Abstract
The transition from quiescence to proliferation is a key regulatory step that can be induced by serum stimulation in cultured fibroblasts. The transcription factor Myc is directly induced by serum mitogens and drives a secondary gene expression program that remains largely unknown. Using mRNA profiling, we identify close to 300 Myc-dependent serum response (MDSR) genes, which are induced by serum in a Myc-dependent manner in mouse fibroblasts. Mapping of genomic Myc-binding sites by ChIP-seq technology revealed that most MDSR genes were directly targeted by Myc, but represented a minor fraction (5.5%) of all Myc-bound promoters (which were 22.4% of all promoters). Other target loci were either induced by serum in a Myc-independent manner, were not significantly regulated or were negatively regulated. MDSR gene products were involved in a variety of processes, including nucleotide biosynthesis, ribosome biogenesis, DNA replication and RNA control. Of the 29 MDSR genes targeted by RNA interference, three showed a requirement for cell-cycle entry upon serum stimulation and 11 for long-term proliferation and/or survival. Hence, proper coordination of key regulatory and biosynthetic pathways following mitogenic stimulation relies upon the concerted regulation of multiple Myc-dependent genes.
Keywords: Myc, chromatin, transcription, serum
Introduction
Quiescent cells can be induced to enter a proliferative state in vitro through exposure to serum mitogens, a process associated with widespread changes in gene expression. Immediate-early serum response genes are induced directly, in a manner that does not depend upon prior protein synthesis. Delayed-early genes, on the other hand, depend upon prior protein synthesis, suggesting a direct role for immediate-early gene products in their activation. Consistent with this concept, immediate-early genes such as c-fos, c-jun or c-myc encode transcription factors (reviewed by Winkles, 1998).
The c-myc gene product, Myc, is a transcription factor that can either activate or repress gene expression. Activation occurs through dimerization with the partner protein Max, and binding to the consensus DNA sequence CACGTG (E-box). Repression occurs through association of Myc/Max dimers with other transcription factors such as Miz-1 or NF-Y, and interference with their function (Izumi et al., 2001; Staller et al., 2001; Seoane et al., 2002; Mao et al., 2003), although it is becoming apparent that Myc may also repress transcription through E-boxes (Adhikary and Eilers, 2005). A number of studies based on chromatin immunoprecipitation (ChIP) have shown that Myc associates with a large (⩾10–20%) fraction of cellular genes in a variety of cell types (Fernandez et al., 2003; Li et al., 2003; Cawley et al., 2004; Zeller et al., 2006; Chen et al., 2008; Eilers and Eisenman, 2008; Kidder et al., 2008; Kim et al., 2008). Myc-binding sites are preferentially found within CpG islands (Fernandez et al., 2003; Zeller et al., 2006) and within chromatin domains bearing the marks characteristic of active promoters, such as tri-methylation of Lys 4 in histone-H3 (H3K4me3) (Guccione et al., 2006; Kim et al., 2008). While Myc itself may favor the maintenance of an active chromatin configuration (Bouchard et al., 2001; Frank et al., 2001; Knoepfler et al., 2006; Cotterman et al., 2008; Martinato et al., 2008), this configuration also acts upstream from Myc, being a pre-requisite for target-site recognition (Guccione et al., 2006). Consistent with this notion, RNA PolII was loaded onto Myc-target promoters prior to—and independently of—Myc binding (Bouchard et al., 2004; Adhikary and Eilers, 2005), and recent data suggested that Myc regulates its target genes mainly at the level of transcriptional elongation in embryonic stem (ES) cells (Rahl et al., 2010). In summary, Myc binds to promoters that pre-exist in a transcriptionally active or poised state, and further modulates their activity.
Several experiments suggested that Myc is required but not sufficient for the activation of its target genes, and that other signals must be involved (Frank et al., 2001; Bouchard et al., 2004). An example is provided by the target gene Snail: on the one hand, Myc bound to the Snail promoter and was required for its activation by transforming growth factor-β signaling in mammary epithelial cells; on the other hand, binding also occurred in fibroblasts, but activation of Snail by serum in those cells did not require Myc (Smith et al., 2009). This dependence upon context for gene regulation—whether cell type or environmental context—may also apply to overexpressed Myc in cancer cells, as suggested for example by the comparison of gene expression profiles in Myc-induced prostate carcinomas in the mouse and a subset of human prostate carcinomas (Ellwood-Yen et al., 2003). These observations might allow understanding why, in spite of multiple gene expression profiles described over the years (see www.myccancergene.org/), Myc activity had not clearly been associated with a universal mRNA signature. Uncovering common denominators in Myc-driven transcription, however, may also depend upon the analytical tools used. For example, linkage and gene-set analysis allowed the identification of Myc as a causal factor in two gene signatures activated in a subset of carcinomas: a serum response/wound-healing program initially discovered in fibroblasts (Chang et al., 2004; Adler et al., 2006), as well as an ‘embryonic stem cell (ESC)-like' expression profile (Wong et al., 2008). A recent re-analysis distinguished the latter as a Myc-driven program common to ES and cancer cells, but distinct from the core pluripotency program in ES cells (Kim et al., 2010). The data described so far did not address whether Myc-associated signatures in cancer cells were related to its action in normal cells, in particular in the serum response.
