Abstract
Cholesterol metabolism is tightly regulated at the cellular level and is essential for cellular growth. MicroRNAs (miRNAs), a class of noncoding RNAs, have emerged as critical regulators of gene expression, acting predominantly at the posttranscriptional level. Recent work from our group and others has shown that hsa-miR-33a and hsa-miR-33b, miRNAs located within intronic sequences of the Srebp genes, regulate cholesterol and fatty acid metabolism in concert with their host genes. Here, we show that hsa-miR-33 family members modulate the expression of genes involved in cell cycle regulation and cell proliferation. MiR-33 inhibits the expression of the cyclin-dependent kinase 6 (CDK6) and cyclin D1 (CCND1), thereby reducing cell proliferation and cell cycle progression. Overexpression of miR-33 induces a significant G1 cell cycle arrest in Huh7 and A549 cell lines. Most importantly, inhibition of miR-33 expression using 2′fluoro/methoxyethyl-modified (2′F/MOE-modified) phosphorothioate backbone antisense oligonucleotides improves liver regeneration after partial hepatectomy (PH) in mice, suggesting an important role for miR-33 in regulating hepatocyte proliferation during liver regeneration. Altogether, these results suggest that Srebp/miR-33 locus may cooperate to regulate cell proliferation and cell cycle progression and may also be relevant to human liver regeneration.
Key words: CDK6, cyclin D1, miR-33, cell cycle, microRNA
Introduction
Intracellular cholesterol is obtained from plasma lipoproteins or synthesized de novo from acetyl-CoA.1 As an essential component of mammalian cell membranes, cells require cholesterol for proliferation.2–5 In addition to its structural requirement, cholesterol is important for other cell functions, such as bile acid and hormone synthesis as well as during embryonic development. The requirement of cholesterol for cell growth and division of mammalian cells has been known for many years,2–4 but whether this is just a consequence of its use for membrane formation or whether it also plays a regulatory role in this process has not been clarified. Cholesterol is also required for cell cycle progression, and its deficiency leads to cell cycle arrest in G2/M.6 Moreover, other non-mevalonate derivatives are essential for the G1-S transition, thus confirming the relationship between the cholesterol synthesis pathway and cell cycle progression.7,8
The intracellular cholesterol level is tightly controlled by feedback mechanisms that operate at both transcriptional and posttranscriptional level. When cells accumulate excess sterols, the activity of 3-hydroxy-3-methylglutaryl coenzyme A reductase (HMGCR), the rate-limiting enzyme of cholesterol biosynthesis, declines by more than 90%, and the cell surface expression of low-density lipoprotein receptor (LDLr) decreases. By contrast, upon depletion of intracellular cholesterol, cells maintain a high activity of HMGCR and high expression of LDLr. This coordinated process is regulated by ER-bound sterol regulatory element-binding proteins (SREBPs).9–12 The SREBP family of basic-helix-loop-helix leucine zipper (bHLH-LZ) transcription factors consist of SRERP-1a, SREBP-1c and SREBP-2.9–12 The SREBPs differ in their tissue-specific expression, target gene selectivity and the relative potencies of their trans-activation domains.9–12 SREBP-1c targets genes that are involved in fatty acid metabolism, such as fatty acid synthase (FASN). SREBP-2 regulates the expression of cholesterol-related genes, such as HMGCR and LDLr. SREBP-1a targets both sets of genes. In addition to regulating lipid metabolism, these transcription factors have also been implicated in the regulation of cell cycle.13 Bengoechea-Alonso et al. have demonstrated that siRNA-mediated silencing of SREBP-1 in human Hela, U2OS and MCF-7 cells leads to an accumulation of cells in late G1 phase, prior to the G1/S transition.13 By contrast, SREBP-1a overexpression, activates the transcription of p21WAF1/CIP1, a universal cyclin-dependent kinase inhibitor, leading to cell growth inhibition and G1 cell cycle arrest.14 In addition to p21, SREBP-1a activation also increases the accumulation of other cyclin-dependent kinase (CDK) inhibitors, including p27, p21 and p16, leading to reduced CDK2 and CDK4 activities and hypophosphorylation of Rb protein.15 Interestingly, SREBP-1a transgenic mice exhibited impaired liver regeneration after partial hepatectomy.15
In addition to classical transcriptional regulators, a class of noncoding RNAs, termed microRNAs (miRNAs), has emerged as critical regulator of gene expression acting predominantly at the post-transcriptional level.16–19 These short (22 nt) double-stranded regulatory noncoding RNAs are mostly processed from primary transcripts by the sequential actions of the endonucleases Drosha and Dicer enzymes. In the cytoplasm, mature miRNAs are incorporated into the cytoplasmatic RNA-induced silencing complex (RISC) and bind to partially complementary target sites in the 3′UTR of specific mRNA. miRNA targeting of mRNAs inhibits their expression through mRNA destabilization, repression of translation or a combination of both processes.16–19
We and others provided the first identification of a highly conserved miRNA family, miR-33, within the intronic sequences of the Srebp genes.20–22 Two miR-33 genes are present in humans: miR-33b, which is encoded within intron 17 of the Srebp-1 gene on chromosome 17, and miR-33a, which is expressed in intron 16 of the Srebp-2 gene on chromosome 22. In mice, however, there is only one miR-33 gene, which is the ortholog of human miR-33a and is located within intron 15 of the mouse Srebp-2 gene. miR-33a and miR-33b are co-transcribed with its host genes, like many intronic miRNAs, and they target genes involved in regulating cholesterol homeostasis (Abca1, Abcg1 and Npc1)20–22 and fatty acid metabolism (Ampk, Cpt1a, Crot, Hadhb and Sirt6).23,24 These findings suggest that Srebp genomic loci, which encode Srebp transcription factors and miR-33a/b may cooperate to regulate lipid metabolism. Of note, the inhibition of miR-33 using different strategies increases plasma high-density lipoproteins (HDL) in mouse and promotes the regression of atherosclerosis in mice.20–22,25,26
Similarly to Srebp/miR-33 locus, the α-myosin heavy chain (alphaMHG) gene, in addition to encoding a major cardiac contractile protein, regulates cardiac growth and gene expression in response to stress and hormonal signaling through miR-208.27 Altogether, these observations suggest that intronic miRNAs work in conjunction with host genes to regulate similar cellular processes.
Since SREBPs regulates cellular proliferation and cell cycle progression, in the current study, we tested the role of miR-33 in regulating these cellular functions. We identify putative binding sites for miR-33 in the 3′UTR of cyclin-dependent kinases (Cdk6, Cdk8 and Cdk19), Ccnd1 and d2, p53, Pten, Myc and mitogen-activated protein kinases (Map3k1, Map3k7, Mapk1, Mapk3, Mapk6, Mapk10 and Mapk14). We show that overexpression of miR-33 inhibits CDK6 and cyclinD1 expression, reduces cell proliferation in different human cell lines and leads to cell cycle arrest in G1 phase. Conversely, endogenous inhibition of miR-33 increases cell proliferation in the human hepatic cell line, Huh7. Furthermore, inhibition of miR-33 expression using 2′fluoro/methoxyethyl-modified (2′F/MOE-modified) phosphorothioate backbone antisense oligonucleotides improves liver regeneration after partial hepatectomy (PH) in mice. Altogether, these results suggest that Srebp/miR-33 may cooperate to regulate cell proliferation and cell cycle progression.
