Summary
Encased within the 280 kb genome in the capsid of the giant myovirus φKZ is an unusual cylindrical proteinaceous “inner body” of highly ordered structure. We present here mass spectrometry, bioinformatic, and biochemical studies that reveal novel information about the φKZ head and the complex inner body. The identification of 39 cleavage sites in 19 φKZ head proteins indicates cleavage of many prohead proteins forms a major morphogenetic step in φKZ head maturation. The φKZ head protease, gp175, is newly identified here by a bioinformatics approach, as confirmed by a protein expression assay. Gp175 is distantly related to T4 gp21 and recognizes and cleaves head precursors at related but distinct S/A/G-X-E recognition sites. Within the φKZ head there are six high copy number proteins that are probable major components of the inner body. The molecular weights of five of these proteins are reduced 35–65% by cleavages making their mature form similar (26–31 kDa), while their precursors are dissimilar (36–88 kDa). Together the six abundant proteins sum to the estimated mass of the inner body (15–20 MDa). The identification of these proteins is important for future studies on the composition and function of the inner body.
Keywords: myovirus, serine protease, cleavage motif, phiKZ, mass spectrometry, abundant capsid proteins, PF12699 paralog family, Hidden Markov Modelling
Introduction
The Pseudomonas aeruginosa phage φKZ is the type virus of a genus of “giant” myoviruses, the φKZ-like phages, whose members have been isolated from diverse geographic locations and are infective for a variety of Pseudomonas species (Krylov et al., 2007). The φKZ-like phages and their gene products are important due to their potential to control pathogenic pseudomonads (Briers et al., 2008, Miroshnikov et al., 2006, Paradis-Bleau et al., 2007, Mesyanzhinov et al., 2002) and genetically manipulate their hosts (Monson et al., 2011). Some, such as φKZ, have already been incorporated into traditional phage therapy cocktails and continue to be examined for novel therapeutic applications [e.g., (Golshahi et al., 2011, Matinkhoo et al., 2011)]. φKZ has a long, 280 kb linear dsDNA genome that encodes numerous proteins that are remarkably diverged from those in other phages, thus making straightforward functional assignments very difficult (Mesyanzhinov et al., 2002).
The φKZ virion also presents a singular research focus as its large T=27 head (Fokine et al., 2005a) contains an unusual immense cylindrical protein “inner body” (IB) encased within the genomic DNA (Krylov et al., 1984). The highly ordered structure of this IB has recently been solved (Wu et al., 2012). The IB is a conserved feature in related phages [(e.g., 201 φ2-1, φPA3, EL, OBP (Krylov et al., 2007)] suggesting it is an essential component of the φKZ-like phages. However, the function, composition, and role in infection of the IB has remained elusive since it was first identified (Krylov et al., 1984). The IB may well prove to be multi-functional and could have roles related to DNA packaging, packaged genome structure, DNA ejection, or it may play a role in all of these aspects of phage development. More specifically, the structurally defined IB may be important for size determination of the T=27 capsid and capsid stability; spooling of DNA tightly around the IB could reduce some of the DNA pressure that might otherwise be exerted on the capsid shell. Spooling of the DNA around the IB may also be necessary to organize such a length of dsDNA at ~500 mg/ml in a sufficiently ordered structure that allows efficient and rapid ejection (Black and Thomas, 2011). The IB may also represent a reservoir of proteins injected into the host cell together with the DNA that are necessary for host takeover. Supporting this hypothesis is that the IB is not detected after infection and injection of the packaged DNA (Krylov et al., 1984). Moreover, in addition to the IB, a number of enzymes [e.g., multi-subunit-β,β′-RNA polymerase (RNAP)] are also found within the φKZ head with an unknown structural relationship to it.
Previous biochemical studies have illustrated the complexity of the φKZ head, containing at least 30 (Lecoutere et al., 2009), but possibly more than 40 (Mesyanzhinov et al., 2002) different proteins. This is in contrast to the head of T4 which contains 13 different proteins (Black et al., 1993). The location of two head proteins (gp120 and gp89) in the complex φKZ SDS-PAGE profile were identified (Mesyanzhinov et al., 2002), however ambiguity regarding the exact locations of other head proteins presented a major hurdle for even simple studies of the head. Also, other than gp89 and gp120, the identity of any other major or high copy number proteins in the φKZ head was unknown. However, the identity of high copy number head proteins is critical for further studies of the IB as any such proteins would become candidates for components of this large structure. Similarly, while gp120 and gp89 were identified as processed (Mesyanzhinov et al., 2002) it was undefined whether other φKZ head proteins were cleaved or not, or which protein represented the prohead protease in any of the φKZ-like phages. Comprehensive mass spectral analyses of P. chlororaphis phage 201 φ2-1 demonstrated that thirteen of its proteins were cleaved at a motif reminiscent of the T4 processing motif (Thomas et al., 2010). However, the exact cleavage sites in all φKZ head proteins could not be inferred from the 201 φ2-1 analyses since these phages are not closely related as illustrated by their homologous virion proteins having ~20–70 percent identity as determined by Blastp (Thomas et al., 2008).
Here we report mass spectrometry (MS) and biochemical analyses of the φKZ tailless full head mutant and a prohead precursor and show that cleavage of numerous head proteins occurs in φKZ. This processing must play a major role in the formation of the IB, in addition to the capsid. Furthermore, using advanced bioinformatic approaches we have identified the φKZ prohead protease which cleaves multiple head proteins at a consensus amino acid recognition motif. This protease is strongly conserved among all known members of this group of phages, reflecting its important role in morphogenesis. We also identify six high copy number proteins (~100–200 copies) within the φKZ head as being IB proteins. This determination was made based on their release from full heads and by their presence in terminase-inhibited protein precursors-processed DNA-free proheads. These characteristics lay the groundwork necessary for a genetic determination of the function(s) of the extraordinary IB structure.
Results and Discussion
Mass spectrometry of φKZ tailless full heads
HPLC-electrospray ionization tandem mass spectrometry (HPLC-ESI-MS/MS) analyses of CsCl purified φKZ tailless mutant (Lavigne et al., 2009, Fokine et al., 2005a, Plotnikova et al., 1982) tailless full heads permitted assignment of peptides to 50 φKZ gene products (Figure 1). Twenty-nine of these proteins have previously been identified in tailless heads (Lecoutere et al., 2009). However, one protein identified by Lecoutere et al., gp128, was not found in our analyses. Conversely, 19 proteins were identified in the current study that were not reported to be present in tailless heads and/or wild type φKZ particles (Lecoutere et al., 2009). Detection of these additional proteins most likely resulted from the approach used here which permitted sampling of low molecular weight proteins and low abundance proteins. Eleven proteins identified in this study (gps 27, 85, 91, 92, 119, 144, 153, 157, 184, 244 and 298) were not identified previously in the tailless heads, although they are φKZ virion proteins (Lecoutere et al., 2009). Eight proteins (gps 42, 81, 96, 129, 177_2, 199, 203 and 228) were identified in the tailless heads in this study that were not previously found in the φKZ virion. These proteins are unlikely to be contaminants as the heads were judged to be highly purified by electron microscopy and the reversion rate of the mutant is about 10−5–10−6. Many of these newly identified proteins are expected to be additional head proteins based on their possessing characteristics indicative of head proteins. For instance, the predicted function of gp129 is that it forms the portal (Cornelissen et al., 2011). Similarly, other proteins have qualities that support them as being head proteins, such as being cleaved at a head prohead protease cleavage motif (e.g., gps 92, 96, 153, 129 and 203), and/or their gene being positioned within, or close to, a major head gene module (e.g., gps 91 and 177_2)(Figure 2).
Figure 1.
SDS-PAGE of proteins in purified φKZ tailless heads. The boundaries of slices that were digested and analyzed by MS are shown to the left of the processed gel lane. Each virion protein is listed next to the slice containing its maximum spectral count. gp refers to φKZ gene product. Virion proteins listed in red/underlined are proteolytically processed by the prohead protease, as confirmed by the detection of a semi-tryptic peptide (see Table 1). In cases where more than one polypeptide segment remained in the virion, the segments have been amended with N, M, or C to indicate N-terminal, middle- (if present) and C-terminal segments, respectively. Newly identified gene products in the tailless mutant are marked with an asterisk. Molecular weight standard was the Fermentas PageRuler Plus Prestained protein ladder.
Figure 2.
Regions of the φKZ genome encoding processed head proteins identified in full heads. Genomic segments from the full GBrowse display of φKZ with peptide coverage from MS of the tailless mutant mapped onto the corresponding genes. Legend: red arrows, proteins detected by MS and determined to be processed; light green arrows, proteins detected by MS, but not known to be processed; black outlined arrows, proteins determined to be abundant (see below); grey arrows, proteins not detected by MS. Boxed regions indicate peptide coverage in purified tailless mutant: lime green, tryptic peptide coverage; red, semi-tryptic peptide ending in glutamate after a processing motif (see Figure 4). Triangles above the peptide coverage boxes indicate prohead protease cleavage sites: dark blue, established by a semi-tryptic peptide in the purified head GeLCMS; purple, established by a semi-tryptic peptide detected in the osmotic shock or acetic acid treated samples that underwent MS analyses.
