Abstract
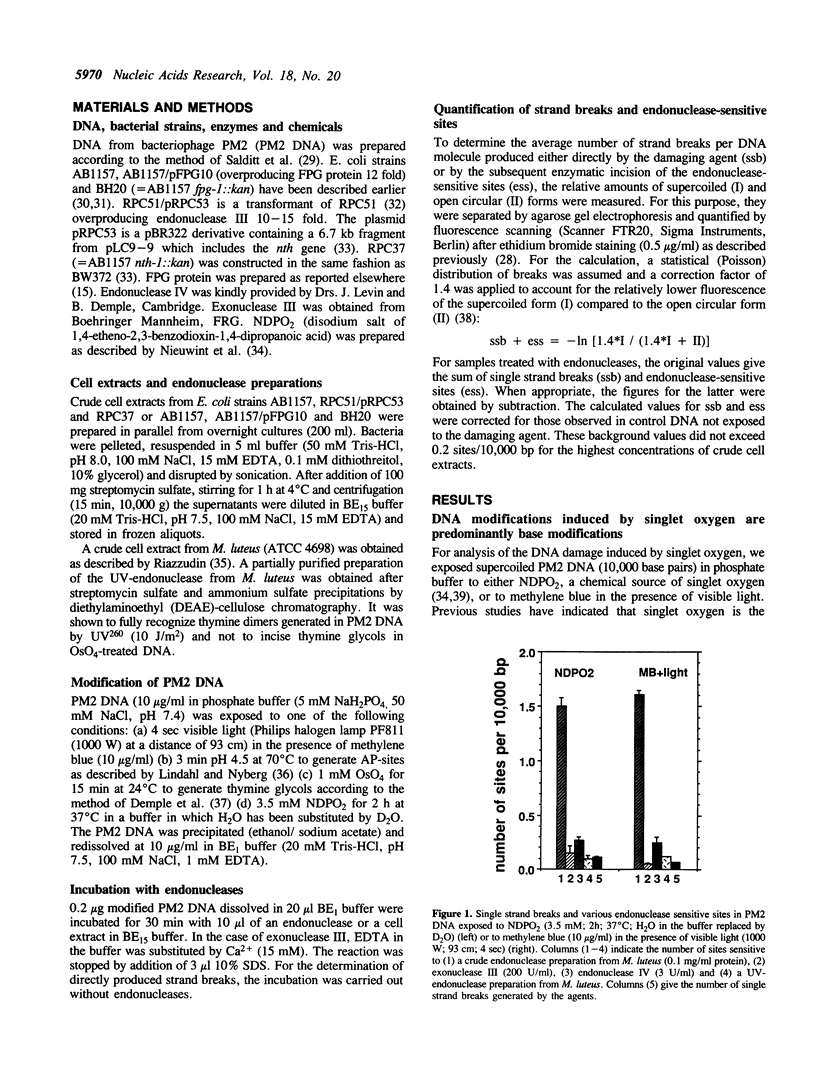
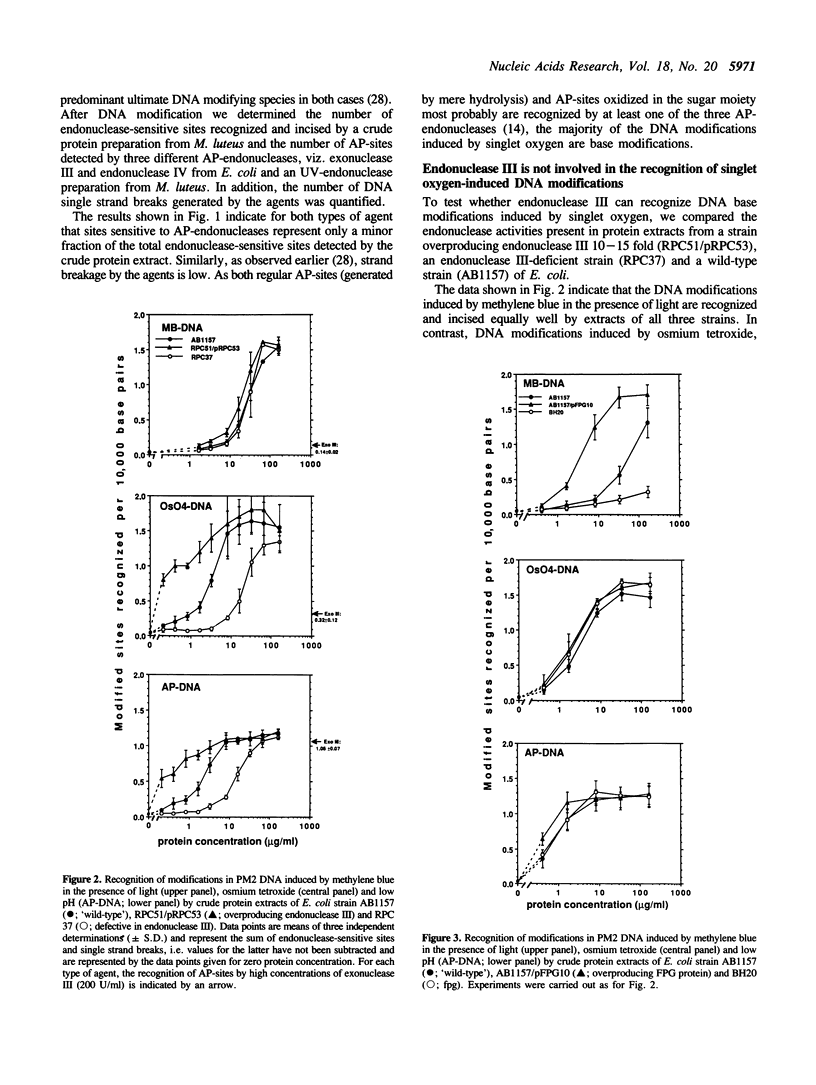
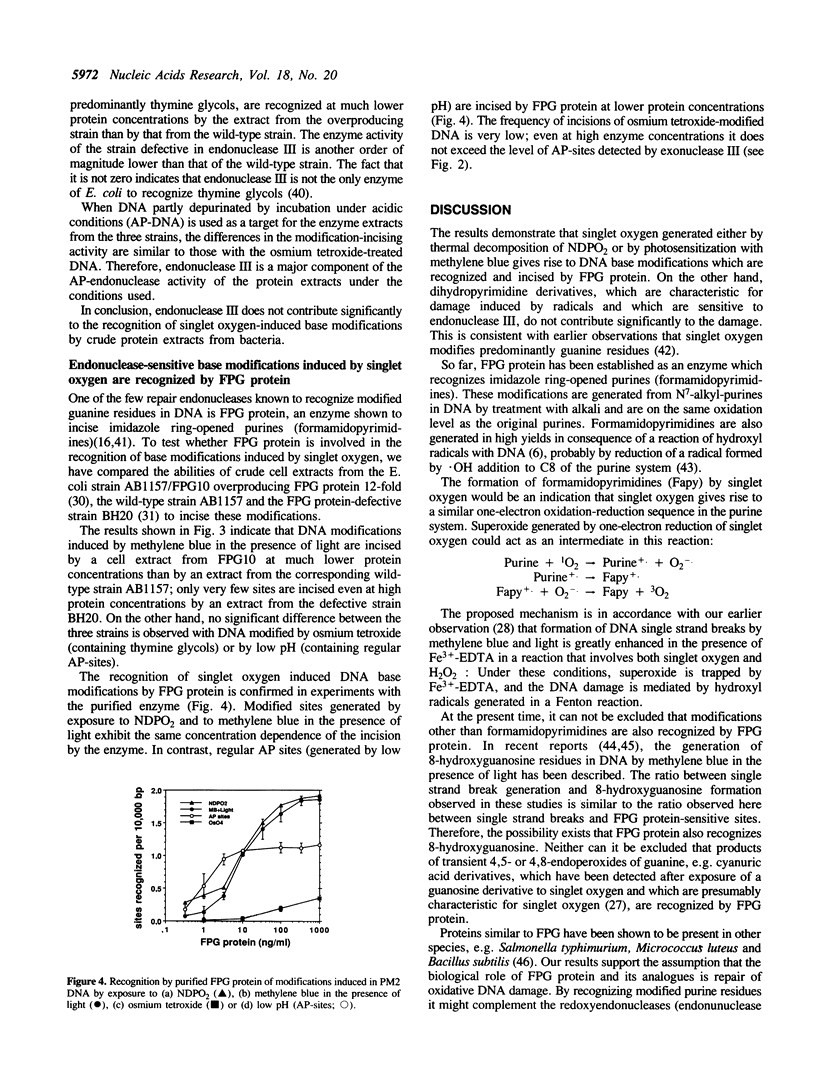
DNA modifications induced either by photosensitization (illumination in the presence of methylene blue) or by chemically generated singlet oxygen (thermal decomposition of an 1,4-etheno-2,3-benzodioxin) are recognized and incised by repair endonucleases present in crude bacterial cell extracts. Only a small fraction of the incised modifications are sites of base loss (AP-sites) sensitive to exonuclease III, endonuclease IV from E. coli or to the UV-endonuclease from M. luteus. Cell extracts from E. coli strains overproducing or defective in endonuclease III recognize the modifications induced by illumination in the presence of methylene blue just as well as do those from wild-type E. coli strains. This indicates that dihydropyrimidine derivatives, which are characteristic of hydroxyl radical-induced DNA modifications, are absent. In contrast, most of the modifications induced are not recognized by a cell extract from a fpg strain defective in formamidopyrimidine-DNA glycosylase FPG protein). Furthermore, incision by a cell extract from an E. coli strain overproducing FPG protein takes place at much lower protein concentration than with the wild-type strain. Experiments with purified FPG protein confirm that this enzyme is responsible for the recognition of singlet oxygen-induced DNA base modifications.
