Abstract
Human telomeres consist of multiple tandem hexameric repeats, each containing a guanine triplet. Guanosine-rich clusters are highly susceptible to oxidative base damage, necessitating base excision repair (BER). Previous demonstration of enhanced strand displacement synthesis by the BER component DNA polymerase β in the presence of telomere protein TRF2 suggests that telomeres employ long-patch (LP) BER. Earlier analyses in vitro showed that efficiency of BER reactions is reduced in the DNA-histone environment of chromatin. Evidence presented here indicates that BER is promoted at telomeres. We found that the three proteins that contact telomere DNA, POT1, TRF1 and TRF2, enhance the rate of individual steps of LP-BER and stimulate the complete reconstituted LP-BER pathway. Thought to protect telomere DNA from degradation, these proteins still apparently evolved to allow selective access of repair proteins.
Key words: telomeres, base excision repair, shelterin complex, oxidative damage, LP-BER
Introduction
The human genome experiences constant genotoxic damage from both normal metabolic byproducts and external environmental factors.1 Generation of reactive oxygen species (ROS) is believed to be the most frequent source of damaging agents within the cell.2 Oxidative damage of bases caused by ROS has been implicated in aging,3–6 neurodegenerative diseases7 and cancer.8–10 Of the four DNA bases, the unique moiety of guanine makes it the most susceptible to electron abstraction.11,12 Further, guanine becomes especially susceptible to oxidation when it is found in tandem or in repeat sequences, i.e., GG or GGG.13–15 Each human telomere consists of 2–15 kilobases16 of double-stranded 5′-TTA GGG-3′ repeats, terminated with 50–500-nts of 3′ single-strand, guanine-rich repeat overhang.17,18 Significantly, recent research has shown that oxidative conditions can cause rapid telomere shortening, known to destabilize the chromosome.19–25
BER is the pathway that removes bases damaged by oxidation. BER can proceed via two sub-pathways, short- and long-patch BER (SP-BER and LP-BER, respectively). Initially, specific DNA glycosylases recognize and excise the damaged base, leaving an intact sugar-phosphate backbone.26 Apurinic/apyrimidinic endonuclease 1 (APE1) recognizes the abasic site and cleaves the sugar-phosphate backbone immediately upstream,27 leaving an upstream 3′ hydroxyl (3′-OH) group and a downstream 5′-deoxyribose-5-phosphate (5′-dRP).28,29 In SP-BER, DNA polymerase β (pol β) uses its lyase activity to remove the 5′-dRP30,31 and its polymerase activity to incorporate the correct nucleotide,32,33 leaving a nick that is sealed by DNA ligase III (LIGIII).34 When the lyase activity of pol β is abrogated, repair proceeds via LP-BER.35,36 Pol β will synthesize into the downstream DNA, displacing the damaged base into a 2–13 nt flap33,37–40 that is cleaved by flap endonuclease 1 (FEN1),41 leaving a nick to be sealed by DNA ligase I (LigI).41
The telomere is a unique environment that has evolved six specialized proteins that coat the repeat DNA, collectively named the shelterin complex.42 The three proteins of interest to our studies directly bind to telomeric repeats. Telomeric repeat binding factors 1 and 2 (TRF1 and TRF2) are telomeric double-stranded DNA (dsDNA) binding proteins.43–46 Protection of telomeres 1 (POT1) is a telomeric single-stranded DNA (ssDNA) binding protein.47 The shelterin complex is primarily thought to play a protective role in the cell through telomerase regulation and inhibition of inappropriate DNA damage response by making the telomere terminus invisible to some repair processes.43,48,49 Whereas nucleotide excision repair (NER) and double-strand break repair (DSBR) were shown to be inhibited by the shelterin components,50–55 there is recent evidence for BER activity in telomeres.56
Earlier, TRF2 was found to interact with and stimulate pol β polymerase activity.57 It was also observed that loss of OGG1, a BER glycoslyase that removes oxidized guanines, led to a disruption in telomere length homeostasis.56,58 Further, loss of FEN1 has been shown to result in telomere dysfunction.59 These results suggest both a role for LP-BER in telomeres and a functional interaction of BER and shelterin proteins.
In the current study, we employed biochemical reconstitution to investigate the interaction of telomere sequences, shelterin proteins and protein components of LP-BER. Our results suggest functional interactions that have evolved to protect telomeric DNA from the instability of oxidative shortening.
Results
POT1, TRF1 and TRF2 interact with BER components.
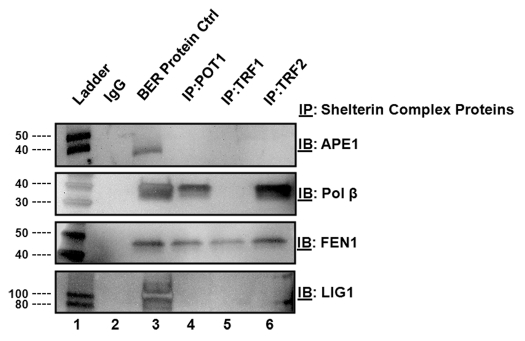
We initially screened for interactions between POT1, TRF1, TRF2 and BER components in vitro by employing a binding assay using purified proteins. We individually incubated the shelterin complex proteins (POT1, TRF1 or TRF2) with each of the LP-BER proteins (APE1, pol β, FEN1 and LigI) in a 1:1 ratio. We then immunoprecitated the bound complex with an antibody recognizing either POT1, TRF1 or TRF2 and probed for interactions using western blot analysis and antibodies against APE1, pol β, FEN1 or LigI. While FEN1 was found to interact with all three proteins of the shelterin complex, (Fig. 1, lanes 4–6), APE1 and LigI did not show stable interactions with any of the shelterin components. Additionally, we detected an interaction between pol β and POT1 (Fig. 1, lane 4). Interactions between TRF2 and pol β and FEN1 were previously reported in references 57 and 60, and confirmed in our assays.
Figure 1.
Shelterin proteins physically interact with BER proteins. Lane 1 indicates a molecular weight marker. Lane 2 contains IgG alone. Lane 3 contains each BER protein probed by western blot analysis. Lanes 4–6 contain immunoprecipitates of either POT1, TRF1 or TRF2, respectively, that were probed by western blot analysis with antibodies against the BER proteins. The specific procedure is included in the Materials and Methods section.
POT1, TRF1 and TRF2 improve APE1 cleavage and binding.
