Abstract
Mutations create the genetic diversity on which selective pressures can act, yet also create structural instability in proteins. How, then, is it possible for organisms to ameliorate mutation-induced perturbations of protein stability while maintaining biological fitness and gaining a selective advantage? Here we used site-specific chromosomal mutagenesis to introduce a selected set of mostly destabilizing mutations into folA—an essential chromosomal gene of Escherichia coli encoding dihydrofolate reductase (DHFR)—to determine how changes in protein stability, activity, and abundance affect fitness. In total, 27 E. coli strains carrying mutant DHFR were created. We found no significant correlation between protein stability and its catalytic activity nor between catalytic activity and fitness in a limited range of variation of catalytic activity observed in mutants. The stability of these mutants is strongly correlated with their intracellular abundance, suggesting that protein homeostatic machinery plays an active role in maintaining intracellular concentrations of proteins. Fitness also shows a significant correlation with intracellular abundance of soluble DHFR in cells growing at 30 °C. At 42 °C, the picture was mixed, yet remarkable: A few strains carrying mutant DHFR proteins aggregated, rendering them nonviable, but, intriguingly, the majority exhibited fitness higher than wild type. We found that mutational destabilization of DHFR proteins in E. coli is counterbalanced at 42 °C by their soluble oligomerization, thereby restoring structural stability and protecting against aggregation.
Keywords: fitness landscape, genotype-phenotype relation, molecular evolution, protein solubility, protein aggregation
In order to evolve, an organism must acquire genetic mutation(s), yet these very mutations can cause structural destabilization of the proteins they encode (1–4), potentially affecting the ability of an organism to survive and reproduce (fitness). This dichotomy of how cells can accommodate evolutionarily beneficial, but structurally destabilizing mutations (the “genotype-phenotype gap”) is central to biology, yet poorly understood.
Past efforts to bridge the genotype-phenotype gap “in one shot” from sequences to phenotype (5) may have fallen foul of the indirect relationship between genomic sequences and phenotypic traits, where many sequence variations lead to the same phenotypic effect. However, the relationship between genomic sequences and phenotypic traits is likely to be indirect, with many sequence variations leading to the same phenotypic effect. A more promising approach, introduced in recent multiscale theoretical models (6–8), is to bridge the scales gap midway by relating coarse-grained molecular traits to an organism’s fitness.
Of all molecular traits, protein stability has been widely recognized as one of the most evolutionarily important (3, 7, 9–11). Indeed, to be functional, almost all proteins must be either stably folded or, in the case of many intrinsically disordered proteins, assume a specific structure upon binding to a partner (12). However, the relationship between stability and fitness, while often postulated (9–11), has not been sufficiently explored experimentally.
As such, we chose to partially close the phenotype-genotype gap by exploring the relationship between stability of an essential enzyme, dihydrofolate reductase (DHFR) encoded by the folA gene in Escherichia coli, and bacterial fitness. DHFR catalyzes an electron transfer reaction from NADPH to 7,8-dihydrofolate (H2F) to form 5,6,7,8-tetrahydrofolate (H4F). DHFR struck us as a good choice for this investigation because tetrahydrofolate is essential for the synthesis of purines, thymidylate, and several amino acids (13), and DHFR is present in relatively low abundance in E. coli (approximately 40 copies/cell) (14), so toxicity from its aggregation should be negligible. It was reasonable therefore to expect that the main effect of perturbations in fitness would be mediated via modulation of the active enzyme copy number.
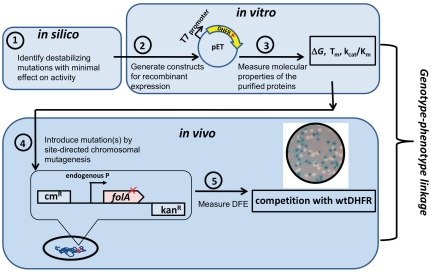
With this in mind, we explored the effects of mutations in DHFR that, theoretically, would have minimal affects on enzyme activity yet provide a broad range of effects on the fitness of our chosen E. coli host organism. Unlike earlier approaches, which considered how overexpression of nonendogenous destabilized proteins from a plasmid affects fitness (15, 16), we aimed to establish the link between the molecular properties of an endogenous essential protein and organismal fitness at maximally realistic conditions through site-specific mutations carried out directly on the chromosome while leaving intact the regulatory region responsible for control of endogenous expression levels (Fig. 1). Through this design, we hoped to determine which particular molecular properties of DHFR have the most pronounced impact on the fitness of E. coli.
Fig. 1.
The experimental approach. (1) 10 DHFR residues, predominantly from the hydrophobic core and at least 4 Å from the NADPH and dihydrofolate binding sites, were chosen for mutagenesis based on the structural and phylogenetic predictions and published biophysical and biochemical data. (2) Sixteen single mutants (Table 1) were generated, cloned into pET vector, expressed, and purified (see SI Materials and Methods). (3) Gibbs free energy difference between folded and unfolded states (ΔG), apparent midtransition temperature of unfolding ( ), and catalytic parameters (kcat, Km) were measured (Table 1 and SI Materials and Methods). (4) A site-directed chromosomal mutagenesis method was developed to introduce in vitro characterized mutations into chromosomal folA gene of E. coli’s MG1655 strain without perturbing the gene’s regulatory region. In addition to 16 single mutant strains, 11 multiple mutant strains were generated by combining exhaustively four most destabilizing mutations (V75H, I91L, W133F, I155A) (Table S1). (5) Fitness effects of the introduced mutations (in total, 27 strains) were measured by growth competition of mutant strains with WT DHFR strain.