Full text
PDF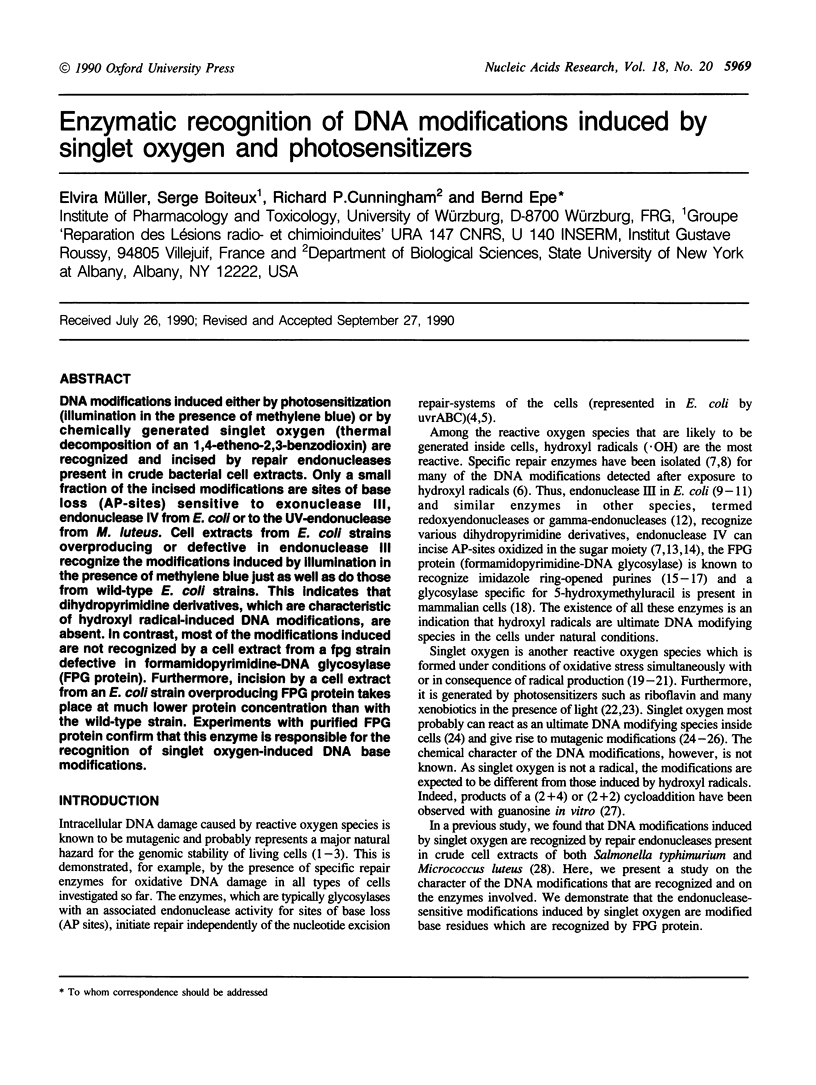

Selected References
These references are in PubMed. This may not be the complete list of references from this article.