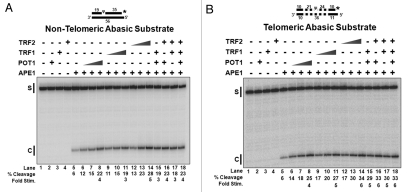
The removal of a damaged base by a DNA glycosylase generates an abasic site that is recognized by APE1. In order to simulate this intermediate in vitro, we designed two duplex DNA substrates, both containing a central tetrahydrofuran (THF) residue as the abasic site. One substrate had a non-telomeric sequence; the other included six telomeric repeat sequences (5′-TTA GGG-3′), three on either side of the THF residue. TRF1 and TRF2 were expected to bind only the telomeric substrate based on their known specificities for telomeric sequences. POT1 was not expected to interact with either substrate, because they did not contain a single-stranded region. Cleavage of substrates labeled at the 3′ end of the THF strand by APE1 created an incised product with distinctive mobility on a denaturing gel. POT1, TRF1 and TRF2 did not show any cleavage activity when incubated alone with the substrate (Fig. 2A and B, lanes 2–4). In the absence of the shelterin components, the limiting APE1 concentration showed ∼6% cleavage activity on both the non-telomeric and telomeric substrates (Fig. 2A and B, lane 5). The substrates were then incubated with APE1 and increasing concentrations of POT1, TRF1 or TRF2 or combinations of these. On the non-telomeric substrate, APE1 cleavage was enhanced ∼4-fold by POT1 (Fig. 2A, lanes 6–8), ∼3-fold by TRF1 (Fig. 2A, lanes 9–11) and ∼5-fold by TRF2 (Fig. 2A, lanes 12–14). APE1 cleavage on a telomeric substrate was improved ∼4-fold by POT1 (Fig. 2B, lanes 6–8), ∼5-fold by TRF1 (Fig. 2B, lanes 9–11) and ∼6-fold by TRF2 (Fig. 2B, lanes 12–14). Combinations of the shelterin components improved APE1 cleavage (Fig. 2A and B, lanes 15–18); however, we did not observe an additive effect of stimulation, even when APE1 was used in sufficiently limiting amounts. This suggests a maximal stimulation of APE1, beyond which cleavage cannot be improved (Fig. S1).
Figure 2.
Shelterin proteins stimulate APE1 endonuclease activity on abasic substrates. (A) APE1 (0.5 fmol/rxn) cleavage on non-telomeric abasic substrate (D6•T3) in the absence/presence of increasing concentrations of individual shelterin proteins, POT1, TRF1 or TRF2, (0, 10, 50, 100 fmol/rxn) or combinations of shelterin proteins (100 fmol/rxn per protein). (B) The same reactions on a telomeric abasic substrate (D8•T2). S, starting substrate; C, Ape1 cleaved product. The asterisk indicates the position of the radiolabel on the substrate and on subsequent substrates in later figures. The dotted lines in the substrate illustrations in this and subsequent figures indicate the telomeric regions.
In our protein interaction assays (Fig. 1), there was no detectable physical interaction between APE1 and the shelterin components. However, because there was improved cleavage by APE1 on both non-telomeric and telomeric substrates in the presence of POT1, TRF1 and TRF2, we examined the mechanism of this stimulation using electrophoretic mobility shift assays (EMSA). Lanes 1–4 of Figure S2A and B show migration of the substrate alone and POT1, TRF1 and TRF2 each alone with substrate, respectively. Because high concentrations of TRF1 and TRF2 interacted with both substrates and could mask APE1 binding, we decreased the concentrations of shelterin components in the presence of APE1. On the non-telomeric substrate, APE1 binding was increased ∼1.4-fold by POT1 (Fig. S2A, lanes 6—8), ∼2-fold by TRF1 (Fig. S2A, lanes 9–11) and ∼2 fold by TRF2 (Fig. S2A, lanes 12–14). APE1 binding was improved on the telomeric substrate ∼2-fold by POT1 (Fig. S2B, lanes 6—8), ∼2-fold by TRF1 (Fig. S2B, lanes 9–11) and ∼2 fold by TRF2 (Fig. S2B, lanes 12–14). These results suggest a transient interaction between the shelterin components and APE1, which increased the local concentration of APE1 on the substrate.
We also tested E. coli single-stranded binding protein, glutathione s-transferase and bovine serum albumin with APE1 and saw no increase in cleavage, indicating a specific relationship between APE1 and these shelterin components (data not shown).
POT1, TRF1 and TRF2 increase FEN1 endonuclease activity on flap substrates.
During LP-BER, a 2–13-nt flap is generated by pol β strand displacement synthesis.40 Moreover, TRF2 stimulates the strand displacement activity of pol β.57 This prompted us to ask whether pol β synthesis and strand displacement activity are also augmented by POT1 and TRF1. Using a substrate containing a primer annealed to a template, we first tested the ability of POT1 and TRF1 to increase primer extension by pol β synthesis. Then, we modified the same substrate by adding a downstream primer containing a short flap to examine whether POT1 and TRF1 can stimulate pol β strand displacement. While TRF2 stimulated pol β synthesis and strand displacement synthesis, as previously reported in reference 57, we did not observe any effect of TRF1 and POT1 on pol β synthesis or strand displacement functions (data not shown).
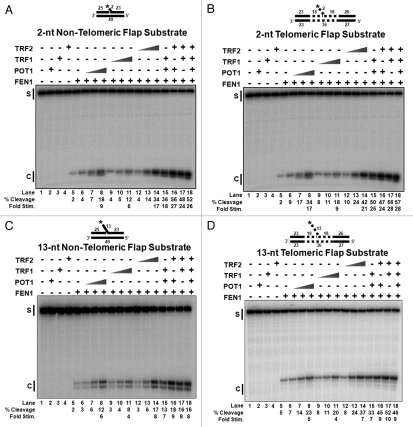
Since strand displacement synthesis by pol β generates a flap during LP-BER, we next examined the ability of the shelterin components to influence cleavage by FEN1. The shortest flap displaced in the LP-BER pathway is 2-nt; therefore, we created two 2-nt flap substrates: one consisting of non-telomeric DNA and the other containing six telomeric repeats (5′-TTA GGG-3′), three on either side of the flap base. LP-BER generates flaps as long as 13-nt,40 so we also created two 13-nt flap substrates: one containing non-telomeric DNA and the other with six telomeric repeats positioned as illustrated in Figure 3D plus two repeats in the 13-nt flap.
Figure 3.
Shelterin proteins stimulate FEN1 activity on flap substrates. (A) FEN1 (0.5 fmol/rxn) cleavage on a 2-nt non-telomeric flap substrate (U1•D1•T1) in the absence/presence of increasing concentrations of individual shelterin proteins, POT1, TRF1 or TRF2, (0, 10, 50, 100 fmol/rxn) or combinations of shelterin proteins (100 fmol/rxn per protein). The same reactions were preformed on (B) a 2-nt telomeric flap substrate (U4•D3•T4), (C) a 13-nt non-telomeric flap substrate (U1•D2•T1) and (D) a 13-nt telomeric flap substrate (U4•D4•T4). S, starting substrate; C, cleaved product.
In the absence of shelterin components, FEN1 cleaved ∼2% of both the non-telomeric and telomeric 2-nt flap substrates (Fig. 3A and B, lanes 5). FEN1 cleavage on the non-telomeric, 2-nt flap substrate was improved ∼9-fold by POT1 (Fig. 3A, lanes 6–8), ∼6-fold by TRF1 (Fig. 3A, lanes 9–11) and ∼17-fold by TRF2 (Fig. 3A, lanes 12–14). FEN1 cleavage on the telomeric, 2-nt flap substrate was increased ∼17-fold by POT1 (Fig. 3B, lanes 6–8), ∼9-fold by TRF1 (Fig. 3B, lanes 9–11) and ∼21-fold by TRF2 (Fig. 3B, lanes 12–14).
On the non-telomeric and telomeric 13-nt flap substrates in the absence of shelterin components, FEN1 cleaved ∼2% and ∼5%, respectively (Fig. 3C and D, lanes 5). FEN1 cleavage on the non-telomeric, 13-nt flap substrate was improved ∼6-fold by POT1 (Fig. 3C, lanes 6–8), ∼4-fold by TRF1 (Fig. 3C, lanes 9–11) and ∼8-fold by TRF2 (Fig. 3C, lanes 12–14). FEN1 cleavage on the telomeric, 13-nt flap substrate was increased ∼5-fold by POT1 (Fig. 3D, lanes 6–8), ∼4-fold by TRF1 (Fig. 3D, lanes 9–11) and ∼7-fold by TRF2 (Fig. 3D, lanes 12–14, and S4).