), and catalytic parameters (kcat, Km) were measured (Table 1 and SI Materials and Methods). (4) A site-directed chromosomal mutagenesis method was developed to introduce in vitro characterized mutations into chromosomal folA gene of E. coli’s MG1655 strain without perturbing the gene’s regulatory region. In addition to 16 single mutant strains, 11 multiple mutant strains were generated by combining exhaustively four most destabilizing mutations (V75H, I91L, W133F, I155A) (Table S1). (5) Fitness effects of the introduced mutations (in total, 27 strains) were measured by growth competition of mutant strains with WT DHFR strain.
Results
Selection and In Vitro Analysis of DHFR Mutants.
The rationale behind choosing DHFR mutations was to cover a broad range of stabilities with minimum effect on activity. To that end, an extensive search of the literature identified 10 loci, all of which were distant (at least 4 Å away) from the enzymatically critical NADPH and H2F binding sites—most being buried in the protein’s hydrophobic core (Table 1). Multiple-sequence alignments at the selected loci were examined, and substitutions with both low and high conservation propensity were identified (Table 1 and Fig. S1).
Table 1.
Thermodynamic, structural, and catalytic properties of in vitro purified DHFR mutants
| Mutant |
 (°C) (°C) |
ΔG(H2O) (kcal/mol) 25 °C |
kcat (s-1) |
kcat/KM (s-1 μM-1) |
ASA* |
Conservation† (%) |
|
| Native |
Substitution |
||||||
| WT | 51.7 | −4.4 | 11.65 | 3.6 | |||
| V40A | 43.2 | −3.31 | 24.05 | 3.22 | 0 | 43.6 | 0.34 |
| I61V | 53.4 | −4.52 | 8.99 | 3.7 | 0 | 19.9 | 79 |
| V75H | 40.6 | −2.18 | 15.5 | 4.57 | 0.05 | 32 | 0 |
| V75I | 39.2 | −2.98 | 18.25 | 5.51 | 0.05 | 32 | 10.3 |
| I91V | 49.1 | −3.14 | 17.87 | 5.13 | 0.07 | 33.3 | 29 |
| I91L | 41.4 | −2.69 | ND‡ | ND‡ | 0.07 | 33.3 | 20 |
| L112V | 47.4 | −2.7 | 11.45 | 2.34 | 0 | 34 | 27.5 |
| W133F | 46.3 | −4.4 | 13.45 | 6.79 | 0.05 | 53.6 | 20.6 |
| W133V | ND‡ | −1.53 | ND‡ | ND‡ | 0.05 | 53.6 | 0.34 |
| I155T | 43.4 | −3.68 | 11.27 | 2.72 | 0.12 | 15.8 | 21 |
| I155L | 45.8 | −2.84 | 12.9 | 5.12 | 0.12 | 15.8 | 1 |
| I155A | 38.5 | −2.42 | 11.95 | 2.71 | 0.12 | 15.8 | 1 |
| I115V | 51.4 | −6 | 10.26 | 2.08 | 0.02 | 55.3 | 34.4 |
| I115A | 46.1 | −3.4 | 7.59 | 0.5 | 0.02 | 55.3 | 0 |
| V88I | 44.3 | −4.22 | 13.8 | 3.12 | 0.34 | 12.4 | 1.4 |
| A145T | 51.3 | −4.13 | 10 | 3.6 | 0.89 | 12.4 | 1.4 |
*ASA (accessible surface area) was calculated by Vadar package (http://vadar.wishartlab.com); cutoff for buried residues is around 0.25.
†Conservation (%) of a native E. coli’s DHFR residue (left column) or a substituted residue (right column) in a given position of 290 aligned mesophylic prokaryotic DHFR sequences retrieved from the Optimal Growth Temperature database (http://pgtdb.csie.ncu.edu.tw).
‡Not determined.
Using site-directed mutagenesis, we generated 16 constructs for recombinant expression of DHFR, each carrying a single unique mutation in the coding region. All DHFR mutants were expressed and purified and their biophysical and catalytic properties assayed initially as follows: (i) Thermal stabilities of the mutants were measured by differential scanning calorimetry (DSC) and the obtained thermograms used to infer apparent thermal transition midpoint temperatures ( ) (Table 1 and Fig. S2); (ii) A two-state folding model was applied to urea denaturation curves to derive urea midtransition concentration (Cm), and the Gibbs free energy difference between folded and unfolded states at 25 °C in water (ΔGH2O) (Table 1 and Fig. S3 and Table S1).
) (Table 1 and Fig. S2); (ii) A two-state folding model was applied to urea denaturation curves to derive urea midtransition concentration (Cm), and the Gibbs free energy difference between folded and unfolded states at 25 °C in water (ΔGH2O) (Table 1 and Fig. S3 and Table S1).