- Ames B. N. Dietary carcinogens and anticarcinogens. Oxygen radicals and degenerative diseases. Science. 1983 Sep 23;221(4617):1256–1264. doi: 10.1126/science.6351251. [DOI] [PubMed] [Google Scholar]
- Aruoma O. I., Halliwell B., Dizdaroglu M. Iron ion-dependent modification of bases in DNA by the superoxide radical-generating system hypoxanthine/xanthine oxidase. J Biol Chem. 1989 Aug 5;264(22):13024–13028. [PubMed] [Google Scholar]
- Asahara H., Wistort P. M., Bank J. F., Bakerian R. H., Cunningham R. P. Purification and characterization of Escherichia coli endonuclease III from the cloned nth gene. Biochemistry. 1989 May 16;28(10):4444–4449. doi: 10.1021/bi00436a048. [DOI] [PubMed] [Google Scholar]
- Boiteux S., Bichara M., Fuchs R. P., Laval J. Excision of the imidazole ring-opened form of N-2-aminofluorene-C(8)-guanine adduct in poly(dG-dC) by Escherichia coli formamidopyrimidine-DNA glycosylase. Carcinogenesis. 1989 Oct;10(10):1905–1909. doi: 10.1093/carcin/10.10.1905. [DOI] [PubMed] [Google Scholar]
- Boiteux S., Huisman O. Isolation of a formamidopyrimidine-DNA glycosylase (fpg) mutant of Escherichia coli K12. Mol Gen Genet. 1989 Jan;215(2):300–305. doi: 10.1007/BF00339732. [DOI] [PubMed] [Google Scholar]
- Boiteux S., O'Connor T. R., Laval J. Formamidopyrimidine-DNA glycosylase of Escherichia coli: cloning and sequencing of the fpg structural gene and overproduction of the protein. EMBO J. 1987 Oct;6(10):3177–3183. doi: 10.1002/j.1460-2075.1987.tb02629.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Boiteux S., O'Connor T. R., Lederer F., Gouyette A., Laval J. Homogeneous Escherichia coli FPG protein. A DNA glycosylase which excises imidazole ring-opened purines and nicks DNA at apurinic/apyrimidinic sites. J Biol Chem. 1990 Mar 5;265(7):3916–3922. [PubMed] [Google Scholar]
- Boorstein R. J., Levy D. D., Teebor G. W. 5-Hydroxymethyluracil-DNA glycosylase activity may be a differentiated mammalian function. Mutat Res. 1987 May;183(3):257–263. doi: 10.1016/0167-8817(87)90008-3. [DOI] [PubMed] [Google Scholar]
- Breimer L. H. Enzymatic excision from gamma-irradiated polydeoxyribonucleotides of adenine residues whose imidazole rings have been ruptured. Nucleic Acids Res. 1984 Aug 24;12(16):6359–6367. doi: 10.1093/nar/12.16.6359. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Breimer L. H., Lindahl T. DNA glycosylase activities for thymine residues damaged by ring saturation, fragmentation, or ring contraction are functions of endonuclease III in Escherichia coli. J Biol Chem. 1984 May 10;259(9):5543–5548. [PubMed] [Google Scholar]
- Cerutti P. A. Prooxidant states and tumor promotion. Science. 1985 Jan 25;227(4685):375–381. doi: 10.1126/science.2981433. [DOI] [PubMed] [Google Scholar]
- Chetsanga C. J., Lindahl T. Release of 7-methylguanine residues whose imidazole rings have been opened from damaged DNA by a DNA glycosylase from Escherichia coli. Nucleic Acids Res. 1979 Aug 10;6(11):3673–3684. doi: 10.1093/nar/6.11.3673. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cunningham R. P., Weiss B. Endonuclease III (nth) mutants of Escherichia coli. Proc Natl Acad Sci U S A. 1985 Jan;82(2):474–478. doi: 10.1073/pnas.82.2.474. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Decuyper-Debergh D., Piette J., Van de Vorst A. Singlet oxygen-induced mutations in M13 lacZ phage DNA. EMBO J. 1987 Oct;6(10):3155–3161. doi: 10.1002/j.1460-2075.1987.tb02626.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Di Mascio P., Menck C. F., Nigro R. G., Sarasin A., Sies H. Singlet molecular oxygen induced mutagenicity in a mammalian SV40-based shuttle vector. Photochem Photobiol. 1990 Mar;51(3):293–298. doi: 10.1111/j.1751-1097.1990.tb01713.x. [DOI] [PubMed] [Google Scholar]
- Doetsch P. W., Henner W. D., Cunningham R. P., Toney J. H., Helland D. E. A highly conserved endonuclease activity present in Escherichia coli, bovine, and human cells recognizes oxidative DNA damage at sites of pyrimidines. Mol Cell Biol. 1987 Jan;7(1):26–32. doi: 10.1128/mcb.7.1.26. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Epe B., Hegler J., Wild D. Singlet oxygen as an ultimately reactive species in Salmonella typhimurium DNA damage induced by methylene blue/visible light. Carcinogenesis. 1989 Nov;10(11):2019–2024. doi: 10.1093/carcin/10.11.2019. [DOI] [PubMed] [Google Scholar]