Cleavage of φKZ head proteins
Information gained by comparing observed and expected MS sequence coverage and 1-D gel migration provided insight into the catalog of φKZ head proteins that had undergone proteolytic cleavage (Table 1). However, the strongest evidence for cleavage was provided by the presence of a “semi-tryptic peptide”, that is, a peptide with a non-tryptic terminus (i.e., no arginine or lysine) in high confidence peptide assignments; these semi-tryptic peptides were detectable by including “semi-tryptic” cleavage as a parameter in the database searches of the tandem MS data. These efforts were facilitated by the detection of peptides from four different φKZ-derived preparations that underwent MS and are reported in this manuscript. High peptide coverage, particularly for the abundant proteins, was frequently observed (see Figure 2). Forty peptides were found that had been produced by cleavages immediately after a glutamic acid, confirming a consensus processing site(s) for 19 different proteins (Figure 2, Supplementary Table 1). Summation of the number of φKZ head proteins and their observed cleavage sites indicates the remarkable amount of processing that must be undertaken by the morphogenetic protease – the cleavage of at least 6200 peptide bonds per prohead, perhaps several fold more.
Table 1.
Prohead protease processed segments of head proteins from the φKZ tailless mutant determined by mass spectrometry. Proteins or segments predicted to be present in the mature virion in more than 100 copies are bolded.
| Residue2 | ||||||
|---|---|---|---|---|---|---|
| gp1 | Slice | Start | Stop | Mw3 | SC4 | SC/Mw5 |
| 84 | 10 | 11* | 413(C) | 45.7 | 34 | 0.7 |
| 86 | 8 | 123* | 428(C) | 35.4 | 47 | 1.3 |
| 89N | 1 | 1 | 122* | 13.1 | 18 | 1.4 |
| 89C | 5 | 156* | 387(C) | 26.1 | 117 | 4.5 |
| 92 | 10 | 86* | 440(C) | 41.1 | 11 | 0.3 |
| 93 | 7 | 157* | 435(C) | 30.9 | 177 | 5.7 |
| 94N | 1 | 12* | 95* | 9.2 | 11 | 1.2 |
| 94M | 1 | 96* | 275* | 20.1 | 15 | 0.7 |
| 94C | 5 | 276* | 505(C) | 26.4 | 14 | 0.5 |
| 95 | 7 | 261* | 547(C) | 31.6 | 177 | 5.6 |
| 96 | 8 | 66* | 371(C) | 34.9 | 6 | 0.2 |
| 97N | 14* | 66* | 5.8 | 24 | 4.1 | |
| 97C | 7 | 533* | 816(C) | 30.7 | 152 | 5.0 |
| 120, capsid | 12 | 164* | 747(C) | 64.9 | 1672 | 25.8 |
| 129N | 3 | 1 | 262* | 29.3 | 5 | 0.2 |
| 129C, portal | 12 | 263* | 896(C) | 70.1 | 89 | 1.3 |
| 153 | 5 | 53* | 304(C) | 28.2 | 42 | 1.5 |
| 162N | 3/4 | 18* | 137* | 13.2 | 41 | 3.1 |
| 162C | 6 | 259* | 522(C) | 29.0 | 156 | 5.4 |
| 163 | 8 | 55* | 395(C) | 38.1 | 45 | 1.2 |
| 175 | 4 | 1 | 210* | 23.7 | 39 | 1.6 |
| 177N | 1 | 1 | 60* | 6.4 | 9 | 1.4 |
| 177C | 11 | 61* | 519(C) | 52.4 | 126 | 2.4 |
| 180_180_2 | 12 | 1 | 505 | 57.8 | 36 | 0.6 |
| 184N | 3 | 1 | 108* | 12.1 | 6 | 0.5 |
| 184C | 1 | 109* | 230 | 13.2 | 21 | 1.6 |
| 203 | 12 | 1 | 653* | 75.3 | 71 | 0.9 |
| 303N | 8 | 61 | 268 | 22.8 | 38 | 1.7 |
| 303M | 1 | 269 | 366(C) | 11.1 | 17 | 1.5 |
| 303C | 7 | 367 | 646(C) | 31.3 | 62 | 2.0 |
The gene product name is appended with N (for N-terminal), M (for middle), or C (for C-terminal) when more than one segment of a gene product were detected in the virion.
A coordinate is appended with an * if confirmed by a semi-tryptic spectrum, or by (C) for an end defined by the stop codon. Semi-tryptic spectrum assignments were above 95% confidence except as noted in Supplementary Table 2.
The molecular weight (MW) was calculated from the sequence.
Peptide spectral counts (SC) were summed over all slices.
SC/MW values were calculated as an approximate indicator of abundance of the polypeptides per virion.
Cleavage of φKZ head proteins occurs at a modified T4-like protease motif
Inspection of the 39 φKZ cleavage sites showed that in addition to cleavage occurring invariably immediately after a glutamic acid there is a strong preference for a serine in the third position upstream of the cleavage site (26 instances), although an alanine or a glycine is also accepted in this position (8 and 4 instances, respectively, Supplementary Table 1). In one instance (gp303) cleavage apparently occurred where phenylalanine is in the third position upstream of the cleavage site. Notably, there is little requirement for any other specific residues, particularly downstream of the cleavage site (Supplementary Figure 2). Overall, the φKZ consensus cleavage sequence (S/A/G-X-E, where X is any amino acid) is similar to, but not the same as that of the T4 prohead protease which cleaves after the motif I/L-X-E (more than 13 cleavages in eight different precursor proteins)(Black et al., 1993). In T4, there is also no requirement for specific residues downstream of the cleavage site. Also, in T4 there is evidence that a shift in the processing motif within a polypeptide results in a retargeting of the cleavage site by the protease (Black et al., 1993, Mullaney and Black, 1996).
Identification of cleavage sites was critical for length determination of the mature polypeptide(s) for each φKZ protein (Table 1). Notably, for some proteins, processing was so extensive that only 30–50% of the precursor protein is retained in the mature head (e.g., gp97, see Table 2). The manner in which several proteins were processed is intriguing, with several proteins cleaved into two or three polypeptides that remain in the mature head (e.g., gps 89, 97, 177, 303). Some of the smaller fragments (e.g., gp89N and gp97N) may be non-functional debris and unable to be cleared from the capsid due to their locale or amino acid composition, similar to the case for the phage T4 “acid soluble peptides” that are found within the mature capsid; alternatively these fragments may have unknown functional significance.
Table 2.
Abundant head proteins in φKZ, in addition to the major capsid protein (gp120, 1560 copies).
| gp | Predicted Mw, kDa | Processed Mw, kDa (%reduction) | Copy number (densitometry) |
|---|---|---|---|
| 89 | 43.2 | ||
| 89C | 26.1 (40) | 60–100 | |
|
| |||
| 90 | 35.9 | 190–230 | |
|
| |||
| 93 | 47.6 | ||
| 93C1 | 30.9 (35) | 110–170 | |
|
| |||
| 95 | 58.5 | ||
| 95C1 | 31.6 (46) | 110–170 | |
|
| |||
| 97 | 87.8 | ||
| 97C | 30.7 (65) | 110–170 | |
|
| |||
| 162 | 57.6 | ||
| 162C1 | 29.0 (50) | 90–160 | |
proteins containing the paralogous domain (PF12699)
Cleavage sites identified in both gps 89 and 120 were consistent with the N-termini identified by Edman degradation (Mesyanzhinov et al., 2002). However, additional processing sites, N-terminal to those found by Edman degradation were also identified in both these proteins, as evidenced by the presence of low abundance semi-tryptic peptides (see Figure 3A for gp120). Similar semi-tryptic peptides that indicated that additional cleavages had occurred upstream of the maturation cleavage site were also identified in a number of other proteins [e.g., gp93 (Figure 4B) and gps 94, 95, 97, 162, 303 (Figure 2)]. The low abundance of these semi-tryptic peptides suggests that they are not present in every particle; the fact that they were identified at all is a consequence of the low detection limits of the MS analyses. Identification of these additional processing sites is important because they demonstrate that there is a multiplicity of processing in the precursor or immature regions of the φKZ head proteins. Examination of the regions in which multiple cleavages have been confirmed by semi-tryptic peptides typically also revealed other potential cleavage motifs in these regions (see Figure 3A and B), suggesting that the detected low abundance cleavages represent a fraction of what actually occurs in the φKZ head during maturation. It seems feasible that there is a redundancy of cleavage sites in these regions that may have evolved as a protective measure to ensure removal of regions of the protein not required in the mature head, raising the question as to whether there is a preferred length for cleaved fragments to ensure optimal removal from the prohead. Observation of sequences in the mature regions of proteins apparently adhering to the cleavage motif but which are apparently never cleaved (e.g., GFE-457 or ALE-723 in gp120, Figure 3A) indicates there are likely to be additional specificity or structural determinants required for cleavage in addition to the S/A/GXE motif.
Figure 3.