Both Cm and ΔGH2O were linearly correlated to  in the studied range of temperatures (30–42 °C), as postulated by protein thermodynamics (17, 18) (Fig. S4). As expected, most mutations (13 out of 16) appeared to be mildly to severely destabilizing with apparent ΔΔG values ranging between 0.18 to 2.87 kcal/mol (Table S1). This initial work led us to choose to use
in the studied range of temperatures (30–42 °C), as postulated by protein thermodynamics (17, 18) (Fig. S4). As expected, most mutations (13 out of 16) appeared to be mildly to severely destabilizing with apparent ΔΔG values ranging between 0.18 to 2.87 kcal/mol (Table S1). This initial work led us to choose to use  to characterize the stability of mutants, because it could be directly measured experimentally, in contrast to ΔGH2O, which is derived under the additional assumption of two-state unfolding.
to characterize the stability of mutants, because it could be directly measured experimentally, in contrast to ΔGH2O, which is derived under the additional assumption of two-state unfolding.
In terms of catalytic activity, the kcat and KM of the DHFR proteins were measured by full progress-curve kinetics (Table 1). As expected, the enzymatic proficiency (kcat/KM) of most mutants was found to be very similar to that of the WT DHFR (with only a twofold difference between all mutants). There was no statistically significant correlation between protein activity and stability (Fig. S4), suggesting a lack of trade-off for this group of mutations. Further analysis also showed no significant correlation between catalytic activity and fitness (Fig. S4), suggesting a catalytic saturation regime for DHFR (as found by Hartl and coworkers for another enzyme) (19).
In addition to 16 single DHFR mutants, we built 11 constructs carrying multiple mutations. This was achieved by exhaustively combining the four most destabilizing single mutations V75H, I91L, W133V, and I155A (Table 1 and Table S2). However, we were unable to purify these mutants at quantities required for in vitro characterization.
Site-Directed Chromosomal Mutagenesis.
Chromosomal mutants were created by a technique developed in this lab that allows incorporation of desired mutations in a controlled manner at any locus within E. coli’s chromosome (see Materials and Methods).
Using this technique, we generated 27 folA mutants in E. coli’s MG1655 strain [16 single mutants with defined molecular properties (Table 1) and 11 multiple mutants (Table S2)].
Fitness Measurements.
Fitness of these strains was determined by competition with WT DHFR strain following the assay developed by Lenski and coworkers (20). To this end, WT and a given mutant strain were mixed in 1∶1 ratio and grown together for 18 h in a range of temperatures. Laboratory fitness of a mutant strain in this assay was defined as the population ratio of mutant strain to WT upon completion of the growth cycle (see Materials and Methods). We next focus on three temperatures: 30 °C as the lower limit of Arrhenius-like dependence of growth rate on temperature (Fig. S5), 42 °C as its highest limit, and 37 °C as a “physiological” temperature for E. coli in humans. We viewed this competition assay as superior to other fitness measurements (such as individually measured growth rates), as it allows the determination of an evolutionarily relevant selective (dis)advantage of a population competing for nutrients. A possible concern might be that fitness variations due to genetic diversity outside the folA locus might contribute to the outcome of the competition experiments. However, our analysis (see Materials and Methods) shows that this is not the case.
Fitness Correlates with Protein Abundance at 30 °C.
We measured intracellular abundances of DHFR proteins for all mutants in both soluble and insoluble fractions of cell lysates. There was no significant correlation between fitness and abundance in the insoluble fraction at all temperatures tested (30 °C, 37 °C, and 42 °C), confirming the notion that for this protein, toxicity of the aggregated fraction does not contribute to fitness (Fig. 2
A and Fig. S6). Soluble protein abundance, however, does appear to be correlated to stability. Correlation of the soluble fraction of the DHFR single mutant proteins with their  grows significantly stronger as temperature increases: R = 0.51 (30 °C), 0.64 (37 °C), and 0.76 (42 °C) (Fig. 2B and Fig. S6). Statistical mechanics of two-state protein folding (21, 22) would predict the relationship between protein stability and intracellular abundance of folded proteins to follow the Boltzmann law:
grows significantly stronger as temperature increases: R = 0.51 (30 °C), 0.64 (37 °C), and 0.76 (42 °C) (Fig. 2B and Fig. S6). Statistical mechanics of two-state protein folding (21, 22) would predict the relationship between protein stability and intracellular abundance of folded proteins to follow the Boltzmann law:
 |
[1] |
where T is temperature, kB is Boltzmann constant, Ctot and Cf are total and folded abundances. The Boltzmann relationship in Eq. 1 predicts a plateau for the observed range of free energies of mutants at ΔG ≤ -2 kcal/mol at 30 °C (Table 1), provided that total abundance is fixed (red circles, Fig. 2B and Fig. S6). Our data, however, show a stronger, linear dependence between abundance and stability (blue circles, Fig. 2B and Fig. S6), suggesting that cytoplasm is an active medium, where stability critically affects total abundance through homeostatic balance between protein production and degradation. Destabilization of a protein apparently shifts this balance toward degradation, as has been observed in vitro (23, 24). In line with this observation is the strong correlation between fitness and protein abundance observed at 30 °C (Fig. 3B). However, we found that this correlation disappears at higher temperature (Fig. S6).
Fig. 2.