Results
miR-33 regulates post-transcriptional CDK6 and CCND1 expression.
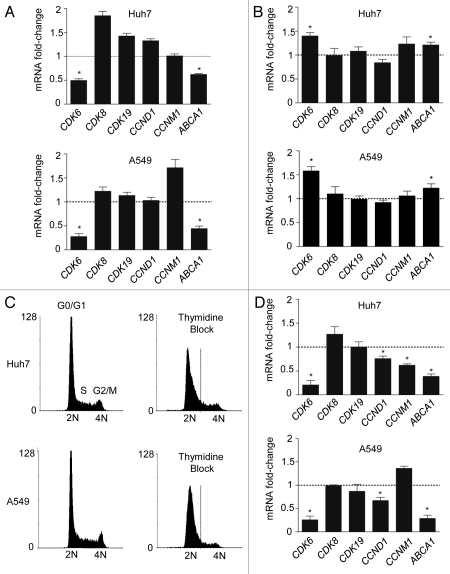
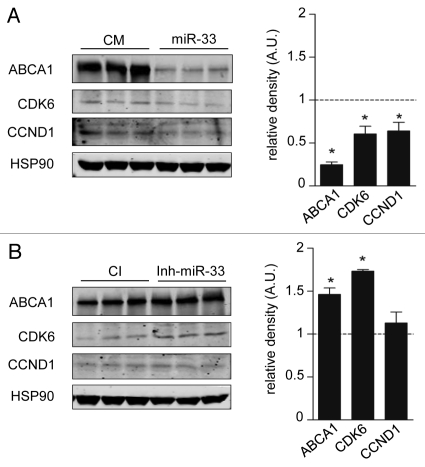
To determine potential miR-33 targets involved in cell proliferation and cell cycle, we used a combination of bio-informatic tools for miRNA target prediction [TargetScan (http:/www.targetscan.org), miRanda (http:/www.microrna.org), DIANAmt (http://diana.cslab.ece.ntua.gr), miRDB (http://mirdb.org), miRWalk (http://www.ma.uni-heidelberg.de/aps/zmf/mirwalk) and PITA (http://genie.weizmann.ac.il)]. Interestingly, the 3′UTR of the cyclin-dependent kinases (Cdk6, Cdk8 and Cdk19), Ccnd1 and d2, p53, Pten, Myc and mitogen-activated protein kinases (Map3k1, Map3k7, Mapk1, Mapk3, Mapk6, Mapk10 and Mapk14) have predicted binding sites for miR-33 (Table 1). Next, we wanted to determine the effect of miR-33 on mRNA expression of CDK6, CDK8, CDK19, CCND1, CCNM1. Transfection of Huh7 cells with miR-33 (40.fold increase expression) significantly inhibited the mRNA levels of CDK6 and ABCA1, a previously identified target of miR-33 (Fig. 1A, upper part). Similar results were observed in A549 cells transfected with miR-33 (Fig. 1A, bottom part). Notably, endogenous inhibition of miR-33 using anti-miR-33 oligonucleotides (2.5 fold decrease) increases mRNA expression of CDK6 and ABCA1 in both cell lines (Fig. 1B). Since the expression level of CCND1 and CCNM1 varies along the cell cycle, we synchronized Huh7 and A549 cells in G1 phase using a double thymidine block, which arrested the cells in early S phase (Fig. 1C). As seen in Figure 1D, transfection of Huh7 (upper part) and A549 (lower part) with miR-33 significantly inhibits CCND1, CDK6 and ABCA1 mRNA levels. Similar regulation of these genes by miR-33 was also seen at the protein level (Fig. 2A and B). Altogether, these results strongly suggest that miR-33 coordinates genes regulating cell cycle progression.
Table 1.
miR-33 predicted target genes
| Gene | DIANAmT | miRanda | miRDB | miRWalk | PITA | Targetscan |
| ABCA1 | X | X | X | X | X | X |
| CDK6 | X | X | X | X | X | |
| CDK8 | X | X | X | X | X | |
| CCND1 | X | X | X | X | ||
| CCND2 | X | X | X | X | ||
| MAP3K1 | X | X | X | X | X | X |
| MAP3K7 | X | X | X | |||
| MAPK1 | X | X | X | |||
| MAPK3 | X | X | X | |||
| MAPK6 | X | X | X | X | X | |
| MAPK10 | X | X | X | X | ||
| MAPK14 | X | X | X | X | X | X |
| p53 | X | X | X | X | X | |
| PIK3R3 | X | X | X | X | X | |
| PTEN | X | X | X | X | ||
| MYC | X | X | X | X |
Gene target prediction based on TargetScan, miRanda, DIANAmt, miRDB and PITA.
Figure 1.
Post-transcriptional regulation of cell cycle genes and ABCA1 by miR-33. (A) Quantitative RT-PCR expression profile of selected miR-33 predicted target in human hepatic Huh7 cell line (upper part) and human lung A549 cell line (bottom part) after overexpressing miR-33 and (B) after endogenous inhibition of miR-33 by using anti-miR-33 oligonucleotides. (C) Flow cytometry analysis of Huh7 cells (upper parts) and A549 cells (bottom parts) synchronized using double thymidine block (right parts). (D) Quantitative RT-PCR analysis of selected miR-33 predicted targets in Huh7 and A549 cells transfected with CM or miR-33 after thymidine block synchronization. Data are the mean ± SEM and are representative of ≥ 3 experiments. *p ≤ 0.05.
Figure 2.
MiR-33 inhibits CDK6 and CCND1 protein expression. Western blot analysis of CDK6, CCND1 and ABCA1 expression from Huh7 cells transfected with CM and miR-33 (A) or CI and anti-miR-33 (B). Data are the mean ± SEM and are representative of more than or equal to three experiments. *p ≤ 0.05.
miR-33 directly targets the 3′UTR of Cdk6 and Ccnd1.