We showed previously that Myc binding to several target promoters was required for their activation by serum (Frank et al., 2001). Here, we profiled the full contribution of Myc to the serum response in immortalized mouse fibroblasts. The c-myc gene was deleted in quiescent 3T9 fibroblasts homozygous for a conditional knockout allele (Trumpp et al., 2001), followed by serum stimulation and mRNA profiling. In parallel, we mapped Myc–DNA interactions at the genome-wide level in serum-treated cells. Altogether, these data provide a comprehensive picture of the core transcriptional program regulated by Myc in response to serum mitogens.
Results
Part of the transcriptional program activated by serum in fibroblasts is Myc-dependent
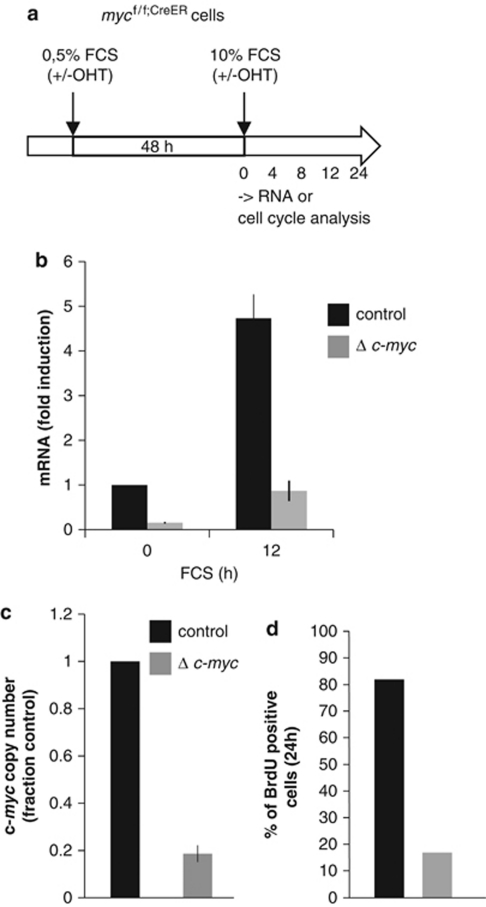
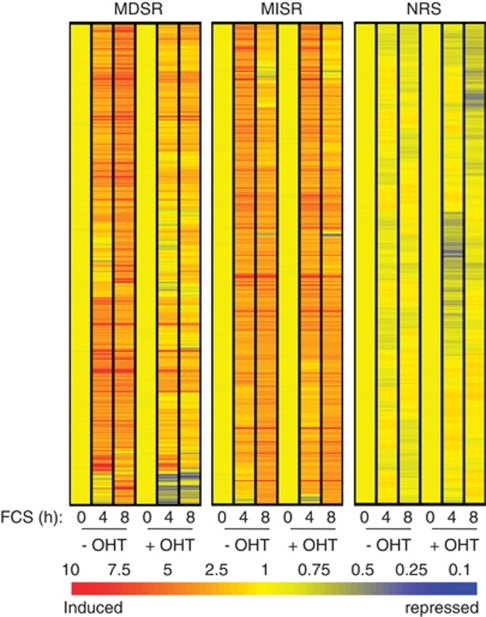
To study the role of Myc in the serum response, we took advantage of immortalized 3T9 fibroblasts derived from homozygous c-mycf/f mice bearing loxP sites in the first intron and in the 3′-untranslated region of the c-myc gene (Trumpp et al., 2001). These cells were transduced with a retroviral vector expressing a chimeric CreER recombinase (hereafter mycf/f;CreER cells), allowing conditional deletion of c-myc upon treatment with 4-hydroxytamoxifen (OHT). To achieve quiescence, near-confluent cultures were serum-starved for 2 days, either with or without OHT: in this manner, the c-myc gene was deleted at the time at which its expression is normally suppressed, interfering minimally with cellular fitness. Cells were then re-seeded in medium containing 10% fetal calf serum and harvested for analysis at the indicated time points (Figure 1a). Serum-induced accumulation of the c-myc mRNA in OHT-treated cells was reduced to 15–20% of that seen in control cells (Figure 1b) (Smith et al., 2009), mirroring the residual levels of the c-myc gene in the treated cells (Figure 1c). Thus, the c-mycf/f allele was efficiently deleted in ⩾80% of the cells upon CreER activation, with residual mRNA levels originating from a minority of undeleted cells. c-myc deletion prevented re-entry in S-phase following serum stimulation (Figure 1d) (Trumpp et al., 2001) and blunted the response of the Myc-target genes Ncl and Ppat (Smith et al., 2009), as observed previously in Rat1 cells (Frank et al., 2001). We thus used oligonucleotide microarrays to profile mRNA expression in control and OHT-treated mycf/f;CreER cells at 0, 4 and 8 h following serum stimulation. Based on a series of criteria outlined under Materials and methods, we identified three groups of genes that followed distinct regulatory patterns (Supplementary Table S1): a Myc-dependent serum response (MDSR, 298 genes), a Myc-independent serum response (MISR, 289 genes) or no response to serum (NRS, 631 genes). The difference among the three regulatory groups was most evident when visualized by hierarchical clustering (Eisen et al., 1998), with the MDSR group clearly composed of mRNAs induced in control, but not—or less—in Myc-deleted cells (Figure 2). The induction of each MDSR mRNA and its reduction without Myc are also plotted in Supplementary Figure S1A. As expected, this group included c-myc itself, Ncl, Ppat and Hspd1 (Frank et al., 2001), as well as 72 genes previously identified as being Myc-regulated (www.myccancergene.org/) (Zeller et al., 2003) (Supplementary Table S1). Other genes that responded to serum in a Myc-dependent manner in Rat1 cells (Frank et al., 2001) were lost from our list, either because of the stringency of our selection criteria, such as Cad, Nme2 (Nm23-H2) and Hspe1, or because they were not Myc-dependent in our experimental system, like Odc1.