Cleavage sites in head proteins of φKZ and their conservation in related phages. Cleavage motifs in (A) the φKZ major head protein, gp120 and (B) φKZ abundant head protein, gp93. Red arrows indicate cleavage sites identified by MS; purple arrows indicate potential cleavage sites that adhere to the cleavage motif, see Figure 4. Blue circles indicate sequences that adhere to the processing motif but are not cleaved. (C) Conservation of cleavage motifs in proteins homologous to processed φKZ proteins. KZ, refers to φKZ, 2-1 refers to 201 φ2-1, and PA3 refers to φPA3. Red underlined residues are identified cleavage sites in φKZ proteins and in (vi) 201 φ2-1 (Thomas et al., 2010). Conserved residues in other phage proteins are also red. Comparison of processing sites in homologs to the φKZ (i) major capsid protein, gp120 (ii) protease, gp175, (iii) gp84, (iv) gp92, (v) gp120 (maturation cleavage), (vi) gp162, and (vii) portal protein, gp129. (D) Conservation of processing motifs in proteins homologous to processed T4 proteins. 44RR refers to 44RRt.8. Red underlined residues are known processing sites in T4 proteins (Black et al., 1993). Conserved residues, representing putative processing sites in homologous proteins are also red. Putative processing motifs in homologs to the T4 processed protein that are not strictly conserved are in bold. Comparison of processing sites in T4 (i) and (ii) major capsid protein, gp23, and (iii) major scaffold protein, gp22 are to their homologs.
Figure 4.
Features of the φKZ morphogenetic protease, gp175. (A) Comparison of the processing of φKZ gp175 compared to that of T4 gp21. Processing of gp21 converts the inactive form into an active serine protease. (B) HHpred alignment of φKZ gp175 HHM with that of T4 gp21. Above the alignment is the predicted secondary structure for gp175 obtained using Psi-Pred. Below the alignment are the structural elements of the herpesvirus fold positioned relative to the T4 sequence and labeled according to (Cheng et al., 2004). Conserved catalytic residues (histidine and serine) are highlighted in red [red stars in (A)]. Candidate third catalytic triad residues are highlighted in light blue and green [light blue and green stars in (A)].
Cleavage of numerous prohead proteins likely represents an essential head morphogenesis step in all φKZ-related phages
The identification of so many cleavage sites in φKZ head proteins enables us to draw conclusions as to the cleavage of head proteins in other phages, particularly 201 φ2-1 and φPA3, phages that are members of the φKZ-like phage genus (Cornelissen et al., 2011). Comparison of the identified cleavage sites in this study in φKZ and those identified in 201 φ2-1 indicates that their respective proteases have very similar cleavage site residue requirements (S/A/G-X-E) and this requirement is likely to be a feature of phages in the φKZ genus. The cleavage sites in 12 φKZ proteins (e.g., gps 86, 89, 96, 153, and 162) are conserved with identified cleavage sites in their respective homolog in 201 φ2-1 (Thomas et al., 2010) and from this work we can infer about another 15 cleavage sites in 201 φ2-1 head proteins (Table S2). From the identified cleavage sites in φKZ and 201 φ2-1 proteins we can also infer cleavages occur at about 27 sites in homologous proteins in φPA3 (Table S2).
We observed that frequently the degree in which a φKZ cleavage site was conserved in its homologs correlated with the overall degree of conservation of the homologs themselves (see Table S4). For instance, the cleavage sites in two highly conserved proteins (major capsid and protease, see below) are the only sites perfectly conserved in their 201 φ2-1 and φPA3 homologs with regard to the three residues upstream of the processing site (Figure 3Ci and ii). However, despite more variations in cleavage site residues with more diverse homologs, we typically observed a conservation of putative cleavage site residues that mirrored the pattern seen in φKZ cleavages. A number of the φKZ cleavage sites are conserved in homologous proteins only with regard to the first and third residues upstream of the processing site, as seen in the homologs to gps 84 and 92 (SXE, Figure 3Ciii and iv). Similarly, in some homologous proteins the third residue upstream of a proposed cleavage site is replaced with an A or G as observed in some φKZ cleavages (e.g., Figure 3Cv and vi). In homologs of other φKZ processed proteins, sequences that adhere to the cleavage motif were frequently found in a position not aligning with, but within a few residues of the actual point of cleavage in the φKZ protein. We believe these motif-like sequences also represent cleavage sites (e.g., Figure 3Cvii).
Notably, we were unable to infer corresponding cleavage sites in the homologs to several φKZ proteins (egs. gps 93, 95, 97 and 303, Table S2). Typically, these are not strongly conserved proteins, however we suspect that numbers of these proteins in φKZ, and most likely their homologs in related phages, have an important role in the mature particle based on their abundance in the φKZ head (see below). Overall, the identification of numerous cleavage sites in φKZ greatly advances our knowledge and expectations of cleavage events in the φKZ-like phages. It also emphasizes that prohead protease cleavages form a major step in capsid maturation for these phages.
Identification of φKZ gp175 as the morphogenetic protease
Sequence-sequence based searches were unable to identify the enzyme responsible for the extensive number of cleavages in φKZ head; hence we sought to identify the φKZ prohead protease using Hidden Markov Model (HMM) based searches. To do this we used local implementations of the Sequence Analysis and Modeling System [SAM (Hughey and Krogh, 1996, Karplus et al., 1998)] and HHpred (Söding, 2005). Initially, Hmmscore (a profile to sequence search) of a custom T4 gp21 model against a library of proteins from four φKZ-like phages (φKZ, EL, φPA3 and 201 φ2-1) identified homologous proteins in the four phages: EL gp192 (E-8), φKZ gp175 (E-6), 201 φ2-1 gp168 (E-5) and φPA3 gp205 (E-3).
To challenge these matches, an alignment of these four protease candidates was constructed using SAM and then converted into a HHM and used for HHpred searches (profile-profile comparisons). An HHsearch of the φKZ gp175 family HHM against all HHpred libraries (from ftp://toolkit.lmb.uni-muenchen.de/HHsearch/databases/) found the best match to be to PHA00911 gp21 prohead core scaffolding protein and protease. The E-value of this match was only 3, however the HHpred P-value, which takes secondary structure into account, was 2.5E-05. This match was solidified using a custom T4 gp21 HHM powerful enough to detect an HHM based on the U35 peptidase family at E-values of 3.5E-05. Members of the U35 peptidase family contain diverged, but known, homologs to the T4 protease, such as the HK97 protease (Liu and Mushegian, 2004, Cheng et al., 2004). When searched against the custom gp21 HHM the φKZ gp175 HHM scored slightly better (2.3E-06, Figure 4) than the U35 family HHM, indicating that φKZ gp175 and its homologs in the φKZ-related phages are homologous, albeit extremely diverged, from T4 gp21.
To confirm the identity of gp175 as the φKZ prohead protease we expressed its predicted mature form in Escherichia coli DH10B. An extract incubation assay of gp175 using expressed full length gp93 as a substrate showed little non-specific proteolysis of other proteins, whereas the majority of gp93 observable by SDS-PAGE was cleaved in a good yield to a length consistent with that found in the mature virion (Figure 5). Protease active site knockout control incubations (data not shown) will be published together with a more detailed enzymatic characterization.
Figure 5.
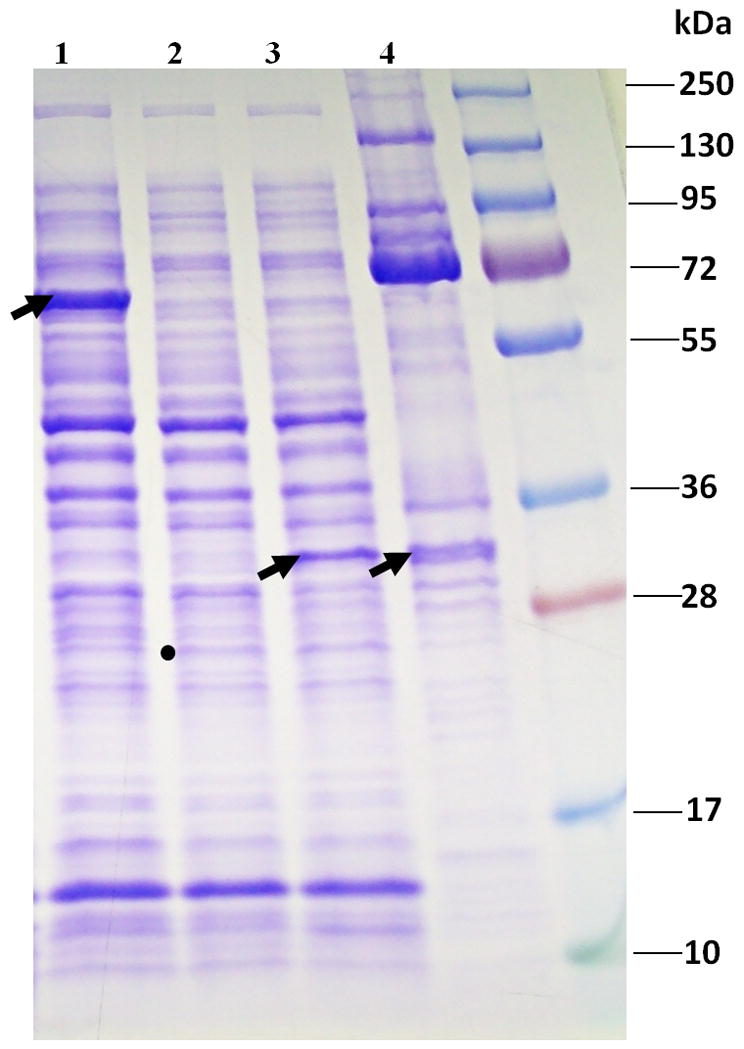
SDS-PAGE of an expression vector assay of the φKZ protease showing cleavage of φKZ head protein gp93. Lane 1, Arrow indicates full length gp93 expressed in DH10B. Lane 2, Dot shows position of mature gp175 in DH10B, much additionally found in the pellet fraction (data not shown). Lane 3, Arrow indicates cleaved gp93 after incubation with mature gp175. Lane 4, Arrow indicates the position of cleaved gp93 in purified wild-type φKZ virions. Lane 5, Molecular weight standard was the Fermentas PageRuler Plus Prestained protein ladder.