Correlating fitness at 30 °C with molecular properties. (A) Fitness measured at 30 °C by competing 27 DHFR mutant strains with a reference WT DHFR strain is plotted against insoluble fraction of DHFR proteins determined in cell lysates by Western blot. Fitness of the WT DHFR strain and its abundance in the insoluble fraction is set to one. (B) Soluble protein abundance (blue circles) determined for single DHFR mutant strains is plotted against apparent midtransition temperature of unfolding ( ) measured in vitro by DSC. Protein abundance predicted at 30 °C by Eq. 1 is depicted in red. Predicted fraction somewhat deviates from a plateau because of the experimental imprecision of ΔG values derived from the urea unfolding under two-state assumption and
) measured in vitro by DSC. Protein abundance predicted at 30 °C by Eq. 1 is depicted in red. Predicted fraction somewhat deviates from a plateau because of the experimental imprecision of ΔG values derived from the urea unfolding under two-state assumption and  values inferred from the DSC thermograms (Fig. S4). (C) Fitness measured at 30 °C by competing 27 DHFR mutant strains with a reference WT DHFR strain is plotted against soluble fraction of DHFR proteins determined in cell lysates by Western blot. Fitness of the WT DHFR strain and its abundance in the soluble fraction is set to one.
values inferred from the DSC thermograms (Fig. S4). (C) Fitness measured at 30 °C by competing 27 DHFR mutant strains with a reference WT DHFR strain is plotted against soluble fraction of DHFR proteins determined in cell lysates by Western blot. Fitness of the WT DHFR strain and its abundance in the soluble fraction is set to one.
Fig. 3.
Distribution of fitness effects and correlation with stability. (A–C) Histograms of distributions of fitness effects at 30 °C, 37 °C, and 42 °C for all mutant DHFR strains (16 single and 11 multiple mutants) as measured by competition with a reference WT DHFR strain. Fitness of the reference strain is set to 1. Individual fitness values can be found at Table S2. (D–F) Correlation of fitness values measured for single mutant DHFR strains by competition at 30 °C, 37 °C, and 42 °C to in vitro measured  .
.
Fitness Inversely Correlates with Stability at 42 °C.
At 30 °C, most (over 55%) of the DHFR mutations were deleterious or nearly neutral (Fig. 3A and Table S2). Very surprisingly, with temperature increase the fraction of strains carrying deleterious mutations dropped to 27% at 37 °C, and to only 14% at 42 °C (Fig. 3
B and C and Table S2). Moreover, the distribution of fitness effects (DFE) became practically bimodal at 42 °C: The effect of DHFR mutations was either nearly lethal or, for most strains, clearly advantageous. We confirmed this unusual observation by competing I155A DHFR mutant strain (that shows highest fitness level at 42 °C among all DHFR mutants, Table S2) with nine independently generated WT DHFR strains. In line with our previous observation, at a low temperature (30 °C), the I155A mutant exhibited reduced fitness (0.92), while at 42 °C its fitness improved remarkably, on average twofold (1.89) compared with WT (Table S3). While at 30 °C and 37 °C no statistically significant correlation between  and fitness can be found, there does appear to be a clear anticorrelation at 42 °C (Fig. 3
D–F), implying an unusual trade-off between fitness and stability (mutant strains encoding less stable DHFR appear to be more fit at the higher temperature) (Fig. 3
D–F).
and fitness can be found, there does appear to be a clear anticorrelation at 42 °C (Fig. 3
D–F), implying an unusual trade-off between fitness and stability (mutant strains encoding less stable DHFR appear to be more fit at the higher temperature) (Fig. 3
D–F).
Soluble Oligomerization at 42 °C.
The results of our fitness experiments presented a paradoxical situation where the main determinants of fitness reverse themselves for strains growing at higher temperature. The observed dependence of fitness on protein abundance and activity at 30 °C is intuitive and consistent with theoretical views based on flux balance theory (19, 25), whereas the data at 42°C where strains carrying less stable mutants appear to be more fit are totally unexpected (and, on the face of it, counterintuitive). What, then, is going on at 42 °C that favors strains carrying less stable mutants? A critical hint comes from the hyperthermophile Thermotoga maritima, whose DHFR exists as a stable homodimer (26). Further, Fernandez and Lynch recently observed that less stable proteomes are enriched in protein complexes (27). This work suggests a possibility that, at higher temperatures, destabilized mutants can form soluble oligomers, preventing their further aggregation and preserving activity. Indeed, such behavior at elevated temperatures close to unfolding transition has been predicted for another protein, SH3 domain (28); furthermore, simulations for several proteins (28–30) and experimental inverstigations (31) have indicated that oligomerization can increase stability by providing additional stabilizing contacts, especially in domain-swapped oligomers. With this in mind, we sought to evaluate, at the protein level, the propensity of DHFR mutants to oligomerize in vitro; initially by solution FRET, then by in vitro cross-linking experiments at “high” (42 °C) and “low” (25 °C) temperatures.