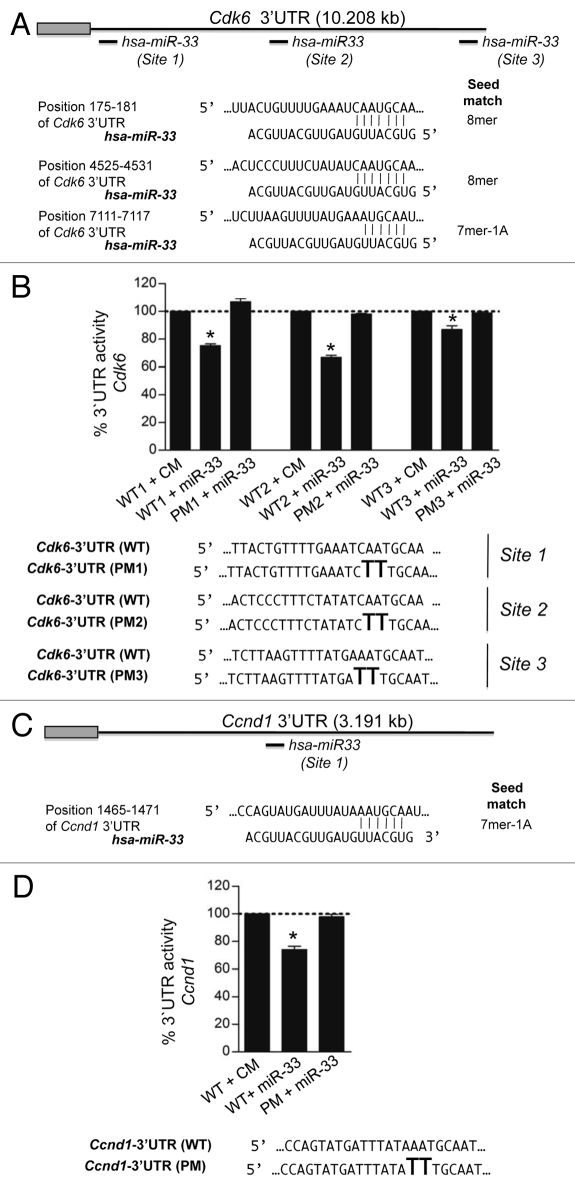
The human Cdk6 and Ccnd1 3′UTR have three and one computationally predicted miR-33 binding sites, respectively (Fig. 3A and C). All the predicted binding sites are evolutionarily conserved (Fig. S1). To directly assess whether miR-33 targets Cdk6 and Ccnd1 directly, we generated reporter constructs with the luciferase coding sequence fused to the 3′UTR of these genes. miR-33 markedly repressed the activity of the Cdk6 and Ccnd1 3′UTR constructs (Fig. 3B and D). Mutation of the miR-33 target sites relieved miR-33 repression of Cdk6 and Ccnd1 3′UTR activity, consistent with the direct interaction of miR-33 with these sequences (Fig. 3B and D).
Figure 3.
MiR-33 specifically targets the 3′UTR of Cdk6 and Ccnd1. (A) Sequence alignment of the human hsa-miR-33 mature sequence with the binding sites of the human Cdk6 3′UTR and (B) human Ccnd1 3′UTR. Luciferase reporter activity in COS-7 cells transfected with CM or miR-33 of the (C) Cdk6 and (D) Ccnd1 3′UTR containing the indicated point mutations (PM) in the miR-33 target sites. Data are expressed as mean % of the 3′UTR activity of control miR ± SEM and are representative of ≥ 3 experiments. *p ≤ 0.05.
miR-33 regulates cell proliferation and cell cycle progression.
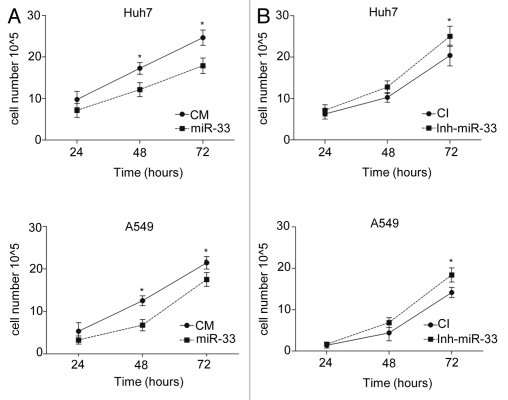
To assess the role of miR-33 in regulating cell proliferation, we transfected Huh7 and A549 cells with miR-33 and measured the viable cell number at different time points. As seen in Figure 4A, overexpression of miR-33 inhibited cell growth in both cell lines. In another set of experiments, we stably overexpressed miR-33 in HeLa and MCF-7 cells using lentivirus and monitored cell proliferation using crystal violet staining and MTT assays. Consistent with our data in Figure 4A, miR-33-transduced cells grew slower compared with control-transduced cells (Fig. S2). In separate experiments, antagonism of endogenous miR-33 increased cell proliferation in Huh7 and A549 cells (Fig. 4B). We also used our stably transfected HeLa cells to study the impact of miR-33 overexpression on the cellular response to mitogenic stimuli. To accomplish this, we serum starved the cells for 72 h and then reintroduced serum, as described in Methods. As expected, the expression of CDK6 and CCND1 was induced when cells were switched from BSA to serum, and the expression of both genes was reduced in cells overexpressing miR-33 (Fig. S3). Intriguingly, miR-33-dependent repression of these targets results in increased mRNA levels of CDK4, CCND2 and CCND3 (Fig. S3). The exact molecular events that lead to these compensatory changes remain obscure, though. Nevertheless, these results demonstrate that miR-33 regulates cell proliferation via targeting CDK6 and CCND1.
Figure 4.
MiR-33 inhibits cell proliferation. Time-course analysis of the effects of (A) overexpression or (B) inhibition of miR-33 on cell proliferation. Huh7 (upper parts) and A549 (bottom parts) cells were cultured in cholesterol-free medium and transfected with CM, miR-33, CI and Inh-miR-33. At the indicated times, the viable cells were counted. Data correspond to means ± SEM and are representative of ≥ 3 experiments. *p ≤ 0.05.
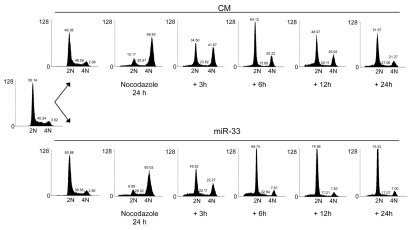
Most of the mitogenic pathways result in the transcriptional induction of D-type cyclins and the subsequent activation of cyclin-dependent kinases (CDKs), such as CDK4 and CDK6. The active CDK4/6-cyclin D complexes then inactivate the retinoblastoma protein (pRB), leading to the EF-dependent transcription of specific cell cycle genes and progression throughout the cell cycle. To assess the effect of miR-33 on cell cycle progression, we synchronized Huh7 cells in mitosis by treating with nocodazole and then transfected with negative control mimic (CM) or miR-33. As seen in Figure 5, cells transfected with miR-33 were arrested in G1 phase after their release from mitosis, suggesting an important role of miR-33 in regulating the G1/S transition.
Figure 5.
MiR-33 induces G1 arrest. Cell cycle distribution of A549 cells transfected with CM or miR-33 and synchronized in G2/M with nocodazole. After 24 h of treatment with nocodazole, cells were washed and released from the G2/M cell cycle arrest. At the indicated times, cells were stained with propidium iodide and analyzed by flow cytometry. 2N, cells have diploid DNA content (G0/G1 phase); 4N (G2/M), cells have tetraploid DNA content. Data correspond to a representative experiment among three that gave similar results. FL2-A corresponds to the fluorescence emitted by propidium iodide bound to DNA, which is measured by cytometry in channel 2 (FL2). To avoid the eventual presence of doublets, among the parameters given by the cytometer area (FL2-A) is preferred in order to quantify the amount of DNA present in the cell.
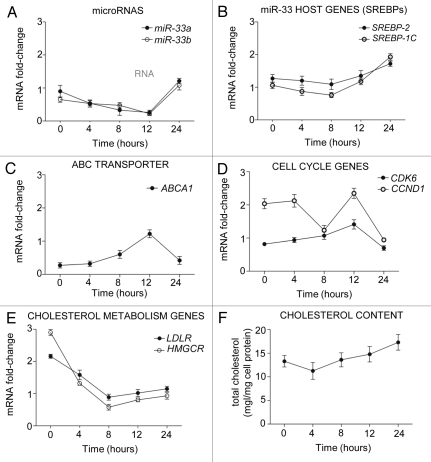
We next analyzed the kinetics of miR-33 mRNA expression during cell cycle progression. To this end, we synchronized Huh7 cells with nocodazole and then analyzed miR-33 expression at 4, 8, 12 and 24 h after releasing them from mitotic arrest. As shown in Figure 6A and B, miR-33a, miR-33b, SREBP1 and SREBP2 levels decreased after releasing the cells from mitosis. Interestingly, miR-33 levels were inversely correlated with the expression of CDK6, CCND1 (Fig. 6D) and ABCA1 (Fig. 6C), suggesting a functional role for miR-33 in regulating the expression of these genes. The inhibition of ABCA1 and the increased expression of genes involved in cellular cholesterol uptake [lowdensity lipoprotein receptor (LDLr)] and cholesterol biosynthesis [3-hydroxy-3-methylglutaryl coenzyme A reductase (HMGCR)] (Fig. 6E) results in an increase in cellular cholesterol levels (Fig. 6F).