Since FEN1 physically interacted with all three proteins in the shelterin complex (Fig. 1), we asked whether the interactions influence FEN1 binding of the flap substrates. Using EMSA, we analyzed the DNA substrate binding efficiency of FEN1 in the presence of POT1, TRF1 and TRF2. On the non-telomeric substrate, FEN1 binding was increased ∼2-fold by POT1 (Fig. S4A, lanes 4–6), ∼4-fold by TRF1 (Fig. S4A, lanes 11–13) and ∼3-fold by TRF2 (Fig. S4A, lanes 18–20). FEN1 binding was improved on the telomeric substrate ∼2-fold by POT1 (Fig. S4B, lanes 4–6), ∼4-fold by TRF1 (Fig. S4B, lanes 11–13) and ∼3-fold by TRF2 (Fig. S4B, lanes 18–20).
Unlike with APE1, there was a more apparent additive effect on FEN1 stimulation, with more than one of the shelterin proteins on all four substrates as observed in lanes Figure 3A–D, lanes 15–18. However, as with APE1, there was a threshold of stimulation by the shelterin proteins, even though the low concentration FEN1 would have allowed detection of very high fold stimulation (Fig. S3).
We also tested E. coli single-stranded binding protein, glutathione s-transferase and bovine serum albumin with FEN1 and saw no increase in cleavage, indicating a specific relationship between FEN1 and these shelterin components (data not shown).
POT1, TRF1 and TRF2 improve LigI binding and ligation.
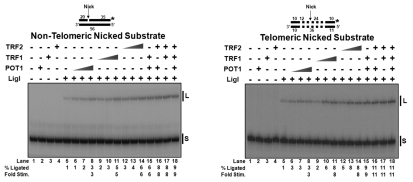
We next tested the effect of shelterin proteins on ligation, the final step in LP-BER. We designed two substrates with a central nick: one consisted of duplex non-telomeric DNA; the other contained two telomeric repeats on the upstream side of the nick and four telomeric repeats downstream of the nick. The nick was located near the center of each substrate.
In the absence of shelterin components, limited LigI sealed ∼1% of both the non-telomeric and telomeric nicked substrates (Fig. 4A and B, lanes 5). Ligation of the non-telomeric nicked substrate was improved ∼3-fold by POT1 (Fig. 4A, lanes 6–8), ∼5-fold by TRF1 (Fig. 4A, lanes 9–11) and ∼6-fold by TRF2 (Fig. 4A, lanes 12–14). Ligation on the telomeric nicked substrate was increased ∼3-fold by POT1 (Fig. 4B, lanes 6–8), ∼8-fold by TRF1 (Fig. 4B, lanes 9–11), and ∼8-fold by TRF2 (Fig. 4B, lanes 12–14). Although there was a small additive effect on LigI stimulation with more than one of the shelterin proteins; as observed in Figure 4A and B, lanes 15–18, there was an upper limit of stimulation observed, as seen with APE1 and FEN1 (Fig. S5).
Figure 4.
Shelterin proteins stimulate LigI ligase activity on nicked substrates. (A) LigI (0.5 fmol/rxn) activity on non-telomeric nicked substrate (U2•D5•T3) in the absence/presence of increasing concentrations of individual shelterin proteins, pOT1, TRF1 or TRF2, (0, 10, 50, 100 fmol/rxn) and combinations of shelterin proteins (100 fmol/rxn per protein). (B) The same reactions on a telomeric nicked substrate (U3•D7•T2). S, starting substrate; L, ligated product.
Results from our protein binding assays (Fig. 1) showed no physical interaction between LigI and the shelterin components. However, since we observed improved joining by LigI on both non-telomeric and telomeric nicked substrates with POT1, TRF1 and TRF2, we examined the mechanism of this stimulation using EMSA analysis. Because high concentrations of TRF1 and TRF2 interacted with the telomeric nick substrate and could mask LigI binding, we decreased the concentrations of shelterin components, so that LigI binding could be evaluated. On the non-telomeric substrate, LigI binding was increased ∼1.3-fold by POT1 (Fig. S6A, lanes 4–6), ∼2-fold by TRF1 (Fig. S6A, lanes 11–13) and ∼2-fold by TRF2 (Fig. S6A, lanes 18–20). LigI binding was improved on the telomeric substrate ∼1.4-fold by POT1 (Fig. S6B, lanes 4–6), ∼2-fold by TRF1 (Fig. S6B, lanes 11–13) and ∼1.3-fold by TRF2 (Fig. S6B, lanes 18–20).
We also tested E. coli single-stranded binding protein, glutathione s-transferase and bovine serum albumin with LigI and saw no increase in cleavage, indicating a specific relationship between LigI and these shelterin components (data not shown).
Reconstitution of LP-BER with POT1, TRF1 and TRF2 in vitro.
The uniqueness of the telomere environment and its specific binding proteins suggests that repair functions have evolved to be integrated with telomere DNA and proteins. In order to better understand repair at the telomeres, we reconstituted LP-BER using essential proteins plus the three shelterin components. The substrates used for these studies are the same as those used for the APE1 cleavage and binding assays (Figs. 2 and S2).
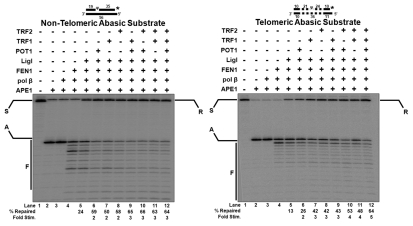
Upon addition of APE1, in lanes 2 of Figure 5A and B, nearly 95% cleavage at the abasic site was observed by the appearance of an APE1 cleaved product (A). The uncleaved substrate was presumably oligos that did not contain THF reside when synthesized. Because the substrate is labeled on the 3′ end of the G-rich strand, there was no change in product appearance in lane 3 (Fig. 5A and B) upon inclusion of pol β. This is expected, because pol β extends from the 3′-OH created by APE1 and displaces into the downstream DNA. Lanes 4 of Figure 5A and B show that addition of FEN1 resulted in short cleavage products (C). This verifies that pol β displaced the downstream DNA, creating various length flaps that became substrates for FEN1. The final LP-BER component, LigI, when added, generated repaired product (R) as seen in lanes 5 of Figure 5A and B. We observed ∼24% and ∼13% repair of the non-telomeric and telomeric substrates, respectively (Fig. 5A and B). Inclusion of POT1, TRF1 or TRF2 significantly increased the amount of repaired product. Specifically, generation of the final ligation product of the non-telomeric substrate was improved ∼2-fold by POT1 (Fig. 5A, lane 6), ∼2-fold by TRF1 (Fig. 5A, lane 7) and ∼2-fold by TRF2 (Fig. 5A, lane 8). Generation of the final ligation product of the telomeric substrate was improved ∼2-fold by POT1 (Fig. 5B, lane 6), ∼3-fold by TRF1 (Fig. 5B, lane 7) and ∼3-fold by TRF2 (Fig. 5B, lane 8).
Figure 5.
