identification of sex chromosome molecular markers using RAPDs and fluorescent in situ hybridization in rainbow trout. Genetica. 1998;101:209–213. doi: 10.1023/A:1018371623919. [DOI] [PubMed] [Google Scholar]
- 35.Kovacs B, Egedi S, Bartfai R, Orban L. Male-specific DNA markers from African catfish (Clarias gariepinus). Genetica. 2001;110:267–276. doi: 10.1023/a:1012739318941. [DOI] [PubMed] [Google Scholar]
- 36.Bello N, Sanchez A. The identification of a sex-specific DNA marker in the ostrich using a random amplified polymorphic DNA (RAPD) assay. Mol Ecol. 1999;8:667–669. doi: 10.1046/j.1365-294x.1999.00549.x. [DOI] [PubMed] [Google Scholar]
- 37.Lessells C, Mateman A. Sexing birds using random amplified polymorphic DNA (RAPD) markers. Mol Ecol. 1998;7:187–195. [Google Scholar]
- 38.Quinn A, Radder R, Sarre S, Georges A, Ezaz T, et al. Isolation and development of a molecular sex marker for Bassiana duperreyi, a lizard with XX/XY sex chromosomes and temperature-induced sex reversal. Mol Genet Genomics. 2009;281:665–672. doi: 10.1007/s00438-009-0437-7. [DOI] [PubMed] [Google Scholar]
- 39.Chang A, Liew WC, Chuah A, Lim Z, Lin Q, et al. FluoMEP: A new genotyping method combining the advantages of randomly amplified polymorphic DNA and amplified fragment length polymorphism. Electrophoresis. 2007;28:525–534. doi: 10.1002/elps.200600715. [DOI] [PubMed] [Google Scholar]
- 40.Conrad DF, Pinto D, Redon R, Feuk L, Gokcumen O, et al. Origins and functional impact of copy number variation in the human genome. Nature. 2010;464:704–712. doi: 10.1038/nature08516. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Redon R, Ishikawa S, Fitch KR, Feuk L, Perry GH, et al. Global variation in copy number in the human genome. Nature. 2006;444:444–454. doi: 10.1038/nature05329. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Albertson D, Segraves R, Sudar D, Chen C, Clark S, et al. Analysis of DNA copy number variation in human papillomavirus-transfected cells during progression toward immortality using comparative genomic hybridization to DNA microarrays. Mol Biol Cell. 1997;8:736–736. [Google Scholar]
- 43.Pinkel D, Seagraves R, Sudar D, Clark S, Poole I, et al. High resolution analysis of DNA copy number variation using comparative genomic hybridization to microarrays. Nat Genet. 1998;20:207–211. doi: 10.1038/2524. [DOI] [PubMed] [Google Scholar]
- 44.Albertson DG, Ylstra B, Segraves R, Collins C, Dairkee SH, et al. Quantitative mapping of amplicon structure by array CGH identifies CYP24 as a candidate oncogene. Nat Genet. 2000;25:144–146. doi: 10.1038/75985. [DOI] [PubMed] [Google Scholar]
- 45.Goidts V, Armengol L, Schempp W, Conroy J, Nowak N, et al. Identification of large-scale human-specific copy number differences by inter-species array comparative genomic hybridization. Hum Genet. 2006;119:185–198. doi: 10.1007/s00439-005-0130-9. [DOI] [PubMed] [Google Scholar]
- 46.Lee A, Gutierrez-Arcelus M, Perry G, Vallender E, Johnson W, et al. Analysis of copy number variation in the rhesus macaque genome identifies candidate loci for evolutionary and human disease studies. Hum Mol Genet. 2008;17:1127–1136. doi: 10.1093/hmg/ddn002. [DOI] [PubMed] [Google Scholar]
- 47.Henrichsen CN, Vinckenbosch N, Zollner S, Chaignat E, Pradervand S, et al. Segmental copy number variation shapes tissue transcriptomes. Nat Genet. 2009;41:424–429. doi: 10.1038/ng.345. [DOI] [PubMed] [Google Scholar]
- 48.Kim H, Cho S, Yu J, Sung S, Kim H. Analysis of copy number variation in 8,842 Korean individuals reveals 39 genes associated with hepatic biomarkers AST and ALT. BMB Rep. 2010;43:547–553. doi: 10.5483/bmbrep.2010.43.8.547. [DOI] [PubMed] [Google Scholar]
- 49.Westerfield M. The zebrafish book. A guide for the laboratory use of zebrafish (Danio rerio). 4th ed. Eugene, OR: University of Oregon Press; 2000. pp. 1.1–11.65. [Google Scholar]