Figure 6.
Mir-33 expression levels correlate inversely with its predicted target genes during cell cycle progression. (A–E) Quantitative RT-PCR analysis of miR-33a/b, SREBP-2, SREBP-1, ABCA1, CDK6, CCND1, LDLR and HMGCR in human hepatic Huh7 cells synchronized with nocodazole for 24 h. Data are the mean ± SEM and are representative of three experiment which gave similar results.
Antagonism of miR-33 in mice promotes liver regeneration.
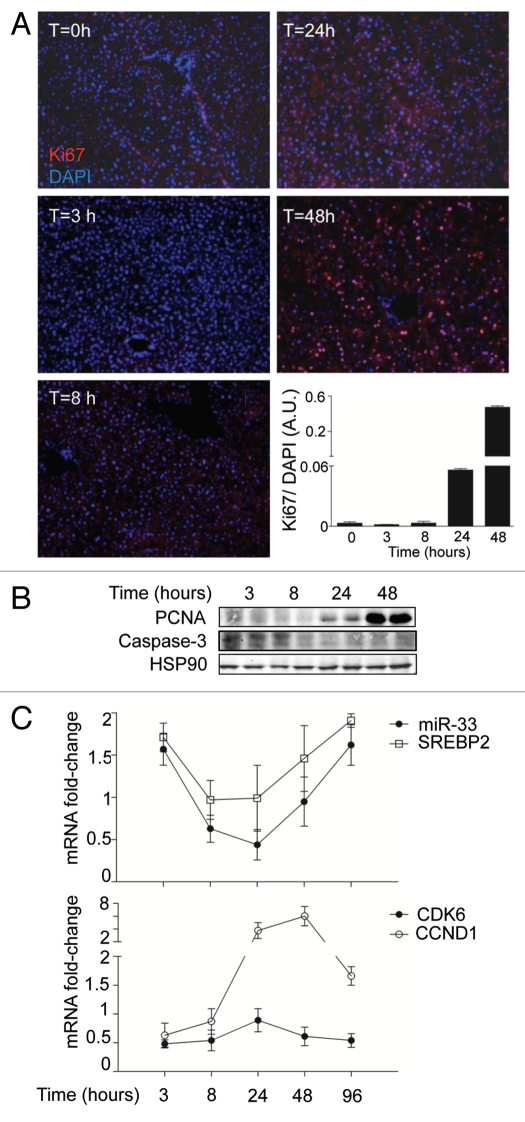
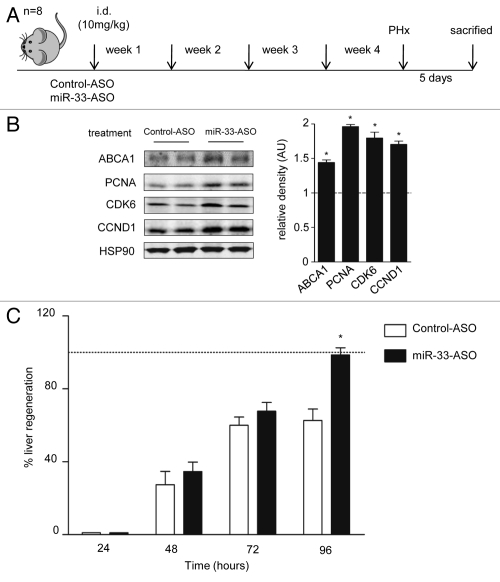
Finally, to analyze the role of miR-33 in a physiological model of cellular proliferation, we studied whether anti-miR-33 therapy could improve liver regeneration in mice. To this end, we performed 2/3 of partial hepatectomy (PH) as a model of liver regeneration and assessed the expression of Ki67, a cellular marker for proliferation. As seen in Figure 7A, a significant number of Ki67-positive cells were observed in sections of livers at 48 h following PH. Similar results were obtained when we analyze the expression of PCNA in liver lysates by western blot (Fig. 7B). Interestingly, the expression of miR-33 correlated inversely with both the proliferative status of the liver and the mRNA expression of CDK6 and CCND1, thus suggesting a role for miR-33 in regulating liver regeneration (Fig. 7C). To assess the effects of inhibiting miR-33 in the PH model, we treated C57/BL/6 mice with 10 mg/Kg of 2′fluoro/methoxyethyl-modified (2′F/MOE-modified) phosphorothioate backbone antisense oligonucleotides every week for one month (Fig. 8A). This antisense therapy has been used successfully in mice to inhibit the function of various miRNAs without apparent toxicity. To determine the efficacy of the anti-miR-33 treatment, we measured the hepatic expression of miR-33 and its target genes after 4 weeks of treatment. Levels of miR-33, detected by qRT-PCR, were decreased by more than 50% in anti-miR-33-treated mice compared with the mice treated with control antimiR- Consistent with these results, the expression of ABCA1, CDK6 and CCND1 were increased in the livers of the mice treated with anti-miR-33 (Fig. 8B). Interestingly, the expression of the proliferative marker PCNA was also increased in mice treated with anti-miR-33 (Fig. 8B). Next, we investigated the effects of anti-miR-33 oligonucleotides in liver regeneration after PH. To this end, we calculated the regeneration index as the increase in the liver-to-body mass ratio, as described in Methods. As seen in Figure 8C, mice treated with anti-miR-33 oligonucleotides showed an accelerated liver regeneration, suggesting that anti-miR-33 therapy might be useful for treating liver disease. Altogether, these results suggest that miR-33 is involved in cell cycle progression and cellular proliferation in coordination with regulating multiple proteins (Fig. S4).
Figure 7.
Liver regeneration after PH causes a rapid induction of CCND1 expression and miR-33 downregulation. (A) Time-course analysis of Ki67-positive cells from liver sections of mice subjected to PH. The computed assisted quantification of the proliferating cells (red) vs. the total number of cells (blue) is shown at the graph. (B) Western blot analysis of PCNA and cleaved caspase 3 expression from liver lysates of mice subjected to PH. (C) Time-course mRNA expression analysis of miR-33, SREBP-2, CDK6 and CCND1 from liver samples of mice subjected to PH. Data are the mean ± SEM (n = 6 mice).
Figure 8.
Inhibition of miR-33 with miR-33-ASO in vivo increased liver regeneration. (A) Experimental outline of miR33-ASO or control-ASO treatment in C57BL/6 mice (n = 8 per group). (B) Representative western blot analysis shows that the inhibition of miR-33 in vivo for 4 weeks increased the expression of PCNA, CDK6, CCND1 and ABCA1. Relative density analysis is shown in the right part. Dotted line positioned at one represents the normalized values of control mice. (C) Effect of miR-33 ASO treatment on liver regeneration after partial hepatectomy. The body weight recovery was used to calculated the percentage of liver regeneration after PHx vs. control animals (dotted line). The data are presented as the mean ± SEM and are representative of eight experiments *p < 0.05.