Shelterin proteins improve LP-BER in vitro. (A) LP-BER reconstituted with a non-telomeric abasic substrate (D6•T3) in the absence/presence of individual shelterin proteins, POT1, TRF1 or TRF2, (100 fmol/rxn) and in combinations. Lanes 1–5 are reactions with substrate alone, APE1 alone (5 fmol/rxn), APE1 plus pol β (10 fmol/rxn), APE1, pol β plus FEN1 (1 fmol/rxn), APE1, pol β, FEN1 plus LigI (0.5 fmol/rxn), respectively. Lanes 6–8 are the minimal LP-BER components plus the addition of POT1, TRF1 and TRF2 (100 fmol/rxn), respectively. Lanes 9–12 are the minimal LP-BER components plus shelterin proteins in combination (100 fmol/rxn each). (B) The same reconstitutions with a telomeric abasic substrate (D8•T2). S, starting substrate; A, APE1 cleavage product; F, FEN1 cleavage products; R, repaired product.
Stimulation by the full complement of shelterin proteins slightly exceeded the highest values obtained with individual telomere proteins, indicating a maximum degree of stimulation as seen earlier measurements with individual LP-BER components. The concentrations of FEN1 and LigI were limited to best display the stimulation of the total repair pathway. When concentrations of FEN1 or LigI were optimized in the absence of shelterin components, nearly complete repair was observed.
Discussion
We show that telomere-specific proteins, POT1, TRF1 and TRF2 stimulate the binding and enzymatic activities of the LP-BER proteins APE1, FEN1 and LigI both individually and when they act together in reconstituted LP-BER using a telomeric substrate. Because telomeres contain a high concentration of guanine repeat sequences, they are more susceptible to oxidative damage than the rest of the genome. The oxidative damage product 8-oxo-guanine (8-oxo-G) is the most abundant base lesion detected at the telomeres,61 indicating that BER must be a frequently required repair pathway. Previous work showed that oxidative damage causes disruption of TRF1 and TRF2 binding, leading to depletion of the shelterin components from the telomeres.62 In addition to maintaining the optimal concentration of the shelterin components at the telomeres, the benefit of minimizing oxidative base damage in telomeres is the prevention of early cell senescence or apoptosis caused by telomere dysfunction. Therefore, we propose that telomere-specific proteins POT1, TRF1 and TRF2 have evolved to enhance BER at telomeres.
Muftuoglu et al. found that pol β and FEN1 both interacted with the telomere-specific protein TRF2. Association with pol β is further supported by the previous observation that TRF2 colocalized with the polymerase outside of the telomeric regions.60 This suggests a physical connection between telomere- and BER-specific proteins. We further asked whether the other telomeric DNA binding proteins interact with the BER pathway, demonstrating that POT1 binds both FEN1 and pol β, and TRF1 binds FEN1. Considering that all three telomeric proteins bind FEN1, it is reasonable to assume that they aid the nuclease in accessing telomeric DNA to find flap intermediates of LP-BER. This concept is consistent with structural evidence for coordinated protein function in BER found by Parikh et al.63 and Mol et al.64 Their results suggest that BER works through a systematic recognition of enzyme complexes formed at each step of LP-BER, presumably diminishing the probability that intermediates could generate further DNA damaging events. Wilson and Kunkel65 referred to this stepwise mechanism as “passing the baton” from APE1 cleavage to pol β lyase and DNA synthesis to ligation during SP-BER. Prasad et al.,66 however, showed that if FEN1 is involved, as in LP-BER, it must be recruited to the enzymatic complex. Our results suggest that one mechanism is by binding shelterin proteins.
These results contrast with analyses of repair protein accessibility to DNA in reconstituted chromatin, where the nucleosome acts as a barrier to the repair proteins, unlike the shelterin components. DNA glycosylase, pol β, FEN1 and LigI were all less efficient catalytically when tested on a reconstituted mononucleosome.67 Moreover, the extent of inhibition of activity depended on both the position of the substrate structure along the histone core and the orientation of the helix with respect to the histones. This behavior is consistent with physical occlusion of the substrate site requiring enzyme activity. Since chromatin hosts BER, additional mechanisms must partially or completely reverse the occlusion during the repair process. Our reconstitutions with telomere proteins show no evidence of occlusion. Moreover, the protein-coated damaged DNA is not simply invisible to the repair complex. Instead, the binding and stimulation properties of the telomeric proteins suggest that BER complexes are specifically recruited to telomeres, and that their repair functions are promoted. The evolution of these mechanisms suggests that efficient BER in telomeres is important for long-term species survival.
All three shelterin proteins stimulated the nuclease activity of FEN1. A more refined view of the results is informative. POT1 exhibited greater stimulation on the 2-nt telomeric flap compared with the non-telomeric analog, suggesting that POT1 has transient binding capacity to this structure, and that binding improves stimulation (Fig. 3). However, binding of POT1 to a 13-nt flap was less stimulatory to FEN1, suggesting that the system has evolved to work on short flaps. In fact, comparing overall stimulation from 2-nt flap cleavage to 13-nt flap cleavage, FEN1 was promoted most by shelterin proteins when the flap was short, implying that the system disfavors formation and processing of long flaps. This conclusion is further supported by the observation that Dna2 nuclease/helicase, which can activate replication protein A-coated flaps for cleavage by FEN1 in Okazaki fragment processing, is ineffective with POT1-coated flaps (unpublished observations). Because telomeric sequences are highly repetitive, they could be subject to sequence disruptions from polymerase slippage by pol β.68 Avoidance of long-flap intermediates may suppress this effect. TRF1 and TRF2 stimulation of FEN1 is also enhanced on a telomeric substrate compared with a nontelomeric substrate. TRF1 and TRF2 interact with and bend the telomeric dsDNA,68–70 a possible mechanism to make the flap base more accessible to FEN1.
We observed moderate stimulation of APE1 cleavage and LigI joining by POT1, TRF1 and TRF2, even though we could detect no physical interaction with the telomere proteins. This resembles the action of APE1, which improves OGG1 glycosylase activity without apparent physical interaction.71–73 POT1 indirectly interacts with dsDNA within the shelterin complex (reviewed in ref. 42). We speculate that POT1 transiently interacts with APE1 and LigI, facilitating a long-lived structural conformation change that leads to an increase in the biological efficiency of BER. Native gel electrophoresis experiments indicate that POT1 stabilizes the interaction of APE1 and LigI for their respective substrates (Figs. S2 and S6).
APE1 binding to an abasic site is critical for prevention of subsequent damage, leading to abortive intermediates.21 Further oxidation or reduction of the 5-dRP moiety formed by APE1 or damage induction not only inhibits lyase activity of pol β but also forms DNA/protein cross-links (DPC) with pol β.21,32,35,74,75 Once bound, APE1 kinks and surrounds the AP strand and cleaves the sugar-phosphate backbone. APE1 then facilitates loading of pol β. POT1 may only weakly interact with APE1, so that it does not disrupt the complex interaction APE1 must have with its substrate. Because POT1 physically interacts with pol β, it may help APE1 direct pol β to its substrate. Similarly, POT1 may have evolved to avoid interference with the LigI mechanism, which involves encircling the nick site.
TRF1 and TRF2 stimulate APE1 and LigI preferentially on telomeric substrates (Fig. 2B, S1B, 4B and S5B). Although TRF1 and TRF2 lose binding capacity if a guanine is converted to an abasic site,62 they may act from an adjacent site. Alternatively, shelterin components may recruit APE1 and LigI indirectly, by exposing damage sites and allowing access for the repair enzymes.