φKZ gp175 conserves catalytic residues and structural elements of serine proteases
φKZ gp175 conserves the proposed serine protease active site residues H-85 and S-140 in T4 gp21 (Figure 4); these residues have been shown to be conserved across the phage U9 and U35 families to the S21 family that contains herpesvirus protease (Liu and Mushegian, 2004, Cheng et al., 2004). The final residue of the serine protease catalytic triad (an aspartate or glutamate in classical serine proteases) is less well conserved throughout the phage/virus phylogenetic spectrum (Cheng et al., 2004). In the U9 family this amino acid position is most often an alanine or glycine (Cheng et al., 2004), while in the φKZ-like phages it is a threonine (Figure 4B). This third residue of the catalytic triad is not considered to play a major role in catalysis and it has been proposed that for the U9 family catalysis might only require a catalytic dyad (Cheng et al., 2004). Alternately, the third residue of the catalytic triad might have migrated—possibly to the well-conserved aspartate two residues downstream of the alanine/glycine residue (Cheng et al., 2004), which is also conserved in the φKZ-like protease candidates (Figure 4B).
The structural elements from a major portion of the herpesvirus protease (β1 – β6), and other members of the peptidase S21 superfamily, also have predicted secondary structure counterparts in phage proteases, with the exception of the herpesvirus αB, which is involved in homodimer formation (Liu and Mushegian, 2004, Cheng et al., 2004). Counterparts to each of the herpesvirus protease elements from β1 – β5, with the exception of β6, are also predicted for φKZ gp175 (Figure 4).
C-terminal cleavage of gp175 may indicate activation by autocleavage
φKZ gp175 is a low copy head protein in the mature head based on an assignment of a relatively low number of spectra to the ~24 kDa protein in our MS experiments (29, compared to 1405 spectra for gp120, the ~65 kDa major capsid protein). This is similar to T4 where only three copies of the T4 protease remain in the mature head, although there are 72 copies of the zymogen present in the unprocessed prohead (Black et al., 1993). Based on the T4 precedent, it is likely to be biological important that φKZ gp175 is C-terminally cleaved at E-210. In T4 the protease is the only head protein known to be C-terminally cleaved, probably at LAE-206 (Keller and Bickle, 1986). Notably, this cleavage of the protease zymogen is an autocleavage event that activates the protease (Showe et al., 1976a, Showe et al., 1976b). The protease auto-activation step is closely controlled by prohead assembly, likely regulated by addition of the vertex proteins (Black et al., 1993). We expect that the processing of φKZ gp175 also activates its zymogen form.
Processed φKZ head proteins of known or inferred function
Only about one quarter of the processed φKZ proteins have either a known function or one that can be inferred by bioinformatics. These are the major capsid protein (gp120), the portal protein (gp129), the protease (gp175), the virion RNAP beta’subunit (gp180), and a helicase (gp203). Several of these proteins have novel or unusual properties. For instance, the fact that the φKZ portal is processed is unusual as processing of the portal protein is an event rarely reported in tailed phages, although it is known that 21 and 26 residues are removed from the N-termini of the λ and P2 portal proteins, respectively (Walker et al., 1982, Rishovd et al., 1994, Rishovd et al., 1998). In fact in T4 the portal is the only early prohead protein that is not processed despite this phage having a relatively high number (13) of processed proteins in its capsid (Black et al., 1993). That the φKZ portal may have such an unusually large propeptide (262 residues) removed suggests it may have more than just a core-targeting function and may contribute significantly to the core itself, possibly connecting the portal to the precursor form of the IB in the prohead.
Helicases and multi-subunit RNAPs have not commonly been found in the virions of characterised phages. In φKZ, presumably the RNAP subunits are injected into the host for transcription of early genes (Thomas et al., 2008). Possibly, they could have an additional role in genome translocation out of the capsid as in T7 (Grayson and Molineux, 2007). Possible functions for gp203 (one of three helicases encoded by φKZ) are more speculative. Gp203, homologous to the SNF-2 helicases, might be injected into the host where it may have a role in DNA transcription or replication, or it might be involved in organization of the DNA within the head. The identification of C-terminal cleavages at a protease motif in the RNAP beta’ subunit and gp203 helicase is a novel finding and supports these enzymes being incorporated into the head based on our knowledge of T4 head morphogenesis (Black et al., 1993).
Abundant φKZ head proteins that are candidates for major IB components
The major capsid protein for φKZ has been reported to be present in 1560 copies per virion (Fokine et al., 2005a). In this study, the relative abundances of the proteins in the φKZ head were estimated from the number of peptide mass spectra assigned with high confidence, after normalization on the basis of calculated molecular weight (SC/Mw), and densitometry. SC/Mw for gps 89, 90, 93, 95, 97 and 162 were substantially higher than those for most of the other head proteins, ranging from 4.5–5.7 (Table 1). The SC/Mw for the major major capsid protein, gp120, was 25.8. However, as a consequence of saturation effects on relative quantification of the major capsid protein using this approach (Thomas et al., 2010), the copy numbers derived from SC/Mw are expected to be over-estimated for gps 89, 90, 93, 95, 97 and 162. In order to obtain an alternative assessment of relative quantities, image analysis was conducted of the Coomassie-stained SDS-PAGE bands containing these six proteins in both purified phage and tailless heads. The densitometry results were in agreement with the MS data, supporting the conclusion that these six proteins are present in high abundance (>100 copies per head, Table 2). This indicates that each φKZ head is composed of more than 2000 protein subunits.
A number of lines of evidence identify these six abundant head proteins as probable major components of the IB. First, the genes encoding five of the abundant head proteins (gps 89, 90, 93, 95 and 97) are located within a single gene module (Figure 2). Although genes with related morphogenetic functions are often observed to be clustered in other phage genomes, the clustered location of these five genes is in φKZ more significant because of the otherwise overall absence of the normal head gene synteny in φKZ (Figure 2). Second, the sum of the observed masses of the IB candidate proteins is consistent with the total estimated mass of the IB (15 – 20 MDa)(Wu et al., 2012). It is important to note that as a consequence of processing, the molecular weights of the mature forms of the six abundant φKZ IB proteins are remarkably similar (Table 2), despite major differences in the sizes of their precursors. Notably, three of the abundant proteins, gps 93, 95 and 162, contain a paralogous domain (PF12699) that is also present in two of the non-abundant head proteins, gp94 and gp163. By analogy to gene duplication events in other phages [e.g., T4 gps 23 and 24 (Fokine et al., 2005b)], the φKZ proteins containing the PF12699 domain are likely to have evolved specialized, although related, functions. Each of the φKZ-related phages has a set of paralogs belonging to this family which we predict to have a major influence on the IB structure.
φKZ internal head proteins
The similarity in the proteolysis of the φKZ head proteins to those in T4 suggests similar overall head morphogenesis. In T4 an immature, uncleaved form of the prohead is first assembled around a structure-determining scaffolding core. The immature prohead is then converted by the gp21 protease to a more mature prohead whose outer shell is composed of the cleaved capsid proteins gp23 and gp24. Additionally, in the mature prohead most of the internal form-determining scaffolding components (gps 22, 21, 67 and 68) have been proteolytically removed. Other scaffolding components nonessential for prohead assembly undergo only limited N-terminal processing and are retained by the mature prohead into which DNA is then packaged by the terminase. Upon phage infection these proteins [the internal proteins (IPs), IPI, IPII, IPIII and Alt] are injected into the host together with the DNA (Black et al., 1993). Addition of the phage T4 terminase inhibitor 9-aminoacridine (9AA) to infected bacteria blocks DNA packaging and leads to the accumulation of fully processed mature T4 proheads that contain the cleaved IPs but lack DNA (Schaerli and Kellenberger, 1980).