Discussion
The most salient feature of this paper is the identification of the role of miR-33 in regulating cell proliferation, cell cycle progression and liver regeneration. We and others provided the first identification of a highly conserved miRNA family, miR-33, within the intronic sequences of the Srebp genes in organisms, ranging from Drosophila to humans. miR-33 targets multiple genes regulating cholesterol homeostasis, including Abca1, Abcg1 and Npc1. In addition to the role in maintaining cholesterol metabolism, we have also demonstrated that miR-33 coordinates genes regulating fatty acid and glucose metabolism. This work opens a new window for the miR-33 family, whose members repress genes involved in cell proliferation and cell cycle progression. Notably, we show that miR-33 negatively regulates CDK6 and CCND1, which results in cell cycle arrest in G1 phase. miR-33 overexpression reduces cell proliferation and retarded cell cycle progression from the late G1 phase into the S phase in human hepatic cells (Huh7). Furthermore, this study also shows that in vivo inhibition of miR-33 using antisense oligonucleotides improves liver regeneration after partial hepatectomy.
The recovery of liver function after partial hepatectomy is promoted by a variety of signals that lead to the prompt re-establishment of liver mass by upregulating cell growth and/or proliferation.28,29 Interestingly, several proteins involved in regulating cell cycle progression play a key role during hepatocyte replication and liver growth.30–32 Indeed, transient expression of CCND1 is sufficient to promote hepatocyte replication and liver growth in vivo.31 D- and E-type cyclins and their CDK partners (CDK4/6) regulate cell cycle progression through G1 to S phase.33 Activation of the CCND1/CDK6 complex phosphorylates and inactivates the retinoblastoma protein (Rb), a transcriptional repressor that, in turn, inhibits the E2F transcription factors and recruits chromatin-remodeling complexes that lead to the repression of specific targeted genes.
In normal adult liver, hepatocytes are highly differentiated and rarely undergo cell division, but they retain a remarkable ability to proliferate in response to acute or chronic injury. Previous studies have suggested that CCND1 is a critical mediator of G1 progression in hepatocytes. Similarly, CDK4/CDK6 regulates cell cycle progression in hepatoma cells. Accordingly, Huh7 and HepG2 cells treated with PD-0332991, a potent inhibitor of these kinases, arrested the cells in G1 phase.34
Here we demonstrate that miR-33 targets CCND1 and CDK6 and regulates hepatocyte proliferation. In addition to CCND1 and CDK6, it has previously been reported that miR-33 also targets p53.35 p53 activates the transcription of genes that induce cell cycle arrest, apoptosis and senescence in response to several stress conditions, including DNA damage. In our work, we did not find differences in the number of apoptotic cells when we overexpressed or inhibited miR-33, suggesting that the effect of miR-33 on p53 and cell viability may be cell type-specific. On the other hand, Thomas M et al. have recently shown that miR-33 also targets the serine/threonine-protein kinase Pim-1 and inhibits proliferation in K562 and LS174T cells.36
In addition to miR-33, many miRNAs have been shown to play a role in regulating proliferation and cell cycle progression.37 The mir-15a-16-1 cluster may induce cell cycle arrest at the G1 phase by targeting critical cell cycle regulators such as CDK1, CDK2 and CDK6 as well as cyclins (D1, D3 and E1).38–40 Indeed, these major cell cycle kinase complexes are regulated by several other miR NAs. Thus, CDK6 mRNA is also targeted by miR-24, miR-34a, miR-124, miR-125b, miR-129, miR-137, miR-195, miR-449 and let-7 family members. The levels of D-type cyclins are downregulated by let-7, miR-15 family, miR-17, miR-19a miR-20a and mir-34.37
The overall impact of miRNAs on liver regeneration has been studied by generating mice with hepatocyte-specific miRNA deficiency. Interestingly, these mice are viable and developed normally into adulthood. However, whereas miRNA-deficient hepatocytes readily exited the G0 phase of the cell cycle, they failed to transition into S phase by 36 h after PH, suggesting a key role for miRNA in regulating liver regeneration.
The whole process of liver regeneration has three clearly distinct phases: an initiation stage, a proliferation stage and a termination stage.41 Most research has focused on the first two stages. However, the participants at the stage of termination have not been well characterized so far. Most of the studies have focused on the anti-proliferative role of TGFβ1. However, the fact that transgenic mice displayed a complete liver regeneration (although delayed) suggests that other mediators must be involved in this process.42 In this context, our results support the idea that miR-33 may have a significant role as a termination signal in the process of liver regeneration. These new findings could open new therapeutic perspectives to stimulate liver regeneration in chronic liver disease or after liver resection in patients affected by hepatocellular carcinoma.
Materials and Methods
Mice.
Six-week-old male C57BL/6 mice were randomized into three groups (n = 20 mice): no treatment (PBS, n = 4), control-ASO (n = 8) or miR-33-ASO (n = 8) (Jackson Laboratory). The mice received a weekly subcutaneous injection of 10 mg/Kg control-ASO, miR-33-ASO or PBS for 4 weeks. All animals were kept under constant temperature and humidity in a 12 h controlled dark/light cycle. Mice were fed ad libitum on a standard pellet diet. All animal experiments were approved by the Institutional Animal Care Use Committee of New York University Medical Center.
Materials.
Chemicals were obtained from Sigma unless otherwise noted. Mouse monoclonal antibodies against CDK6 (1:1,000) and PCNA (1:1,000), rabbit monoclonal antibody against CCND1 (1:1,000) and cleaved caspase 3 (Asp 175) (1:500) were purchased from Cell Signaling. Mouse monoclonal HSP-90 (1:1,000) antibody was from BD Bioscience. Mouse monoclonal ABCA1 (1:1,000) antibody was purchased from Abcam. Secondary fluorescently-labeled antibodies were from Molecular Probes (Invitrogen).
Cell culture.
A549, Hela, Huh7 and COS-7 cells were obtained from American Type Tissue Collection. A549, Hela, Huh7 and COS-7 cells were maintained in Dulbecco's modified Eagle medium (DMEM) containing 10% FBS, 100 U/ml penicillin and 100 µg/ml streptomycin. Cells were treated with Nocodazole (250 ng/ml, Sigma) 24 h post-transfection or with 2 mM Thymidine for 18 h as indicated in figure legends. Stable cell lines were generated upon transduction with lentiviral vectors and selection with puromycin (2 ug/mL). Briefly, murine miR-33 was amplified from genomic DNA and cloned into pSicoR, as described in Marquart el al. (PNAS 2010); the GFP cassette in pSicoR was then swapped with a puromycin resistance cassette. Lentiviruses were obtained by cotransfection in HEK293 cells of empty or miR-33 pSicoR-Puro, with vectors providing gag, pol and vsv-G. Supernatants were collected 48 h after transfection and used to transduce a variety of cell lines.
Cell cycle analysis.