Solution FRET was carried out with Cy3 as a donor label and Cy5 as an acceptor to determine whether mutant DHFR associates in vitro at 42 °C. We found that the highly destabilized mutant I155A, which gives rise to the most fit (at 42 °C) strain, shows pronounced protein-protein interaction at 42 °C in contrast to WT (also at 42 °C) and the same mutant at 25 °C, matching our observations on fitness (Fig. S7). However, FRET data for another destabilized mutant, W133V, whose strain has very low fitness at 42 °C, was qualitatively similar to I155A and different from WT (Fig. S7). This latter anomaly was thought to be due to a limitation of FRET in its inability to distinguish between oligomers and aggregates as FRET merely monitors a close proximity between donor and acceptor dyes attached to different protein molecules. We hypothesized that two distinctly different types of fitness effects as seen in Fig. 3C at 42 °C may be due to the differences in the association behavior of different mutants (an effect FRET is incapable of identifying). To test this hypothesis, we carried out in vitro cross-linking experiments at room temperature (25 °C) and at 42 °C on the purified DHFR proteins (Fig. 4 and Fig. S8). Strikingly, we found that many mutants that exhibit higher fitness at 42 °C (e.g., I155A) have a strong tendency to form oligomers at 42 °C but not at room temperature (Fig. 4A). Furthermore, oligomer-forming mutants did not exhibit high molecular weight bands typical of aggregated proteins, whereas mutants of low fitness (at 42 °C) strains showed a pronounced aggregation and a much weaker homodimer band (Fig. 4 and Fig. S8). Quantitatively, we found a significant correlation between the propensity to oligomerize (assessed by density of all DHFR oligomerized species bands in the gels) with both fitness at 42 °C (Fig. 4B) and unfolding transition temperature Tm (Fig. 4C). Our postulation of soluble oligomerization is further supported by native gel electrophoresis (Fig. S9), where the destabilized I155A mutant shows the same pattern of dimers at 42 °C but not at lower temperatures, a behavior not observed for WT proteins.
Fig. 4.
In vitro oligomerization assay. (A) A representative cross-linking experiment. 11 μM of WT DHFR, W133V, or I155A purified proteins in 25 mM potassium-phosphate buffer (pH 7.8) and 100 μM NADPH were incubated for 45 min at room temperature (RT) or 42 °C with or without 2 mM of cross-linking (c-l) agent (glutaraldehyde). Shown is the SDS/PAGE analysis of 10 μL of protein samples after Coomassie staining. Red arrows indicate molecular weight equivalent of monomeric and oligomeric protein species. Smears at high molecular weight seen for W133V protein at 37 °C and 42 °C in the presence of c-l are due to extensive aggregation. (B) The in vitro propensity to oligomerize is correlated to fitness values measured by competition at 42 °C. Oligomerization propensity was determined by measuring the relative density of the SDS/PAGE bands corresponding to all DHFR oligomerized species for each of the purified mutant (Fig. S8). The degree of oligomerization for every mutant is normalized to WT DHFR. (C) Correlation of oligomerization propensity (as measured in B) to mutant’s  .
.
Discussion
A key aspect of this study is that it introduces mutations in the gene of interest directly on the chromosome leaving the upstream region intact to keep protein abundance at its endogenous level, including possible feedback control mechanisms. This contrasts with the more common approach where DHFR mutant genes are expressed from a plasmid. This is problematic as effects observed in this work are strongly concentration-dependent so that nonendogenous abundance of mutant proteins would have concealed or distorted the relevant phenomenology.
A common expectation is that destabilization should be detrimental to fitness, first, because active proteins are thought to be lost to unfolding (see Eq. 1) and, second, because destabilized proteins are believed to aggregate causing both irreversible loss of function and possible toxicity (8, 32). Our results suggest that loss of function rather than toxicity from aggregation is responsible for most fitness effects at 30 °C (Fig. 2). This finding might appear at variance with recent theoretical postulates that posit that misfolding-induced aggregation is a major source of fitness effects (8), but we note that DHFR is a low copy number enzyme. As such it is possible that the effect of misfolding-induced aggregation may be more pronounced for highly expressed proteins as it was indeed observed in recent experiments where a nonendogenous protein was greatly overexpressed from a plasmid in yeast (16). However, the behavior, which we report at a higher temperature (42 °C), goes against the conventional wisdom because an unexpected physical factor interferes. Our work shows that less stable DHFR proteins tend to escape aggregation by forming soluble oligomers, and this phenomenon is responsible for a surprising anticorrelation between stability and fitness at 42 °C.
Dimer stabilization through domain swapping has been observed in silico and in vitro for many proteins (28, 33, 34). Recent observations that homodimers are prevalent in proteomes (35) may indicate that soluble oligomerization serves as a common evolutionary mechanism to evolve stable functional proteins (36, 37). It appears that this mechanism may provide a route to escape the detrimental effects of destabilizing mutations by opening sequence space for a broader exploration at higher temperatures. It is tempting to suggest that it can serve as a universal route to “sequence-based” (38) thermal adaptation for many proteins of originally mesophilic organisms which colonize warmer environments. While the structural aspects of mutant DHFR dimerization remain to be discovered, our findings provide a mechanistic support to this view.
We found here a peculiar temperature-dependent trade-off between stability and fitness, which is very different from the postulated trade-off between stability and activity (1). For example, the destabilized mutant I155A is less fit than WT at 30 °C when it cannot dimerize via domain swapping, and the effect of destabilization is in a decreased copy number of active proteins. However, the same mutant is much more fit than WT at 42 °C where soluble oligomerization of the same I155A mutant prevents its aggregation while WT DHFR partly aggregates.