Host uptake of 9-aminoacridine has a strikingly similar effect on φKZ head morphogenesis enabling us to isolate a comparable packaging-inhibited φKZ prohead in which the abundant, candidate IB processed head proteins are components (Figure 6). The 9AA particles isolated by glycerol gradient centrifugation (Figure 6A) contained the mature capsid protein and a number of other abundant inner body proteins in proportions comparable to those found in the mature capsid (Figure 6B). The particles contained little, if any DNA (Figure 6C). Significantly, electron microscopy of these particles revealed a kernel-like object within the DNA-free prohead which apparently represents a precursor or degraded form of the IB that evidently can assemble in the absence of DNA packaging (Figure 7, Supplemental Figures 3 and 4). Isolation of these processed mature proheads shows that the φKZ protease operates like the T4 protease, prior to and independent of DNA packaging, and unlike the adenovirus protease that translocates along the packaged DNA to access its cleavage sites in the capsid (Mangel et al., 2003).
Figure 6.

Purification of processed DNA-free IB containing proheads produced by terminase inhibitor 9AA. (A) SDS-PAGE of fractions from a glycerol gradient of 9AA-treated φKZ. Fraction numbers are given above each lane and are numbered from the bottom to the top of the gradient. P refers to the resuspended pellet fraction. (B) SDS-PAGE of purified whole φKZ (KZ; KZ2 refers to 2-fold dilution) and Fraction 3 (F3). Arrow indicates the band containing the mature form of the major capsid protein. Note the similar proportions of cleaved IB proteins in proheads and phages (bracketed). (C) Agarose gel stained with ethidium bromide showing DNA content of purified whole phage (KZ); pellet fraction (P); and Fraction 3 (F3).
Figure 7.

(A) Cryo-electron micrograph of purified φKZ capsid-related particles produced in the presence of 9AA. The thin-walled polyhedral shape suggests that the capsids have the mature conformation. Most particles have an internal structure that appears to be related to the inner body but is shorter and more variable in appearance and may represent breakdown products. (B) Central 1.6 nm-thick slices through cryo-electron tomographic reconstruction of four 9AA-treated φKZ capsids. White arrows point to cylindrical internal densities that in some cases have about the same diameter as the inner body but are shorter and do not pass through the center of the capsid. Space bars represent 100 nm.
Proteins released from the T4 capsid shell by osmotic shock were observed to be either scaffolding core-related or IP-related (Black et al., 1993). Osmotic shock treatment of φKZ purified tailless heads followed by separation of the empty shells (Supplemental Figure 5) from the released proteins, enabled the identification of proteins released from the capsid, and therefore, most likely packaged within the capsid itself and not tightly associated with the capsid shell (Figure 8). The released proteins included the abundant IB proteins listed in Table 2 (Figure 8) and based on the T4 precedent are likely to have scaffolding core- and/or IP-related functions.
Figure 8.
Osmotic shock treatment of φKZ tailless heads. (A) SDS-PAGE of glycerol gradient fractions of the tailless mutant after osmotic shock treatment. P, refers to resuspended pellet. Fractions are numbered from the bottom of the gradient (1) to the top of the gradient (14). Fractions 1 and 2 were combined. (B) DNA gel of the glycerol fractions, showing no or little DNA in fraction 6 that contained empty shells (see Supplemental Figure 3). H, refers to heads from purified φKZ tailless mutant. The standard used was the Fermentas GeneRuler 1 kb DNA ladder. (C) SDS-PAGE of fraction 14 showing the slices excised for MS identification of proteins released from within the capsid. Virion proteins in red/underlined are proteolytically processed as determined by the presence of a semi-tryptic peptide (see Table 1).
An additional experiment also indicates that most of the abundant φKZ IB proteins do not have one T4 IP-like signature characteristic. The T4 IPs and Alt precipitate with the still-condensed genomic DNA released from full heads by disruption of the capsid by treatment with 70% acetic acid (generally a protein solubilizing solvent) whereas these proteins are soluble as isolated from heads or proheads lacking DNA (Zachary and Black, 1991). The other T4 head proteins, with the exception of the portal protein, are found in the supernatant (Zachary and Black, 1991). The same treatment of φKZ tailless heads also resulted in most proteins being enriched in the supernatant as assessed by SDS-PAGE and the SC/Mw values for each protein in both fractions (Figure 9). As expected, most of the DNA was found in the pellet. Only two processed, encapsidated proteins – the abundant protein (gp93) and the low abundance helicase (gp203) – were enriched in the pellet fraction. By this criterion therefore the soluble gp93 IB protein has a unique T4 IP-like association with the DNA unlike the remainder of the IB proteins. This suggests that most of the IB proteins are segregated away from the DNA in the protein rich cylinder (Wu et al., 2012), whereas the T4 IPs are dispersed within the DNA (Black and Thomas, 2011).
Figure 9.
SDS-PAGE of pellet and supernatant fractions after acetic acid treatment of φKZ tailless heads (A) 10% SDS-PAGE gel stained with Coomassie Blue stain. Lanes: (1) purified whole φKZ, (2) buoyant density purified tailless mutant, (3) step CsCl purified tailless mutant, (4) tailless mutant purified by differential centrifugation, (5) Fermentas Page Ruler, (6) pellet fraction from acetic acid treated sample from Lane 4, (7) 0.5 volume of sample in Lane 6, (8) supernatant fraction from acetic acid treated sample from Lane 4, and (9) 0.5 volume of sample loaded in Lane 8. (B) 12% SDS-PAGE gel stained with blue silver stain of pellet and supernatant fractions of acetic acid treated φKZ tailless heads. The boundaries of slices excised for GeLCMS are marked. Head proteins identified in the pellet fraction are provided and the ratio of their enrichment included in parentheses [(SC/Mw in pellet fraction)/(SC/Mw in supernatant fraction)]. Processed head proteins enriched in the pellet fraction are underlined.
Conservation of many φKZ internal head proteins is analogous to the degree of conservation of T4 proteins with scaffolding/core functions
Comparison of the conservation of the homologs to the processed proteins in T4 and φKZ among the members of their respective families supports that the candidate φKZ IB proteins, and in fact the majority of φKZ processed proteins, do not behave like T4 IP proteins, but more like the T4 scaffolding core proteins. Based on the degree by which the T4 processed proteins are conserved through the T4-like phages (from closely related phages, to more diverged members, Table S3) these proteins can be roughly be divided into three groups: (1) proteins that are highly conserved to approximately the same degree as known essential proteins (e.g., terminase, portal and tail sheath proteins), and include the major capsid protein, gp21 and the core-related protein, gp68; (2) proteins that are more diverged than the highly conserved proteins, but for most of which homologs could still be identified in the more distantly related T4-related phages (e.g., gp67 and gp22); and (3) proteins that are poorly conserved, and for which counterparts were not identifiable in many phages (e.g., IP and Alt proteins). Notably, the genes encoding the proteins in categories (1) and (2) are all located in one major region, however the genes encoding the IPs and Alt are in three different locales in the T4 genome.
A comparison between the processed proteins in φKZ and their homologs in the related phages 201 φ2-1, φPA3 and EL (Table S4) revealed that all processed φKZ proteins have identifiable homologs in 201 φ2-1 and φPA3, and the more highly conserved of these proteins have very diverged counterparts in EL [recently proposed to belong to a separate genus, the EL-like phages (Cornelissen et al., 2011)]. Generally, the proteins of the phages 201 φ2-1 (and φPA3) have similar degrees of divergence from their φKZ homologs as KVP40 proteins have from their T4 homologs (Thomas et al., 2008). Therefore the very existence of identifiable homologs to the φKZ processed proteins in 201 φ2-1 and φPA3 illustrates that their conservation is very dissimilar to the poor conservation of the IP proteins in the T4-like phages.
Several of the φKZ processed proteins are conserved to a comparable degree as the major morphogenetic proteins terminase and sheath, including the major capsid protein, portal, protease, virion RNAP subunits, helicase-gp203, gp89 (an abundant protein) and gp184 (Table S4). φKZ processed proteins more moderately conserved (divergence of 50–70%) include gps 84, 92, 163, 177 and 90, the latter an abundant head protein for which processing has not been observed. Lesser conserved processed φKZ proteins (70–80% divergence) include gps 86, 93, 94, 95, 96, 97 and 162. The degree of conservation of these more moderately and lesser conserved φKZ processed proteins is reminiscent of the conservation of the various T4 proteins with scaffolding core-related functions to their homologs in KVP40.
Concluding Remarks
Proteolysis of head proteins is a critical step in the morphogenesis of many tailed phages, facilitating the conversion of the prohead to the mature capsid. In this study we show sequences of the cleavage sites in nineteen head proteins in φKZ. We identify informatically and directly by enzyme activity the protease likely responsible for all of these cleavages as judged by action at consensus S/A/G-X-E recognition sequences at 39 cleavage sites. Processing of φKZ head proteins thus occurs at a sequence similar but different from the I/L-X-E recognition sequence of the distantly related T4 phage protease. We found evidence of multiple processing sites in nine φKZ proteins. This appears to represent a built-in mechanism by which the phage ensures that the majority of the propeptide regions are removed and emphasizes the essential nature of processing in φKZ head morphogenesis.