- Epe B., Mützel P., Adam W. DNA damage by oxygen radicals and excited state species: a comparative study using enzymatic probes in vitro. Chem Biol Interact. 1988;67(1-2):149–165. doi: 10.1016/0009-2797(88)90094-4. [DOI] [PubMed] [Google Scholar]
- Floyd R. A., West M. S., Eneff K. L., Schneider J. E. Methylene blue plus light mediates 8-hydroxyguanine formation in DNA. Arch Biochem Biophys. 1989 Aug 15;273(1):106–111. doi: 10.1016/0003-9861(89)90167-7. [DOI] [PubMed] [Google Scholar]
- Friedmann T., Brown D. M. Base-specific reactions useful for DNA sequencing: methylene blue--sensitized photooxidation of guanine and osmium tetraoxide modification of thymine. Nucleic Acids Res. 1978 Feb;5(2):615–622. doi: 10.1093/nar/5.2.615. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ito T. Cellular and subcellular mechanisms of photodynamic action: the 1O2 hypothesis as a driving force in recent research. Photochem Photobiol. 1978 Oct-Nov;28(4-5):493–508. doi: 10.1111/j.1751-1097.1978.tb06957.x. [DOI] [PubMed] [Google Scholar]
- Joshi P. C. Comparison of the DNA-damaging property of photosensitised riboflavin via singlet oxygen (1O2) and superoxide radical O2-. mechanisms. Toxicol Lett. 1985 Aug;26(2-3):211–217. doi: 10.1016/0378-4274(85)90169-9. [DOI] [PubMed] [Google Scholar]
- Katcher H. L., Wallace S. S. Characterization of the Escherichia coli X-ray endonuclease, endonuclease III. Biochemistry. 1983 Aug 16;22(17):4071–4081. doi: 10.1021/bi00286a013. [DOI] [PubMed] [Google Scholar]
- Levin J. D., Johnson A. W., Demple B. Homogeneous Escherichia coli endonuclease IV. Characterization of an enzyme that recognizes oxidative damage in DNA. J Biol Chem. 1988 Jun 15;263(17):8066–8071. [PubMed] [Google Scholar]
- Lindahl T., Nyberg B. Rate of depurination of native deoxyribonucleic acid. Biochemistry. 1972 Sep 12;11(19):3610–3618. doi: 10.1021/bi00769a018. [DOI] [PubMed] [Google Scholar]
- Lloyd R. S., Haidle C. W., Robberson D. L. Bleomycin-specific fragmentation of double-stranded DNA. Biochemistry. 1978 May 16;17(10):1890–1896. doi: 10.1021/bi00603a014. [DOI] [PubMed] [Google Scholar]
- Menghini R. Genotoxicity of active oxygen species in mammalian cells. Mutat Res. 1988 May;195(3):215–230. [PubMed] [Google Scholar]
- Povirk L. F., Steighner R. J. Oxidized apurinic/apyrimidinic sites formed in DNA by oxidative mutagens. Mutat Res. 1989 Sep;214(1):13–22. doi: 10.1016/0027-5107(89)90193-0. [DOI] [PubMed] [Google Scholar]
- Salditt M., Braunstein S. N., Camerini-Otero R. D., Franklin R. M. Structure and synthesis of a lipid-containing bacteriophage. X. Improved techniques for the purification of bacteriophage PM2. Virology. 1972 Apr;48(1):259–262. doi: 10.1016/0042-6822(72)90133-x. [DOI] [PubMed] [Google Scholar]
- Sancar A., Sancar G. B. DNA repair enzymes. Annu Rev Biochem. 1988;57:29–67. doi: 10.1146/annurev.bi.57.070188.000333. [DOI] [PubMed] [Google Scholar]
- Saporito S. M., Cunningham R. P. Nucleotide sequence of the nfo gene of Escherichia coli K-12. J Bacteriol. 1988 Nov;170(11):5141–5145. doi: 10.1128/jb.170.11.5141-5145.1988. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Schneider J. E., Price S., Maidt L., Gutteridge J. M., Floyd R. A. Methylene blue plus light mediates 8-hydroxy 2'-deoxyguanosine formation in DNA preferentially over strand breakage. Nucleic Acids Res. 1990 Feb 11;18(3):631–635. doi: 10.1093/nar/18.3.631. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Teebor G. W., Boorstein R. J., Cadet J. The repairability of oxidative free radical mediated damage to DNA: a review. Int J Radiat Biol. 1988 Aug;54(2):131–150. doi: 10.1080/09553008814551591. [DOI] [PubMed] [Google Scholar]
- Wallace S. S. AP endonucleases and DNA glycosylases that recognize oxidative DNA damage. Environ Mol Mutagen. 1988;12(4):431–477. doi: 10.1002/em.2860120411. [DOI] [PubMed] [Google Scholar]