This study highlights the complexity of the concept of fitness, which is central to population genetics. Mutations that provide higher fitness under one set of conditions can be detrimental under another. It seems that the fate of a mutation is determined not by its fitness effect under fixed conditions but rather by an organism’s lifestyle (e.g., generalist versus specialist). Indeed some DHFR mutations, which provide high fitness to MG1655 E.coli at 42 °C, are wild type in other bacterial species.
The metaphor of a “rugged fitness landscape” is often invoked to reflect the notion that fitness effects cannot be predicted from sequence variation. This study shows that fitness effects of mutations, while difficult to rationalize at the level of sequence variation, can be “projected” on a small number of “axes” that reflect coarse-grained Biophysical properties of proteins such as stability or intracellular abundance. Organismal fitness in the space of such coarse-grained properties can then appear more “smooth.” Indeed, a significant correlation exists between fitness and several coarse-grained quantities. This is good news for biophysics-based multiscale modeling of evolution. The bad news is that it is still challenging to postulate such correlations a priori—for example, simple equilibrium statistical mechanics considerations may be not applicable because cellular environments represent an active medium where energy consuming machinery acts on proteins. Further issues may be encountered where unexpected equilibrium mechanisms, such as soluble oligomerization, may intervene in organismal fitness.
Materials and Methods
Site-Directed Chromosomal Mutagenesis.
The method is a modification of a chromosomal gene knock-out protocol (39). Briefly, the folA gene carrying the desired mutation(s) with an entire endogenous regulatory region (191 bp separating the stop codon of the upstream kefC gene and start codon of the folA gene) was placed on a pKD13 plasmid flanked by two different antibiotic markers (genes encoding kanamycin (kanR) and chloramphenicol (cmR) resistances). The entire cassette was then amplified with two primers tailed with 50 nucleotides homologous to the region of the chromosome intended for recombination (kefC gene upstream and apaH gene downstream to folA). The amplified product was transformed into BW25113 strain with an induced Red helper plasmid. The recombinants were selected on plates carrying both antibiotics. Strains carrying the desired mutation in the chromosome were verified by sequencing. Identified chromosomal mutations were then retransformed into MG1655 strain by P1 transduction and double antibiotic selection (kan and cm) and again verified by sequencing.
Competition Assay.
Laboratory fitness was determined by competing each of the strains expressing mutant DHFR protein with WT DHFR strain in M9 minimal media supplemented with 0.2% glucose, LmM MgSO4, 0.1% casamino acids, and 0.5 μg/mL thiamine. To this end, MG1655 E. coli strain carrying WT folA gene flanked by cmR and kanR genes was mixed with one of the MG1655 DHFR mutant strains in a 1∶1 ratio (≈104 cells each) in 50 mL of medium. Prior to mixing, cells were grown separately overnight from a single colony, diluted 1/100 and regrown to early exponential phase (OD600 ≈ 0.2) at 30 °C. The competition assay was performed for 18 h at 30 °C, 37 °C, and 42 °C. To distinguish between the competing strains, a knock-out mutation was introduced in the lacZ gene of each strain. Two identical competition experiments were performed with the lacZ knockout (a neutral marker under the condition of the competition experiment) always present in one of the competing strains. The ratio before and after competition was determined by plating the culture on LB agar plates supplemented with X-gal and IPTG (lacZ- strain generates white colonies, whereas lacZ+ strain generates blue colonies, hence “blue-white” swap). The swap was performed to ensure the neutrality of the lacZ marker. Around 3,000 to 5,000 colonies were counted for each competition experiment.
Estimating Fitness Errors.
Two steps were taken to establish the error bars for fitness measurements.
First, experimental noise was measured for any competing pair of strains by repeating the competition experiments 2–6 times. “Noise” levels were found to be within 13% for measurements performed at 30 °C and 17% at 42 °C (Figs. 2 and 3). Second, although WT DHFR strains, just as the WT MG1655 strain, do not carry mutation in the folA gene, they were subjected to double antibiotic selection, and, therefore, could carry unrelated hitchhiked mutations in other parts of the genome. It is possible, therefore, that fitness variations could arise due to genetic variation outside of the folA locus in competing strains. This was addressed by using our chromosomal insertion technique to generate additional 12 control WT strains whose folA gene encodes WT DHFR to measure any unintended genetic variability that this technique might introduce elsewhere in the genome. Twelve individual competition assays were carried out by competing each of the new control WT DHFR strains with our original WT DHFR reference strain. We found that the experimental error for these competitions was comparable to the error found for any repeatedly competed pair of strains (10% for competitions at 30 °C and 18% at 42 °C) (Table S4). This analysis establishes the error bars for fitness measurements.
Intracellular Protein Abundance.
Cells were grown in supplemented M9 medium for 6 h at 30 °C, chilled on iced for 30 min and lysed with BugBuster (Novagen). The insoluble fraction was separated by centrifugation, then solubilized by Inclusion Body Solubilization Reagent (Thermo Scientific), and dialyzed against PBS. DHFR amounts in the soluble and insoluble fractions were determined by SDS/PAGE followed by Western blot using rabbit-anti E. coli’s DHFR polyclonal antibodies (custom raised by Pacific Immunology) by measuring densities of the DHFR bands. All protein abundances are normalized to the amounts detected for WT DHFR then arbitrarily set to 1. The overall amount of protein in the lysates was estimated with BCA protein assay kits (Pierce).