Synchro=nous cells were cultured in DMEM supplemented with antibiotics (100 units of penicillin/ml and 100 µg/ml of streptomycin) at 37°C in a humidified atmosphere containing 5% CO2. At the end of the incubation the cells were washed twice with ice-cold phosphate-buffered saline (PBS), fixed in 70% cold ethanol, treated with 100 µg/ mL ribonuclease A (Roche) and labeled with 50 µg/mL propidium iodide (PI) for 1 h at 37°C. The cells were analyzed by flow cytometry (FACScalibur, Becton-Dickinson) using selective gating to exclude the doublets of cells and subjected to MODFIT analysis (Verity Software House, Inc.). Cells were treated with 250 ng/ml Nocodazole (Sigma) 24 h post-transfection.
Surgical procedure.
All operations were performed under Isoflurane anesthesia. A 70% partial hepatectomy (PH) was performed according to the technique described by Higgins and Anderson. The abdomen was opened via a midline incision. Two thirds of the liver (median and left lobes) were removed. After the PH, the weights of regenerating liver were measured and the hepatic tissue was preserved for future processing at the following time points: 3, 8, 24 and 48 h in the case of untreated mice and and 5 d in the case of treated mice. The percentage of liver regeneration was calculated following the formula (weight of non removed lobes/weight of total liver) and normalized by total body weigh of mice.
Western blot analysis.
Cells were lysed in ice-cold buffer containing 50 mM TRIS-HCl, pH 7.5, 125 mM NaCl, 1% NP-40, 5.3 mM NaF, 1.5 mM Na4P2O7 and 1 mM orthovanadate, 175 mg/ml octylglucopyranoside and 1 mg/ml of protease inhibitor cocktail (Roche) and 0.25 mg/ml AEBSF (Roche). Cell lysates were rotated at 4°C for 1 h before the insoluble material was removed by centrifugation at 12,000x g for 10 min. After normalizing for equal protein concentration, cell lysates were resuspended in SDS sample buffer before separation by SDS-PAGE. Following transfer of the proteins onto nitrocellulose membranes, the membranes were probed with the indicated antibodies, and protein bands were visualized using the Odyssey Infrared Imaging System (LI-COR Biotechnology). Densitometry analysis of the gels was performed using ImageJ software.
RNA isolation and quantitative real-time PCR.
Total RNA was isolated using TRIzol reagent (Invitrogen) according to the manufacturer's protocol. from the NIH (http://rsbweb.nih.gov/ij/). For mRNA quantification, cDNA was synthesized using Taqman RT reagents (Applied Biosystems), following the manufacturer's protocol. Quantitative real-time PCR was performed in triplicate using iQ SYBR green Supermix (BioRad) on iCycler Real-Time Detection System (BioRad). mRNA levels were normalized to GAPDH as a housekeeping gene. The primer sequences used were:
ABCA1, 5′-GGT TTG GAG ATG GTT ATA CAA TAG TTG T-3′ and 5′-CCC GGA AAC GCA AGT CC-3′;
ABCG1, 5′-TCA CCC AGT TCT GCA TCC TCT T-3′ and 5′-GCA GAT GTG TCA GGA CCG AGT-3′;
GAPDH, 5′-AAC TTT GGC ATT GTG GAA GG-3′ and 5′-ACA CAT TGG GGG TAG GAA CA-3′;
SREBP2, 5′-GCG TTC TGG AGA CCA TGG A-3′ and 5′-ACA AAG TTG CTC TGA AAA CAA ATC A-3′;
SREBP1, 5′-ACT TCC CTG GCC TAT TTG ACC-3′ and 5′-GGC ATG GAC GGG TAC ATC TT-3′;
HMGCR 5′-CTT GTG GAA TGC CTT GTG ATT G-3′ and 5′-AGC CGA AGC AGC ACA TGA T-3′;
CDK6, 5′-ACC TTC GAG CAC CCC AAC GT-3′ and 5′-ACC ACA GCG TGA CGA CCA CT-3′;
CDK8, 5′-GCC AGT TCA GTT ACC TCG GGG A-3′ and 5′-GGG CAA AGC CCA TGT CAG CAA T-3′;
CDK19, 5′-TTT GCC GGC TGCCAG ATT CC-3′ and 5′-AGG CGA GAACTG GAG TGC TGA-3′;
CyclinD1, 5′-TCG TTG CCC TCT GTG CCA CA-3′ and 5′-AGG CAG TCC GGG TCA CAC TT-3′:
CyclinM1, 5′-CAC CCG CTT CTA CAA CCG GC-3′ and 5′-AGG GCT CCA CTT CTG TGG CC-3′.
For miRNA quantification, total RNA was reverse transcribed using the RT2 miRNA First Strand Kit (SABiosciences). Primers specific for human miR-33a, miR-33b, miR-16 (SABiosciences) were used and values normalized to human SNORD38b as housekeeping gene.
miR-33 and anti-miR-33 transfection.
A549, Hela and Huh7 cells were transfected with 40 nM miRIDIAN miRNA mimics (miR-33) or with 60 nM miRIDIAN miRNA inhibitors (inh-miR-33) (Dharmacon) utilizing Oligofectamine (Invitrogen). All experimental control samples were treated with an equal concentration of a non-targeting control mimic sequence (CM) or an inhibitor negative control sequence (CI) to control for non-sequence-specific effects in miRNA experiments.
3′UTR luciferase reporter assays.
cDNA fragments corresponding to the entire 3′UTR of Ccnd1 and the three predicted miRNA target sites of Cdk6 were amplified by RT-PCR from total RNA extracted from A549 cells with XhoI and NotI linkers. The PCR products were directionally cloned downstream of the Renilla luciferase open reading frame of the psiCHECK2™ vector (Promega) that also contained a constitutively expressed firefly luciferase gene, which is used to normalize transfections. Point mutations in the seed region of predicted miR-33 sites within the 3′UTR of Ccnd1 and Cdk6 were generated using Multisite-Quickchange (Stratagene) according to the manufacturer's protocol. All constructs were confirmed by sequencing. COS-7 cells were plated into 12-well plates (Costar) and cotransfected with 0.5 g of the indicated 3′UTR luciferase reporter vectors and the miR-33 mimic or negative control miRNA mimic (CM) (Dharmacon) using Lipofectamine 2000 (Invitrogen). Luciferase activity was measured using the Dual-Glo Luciferase Assay System (Promega). Renilla luciferase activity was normalized to the corresponding firefly luciferase activity and plotted as a percentage of the control (cells co-transfected with the corresponding concentration of control mimic). Experiments were performed in triplicate wells of a 12-well plate and repeated at least three times.
Cell proliferation assay.
Huh7 and A549 cells were grown in DMEM supplemented with 10% lipoprotein-deficient serum (LPDS) and transfected with CM, miR-33, control miRNA inhibitor (CI) and antisense inhibitor of miR-33 (inh-miR-33). At the indicated times, the viable cells were counted by trypan blue dye exclusion using a hemocytometer.
Crystal violet method.
After treatment, the medium was removed, and 24-well dishes were washed with PBS, fixed with 1% glutaraldehyde for 15 min, washed twice with PBS and stained with 100 µl of 0.1% aqueous crystal violet for 20 min. Dishes were rinsed four times in tap water and allowed to dry. If cell mass estimation was desired, 100 µl of 10% acetic acid was added, and the content of each well was mixed before reading the absorbance at 595 nm.
Immunohistochemistry.