The other main conclusion of our study is the identification of six similar size, similar abundance internal head proteins that provide the biochemical basis for the regular multi-tiered IB cylinder structure (Wu et al., 2012). Potentially, these six proteins could be segregated within individual tiers, or mixed in few or many combinations in several or more of these tiers. Further work will be required to determine their structural organization. Further study is also required on the IB precursor; the extensive cleavages in the IB proteins suggest that this structure may be quite different and not readily inferred from its mature form. Studies of related phages with differences in IB protein composition will enhance our understanding of the φKZ IB, and differences in IB structure among these phages may also help explain the major differences in genome length (280 kb, φKZ; 316 kb, 201 φ2-1; 308 kb, φPA3; 211 kb, EL) found among these phages whose capsids are of similar size (Krylov et al., 2007). The determination of whether or not the (thus far) unique IB of the PhiKZ-like phages is essential and, if so, the nature of its function(s) may be answered by a combined genetic, biochemical and structural approach.
Experimental procedures
Bacteria and phages
The Lory strain of Pseudomonas aeruginosa PAO1 (BEO3) was kindly provided by Dr Robert Ernst. Phage φKZ (Mesyanzhinov et al., 2002, Fokine et al., 2007) and its tailless mutant (Lavigne et al., 2009, Fokine et al., 2005a, Plotnilova, et al., 1982) were generous gifts of Drs Andrei Fokine and Konstantin Miroshnikov, respectively. We also thank Dr Miroshnikov for advice on propagation of the tailless mutant.
Purification of phages
φKZ was propagated as described previously (Wu et al., 2012). Purification of the tailless mutant was by step CsCl gradient as described previously (Fokine et al., 2005a), with the exception that particles were grown in liquid culture and concentrated by PEG precipitation. An overnight culture of P. aeruginosa was diluted 1:1000 into 1L of LB and grown with shaking at 37°C until OD600 was 0.33. The culture was then transferred to 40°C and infected with the tailless mutant phage at a MOI of 5 and grown for 2 hr, 15 min until lysis was observed. Bacterial debris was removed by a low speed spin (10,400 x g, 5 min) and the sample treated with Pancreatic DNAase I (Roche)(1500 U, 37°C, 30 min). Heads were concentrated by slow stirring overnight at 4°C in 1M NaCl and 10% PEG (final concentration) and then pelleted (11,300 x g, 4°C, 30 min). Pellets were resuspended in 20 ml SM buffer [50 mM Tris-HCl (pH 7.5), 100 mM NaCl, 8 mM MgSO4, 0.002% gelatin] containing Complete Protease Inhibitor (Roche) and then layered (5.8 ml/tube) onto CsCl step gradients composed of the following concentrations of CsCl: 1.59 g ml−1 (1.0 ml), 1.52 g ml−1 (0.75 ml), 1.5 g ml−1 (1.5 ml), 1.30 g ml−1 (1.5 ml) and 1.21 g ml−1 (1.5 ml). The buffer used throughout the gradient was 10 mM Tris-HCl (pH 7.5) and 1 mM MgCl2. Tubes were spun (108,000 x g, 10°C, 3 h) in a Beckman Coulter SW41 rotor. The band judged to contain only heads by SDS-PAGE and transmission electron microscopy had a buoyant density of 1.46 g/ml. This is higher than the buoyant density of purified intact phage (1.36 g ml−1). Heads were found to be unstable upon dialysis unless the CsCl was removed by dialyzing initially against a buffer composed of 50 mM Tris-HCl (pH 7.5), 5M NaCl and 100 mM MgCl2 (4°C), followed by a series of buffers composed of reducing concentrations of salts, concluding at 50 mM Tris-HCl (pH 7.5) and 10 mM MgCl2.
Production of DNA-deficient prohead particles
Treatment of φKZ with the T4 terminase inhibitor 9-aminoacridine (9AA) was generally as described for T4 (Schaerli and Kellenberger, 1980), with the exception that preliminary experiments were undertaken to determine the concentration of 9AA required to inhibit phage particle production (15 μg ml−1); the concentration that was subsequently used was10-fold higher than required for the same effect on T4. This can be attributed, in part, to the higher tolerance of P. aeruginosa to 9AA than E. coli (Wainwright et al., 1997). P. aeruginosa was propagated in 100 ml of M9S media at 37°C with shaking until OD600 0.45. Purified φKZ was used to infect at a MOI of 5; 9AA was added 30 seconds after infection. At OD600 0.68 the cells were harvested by centrifugation at 10,400 x g for 5 min. The pellet was resuspended in 800 μl of SM buffer, DNAase (100 units) and CHCl3 (40 μl) were added and the mixture was incubated for 10 min (37°C) and then centrifuged for 5 min (4,300 x g, 4°C). The supernatant (200 μl) was immediately loaded onto a pre-formed 15–45% glycerol gradient (50 mM Tris-HCl, pH7.5; 5 mM MgCl2) and centrifuged in a Beckman Coulter SW50.1 rotor (147,000 x g, 1hr, 4°C). Fractions (~350 μl) were collected from the bottom of the tube and run on SDS-PAGE (10% polyacrylamide) and DNA agarose gels (0.7 – 1%). Non-dialysed fractions of interest were concentrated approximately 10-fold, by initially diluting the sample with 1280 μl of SM and then centrifugation (38,700 x g, 30 min, 4°C) in a SS34 rotor. The pellet was resuspended in SM buffer.
Preparation of disrupted φKZ tailless particles
Purified heads in CsCl were osmotically shocked by dialysis from CsCl to a buffer containing 50 mM Tris-Cl (pH 7.5); 10 mM Mg Cl2. Particles were determined to be disrupted by a large increase in the viscosity of the sample such that it prevented it from being pipetted. This viscosity was reduced by treatment with DNAase (10U/200 μl of sample) for 10 min (37°C) and the sample was then loaded onto a pre-poured, chilled glycerol gradient and treated as described for the 9AA samples above. SDS-PAGE bands containing proteins released from the capsid underwent MS analyses as described above.
Acetic acid treatment of φKZ tailless heads
Acetic acid treatment of φKZ tailless heads was as previously described (Zachary and Black, 1991). Heads were propagated as described above but instead of purification by CsCl gradient underwent differential centrifugation (2x 7,600 x g, 10 min; 38,700 x g, 30 min, 4°C). Pellets were resuspended in SM buffer and judged to contain only a small percentage of intact phage as judged by comparison of its SDS-PAGE profile with those of intact phage and purified heads. Acetic acid (70% final v/v) was added to the sample, vortexed, left at room temperature for 20 min, and then centrifuged in a benchtop centrifuge (16,000 x g, 4°C, 10 min). Pellets were dried by speedivac centrifugation (20 min, 60°C) then resuspended in 10 mM Tris-HCl (pH 7.5). The supernatant was dialyzed against water (1hr, 4°C), underwent Speedivac centrifugation, and was resuspended in an equal volume of 10 mM Tris-HCl (pH 7.5) as to the pellet fraction. Samples underwent SDS-PAGE, DNA gels and MS analyses as described above. For the MS analyses double the volume of pellet fraction was loaded on the SDS-PAGE gel as for the supernatant fraction. As a consequence, the number of spectra identified for each protein in the pellet fraction was halved for calculations of SC/Mw to enable a more realistic comparison of the abundance of each protein species in pellet versus supernatant fractions.
Mass spectrometry
Samples were boiled for 10 min in SDS sample buffer (Bio-Rad) prior to electrophoresis on Criterion XT 12% SDS-PAGE reducing gels (Bio-Rad) in the presence of MOPS buffer (Bio-Rad). Proteins were visualized by staining with Coomassie blue. Gel lanes were divided into the following number of slices: acetic acid treatment of φKZ tailless heads, 7 slices; purified heads, 17 slices; disrupted φKZ tailless particles, 7 slices. Efforts were made to avoid transecting visible stained bands. Proteins in the gel slices were digested in situ with trypsin (Promega) and analyzed by HPLC-ESI-tandem mass spectrometry on either a Thermo Fisher LTQ mass spectrometer (acetic acid treatment of φKZ tailless heads) or a Thermo Fisher LTQ Orbitrap Velos mass spectrometer (purified heads and disrupted φKZ tailless particles). For the LTQ analyses, on-line HPLC separation of the digests was accomplished with an Eksigent NanoLC fitted with a PicoFrit™ (New Objective; 75 μm i.d.) column packed to 11 cm with C18 adsorbent (Vydac; 218MS 5 μm, 300 Å). A strategy was employed in which a survey scan was acquired followed by data-dependent collision-induced dissociation (CID) spectra of the seven most intense ions in the survey scan above a set threshold. For analyses on the Orbitrap, an Eksigent NanoLC-Ultra 2-D HPLC system was used with separation accomplished by a PicoFrit™ (New Objective; 75 μm i.d.) column packed to 15 cm with C18 adsorbent (Vydac; 218MS 5 μm, 300 Å). Precursor ions were acquired on the Orbitrap at a resolution of 60,000 (m/z 400). Data-dependent CID spectra of the six most intense ions in the survey scan were acquired from the linear trap while the precursor ion spectra were being collected. Mascot (Matrix Science; London, UK) was used to search the uninterpreted CID spectra searched against a locally-generated φKZ protein database that had been concatenated with the SwissProt (version 51.6) database. Methionine was considered as a variable modification and semi-trypsin was specified as the proteolytic agent. Determination of probabilities of protein identifications and cross correlation of the Mascot results with X! Tandem were accomplished by Scaffold (Proteome Software). The tandem MS results obtained from the digest of each gel slice were searched separately using Mascot, and the data files were either evaluated individually or combined into datasets for processing by Scaffold (using the “MudPIT” option).