Statistics.
R and P—values for linear correlations were determined by ANOVA (analysis of variation) test (40).
Supplementary Material
Acknowledgments.
We acknowledge Roy Kishony’s help at the early stages of this project and Dan Tawfik, Art Horwich, Bill Eaton, Maxim Frank-Kamenetskii, Sergey Maslov, and Stephen MQc. Gould for commenting on the manuscript. We acknowledge Xiaowei Zhuang for helping with FRET measurements. We thank Adrian Serohijos for the bioinformatics analysis and Yakov Pechersky, Abhishek Chintapalli, and Phil Snyder for technical assistance. This work was supported by National Institutes of Health Grant GM 068670 and long-term postdoctoral fellowship from the Human Frontier Science Program (S.B.).
Footnotes
The authors declare no conflict of interest.
*This Direct Submission article had a prearranged editor.
This article contains supporting information online at www.pnas.org/lookup/suppl/doi:10.1073/pnas.1118157109/-/DCSupplemental.
References
- 1.DePristo MA, Weinreich DM, Hartl DL. Missense meanderings in sequence space: A biophysical view of protein evolution. Nat Rev Genet. 2005;6:678–687. doi: 10.1038/nrg1672. [DOI] [PubMed] [Google Scholar]
- 2.Bloom JD, Labthavikul ST, Otey CR, Arnold FH. Protein stability promotes evolvability. Proc Natl Acad Sci USA. 2006;103:5869–5874. doi: 10.1073/pnas.0510098103. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Zeldovich KB, Chen P, Shakhnovich EI. Protein stability imposes limits on organism complexity and speed of molecular evolution. Proc Natl Acad Sci USA. 2007;104:16152–16157. doi: 10.1073/pnas.0705366104. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Soskine M, Tawfik DS. Mutational effects and the evolution of new protein functions. Nat Rev Genet. 2010;11:572–582. doi: 10.1038/nrg2808. [DOI] [PubMed] [Google Scholar]
- 5.Weinreich DM, Delaney NF, DePristo MA, Hartl DL. Darwinian evolution can follow only very few mutational paths to fitter proteins. Science. 2006;312:111–114. doi: 10.1126/science.1123539. [DOI] [PubMed] [Google Scholar]
- 6.Zeldovich KB, Chen P, Shakhnovich BE, Shakhnovich EI. A first-principles model of early evolution: Emergence of gene families, species, and preferred protein folds. PLoS Comput Biol. 2007;3:1224–1238. doi: 10.1371/journal.pcbi.0030139. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Taverna DM, Goldstein RA. Why are proteins marginally stable? Proteins. 2002;46:105–109. doi: 10.1002/prot.10016. [DOI] [PubMed] [Google Scholar]
- 8.Drummond DA, Wilke CO. Mistranslation-induced protein misfolding as a dominant constraint on coding-sequence evolution. Cell. 2008;134:341–352. doi: 10.1016/j.cell.2008.05.042. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Bloom JD, Raval A, Wilke CO. Thermodynamics of neutral protein evolution. Genetics. 2007;175:255–266. doi: 10.1534/genetics.106.061754. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Pena MI, Davlieva M, Bennett MR, Olson JS, Shamoo Y. Evolutionary fates within a microbial population highlight an essential role for protein folding during natural selection. Mol Sys Biol. 2010;6:1–12. doi: 10.1038/msb.2010.43. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Wylie CS, Shakhnovich EI. A biophysical protein folding model accounts for most mutational fitness effects in viruses. Proc Natl Acad Sci USA. 2011;108:9916–9921. doi: 10.1073/pnas.1017572108. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Wright PE, Dyson HJ. Linking folding and binding. Curr Opin Struct Biol. 2009;19:31–38. doi: 10.1016/j.sbi.2008.12.003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Benkovic SJ, Fierke CA, Naylor AM. Insights into enzyme function from studies on mutants of dihydrofolate reductase. Science. 1988;239:1105–1110. doi: 10.1126/science.3125607. [DOI] [PubMed] [Google Scholar]
- 14.Taniguchi Y, et al. Quantifying E. coli proteome and transcriptome with single-molecule sensitivity in single cells. Science. 2010;329:533–538. doi: 10.1126/science.1188308. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Bershtein S, Segal M, Bekerman R, Tokuriki N, Tawfik DS. Robustness-epistasis link shapes the fitness landscape of a randomly drifting protein. Nature. 2006;444:929–932. doi: 10.1038/nature05385. [DOI] [PubMed] [Google Scholar]
- 16.Geiler-Samerotte KA, et al. Misfolded proteins impose a dosage-dependent fitness cost and trigger a cytosolic unfolded protein response in yeast. Proc Natl Acad Sci USA. 2011;108:680–685. doi: 10.1073/pnas.1017570108. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Privalov PL. Stability of proteins: Small globular proteins. Adv Protein Chem. 1979;33:167–241. doi: 10.1016/s0065-3233(08)60460-x. [DOI] [PubMed] [Google Scholar]
- 18.Robertson AD, Murphy KP. Protein structure and the energetics of protein stability. Chem Rev. 1997;97:1251–1268. doi: 10.1021/cr960383c. [DOI] [PubMed] [Google Scholar]