For the histological and the immunohistochemistry experiments, liver samples were fixed in 4% paraformaldehyde (PFA) at 4°C, cryoprotected overnight in 30% sucrose solution and embedded in OCT (Tissue-Tek® O.C.T™ Compound, SAKURA) embedding compound and frozen on dry ice. Next, 8-µm frozen sections were rehydrated and lipid droplets deposition was detected by Oil Red-O (Sigma) staining. Sections were rinsed with 60% isopropanol and stained for 20 min with filtered Oil Red O solution (0.5% in isopropanol followed by a 60% dilution in distilled water). After two rinses with 60% isopropanol and distilled water, slides were counterstained with hematoxylin for 4 min, rinsed with water and mounted. Digital images were taken with a Leica DM 4000B microscope.
Immunohistofluorescence.
Tissues were fixed with 4% PFA, cryoprotected overnight in a 30% sucrose solution and embedded in OCT. Frozen sections of 8 µm were rehydrated, blocked with 5% normal goat serum (NGS) and incubated with mouse anti-Ki67 (1:100, Abcam). Controls without primary antibodies were used as negative controls. Binding sites of the primary antibodies were revealed with Alexa-597 goat-anti-mouse IgG (1:1,000, Invitrogen). Samples were analyzed with a fluorescence microscope (Nikon Eclipse E600).
Statistics.
Data are presented as the mean ± standard error of the mean (SEM) (n is noted in the figure legends). The statistical significance of differences was evaluated with the Student t-test. Significance was accepted at the level of p < 0.05.
Acknowledgements
We thank Christine Esau from Regulus Therapeutics for providing us the 2′fluoro/methoxyethyl-modified (2′F/MOE-modified) phosphorothioate backbone antisense oligonucleotides.
Financial Support
This work was supported by grants from the National Institutes of Health R01HL107953 and R01HL106063 (to C.F.H.), R01HL105945 (to Y.S.) and R01HL107794 (to A.B.), the Ministerio de Ciencia e Innovación-Plan Nacional de I+D+I (SAF2010-19025 to M.M.R.), the Deutsche Forschungsgemeinschaft DFG EXC. 257 (D.C.S. and E.E.). M.P.P. by MICINN (contract number BES 2007-16909). A.G.S. and A.C.W. by Capes Foundation, Ministry of Education of Brazil, Brazil. CIBERehd is financed by Instituto de Salud Carlos III. CIBERehd and CIBERobn are financed by Instituto de Salud Carlos III, Spain.
Financial Disclosure
C.F.H. and Y.S. have patents on the use of miR-33 inhibitors.
Supplementary Material
References
- 1.Goldstein JL, Brown MS. Regulation of the mevalonate pathway. Nature. 1990;343:425–430. doi: 10.1038/343425a0. [DOI] [PubMed] [Google Scholar]
- 2.Brown MS, Goldstein JL. Suppression of 3-hydroxy-3-methylglutaryl coenzyme A reductase activity and inhibition of growth of human fibroblasts by 7-ketocholesterol. J Biol Chem. 1974;249:7306–7314. [PubMed] [Google Scholar]
- 3.Chen HW, Heiniger HJ, Kandutsch AA. Relationship between sterol synthesis and DNA synthesis in phytohemagglutinin-stimulated mouse lymphocytes. Proc Natl Acad Sci USA. 1975;72:1950–1954. doi: 10.1073/pnas.72.5.1950. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Chen HW, Kandutsch AA, Waymouth C. Inhibition of cell growth by oxygenated derivatives of cholesterol. Nature. 1974;251:419–421. doi: 10.1038/251419a0. [DOI] [PubMed] [Google Scholar]
- 5.Fernández C, Lobo Md Mdel V, Gómez-Coronado D, Lasunción MA. Cholesterol is essential for mitosis progression and its deficiency induces polyploid cell formation. Exp Cell Res. 2004;300:109–120. doi: 10.1016/j.yexcr.2004.06.029. [DOI] [PubMed] [Google Scholar]
- 6.Martínez-Botas J, Suárez Y, Ferruelo AJ, Gómez-Coronado D, Lasuncion MA. Cholesterol starvation decreases p34(cdc2) kinase activity and arrests the cell cycle at G2. FASEB J. 1999;13:1359–1370. doi: 10.1096/fasebj.13.11.1359. [DOI] [PubMed] [Google Scholar]
- 7.Crick DC, Andres DA, Waechter CJ. Geranylgeraniol promotes entry of UT-2 cells into the cell cycle in the absence of mevalonate. Exp Cell Res. 1997;231:302–307. doi: 10.1006/excr.1997.3480. [DOI] [PubMed] [Google Scholar]
- 8.Martínez-Botas J, Ferruelo AJ, Suárez Y, Fernández C, Gómez-Coronado D, Lasunción MA. Dose-dependent effects of lovastatin on cell cycle progression. Distinct requirement of cholesterol and non-sterol mevalonate derivatives. Biochim Biophys Acta. 2001;1532:185–194. doi: 10.1016/S1388-1981(01)00125-1. [DOI] [PubMed] [Google Scholar]
- 9.Brown MS, Goldstein JL. The SREBP pathway: regulation of cholesterol metabolism by proteolysis of a membrane-bound transcription factor. Cell. 1997;89:331–340. doi: 10.1016/S0092-8674(00)80213-5. [DOI] [PubMed] [Google Scholar]
- 10.Brown MS, Goldstein JL. A proteolytic pathway that controls the cholesterol content of membranes, cells and blood. Proc Natl Acad Sci USA. 1999;96:11041–11048. doi: 10.1073/pnas.96.20.11041. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Espenshade PJ. SREBPs: sterol-regulated transcription factors. J Cell Sci. 2006;119:973–976. doi: 10.1242/jcs02866. [DOI] [PubMed] [Google Scholar]
- 12.Osborne TF. Sterol regulatory element-binding proteins (SREBPs): key regulators of nutritional homeostasis and insulin action. J Biol Chem. 2000;275:32379–32382. doi: 10.1074/jbc.R000017200. [DOI] [PubMed] [Google Scholar]
- 13.Bengoechea-Alonso MT, Ericsson J. Cdk1/cyclin B-mediated phosphorylation stabilizes SREBP1 during mitosis. Cell Cycle. 2006;5:1708–1718. doi: 10.4161/cc.5.15.3131. [DOI] [PubMed] [Google Scholar]
- 14.Inoue N, Shimano H, Nakakuki M, Matsuzaka T, Nakagawa Y, Yamamoto T, et al. Lipid synthetic transcription factor SREBP-1a activates p21WAF1/CIP1, a universal cyclin-dependent kinase inhibitor. Mol Cell Biol. 2005;25:8938–8947. doi: 10.1128/MCB.25.20.8938-47.2005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Nakakuki M, Shimano H, Inoue N, Tamura M, Matsuzaka T, Nakagawa Y, et al. A transcription factor of lipid synthesis, sterol regulatory element-binding protein (SREBP)-1a causes G(1) cell cycle arrest after accumulation of cyclin-dependent kinase (cdk) inhibitors. FEBS J. 2007;274:4440–4452. doi: 10.1111/j.1742-4658.2007.05973.x. [DOI] [PubMed] [Google Scholar]
- 16.Ambros V. MicroRNA pathways in flies and worms: growth, death, fat, stress and timing. Cell. 2003;113:673–676. doi: 10.1016/S0092-8674(03)00428-8. [DOI] [PubMed] [Google Scholar]