While the shelterin proteins stimulate BER on both telomeric and non-telomeric substrates, the localization of shelterin to telomeres is consistent with the conclusion that it exerts its effects on DNA repair in the telomere environment. Our results show stimulation of LP-BER. In stark contrast, Rochette and Brash50 showed that the inhibition of NER in telomeres led to higher incidence of cyclobutane pyrimidine dimers. Furthermore, TRF2 inhibits the XPF-ERCC1 endonuclease in vivo, which is required for NER.76,77 The types of damage NER repairs are recognized by distortion of helical structure caused by the presence of bulky adducts or unnatural covalent bonds. TRF2 also interacts with and inhibits the ATM kinase to suppress DNA double-strand break repair at ends of telomeres.78 Furthermore, Fink et al. discovered that targeting of a key NHEJ protein, Ku80, in the absence of apparent DNA damage, contributes to telomere dysfunction through TRF2 abstraction from the telomere.79 A reasonable set of conclusions is that base damage is a very prevalent lesion in telomeres and NER substrates, and double-strand breaks are less common.
Although telomeres are non-coding DNA, their integrity is vital to cell health. However, their triplet guanine-containing repeat sequence makes them particularly susceptible to dangerous shortening after oxidative damage. Our results indicate that they have evolved a way to promote efficient repair of this damage by recruitment of LP-BER proteins.
Materials and Methods
Substrates.
All synthetic oligonucleotides were purchased from Integrated DNA Technologies. Radioactive nucleotides [α-32P] dCTP and [γ-32P] ATP were purchased from Perkin Elmer Life Sciences. Klenow fragment of E. coli DNA polymerase I (KF) and T4 polynucleotide kinase (PNK) for 3′ and 5′ labeling, respectively, were purchased from Roche Applied Science. All remaining reagents were purchased from the best commercially available products.
Purified proteins.
Human FEN1 was purified as described in reference 80. Human APE1 was purified as described in reference 81. Human pol β was purified as described in reference 82. Human LigI was generously provided by Dr. Jeffrey Hayes and purified as described in reference 83. Recombinant human glutatione s-transferase-tagged POT1 and histidine-tagged TRF1 and TRF2 were purified as described in references 62, 84 and 85.
Oligonucleotides.
All experimental substrates are listed in Table S1. A “ψ” in the nucleotide sequence represents a tetrahydrofuran (THF) residue. Labels were incorporated at the 5′ end of the DNA primer by incubation with PNK and [γ-32P] ATP. Labels at the 3′ end were added by first annealing 20 pmol of primer to 50 pmol of template containing a 5′ guanine-overhang. The annealed complex was incubated with the KF and [α-32P] dCTP. Urea-PAGE was used to purify labeled primers. Experimental substrates were created by combining primers in annealing buffer containing 10 mM TRIS-HCl (pH 8.0), 50 mM NaCl and 1 mM DTT, heated to 95°C for 5 min. and slowly cooled to room temp. Substrates were annealed in a 1:2:4 ratio (labeled primer:template:upstream primer). Substrates containing an internal THF residue were annealed in a 1:2 ratio (labeled primer:template).
Enzyme assays.
To analyze how POT1, TRF1 and TRF2 affected APE1, FEN1 and LigI, 20 µl reaction mixtures containing the indicated enzyme quantities and 5 fmol of 32P radio-labeled DNA substrate were performed at 37°C and sampled for analysis at 10 min unless otherwise noted. Reactions were stopped by addition of 2X termination dye [90% formamide (v/v), 10 mM EDTA, 0.1% bromophenol blue and 0.1% xylene cyanole]. Unless stated otherwise, the reaction buffer contained 50 mM TRIS-HCl (pH 8.0), 2 mM DTT, 30 mM NaCl, 0.1 mg/ml bovine serum albumin (BSA), 5% glycerol, 8 mM MgCl2 and 2 mM ATP. All results are from at least three independent experiments.
Electrophoretic mobility gel shift assay (EMSA).
To analyze APE1, FEN1 and LigI binding with POT1, TRF1 and TRF2, 20 µl reaction mixtures containing the indicated quantities of enzymes and 5 fmol of 32P-radiolabeled DNA substrate were incubated at 37°C for 10 min. unless indicated otherwise. All results are from at least three independent experiments.
Gel analysis.
After electrophoresis, gels were dried, exposed on a phosphor screen and scanned using a PhosphorImager (GE Healthcare). Scanned images were analyzed using ImageQuant Version 5.0.
Binding assay.
Protein interactions were analyzed by incubating 1 ng of purified proteins of interest (in a 1:1 ratio) in coupling buffer (25 mM HEPES (pH 7.5), 100 mM NaCl, 1 mM EDTA, 1 mM DTT and 10% glycerol) for 1 h at 4°C. Concurrently, protein agarose A was incubated with antibody for the bait protein for 1 h at 4°C. The protein/coupling buffer mixture was washed and added to the protein agarose A/antibody slurry then incubated overnight in a rotator at 4°C. The mixture was spun down, washed, and the supernatant was removed. The slurry was incubated with 20 µl SDS/reduction solution for 5 min at 94°C. The supernatant was removed, and protein was separated by SDS-PAGE (7–14%). Analysis was performed using standard western blotting procedure with antibody to prey protein and HRP-linked 2° antibody. All results are from at least three independent experiments.
Acknowledgements
We would like to thank members of the Bambara and Opresko laboratories for helpful suggestions and discussions. This work was funded by the National Institutes of Health grant GM024441 to R.A.B., with additional support from NIH grant ES015052 to P.L.O.
Disclosure of Potential Conflicts of Interest
No potential conflicts of interest were disclosed.