- 19.Dykhuizen DE, Dean AM, Hartl DL. Metabolic flux and fitness. Genetics. 1987;115:25–31. doi: 10.1093/genetics/115.1.25. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Elena SF, Lenski RE. Evolution experiments with microorganisms: The dynamics and genetic bases of adaptation. Nat Rev Genet. 2003;4:457–469. doi: 10.1038/nrg1088. [DOI] [PubMed] [Google Scholar]
- 21.Privalov PL, Khechinashvili NN. A thermodynamic approach to the problem of stabilization of globular protein structure: A calorimetric study. J Mol Biol. 1974;86:665–684. doi: 10.1016/0022-2836(74)90188-0. [DOI] [PubMed] [Google Scholar]
- 22.Shakhnovich E. Protein folding thermodynamics and dynamics: Where physics, chemistry, and biology meet. Chem Rev. 2006;106:1559–1588. doi: 10.1021/cr040425u. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Parsell DA, Sauer RT. The structural stability of a protein is an important determinant of its proteolytic susceptibility in Escherichia coli. J Biol Chem. 1989;264:7590–7595. [PubMed] [Google Scholar]
- 24.Park C, Marqusee S. Pulse proteolysis: A simple method for quantitative determination of protein stability and ligand binding. Nat Methods. 2005;2:207–212. doi: 10.1038/nmeth740. [DOI] [PubMed] [Google Scholar]
- 25.Wang Z, Zhang J. Impact of gene expression noise on organismal fitness and the efficacy of natural selection. Proc Natl Acad Sci USA. 2011;108:E67–E76. doi: 10.1073/pnas.1100059108. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Dams T, et al. The crystal structure of dihydrofolate reductase from Thermotoga maritima: Molecular features of thermostability. J Mol Biol. 2000;297:659–672. doi: 10.1006/jmbi.2000.3570. [DOI] [PubMed] [Google Scholar]
- 27.Fernandez A, Lynch M. Non-adaptive origins of interactome complexity. Nature. 2011;474:502–505. doi: 10.1038/nature09992. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Ding F, Dokholyan NV, Buldyrev SV, Stanley HE, Shakhnovich EI. Molecular dynamics simulation of the SH3 domain aggregation suggests a generic amyloidogenesis mechanism. J Mol Biol. 2002;324:851–857. doi: 10.1016/s0022-2836(02)01112-9. [DOI] [PubMed] [Google Scholar]
- 29.Malevanets A, Sirota FL, Wodak SJ. Mechanism and energy landscape of domain swapping in the B1 domain of protein G. J Mol Biol. 2008;382:223–235. doi: 10.1016/j.jmb.2008.06.025. [DOI] [PubMed] [Google Scholar]
- 30.Yang S, Levine H, Onuchic JN. Protein oligomerization through domain swapping: Role of inter-molecular interactions and protein concentration. J Mol Biol. 2005;352:202–211. doi: 10.1016/j.jmb.2005.06.062. [DOI] [PubMed] [Google Scholar]
- 31.Rousseau F, Schymkowitz JW, Wilkinson HR, Itzhaki LS. Three-dimensional domain swapping in p13suc1 occurs in the unfolded state and is controlled by conserved proline residues. Proc Natl Acad Sci USA. 2001;98:5596–5601. doi: 10.1073/pnas.101542098. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Dobson CM. Protein aggregation and its consequences for human disease. Protein Pept Lett. 2006;13:219–227. doi: 10.2174/092986606775338362. [DOI] [PubMed] [Google Scholar]
- 33.Gronenborn AM. Protein acrobatics in pairs—dimerization via domain swapping. Curr Opin Struct Biol. 2009;19:39–49. doi: 10.1016/j.sbi.2008.12.002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Rousseau F, et al. Domain swapping in p13suc1 results in formation of native-like, cytotoxic aggregates. J Mol Biol. 2006;363:496–505. doi: 10.1016/j.jmb.2006.07.061. [DOI] [PubMed] [Google Scholar]
- 35.Ispolatov I, Yuryev A, Mazo I, Maslov S. Binding properties and evolution of homodimers in protein—protein interaction networks. Nucleic Acids Res. 2005;33:3629–3635. doi: 10.1093/nar/gki678. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Pereira-Leal JB, Levy ED, Kamp C, Teichmann SA. Evolution of protein complexes by duplication of homomeric interactions. Genome Biol. 2007;8:R51.1–R51.12. doi: 10.1186/gb-2007-8-4-r51. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Lukatsky DB, Shakhnovich BE, Mintseris J, Shakhnovich EI. Structural similarity enhances interaction propensity of proteins. J Mol Biol. 2007;365:1596–1606. doi: 10.1016/j.jmb.2006.11.020. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Berezovsky IN, Shakhnovich EI. Physics and evolution of thermophilic adaptation. Proc Natl Acad Sci USA. 2005;102:12742–12747. doi: 10.1073/pnas.0503890102. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Datsenko KA, Wanner BL. One-step inactivation of chromosomal genes in Escherichia coli K-12 using PCR products. Proc Natl Acad Sci USA. 2000;97:6640–6645. doi: 10.1073/pnas.120163297. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Freedman D, Pisani R, Purves R. Statistics. 4th Ed. New York: W.W. Norton; 2007. [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.