- 50.Parichy D, Elizondo M, Mills M, Gordon T, Engeszer R. Normal table of postembryonic zebrafish development: staging by externally visible anatomy of the living fish. Dev Dyn. 2009;238:2975–3015. doi: 10.1002/dvdy.22113. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Sambrook J, Russell D. Purification of Nucleic Acids by Extraction with Phenol:Chloroform. Cold Spring Harb Protoc 2006: pdb.prot4455- Cold Spring Harbor, NY: Cold Spring Harbor Laboratory Press; 2006. [DOI] [PubMed] [Google Scholar]
- 52.Michelmore R, Paran I, Kesseli R. Identification of markers linked to disease-resistance genes by bulked segregant analysis: a rapid method to detect markers in specific genomic regions by using segregating populations. P Natl Acad Sci USA. 1991;88:9828–9832. doi: 10.1073/pnas.88.21.9828. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Rozen S, Skaletsky H. Primer3 on the WWW for General Users and for Biologist Programmers. In: Krawetz S, Misener S, editors. Bioinformatics Methods and Protocols: Methods in Molecular Biology. Totowa, NJ: Humana Press; 1999. pp. 365–386. [DOI] [PubMed] [Google Scholar]
- 54.Hoebeeck J, Speleman F, Vandesompele J. Real-Time Quantitative PCR as an Alternative to Southern Blot or Fluorescence In Situ Hybridization for Detection of Gene Copy Number Changes. In: Hilario E, Mackay J, editors. Protocols for Nucleic Acid Analysis by Nonradioactive Probes. Totowa, NJ: Humana Press; 2007. pp. 205–226. [DOI] [PubMed] [Google Scholar]
- 55.Li C, Orti G, Zhang G, Lu G. A practical approach to phylogenomics: the phylogeny of ray-finned fish (Actinopterygii) as a case study. BMC Evol Biol. 2007;7:44. doi: 10.1186/1471-2148-7-44. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.Nanda I, Feichtinger W, Schmid M, Schröder J, Zischler H, et al. Simple repetitive sequences are associated with differentiation of the sex chromosomes in the guppy fish. J Mol Evol. 1990;30:456–462. [Google Scholar]
- 57.Kirankumar S, Anathy V, Pandian T. Hormonal induction of supermale golden rosy barb and isolation of Y-chromosome specific markers. Gen Com Endocrinol. 2003;134:62–71. doi: 10.1016/s0016-6480(03)00218-1. [DOI] [PubMed] [Google Scholar]
- 58.Kirankumar S, Pandian T. Production and progeny testing of androgenetic rosy barb (Puntius conchonius). J Exp Zool Part A. 2004;301A:938–951. doi: 10.1002/jez.a.117. [DOI] [PubMed] [Google Scholar]
- 59.Baroiller JF, D'Cotta H. Environment and sex determination in farmed fish. Comp Biochem Physiol C Toxicol Pharmacol. 2001;130:399–409. doi: 10.1016/s1532-0456(01)00267-8. [DOI] [PubMed] [Google Scholar]
- 60.Amores A, Postlethwait JH. Banded chromosomes and the zebrafish karyotype. In: Detrich HW, Westerfield M, Zon LI, editors. The Zebrafish: Genetics and Genomics. San Diego, CA: Academic Press; 1999. pp. 323–338. [DOI] [PubMed] [Google Scholar]
- 61.Daga R, Thode G, Amores A. Chromosome complement, C-banding, Ag-NOR and replication banding in the zebrafish Danio rerio. Chromosome Res. 1996;4:29–32. doi: 10.1007/BF02254941. [DOI] [PubMed] [Google Scholar]
- 62.Endo A, Ingalls TH. Chromosomes of the zebra fish. A model for cytogenetic, embryologic, and ecologic study. J Hered. 1968;59:382–384. doi: 10.1093/oxfordjournals.jhered.a107755. [DOI] [PubMed] [Google Scholar]
- 63.Gornung E, Gabrielli I, Cataudella S, Sola L. CMA(3)-banding pattern and fluorescence in situ hybridization with 18S rRNA genes in zebrafish chromosomes. Chromosome Res. 1997;5:40–46. doi: 10.1023/a:1018441402370. [DOI] [PubMed] [Google Scholar]
- 64.Phillips B, Reed M. Localization of repetitive DNAs to zebrafish (Danio rerio) chromosomes by fluorescence in situ hybridization (FISH). Chromosome Res. 2000;8:27–35. doi: 10.1023/a:1009271017998. [DOI] [PubMed] [Google Scholar]
- 65.Pijnacker LP, Ferwerda MA. Zebrafish chromosome banding. Genome. 1995;38:1052–1055. doi: 10.1139/g95-140. [DOI] [PubMed] [Google Scholar]
- 66.Post A. Vergleichende Untersuchungen der Chromosomenzahlen bei Susswassernteleosteern. Z Zool Syst Evol. 1964;3:47–93. [Google Scholar]