Identification of abundant proteins in the φKZ head
An estimate of abundance for each protein in the purified head sample was made by dividing its spectral counts by its molecular weight (SC/Mw). To obtain the number of spectra assigned to the mature region(s) of the processed proteins, a “Spectrum report” was exported from Scaffold into Microsoft Excel, which was then saved as tab-delineated text. Spectra over defined regions of each protein were then summed using the perl script MSsort (Thomas et al., 2010). The program may be obtained from S.C. Hardies through the website at http://biochem.uthscsa.edu/~hs_lab/scripting/MSprogs.html. Estimates of the copy number of each protein identified as abundant by SC/Mw were made by densitometry of the SDS-PAGE bands containing these proteins. Gels prepared with serially-diluted samples of purified φKZ and the tailless mutant were analyzed using ImageJ software (Rasband, 1997–2011). Controls used for copy number estimation of the abundant proteins were the bands containing the major capsid (gp120) and the major sheath (gp29).
Cryo-electron microscopy and tomography of 9-aminoacridine treated φKZ proheads
Cryo-EM was performed using a Philips CM20-FEG electron microscope as described (Cheng et al., 2002).
Tomographic data collection and reconstruction were carried out as described (Harris et al., 2006). Briefly, a sample of 9AA treated particles was mixed 50:50 with a BSA gold suspension (Aurion, Electron Microscopy Sciences, Hatfield, PA), applied to a Quantifoil R2/2 holey carbon grid (Quantifoil, SPI, West Chester, PA) and plunge-frozen using a Vitrobot (FEI, Hillsboro, OR). Tilt series covering the range from −60° to +60° in 2° increments were collected at −4 μm nominal defocus, 38,500x magnification (7.8 Å pixel size), and a total electron dose per series of ~ 70 e- Å-2 using SerialEM (David N, 2005) on a Tecnai T12 electron microscope (FEI) equipped with a GIF 2002 energy filter (Gatan, Pleasanton, CA) operating in zero-loss mode. Reconstructions were calculated with IMOD (Kremer et al., 1996). Capsids were extracted from the tomogram as subvolumes and denoised by 30 iterations of anisotropic non-linear diffusion (Frangakis and Hegerl, 2001) as implemented in Bsoft (Heymann et al., 2008).
Expression vector assay of the φKZ protease
The gene encoding the full length form of gp93 was amplified by PCR using the primers 5′-GCGCCATGGCTCTACTTAAAATGCTAGACA-3′ and 5′-GCGGGTACCTTACTTCTCATCATTCTCAGG-3′. The gene encoding the mature form of gp175 was amplified with the primers 5′-GCGCCATGGAACCATATTCTAATTTTTCTGGT-3′ and 5′-GCGTCTAGATTACTCTAATGCAGGGGAATTCCATTTA-3′. Both genes were cloned using the NcoI and XbaI sites of the vector pHERD20T (Qiu et al., 2008), kindly provided by Dr Hongwei Yu, and transformed into Escherichia coli DH10B. Full length gp93 and mature gp175 were induced individually and the cells disrupted by freeze-thaw and lysozyme treatment. Cell lysates containing either gp93, gp175, or the two mixed together, were incubated for 30 minutes at 37°C. Samples were then spun in a benchtop centrifuge to separate soluble and insoluble fractions and the fractions assayed by SDS-PAGE. Only the soluble fractions are shown in Figure 5.
Supplementary Material
Acknowledgments
φKZ and the φKZ tailless mutant were generously provided by Dr. A. Fokine (Purdue University) and Dr. K. Miroshnikov (Russian Academy of Sciences), respectively. Pseudomonas aeruginosa PAO1 was kindly provided by Dr R. Ernst (Univ. of Maryland Baltimore). The pHERD20T vector was provided by Dr Yu (Marshall University). We thank Kevin Hakala and Sam Pardo for the excellent MS analyses that were conducted in the UTHSCSA Institutional Mass Spectrometry Laboratory. We thank Ru-ching Hsia (Univ. of Maryland Baltimore) for her help with TEM and Qin Dan for her technical support. We are extremely grateful to Dr. S.C. Hardies for allowing access to his collection of bioinformatics software and the UTHSCSA Bioinformatics Center for assistance with computational aspects of the project. This work was supported by NIH grant AI11676 to L.W.B. and by the Intramural Research Program of NIAMS.
References
- Black LW, Showe MK, Steven AC. Morphogenesis of the T4 head. In: Karam JD, editor. Molecular Biology of Bacteriophage T4. Washington, DC: ASM Press; 1993. pp. 218–258. [Google Scholar]
- Black LW, Thomas JA. Condensed genome structure. In: Rossman M, Rao VB, editors. Viral Molecular Machines. New York: Springer; 2012. pp. 469–488. [Google Scholar]
- Briers Y, Miroshnikov K, Chertkov O, Nekrasov A, Mesyanzhinov V, Volckaert G, Lavigne R. The structural peptidoglycan hydrolase gp181 of bacteriophage φKZ. Biochem Biophys Res Commun. 2008;374:747–751. doi: 10.1016/j.bbrc.2008.07.102. [DOI] [PubMed] [Google Scholar]
- Cheng H, Shen N, Pei J, Grishin NV. Double-stranded DNA bacteriophage prohead protease is homologous to herpesvirus protease. Protein Science. 2004;13:2260–2269. doi: 10.1110/ps.04726004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cheng N, Trus BL, Belnap DM, Newcomb WW, Brown JC, Steven AC. Handedness of the Herpes Simplex Virus capsid and procapsid. J Virol. 2002;76:7855–7859. doi: 10.1128/JVI.76.15.7855-7859.2002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cornelissen A, Hardies SC, Shaburova OV, Krylov VN, Mattheus W, Kropinski AM, Lavigne R. Complete genome sequence of the giant virus OBP and comparitive genome analysis of the diverse φKZ-related phages. J Virol. 2011;86:1844–52. doi: 10.1128/JVI.06330-11. [DOI] [PMC free article] [PubMed] [Google Scholar]
- David NM. Automated electron microscope tomography using robust prediction of specimen movements. J Struct Biol. 2005;152:36–51. doi: 10.1016/j.jsb.2005.07.007. [DOI] [PubMed] [Google Scholar]
- Fokine A, Battisti A, Bowman V, Efimov A, Kurochkina L, Chipman P, et al. CryoEM Study of the Pseudomonas bcacteriophage φKZ. Structure. 2007;15:1099–1104. doi: 10.1016/j.str.2007.07.008. [DOI] [PubMed] [Google Scholar]
- Fokine A, Kostyuchenko VA, Efimov AV, Kurochkina LP, Sykilinda NN, Robben J, et al. A three-dimensional cryo-electron microscopy structure of the bacteriophage φKZ head. J Mol Biol. 2005a;352:117–124. doi: 10.1016/j.jmb.2005.07.018. [DOI] [PubMed] [Google Scholar]
- Fokine A, Leiman PG, Shneider MM, Ahvazi B, Boeshans KM, Steven AC, et al. Structural and functional similarities between the capsid proteins of bacteriophages T4 and HK97 point to a common ancestry. Proc Natl Acad Sci. 2005b;102:7163–7168. doi: 10.1073/pnas.0502164102. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Frangakis AS, Hegerl R. Noise reduction in electron tomographic reconstructions using nonlinear anisotropic diffusion. J Struct Biol. 2001;135:239–250. doi: 10.1006/jsbi.2001.4406. [DOI] [PubMed] [Google Scholar]
- Golshahi L, Lynch KH, Dennis JJ, Finlay WH. In vitro lung delivery of bacteriophages KS4-M and φKZ using dry powder inhalers for treatment of Burkholderia cepacia complex and Pseudomonas aeruginosa infections in cystic fibrosis. Journal of Applied Microbiology. 2011;110:106–117. doi: 10.1111/j.1365-2672.2010.04863.x. [DOI] [PubMed] [Google Scholar]
- Grayson P, Molineux IJ. Is phage DNA ‘injected’ into cells—biologists and physicists can agree. Curr Opin Microbiol. 2007;10:401–409. doi: 10.1016/j.mib.2007.04.004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Harris A, Cardone G, Winkler DC, Heymann JB, Brecher M, White JM, Steven AC. Influenza virus pleiomorphy characterized by cryoelectron tomography. Proc Natl Acad Sci. 2006;103:19123–19127. doi: 10.1073/pnas.0607614103. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Heymann BJ, Cardone G, Winkler DC, Steven AC. Computational resources for cryo-electron tomography in Bsoft. J Struct Biol. 2008;161:232–242. doi: 10.1016/j.jsb.2007.08.002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hughey R, Krogh A. Hidden Markov models for sequence analysis: Extension and analysis of the basic method. Comput Appl Biosci. 1996;12:95–107. doi: 10.1093/bioinformatics/12.2.95. [DOI] [PubMed] [Google Scholar]