Supplementary Material
References
- 1.Dawson TL, Gores GJ, Nieminen AL, Herman B, Lemasters JJ. Mitochondria as a source of reactive oxygen species during reductive stress in rat hepatocytes. Am J Physiol. 1993;264:961–967. doi: 10.1152/ajpcell.1993.264.4.C961. [DOI] [PubMed] [Google Scholar]
- 2.Lindahl T. Instability and decay of the primary structure of DNA. Nature. 1993;362:709–715. doi: 10.1038/362709a0. [DOI] [PubMed] [Google Scholar]
- 3.Beckman KB, Ames BN. The free radical theory of aging matures. Physiol Rev. 1998;78:547–581. doi: 10.1152/physrev.1998.78.2.547. [DOI] [PubMed] [Google Scholar]
- 4.Gil Del Valle L. Oxidative stress in aging: Theoretical outcomes and clinical evidences in humans. Biomed Pharmacother. 2010 doi: 10.1016/j.biopha.2010.09.010. Epub Ahead of Print. [DOI] [PubMed] [Google Scholar]
- 5.Jomova K, Vondrakova D, Lawson M, Valko M. Metals, oxidative stress and neurodegenerative disorders. Mol Cell Biochem. 2010;345:91–104. doi: 10.1007/s11010-010-0563-x. [DOI] [PubMed] [Google Scholar]
- 6.Sedelnikova OA, Redon CE, Dickey JS, Nakamura AJ, Georgakilas AG, Bonner WM. Role of oxidatively induced DNA lesions in human pathogenesis. Mutat Res. 2010;704:152–159. doi: 10.1016/j.mrrev.2009.12.005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Essick EE, Sam F. Oxidative stress and autophagy in cardiac disease, neurological disorders, aging and cancer. Oxid Med Cell Longev. 2010;3:168–177. doi: 10.4161/oxim.3.3.12106. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Tudek B, Winczura A, Janik J, Siomek A, Foksinski M, Oliski R. Involvement of oxidatively damaged DNA and repair in cancer development and aging. Am J Transl Res. 2010;2:254–284. [PMC free article] [PubMed] [Google Scholar]
- 9.Halliwell B. Oxidative stress and cancer: have we moved forward? Biochem J. 2007;401:1–11. doi: 10.1042/BJ20061131. [DOI] [PubMed] [Google Scholar]
- 10.Klaunig JE, Kamendulis LM, Hocevar BA. Oxidative stress and oxidative damage in carcinogenesis. Toxicol Pathol. 2010;38:96–109. doi: 10.1177/0192623309356453. [DOI] [PubMed] [Google Scholar]
- 11.Steenken S, aJSV How easily oxidizable is DNA? One-electron reduction potentials of adenosine and guanosine radicals in aqueous solution. J Am Chem Soc. 1997;119:617–618. doi: 10.1021/ja962255b. [DOI] [Google Scholar]
- 12.Neeley WL, Essigmann JM. Mechanisms of formation, genotoxicity and mutation of guanine oxidation products. Chem Res Toxicol. 2006;19:491–505. doi: 10.1021/tx0600043. [DOI] [PubMed] [Google Scholar]
- 13.Saito I, Takayama M, Nakamura T, Sugiyama H, Komeda Y, Iwasaki M. The most electron-donating sites in duplex DNA: guanine-guanine stacking rule. Nucleic Acids Symp Ser. 1995;34:191–192. [PubMed] [Google Scholar]
- 14.Saito I, Masami T, Hiroshi S, Kazuhiko N, Akira T, Masahide Y. Photoinduced DNA Cleavage via Electron Transfer: Demonstration That Guanine Residues Located 5′ to Guanine Are the Most Electron-Donating Sites. J Am Chem Soc. 1995;117:6406–6407. doi: 10.1021/ja00128a050. [DOI] [Google Scholar]
- 15.Sugiyama HS. Theoretical Studies of GG-Specific Photocleavage of DNA via Electron Transfer: Significant Lowering of Ionization Potential and 5′-Localization of HOMO of Stacked GG Bases in B-Form DNA. J Am Chem Soc. 1996;118:7063–7068. doi: 10.1021/ja9609821. [DOI] [Google Scholar]
- 16.de Lange T, Shiue L, Myers RM, Cox DR, Naylor SL, Killery AM, et al. Structure and variability of human chromosome ends. Mol Cell Biol. 1990;10:518–527. doi: 10.1128/mcb.10.2.518. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Makarov VL, Hirose Y, Langmore JP. Long G tails at both ends of human chromosomes suggest a C strand degradation mechanism for telomere shortening. Cell. 1997;88:657–666. doi: 10.1016/s0092-8674(00)81908-x. [DOI] [PubMed] [Google Scholar]
- 18.McElligott R, Wellinger RJ. The terminal DNA structure of mammalian chromosomes. EMBO J. 1997;16:3705–3714. doi: 10.1093/emboj/16.12.3705. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Kawanishi S, Oikawa S. Mechanism of telomere shortening by oxidative stress. Ann NY Acad Sci. 2004;1019:278–284. doi: 10.1196/annals.1297.047. [DOI] [PubMed] [Google Scholar]
- 20.von Zglinicki T. Oxidative stress shortens telomeres. Trends Biochem Sci. 2002;27:339–344. doi: 10.1016/S0968-0004(02)02110-2. [DOI] [PubMed] [Google Scholar]
- 21.Dedon PC. The chemical toxicology of 2-deoxyribose oxidation in DNA. Chem Res Toxicol. 2008;21:206–219. doi: 10.1021/tx700283c. [DOI] [PubMed] [Google Scholar]
- 22.Henle ES, Han Z, Tang N, Rai P, Luo Y, Linn S. Sequence-specific DNA cleavage by Fe2+-mediated fenton reactions has possible biological implications. J Biol Chem. 1999;274:962–971. doi: 10.1074/jbc.274.2.962. [DOI] [PubMed] [Google Scholar]
- 23.Oikawa S, Tada-Oikawa S, Kawanishi S. Site-specific DNA damage at the GGG sequence by UVA involves acceleration of telomere shortening. Biochemistry. 2001;40:4763–4768. doi: 10.1021/bi002721g. [DOI] [PubMed] [Google Scholar]
- 24.Ayouaz A, Raynaud C, Heride C, Revaud D, Sabatier L. Telomeres: hallmarks of radiosensitivity. Biochimie. 2008;90:60–72. doi: 10.1016/j.biochi.2007.09.011. [DOI] [PubMed] [Google Scholar]
- 25.Rhee DB, Ghosh A, Lu J, Bohr VA, Liu Y. Factors that influence telomeric oxidative base damage and repair by DNA glycosylase OGG1. DNA Repair (Amst) 2011;10:34–44. doi: 10.1016/j.dnarep.2010.09.008. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Scharer OD, Jiricny J. Recent progress in the biology, chemistry and structural biology of DNA glycosylases. Bioessays. 2001;23:270–281. doi: 10.1002/1521-1878(200103)23:3<270::AID-BIES1037>3.0.CO;2-J. [DOI] [PubMed] [Google Scholar]
- 27.Demple B, Herman T, Chen DS. Cloning and expression of APE, the cDNA encoding the major human apurinic endonuclease: definition of a family of DNA repair enzymes. Proc Natl Acad Sci USA. 1991;88:11450–11454. doi: 10.1073/pnas.88.24.11450. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Chen DS, Herman T, Demple B. Two distinct human DNA diesterases that hydrolyze 3′-blocking deoxyribose fragments from oxidized DNA. Nucleic Acids Res. 1991;19:5907–5914. doi: 10.1093/nar/19.21.5907. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Wilson DM, 3rd, Takeshita M, Grollman AP, Demple B. Incision activity of human apurinic endonuclease (Ape) at abasic site analogs in DNA. J Biol Chem. 1995;270:16002–16007. doi: 10.1074/jbc.270.27.16002. [DOI] [PubMed] [Google Scholar]
- 30.Matsumoto Y, Kim K. Excision of deoxyribose phosphate residues by DNA polymerase beta during DNA repair. Science. 1995;269:699–702. doi: 10.1126/science.7624801. [DOI] [PubMed] [Google Scholar]