- 67.Rishi KK. Karyotypic studies on four species of fishes. Nucleus. 1976;19:95–98. [Google Scholar]
- 68.Schreeb K, Groth G, Sachsse W, Freundt K. The karyotype of the zebrafish (Brachydanio rerio). J Exp Anim Sci. 1993;36:27–31. [PubMed] [Google Scholar]
- 69.Ueda T, Naoi H. BrdU-4Na-EDTA-Giemsa band karyotypes of 3 small freshwater fish, Danio rerio, Oryzias latipes, and Rhodeus ocellatus. Genome. 1999;42:531. [Google Scholar]
- 70.Fontana F, Chiarelli B, Rossi AC. Il cariotipo di alcune specie di Cyprinidae, Centrarchidae, Characidae studiate mediante culture in vivo. Caryologia. 1970;23:549–564. [Google Scholar]
- 71.Sharma K, Sharma O, Tripathi M. Female heterogamety in Danio rerio (Cypriniformes: Cyprinidae). P Natl Acad Sci India B. 1998;68:123–126. [Google Scholar]
- 72.Wallace BMN, Wallace H. Synaptonemal complex karyotype of zebrafish. Heredity. 2003;90:136–140. doi: 10.1038/sj.hdy.6800184. [DOI] [PubMed] [Google Scholar]
- 73.Traut W, Winking H. Meiotic chromosomes and stages of sex chromosome evolution in fish: zebrafish, platyfish and guppy. Chromosome Res. 2001;9:659–672. doi: 10.1023/a:1012956324417. [DOI] [PubMed] [Google Scholar]
- 74.Magerhans A, Müller-Belecke A, Hörstgen-Schwark G. Effect of rearing temperatures post hatching on sex ratios of rainbow trout (Oncorhynchus mykiss) populations. Aquaculture. 2009;294:25–29. [Google Scholar]
- 75.Wellcome Trust Sanger Institute. Zv8, the 8th intergrated whole genome assembly of the zebrafish genome has been released. 2008. The Danio rerio Sequencing Project Available: http://www.sanger.ac.uk/Projects/D_rerio/Zv8_assembly_information.shtml. Accessed 2011 Jun 22.
- 76.Matsuda M, Nagahama Y, Shinomiya A, Sato T, Matsuda C, et al. DMY is a Y-specific DM-domain gene required for male development in the medaka fish. Nature. 2002;417:559–563. doi: 10.1038/nature751. [DOI] [PubMed] [Google Scholar]
- 77.Brown KH, Dobrinski KP, Lee AS, Gokcumen O, Mills RE, et al. Extensive genetic diversity and substructuring among zebrafish strains revealed through copy number variant analysis. P Natl Acad Sci USA. 2012;109:529–534. doi: 10.1073/pnas.1112163109. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 78.Bradley KM, Breyer JP, Melville DB, Broman KW, Knapik EW, et al. A SNP-based linkage map for zebrafish reveals sex determination loci. G3. 2011;1:3–9. doi: 10.1534/g3.111.000190. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 79.Tong S, Hsu H, Chung B. Zebrafish monosex population reveals female dominance in sex determination and earliest events of gonad differentiation. Dev Biol. 2010;344:849–856. doi: 10.1016/j.ydbio.2010.05.515. [DOI] [PubMed] [Google Scholar]
- 80.Arnold AP. The end of gonad-centric sex determination in mammals. Trends Genet. 2012;28:55–61. doi: 10.1016/j.tig.2011.10.004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 81.Ewert M, Nelson C. Sex determination in turtles: Diverse patterns and some possible adaptive values. Copeia. 1991;1991:50–69. [Google Scholar]
- 82.Harvey PH, Slatkin M. Some like it hot: temperature-determined sex. Nature. 1982;296:807–808. doi: 10.1038/296807a0. [DOI] [PubMed] [Google Scholar]
- 83.Ferguson M, Joanen T. Temperature of egg incubation determines sex in Alligator mississippiensis. Nature. 1982;296:850–853. doi: 10.1038/296850a0. [DOI] [PubMed] [Google Scholar]
- 84.Tessema M, Müller-Belecke A, Hörstgen-Schwark G. Effect of rearing temperatures on the sex ratios of Oreochromis niloticus populations. Aquaculture. 2006;258:270–277. [Google Scholar]
- 85.Ospina-Alvarez N, Piferrer F. Temperature-dependent sex determination in fish revisited: Prevalence, a single sex ratio response pattern, and possible effects of climate change. PLoS ONE. 2008;3:e2837. doi: 10.1371/journal.pone.0002837. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 86.Engeszer RE, Patterson LB, Rao AA, Parichy DM. Zebrafish in the wild: A review of natural history and new notes from the field. Zebrafish. 2007;4:21–40. doi: 10.1089/zeb.2006.9997. [DOI] [PubMed] [Google Scholar]