- Karplus K, Barrett C, Hughey R. Hidden markov models for detecting remote protein homologies. Bioinformatics. 1998;14:846–856. doi: 10.1093/bioinformatics/14.10.846. [DOI] [PubMed] [Google Scholar]
- Keller B, Bickle TA. The nucleotide sequence of gene 21 of bacteriophage T4 coding for the prohead protease. Gene. 1986;49:245–251. doi: 10.1016/0378-1119(86)90285-4. [DOI] [PubMed] [Google Scholar]
- Kremer JR, Mastronarde DN, McIntosh JR. Computer visualization of three-dimensional image data using IMOD. J Struct Biol. 1996;116:71–76. doi: 10.1006/jsbi.1996.0013. [DOI] [PubMed] [Google Scholar]
- Krylov VN, Dela Cruz DM, Hertveldt K, Ackermann HW. “φKZ-like viruses”, a proposed new genus of myovirus bacteriophages. Arch Virol. 2007;152:1955–1959. doi: 10.1007/s00705-007-1037-7. [DOI] [PubMed] [Google Scholar]
- Krylov VN, Smirnova TA, Minenkova IB, Plotnilova TG, Zhazikov IZ, Khrenova EA. Pseudomonas bacteriophage φKZ contains an inner body in its capsid. Can J Microbiol. 1984;30:758–762. doi: 10.1139/m84-116. [DOI] [PubMed] [Google Scholar]
- Lavigne R, Darius P, Summer EJ, Seto D, Mahadevan P, Nilsson AS, et al. Classification of Myoviridae bacteriophages using protein sequence similarity. Bmc Microbiol. 2009;9:224. doi: 10.1186/1471-2180-9-224. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lecoutere E, Ceyssens PJ, Miroshnikov K, Mesyanzhinov V, Krylov VN, Noben JP, et al. Identification and comparative analysis of the structural proteomes of φKZ and EL, two giant Pseudomonas aeruginosa bacteriophages. Proteomics. 2009;9:3215–3219. doi: 10.1002/pmic.200800727. [DOI] [PubMed] [Google Scholar]
- Liu J, Mushegian A. Displacements of prohead protease genes in the late operons of double-stranded-DNA bacteriophages. J Bacteriol. 2004;186:4369–4375. doi: 10.1128/JB.186.13.4369-4375.2004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mangel WF, Baniecki ML, McGrath WJ. Specific interactions of the adenovirus proteinase with the viral DNA, an 11-amino-acid viral peptide, and the cellular protein actin. Cellular and Molecular Life Sciences. 2003;60:2347–2355. doi: 10.1007/s00018-003-2318-2. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Matinkhoo S, Lynch KH, Dennis JJ, Finlay WH, Vehring R. Spray-dried respirable powders containing bacteriophages for the treatment of pulmonary infections. Journal of Pharmaceutical Sciences. 2011;100:5197–5205. doi: 10.1002/jps.22715. [DOI] [PubMed] [Google Scholar]
- Mesyanzhinov VV, Robben J, Grymonprez B, Kostyuchenko VA, Bourkaltseva MV, Sykilinda NN, et al. The genome of bacteriophage φKZ of Pseudomonas aeruginosa. J Mol Biol. 2002;317:1–19. doi: 10.1006/jmbi.2001.5396. [DOI] [PubMed] [Google Scholar]
- Miroshnikov KA, Faizullina NM, Sykilinda NN, Mesyanzhinov VV. Properties of the endolytic transglycosylase encoded by gene 144 of Pseudomonas aeruginosa bacteriophage φKZ. Biochemistry (Moscow) 2006;71:300–305. doi: 10.1134/s0006297906030102. [DOI] [PubMed] [Google Scholar]
- Monson R, Foulds I, Foweraker J, Welch M, Salmond GPC. The Pseudomonas aeruginosa generalized transducing phage φPA3 is a new member of the φKZ-like group of ‘jumbo’ phages, and infects model laboratory strains and clinical isolates from cystic fibrosis patients. Microbiology. 2011;157:859–867. doi: 10.1099/mic.0.044701-0. [DOI] [PubMed] [Google Scholar]
- Mullaney JM, Black LW. Capsid targeting sequence targets foreign proteins into bacteriophage T4 and permits proteolytic processing. J Mol Biol. 1996;261:372–385. doi: 10.1006/jmbi.1996.0470. [DOI] [PubMed] [Google Scholar]
- Paradis-Bleau C, Cloutier I, Lemieux L, Sanschagrin F, Laroche J, Auger M, et al. Peptidoglycan lytic activity of the Pseudomonas aeruginosa phage φKZ gp144 lytic transglycosylase. FEMS Microbiol Lett. 2007;266:201–209. doi: 10.1111/j.1574-6968.2006.00523.x. [DOI] [PubMed] [Google Scholar]
- Plotnikova TG, Dzhusupova AB, Khrenova EA, Krylov VN. Genetic and phenogenetic study of the ts-mutant group of Pseudomonas aeruginosa PA01 phage φKZ. Genetika. 1292;18:1793–1798. [Russian] [PubMed] [Google Scholar]
- Qiu D, Damron FH, Mima T, Schweizer HP, Yu HD. PBAD-based shuttle vectors for functional analysis of toxic and highly regulated genes in Pseudomonas and Burkholderia spp. and other bacteria. Appl Environ Micro. 2008;74:7422–7426. doi: 10.1128/AEM.01369-08. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rasband WS. ImageJ. U. S. National Institutes of Health; Bethesda, Maryland, USA: 1997–2011. http://imagej.nih.gov/ij/ [Google Scholar]
- Rishovd S, Holzenburg A, Johansen BV, Lindqvist BH. Bacteriophage P2 and P4 morphogenesis: Structure and function of the connector. Virology. 1998;245:11–17. doi: 10.1006/viro.1998.9153. [DOI] [PubMed] [Google Scholar]
- Rishovd S, Marvik OJ, Jacobsen E, Lindqvist BH. Bacteriophage P2 and P4 morphogenesis: Identification and characterization of the portal protein. Virology. 1994;200:744–751. doi: 10.1006/viro.1994.1238. [DOI] [PubMed] [Google Scholar]
- Schaerli C, Kellenberger Head maturation pathway of bacteriophages T4 and T2. V Maturable epsilon-particle accumulating an acridine-treated bacteriophage T4-infected cells. J Virol. 1980;33:830–844. doi: 10.1128/jvi.33.2.830-844.1980. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Showe MK, Isobe E, Onorato L. Bacteriophage T4 prehead proteinase: I. Purification and properties of a bacteriophage enzyme which cleaves the capsid precursor proteins. J Mol Biol. 1976a;107:35–54. doi: 10.1016/s0022-2836(76)80016-2. [DOI] [PubMed] [Google Scholar]
- Showe MK, Isobe E, Onorato L. Bacteriophage T4 prehead proteinase: II. Its cleavage from the product of gene 21 and regulation in phage-infected cells. J Mol Biol. 1976b;107:55–69. doi: 10.1016/s0022-2836(76)80017-4. [DOI] [PubMed] [Google Scholar]
- Söding J. Protein homology detection by HMM-HMM comparison. Bioinformatics. 2005;21:951–960. doi: 10.1093/bioinformatics/bti125. [DOI] [PubMed] [Google Scholar]
- Thomas JA, Rolando MR, Carroll CA, Shen PS, Belnap DM, Weintraub ST, et al. Characterization of Pseudomonas chlororaphis myovirus 201 φ2-1 via genomic sequencing, mass spectrometry, and electron microscopy. Virology. 2008;376:330–338. doi: 10.1016/j.virol.2008.04.004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Thomas JA, Weintraub ST, Hakala K, Serwer P, Hardies SC. Proteome of the large Pseudomonas myovirus 201 φ2-1: Delineation of proteolytically processed virion proteins. Molecular & Cellular Proteomics. 2010;9:940–951. doi: 10.1074/mcp.M900488-MCP200. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wainwright M, Phoenix DA, Marland J, Wareing DR, Bolton FJ. In-vitro photobactericidal activity of aminoacridines. J Antimicrob Chemother. 1997;40:587–589. doi: 10.1093/jac/40.4.587. [DOI] [PubMed] [Google Scholar]
- Walker JE, Auffret AD, Carne A, Gurnett A, Hanisch P, Hill D, Saraste M. Solid-phase sequence analysis of polypeptides eluted from polyacrylamide gels. Eur J Biochem. 1982;123:253–260. doi: 10.1111/j.1432-1033.1982.tb19761.x. [DOI] [PubMed] [Google Scholar]
- Wu W, Thomas JA, Cheng N, Black LW, Steven AC. Bubblegrams reveal the inner body structure of φKZ. Science. 2012;335:182. doi: 10.1126/science.1214120. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zachary A, Black LW. Isolation and characterization of a portal protein-DNA complex from dsDNA bacteriophage. Intervirol. 1991;33:6–16. doi: 10.1159/000150225. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.