- 31.Piersen CE, Prasad R, Wilson SH, Lloyd RS. Evidence for an imino intermediate in the DNA polymerase beta deoxyribose phosphate excision reaction. J Biol Chem. 1996;271:17811–17815. doi: 10.1074/jbc.271.30.17811. [DOI] [PubMed] [Google Scholar]
- 32.Sobol RW, Horton JK, Kuhn R, Gu H, Singhal RK, Prasad R, et al. Requirement of mammalian DNA polymerase-beta in base-excision repair. Nature. 1996;379:183–186. doi: 10.1038/379183a0. [DOI] [PubMed] [Google Scholar]
- 33.Wilson SH. Mammalian base excision repair and DNA polymerase beta. Mutat Res. 1998;407:203–215. doi: 10.1016/s0921-8777(98)00002-0. [DOI] [PubMed] [Google Scholar]
- 34.Demple B, Harrison L. Repair of oxidative damage to DNA: enzymology and biology. Annu Rev Biochem. 1994;63:915–948. doi: 10.1146/annurev.bi.63.070194.004411. [DOI] [PubMed] [Google Scholar]
- 35.Guan L, Bebenek K, Kunkel TA, Greenberg MM. Inhibition of short patch and long patch base excision repair by an oxidized abasic site. Biochemistry. 2010;49:9904–9910. doi: 10.1021/bi101533a. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Jacobs AC, Kreller CR, Greenberg MM. Long Patch Base Excision Repair Compensates for DNA Polymerase β Inactivation by the C4'-Oxidized Abasic Site. Biochemistry. 2010 doi: 10.1021/bi1017667. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Memisoglu A, Samson L. Base excision repair in yeast and mammals. Mutat Res. 2000;451:39–51. doi: 10.1016/S0027-5107(00)00039-7. [DOI] [PubMed] [Google Scholar]
- 38.Sung JS, Demple B. Roles of base excision repair subpathways in correcting oxidized abasic sites in DNA. FEBS J. 2006;273:1620–1629. doi: 10.1111/j.1742-4658.2006.05192.x. [DOI] [PubMed] [Google Scholar]
- 39.Wilson DM, 3rd, Barsky D. The major human abasic endonuclease: formation, consequences and repair of abasic lesions in DNA. Mutat Res. 2001;485:283–307. doi: 10.1016/s0921-8777(01)00063-5. [DOI] [PubMed] [Google Scholar]
- 40.Sattler U, Frit P, Salles B, Calsou P. Long-patch DNA repair synthesis during base excision repair in mammalian cells. EMBO Rep. 2003;4:363–367. doi: 10.1038/sj.embor.embor796. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Klungland A, Lindahl T. Second pathway for completion of human DNA base excision-repair: reconstitution with purified proteins and requirement for DNase IV (FEN1) EMBO J. 1997;16:3341–33418. doi: 10.1093/emboj/16.11.3341. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Palm W, de Lange T. How shelterin protects mammalian telomeres. Annu Rev Genet. 2008;42:301–334. doi: 10.1146/annurev.genet.41.110306.130350. [DOI] [PubMed] [Google Scholar]
- 43.de Lange T. Shelterin: the protein complex that shapes and safeguards human telomeres. Genes Dev. 2005;19:2100–2110. doi: 10.1101/gad.1346005. [DOI] [PubMed] [Google Scholar]
- 44.Bianchi A, Stansel RM, Fairall L, Griffith JD, Rhodes D, de Lange T. TRF1 binds a bipartite telomeric site with extreme spatial flexibility. EMBO J. 1999;18:5735–5744. doi: 10.1093/emboj/18.20.5735. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Court R, Chapman L, Fairall L, Rhodes D. How the human telomeric proteins TRF1 and TRF2 recognize telomeric DNA: a view from high-resolution crystal structures. EMBO Rep. 2005;6:39–45. doi: 10.1038/sj.embor.7400314. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Hanaoka S, Nagadoi A, Nishimura Y. Comparison between TRF2 and TRF1 of their telomeric DNA-bound structures and DNA-binding activities. Protein Sci. 2005;14:119–130. doi: 10.1110/ps.04983705. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Baumann P, Cech TR. Pot1, the putative telomere endbinding protein in fission yeast and humans. Science. 2001;292:1171–1175. doi: 10.1126/science.1060036. [DOI] [PubMed] [Google Scholar]
- 48.Martinez P, Blasco MA. Telomeric and extra-telomeric roles for telomerase and the telomere-binding proteins. Nat Rev Cancer. 2011;11:161–176. doi: 10.1038/nrc3025. [DOI] [PubMed] [Google Scholar]
- 49.Sheppard SA, Loayza D. LIM-domain proteins TRIP6 and LPP associate with shelterin to mediate telomere protection. Aging (Albany NY) 2010;2:432–444. doi: 10.18632/aging.100170. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Rochette PJ, Brash DE. Human telomeres are hypersensitive to UV-induced DNA Damage and refractory to repair. PLoS Genet. 2010;6:1000926. doi: 10.1371/journal.pgen.1000926. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Muñoz P, Blanco R, de Carcer G, Schoeftner S, Benetti R, Flores JM, et al. TRF1 controls telomere length and mitotic fidelity in epithelial homeostasis. Mol Cell Biol. 2009;29:1608–1625. doi: 10.1128/MCB.01339-08. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Muñoz P, Blanco R, Flores JM, Blasco MA. XPF nuclease-dependent telomere loss and increased DNA damage in mice overexpressing TRF2 result in premature aging and cancer. Nat Genet. 2005;37:1063–1071. doi: 10.1038/ng1633. [DOI] [PubMed] [Google Scholar]
- 53.Kabir S, Sfeir A, de Lange T. Taking apart Rap1: an adaptor protein with telomeric and non-telomeric functions. Cell Cycle. 2010;9:4061–4067. doi: 10.4161/cc.9.20.13579. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Horikawa I, Fujita K, Harris CC. p53 governs telomere regulation feedback too, via TRF2. Aging (Albany NY) 2011;3:26–32. doi: 10.18632/aging.100271. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Dregalla RC, Zhou J, Idate RR, Battaglia CL, Liber HL, Bailey SM. Regulatory roles of tankyrase 1 at telomeres and in DNA repair: suppression of T-SCE and stabilization of DNA-PKcs. Aging (Albany NY) 2010;2:691–708. doi: 10.18632/aging.100210. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.Wang Z, Rhee DB, Lu J, Bohr CT, Zhou F, Vallabhaneni H, et al. Characterization of oxidative guanine damage and repair in mammalian telomeres. PLoS Genet. 2010;6:1000951. doi: 10.1371/journal.pgen.1000951. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57.Muftuoglu M, Wong HK, Imam SZ, Wilson DM, 3rd, Bohr VA, Opresko PL. Telomere repeat binding factor 2 interacts with base excision repair proteins and stimulates DNA synthesis by DNA polymerase beta. Cancer Res. 2006;66:113–124. doi: 10.1158/0008-5472.CAN-05-2742. [DOI] [PubMed] [Google Scholar]
- 58.Lu J, Liu Y. Deletion of Ogg1 DNA glycosylase results in telomere base damage and length alteration in yeast. EMBO J. 2010;29:398–409. doi: 10.1038/emboj.2009.355. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59.Saharia A, Guittat L, Crocker S, Lim A, Steffen M, Kulkarni S, et al. Flap endonuclease 1 contributes to telomere stability. Curr Biol. 2008;18:496500. doi: 10.1016/j.cub.2008.02.071. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60.Fotiadou P, Henegariu O, Sweasy JB. DNA polymerase beta interacts with TRF2 and induces telomere dysfunction in a murine mammary cell line. Cancer Res. 2004;64:3830–3837. doi: 10.1158/0008-5472.CAN-04-0136. [DOI] [PubMed] [Google Scholar]