- 87.Spence R, Gerlach G, Lawrence C, Smith C. The behaviour and ecology of the zebrafish, Danio rerio. Biol Rev. 2008;83:13–34. doi: 10.1111/j.1469-185X.2007.00030.x. [DOI] [PubMed] [Google Scholar]
- 88.Abozaid H, Wessels S, Hörstgen-Schwark G. Effect of Rearing Temperatures during Embryonic Development on the Phenotypic Sex in Zebrafish (Danio rerio). Sex Dev. 2011;5:259–265. doi: 10.1159/000330120. [DOI] [PubMed] [Google Scholar]
- 89.Uchida D, Yamashita M, Kitano T, Iguchi T. An aromatase inhibitor or high water temperature induce oocyte apoptosis and depletion of P450 aromatase activity in the gonads of genetic female zebrafish during sex-reversal. Comp Biochem Physiol A Mol Integr Physiol. 2004;137:11–20. doi: 10.1016/s1095-6433(03)00178-8. [DOI] [PubMed] [Google Scholar]
- 90.Krueger W, Oliveira K. Evidence for environmental sex determination in the American eel, Anguilla rostrata. Environ Biol Fish. 1999;55:381–389. [Google Scholar]
- 91.Lawrence C, Ebersole J, Kesseli R. Rapid growth and out-crossing promote female development in zebrafish (Danio rerio). Environ Biol Fish. 2008;81:239–246. [Google Scholar]
- 92.Yu RMK, Wu RSS. Hypoxia affects sex differentiation and development, leading to a male-dominated population in zebrafish (Danio rerio). Environ Sci Technol. 2006;40:3118–3122. doi: 10.1021/es0522579. [DOI] [PubMed] [Google Scholar]
- 93.Shang E, Yu R, Wu R. Hypoxia alters sex hormones, sex ratio and favors a male-dominated population in fish. Comp Biochem Physiol A Mol Integr Physiol. 2007;148:S118–S118. [Google Scholar]
- 94.Pelegri F, Schulte-Merker S. A gynogenesis-based screen for maternal-effect genes in the zebrafish Danio rerio. In: Detrich HW, Westerfield M, Leonard IZ, editors. Methods in Cell Biology. Waltham, MA: Academic Press; 1998. pp. 1–20. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Multifactorial selective breeding was carried out over a few generations for five lines of zebrafish to select for pairs that produced a sex-biased family. All multifactorial crosses were set up using full siblings. The selective breeding process involved selecting for pairs that produce highly biased sex ratio (highlighted by different colour box) then offspring from the selected pairs were used to set up multifactorial crosses in the next generation. This was repeated for a few generations. The two all-male families were from the D8 line (D8F3_1 and D8F3_2). The control family D4F3 had a mass cross performed in the F2 generation without selection.
(XLSX)
Plots of the number of fish present in each population over time. Each line represents one population, housed in a single tank. Data points of fish counts are represented by diamonds. Populations with higher than the overall average percentage of males are colored orange while those populations with a lower than average male percentage are colored blue. These data are from experiment 1 and 2 shown in Table S1.
(PPTX)
Sex-linked FluoMEP markers obtained by bulk segregant analysis performed on pooled male and female samples of guppy ( Poecilia reticulata). A) and rosy barb (Puntius conchonius; B). Primer combinations are indicated on the top right corner of the peak profiles. Red boxes indicate the sex-linked markers that were confirmed through individual testing. The remaining differences are false positives that have occurred with similar frequency in both species depicted here, as well as in the zebrafish (not shown).
(PPTX)
Inheritance pattern of the five CNVRs universal to all four families. None of them showed sex-linked inheritance pattern. Gain in copy number is indicated by blue box while loss in copy number is indicated by red box.
(PPTX)
Data tables for rearing density experiment 1 and 2.
(XLSX)
Sequences of all primers used.
(XLSX)