- 17.Ambros V. The functions of animal microRNAs. Nature. 2004;431:350–355. doi: 10.1038/nature02871. [DOI] [PubMed] [Google Scholar]
- 18.Bartel DP. MicroRNAs: target recognition and regulatory functions. Cell. 2009;136:215–233. doi: 10.1016/j.cell.2009.01.002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Filipowicz W, Bhattacharyya SN, Sonenberg N. Mechanisms of post-transcriptional regulation by microRNAs: are the answers in sight? Nat Rev Genet. 2008;9:102–114. doi: 10.1038/nrg2290. [DOI] [PubMed] [Google Scholar]
- 20.Marquart TJ, Allen RM, Ory DS, Baldán A. miR-33 links SREBP-2 induction to repression of sterol transporters. Proc Natl Acad Sci USA. 2010;107:12228–12232. doi: 10.1073/pnas.1005191107. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Najafi-Shoushtari SH, Kristo F, Li Y, Shioda T, Cohen DE, Gerszten RE, et al. MicroRNA-33 and the SREBP host genes cooperate to control cholesterol homeostasis. Science. 2010;328:1566–1569. doi: 10.1126/science.1189123. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Rayner KJ, Suárez Y, Dávalos A, Parathath S, Fitzgerald ML, Tamehiro N, et al. MiR-33 contributes to the regulation of cholesterol homeostasis. Science. 2010;328:1570–1573. doi: 10.1126/science.1189862. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Dávalos A, Goedeke L, Smibert P, Ramírez CM, Warrier NP, Andreo U, et al. miR-33a/b contribute to the regulation of fatty acid metabolism and insulin signaling. Proc Natl Acad Sci USA. 2011;108:9232–9237. doi: 10.1073/pnas.1102281108. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Gerin I, Clerbaux LA, Haumont O, Lanthier N, Das AK, Burant CF, et al. Expression of miR-33 from an SREBP2 intron inhibits cholesterol export and fatty acid oxidation. J Biol Chem. 2010;285:33652–33661. doi: 10.1074/jbc.M110.152090. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Horie T, Ono K, Horiguchi M, Nishi H, Nakamura T, Nagao K, et al. MicroRNA-33 encoded by an intron of sterol regulatory element-binding protein 2 (Srebp2) regulates HDL in vivo. Proc Natl Acad Sci USA. 2010;107:17321–17326. doi: 10.1073/pnas.1008499107. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Rayner KJ, Sheedy FJ, Esau CC, Hussain FN, Temel RE, Parathath S, et al. Antagonism of miR-33 in mice promotes reverse cholesterol transport and regression of atherosclerosis. J Clin Invest. 2011;121:2921–2931. doi: 10.1172/JCI57275. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.van Rooij E, Sutherland LB, Qi X, Richardson JA, Hill J, Olson EN. Control of stress-dependent cardiac growth and gene expression by a microRNA. Science. 2007;316:575–579. doi: 10.1126/science.1139089. [DOI] [PubMed] [Google Scholar]
- 28.Michalopoulos GK. Liver regeneration. J Cell Physiol. 2007;213:286–300. doi: 10.1002/jcp.21172. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Overturf K, al-Dhalimy M, Ou CN, Finegold M, Grompe M. Serial transplantation reveals the stem-cell-like regenerative potential of adult mouse hepatocytes. Am J Pathol. 1997;151:1273–1280. [PMC free article] [PubMed] [Google Scholar]
- 30.Boylan JM, Gruppuso PA. D-type cyclins and G1 progression during liver development in the rat. Biochem Biophys Res Commun. 2005;330:722–730. doi: 10.1016/j.bbrc.2005.03.042. [DOI] [PubMed] [Google Scholar]
- 31.Hanse EA, Nelsen CJ, Goggin MM, Anttila CK, Mullany LK, Berthet C, et al. Cdk2 plays a critical role in hepatocyte cell cycle progression and survival in the setting of cyclin D1 expression in vivo. Cell Cycle. 2009;8:2802–2809. doi: 10.4161/cc.8.17.9465. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Kato A, Bamba H, Shinohara M, Yamauchi A, Ota S, Kawamoto C, et al. Relationship between expression of cyclin D1 and impaired liver regeneration observed in fibrotic or cirrhotic rats. J Gastroenterol Hepatol. 2005;20:1198–1205. doi: 10.1111/j.1440-746.2005.03829.x. [DOI] [PubMed] [Google Scholar]
- 33.Nurse P. Checkpoint pathways come of age. Cell. 1997;91:865–867. doi: 10.1016/S0092-8674(00)80476-6. [DOI] [PubMed] [Google Scholar]
- 34.Rivadeneira DB, Mayhew CN, Thangavel C, Sotillo E, Reed CA, Graña X, et al. Proliferative suppression by CDK4/6 inhibition: complex function of the retinoblastoma pathway in liver tissue and hepatoma cells. Gastroenterology. 2010;138:1920–1930. doi: 10.1053/j.gastro.2010.01.007. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Herrera-Merchan A, Cerrato C, Luengo G, Dominguez O, Piris MA, Serrano M, et al. miR-33-mediated downregulation of p53 controls hematopoietic stem cell self-renewal. Cell Cycle. 2010;9:3277–3285. doi: 10.4161/cc.9.16.12598. [DOI] [PubMed] [Google Scholar]
- 36.Thomas M, Lange-Grunweller K, Weirauch U, Gutsch D, Aigner A, Grunweller A, et al. The proto-oncogene Pim-1 is a target of miR-33a. Oncogene. doi: 10.1038/onc.2011.278. [DOI] [PubMed] [Google Scholar]
- 37.Bueno MJ, Malumbres M. MicroRNAs and the cell cycle. Biochim Biophys Acta. 2011;1812:592–601. doi: 10.1016/j.bbadis.2011.02.002. [DOI] [PubMed] [Google Scholar]
- 38.Linsley PS, Schelter J, Burchard J, Kibukawa M, Martin MM, Bartz SR, et al. Transcripts targeted by the microRNA-16 family cooperatively regulate cell cycle progression. Mol Cell Biol. 2007;27:2240–2252. doi: 10.1128/MCB.02005-06. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Takeshita F, Patrawala L, Osaki M, Takahashi RU, Yamamoto Y, Kosaka N, et al. Systemic delivery of synthetic microRNA-16 inhibits the growth of metastatic prostate tumors via downregulation of multiple cell cycle genes. Mol Ther. 2010;18:181–187. doi: 10.1038/mt.2009.207. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Wang F, Fu XD, Zhou Y, Zhang Y. Downregulation of the cyclin E1 oncogene expression by microRNA-16-1 induces cell cycle arrest in human cancer cells. BMB Rep. 2009;42:725–730. doi: 10.5483/BMBRep.2009.42.11.725. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Michalopoulos GK, DeFrances MC. Liver regeneration. Science. 1997;276:60–66. doi: 10.1126/science.276.5309.60. [DOI] [PubMed] [Google Scholar]
- 42.Sanderson N, Factor V, Nagy P, Kopp J, Kondaiah P, Wakefield L, et al. Hepatic expression of mature transforming growth factor beta1 in transgenic mice results in multiple tissue lesions. Proc Natl Acad Sci USA. 1995;92:2572–2576. doi: 10.1073/pnas.92.7.2572. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.