- 61.Oikawa S, Kawanishi S. Site-specific DNA damage at GGG sequence by oxidative stress may accelerate telomere shortening. FEBS Lett. 1999;453:365–368. doi: 10.1016/S0014-5793(99)00748-6. [DOI] [PubMed] [Google Scholar]
- 62.Opresko PL, Fan J, Danzy S, Wilson DM, 3rd, Bohr VA. Oxidative damage in telomeric DNA disrupts recognition by TRF1 and TRF2. Nucleic Acids Res. 2005;33:1230–1239. doi: 10.1093/nar/gki273. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 63.Parikh SS, Mol CD, Hosfield DJ, Tainer JA. Envisioning the molecular choreography of DNA base excision repair. Curr Opin Struct Biol. 1999;9:37–47. doi: 10.1016/S0959440X(99)80006-2. [DOI] [PubMed] [Google Scholar]
- 64.Mol CD, Izumi T, Mitra S, Tainer JA. DNA-bound structures and mutants reveal abasic DNA binding by APE1 and DNA repair coordination [corrected] Nature. 2000;403:451–456. doi: 10.1038/35000249. [DOI] [PubMed] [Google Scholar]
- 65.Wilson SH, Kunkel TA. Passing the baton in base excision repair. Nat Struct Biol. 2000;7:176–178. doi: 10.1038/82818. [DOI] [PubMed] [Google Scholar]
- 66.Prasad R, Shock DD, Beard WA, Wilson SH. Substrate channeling in mammalian base excision repair pathways: passing the baton. J Biol Chem. 2010;285:40479–40488. doi: 10.1074/jbc.M110.155267. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 67.Jagannathan I, Cole HA, Hayes JJ. Base excision repair in nucleosome substrates. Chromosome Res. 2006;14:27–37. doi: 10.1007/s10577-005-1020-7. [DOI] [PubMed] [Google Scholar]
- 68.Bianchi A, Smith S, Chong L, Elias P, de Lange T. TRF1 is a dimer and bends telomeric DNA. EMBO J. 1997;16:1785–1794. doi: 10.1093/emboj/16.7.1785. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 69.Griffith JD, Comeau L, Rosenfield S, Stansel RM, Bianchi A, Moss H, et al. Mammalian telomeres end in a large duplex loop. Cell. 1999;97:503–514. doi: 10.1016/S0092-8674(00)80760-6. [DOI] [PubMed] [Google Scholar]
- 70.Stansel RM, de Lange T, Griffith JD. T-loop assembly in vitro involves binding of TRF2 near the 3′ telomeric overhang. EMBO J. 2001;20:5532–5540. doi: 10.1093/emboj/20.19.5532. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 71.Vidal AE, Hickson ID, Boiteux S, Radicella JP. Mechanism of stimulation of the DNA glycosylase activity of hOGG1 by the major human AP endonuclease: bypass of the AP lyase activity step. Nucleic Acids Res. 2001;29:1285–1292. doi: 10.1093/nar/29.6.1285. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 72.Saitoh T, Shinmura K, Yamaguchi S, Tani M, Seki S, Murakami H, et al. Enhancement of OGG1 protein AP lyase activity by increase of APEX protein. Mutat Res. 2001;486:31–40. doi: 10.1016/s0921-8777(01)00078-7. [DOI] [PubMed] [Google Scholar]
- 73.Hill JW, Hazra TK, Izumi T, Mitra S. Stimulation of human 8-oxoguanine-DNA glycosylase by AP-endonuclease: potential coordination of the initial steps in base excision repair. Nucleic Acids Res. 2001;29:430–438. doi: 10.1093/nar/29.2.430. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 74.Sung JS, DeMott MS, Demple B. Long-patch base excision DNA repair of 2-deoxyribonolactone prevents the formation of DNA-protein cross-links with DNA polymerase beta. J Biol Chem. 2005;280:39095–39103. doi: 10.1074/jbc.M506480200. [DOI] [PubMed] [Google Scholar]
- 75.Liu P, Qian L, Sung JS, de Souza-Pinto NC, Zheng L, Bogenhagen DF, et al. Removal of oxidative DNA damage via FEN1-dependent long-patch base excision repair in human cell mitochondria. Mol Cell Biol. 2008;28:4975–4987. doi: 10.1128/MCB.00457-08. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 76.Zhu XD, Niedernhofer L, Kuster B, Mann M, Hoeijmakers JH, de Lange T. ERCC1/XPF removes the 3′ overhang from uncapped telomeres and represses formation of telomeric DNA-containing double minute chromosomes. Mol Cell. 2003;12:1489–198. doi: 10.1016/S10972765(03)00478-7. [DOI] [PubMed] [Google Scholar]
- 77.Miller D, Reynolds GE, Mejia R, Stark JM, Murnane JP. Subtelomeric regions in mammalian cells are deficient in DNA double-strand break repair. DNA Repair (Amst) 2011;10:536–544. doi: 10.1016/j.dnarep.2011.03.001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 78.Denchi EL, de Lange T. Protection of telomeres through independent control of ATM and ATR by TRF2 and POT1. Nature. 2007;448:1068–1071. doi: 10.1038/nature06065. [DOI] [PubMed] [Google Scholar]
- 79.Fink LS, Lerner CA, Torres PF, Sell C. Ku80 facilitates chromatin binding of the telomere binding protein, TRF2. Cell Cycle. 2010;9:3798–3806. doi: 10.4161/cc.9.18.13129. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 80.Bornarth CJ, Ranalli TA, Henricksen LA, Wahl AF, Bambara RA. Effect of flap modifications on human FEN1 cleavage. Biochemistry. 1999;38:13347–13354. doi: 10.1021/bi991321u. [DOI] [PubMed] [Google Scholar]
- 81.Tom S, Ranalli TA, Podust VN, Bambara RA. Regulatory roles of p21 and apurinic/apyrimidinic endonuclease 1 in base excision repair. J Biol Chem. 2001;276:48781–48789. doi: 10.1074/jbc.M109626200. [DOI] [PubMed] [Google Scholar]
- 82.Menge KL, Hostomsky Z, Nodes BR, Hudson GO, Rahmati S, Moomaw EW, et al. Structure-function analysis of the mammalian DNA polymerase beta active site: role of aspartic acid 256, arginine 254 and arginine 258 in nucleotidyl transfer. Biochemistry. 1995;34:15934–15942. doi: 10.1021/bi00049a008. [DOI] [PubMed] [Google Scholar]
- 83.Henricksen LA, Veeraraghavan J, Chafin DR, Bambara RA. DNA ligase I competes with FEN1 to expand repetitive DNA sequences in vitro. J Biol Chem. 2002;277:22361–22369. doi: 10.1074/jbc.M201765200. [DOI] [PubMed] [Google Scholar]
- 84.Sowd G, Lei M, Opresko PL. Mechanism and substrate specificity of telomeric protein POT1 stimulation of the Werner syndrome helicase. Nucleic Acids Res. 2008;36:4242–4256. doi: 10.1093/nar/gkn385. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 85.Opresko PL, von Kobbe C, Laine JP, Harrigan J, Hickson ID, Bohr VA. Telomere-binding protein TRF2 binds to and stimulates the Werner and Bloom syndrome helicases. J Biol Chem. 2002;277:41110–411109. doi: 10.1074/jbc.M205396200. [DOI] [PubMed] [Google Scholar]
- 86.Chan K, Houlbrook S, Zhang QM, Harrison M, Hickson ID, Dianov GL. Overexpression of DNA polymerase beta results in an increased rate of frame-shift mutations during base excision repair. Mutagenesis. 2007;22:183–188. doi: 10.1093/mutage/gel070. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.