Figure 1.
Conditional deletion of the c-myc gene in quiescent cells impairs cell-cycle re-entry. (a) A schematic representation of the experimental approach. Near-confluent cultures of mycf/f;CreER fibroblasts were serum-starved for 48 h with or without OHT to induce c-myc deletion. Following stimulation with 10% fetal calf serum, RNA was isolated at the indicated time points for profiling (Figure 2). (b) c-myc mRNA levels were measured in mycf/f;CreER cells, either without (control) or with OHT to induce c-myc deletion (Δ c-myc), followed by serum stimulation, as indicated. The data were normalized to 36B4 mRNA. The average±s.d. of three independent experiments is shown. (c) Relative amount of c-myc genomic DNA measured by quantitative RT–PCR in mycf/f;CreER cells left untreated (black bars) or treated with OHT (gray bars). Values were normalized to a PCR amplicon in the Nucleolin locus, and represent the average±s.d. from three independent experiments. (d) Cumulative percentages of cells traversing S-phase over 24 h of fetal calf serum stimulation in cultures treated as above. BrdU was added to the culture medium at time 0, and incorporation was assayed by flow cytometry.
Figure 2.
Identification of regulatory patterns following Myc deletion and serum stimulation. The heatmaps show the different classes of gene identified by their expression patterns: MDSR, MISR and NRS. The genes in each group were clustered hierarchically on the basis of their microarray-based expression profiles. Changes in mRNA levels were expressed as fold-induction relative to time 0 in each culture, and color-coded as indicated.
We also addressed the behavior in our experimental system of 230 genes reported previously to be upregulated upon Myc overexpression in Rat1 cells (list ‘Up in rat' Supplementary Table S2), identified mainly in profiling studies compiled in the Myc database (www.myccancergene.org) (Zeller et al., 2003). As shown in Supplementary Figure S2A and Supplementary Table S2, the majority of these genes responded positively to serum in our cells, but only few showed overlap with MDSR genes (37/230 or 16%). Limiting our analysis to the subset of genes identified in multiple studies improved the overlap with MDSR genes to up to 32% (Supplementary Table S3). Altogether, our data indicate that those genes that depend upon endogenous Myc (that is, our MDSR list) show only a partial overlap with genes that are induced upon ectopic Myc activation. A screen for Myc-inducible genes in mouse fibroblasts (our unpublished data) yielded the same conclusion.
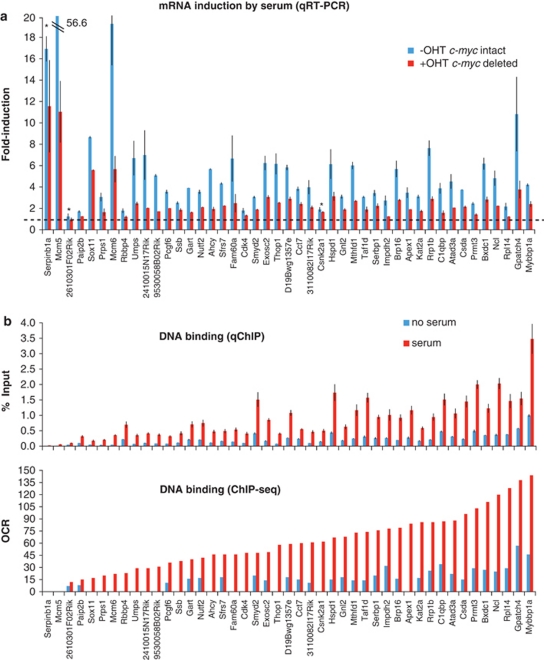
To validate our microarray expression data, we repeated three independent experiments and measured mRNA levels for 45 MDSR genes by quantitative reverse transcription (RT)–PCR at 0, 4, 8 and 12 h following serum stimulation. Figure 3a reports fold-activation for each mRNA at its peak time: 43 mRNAs (93.5%) showed a ⩾1.5 × serum response in control cells, with a significant reduction in Myc-deleted cells (P⩽0.01). Only two mRNAs failed this test, lacking either induction or significant Myc dependence (asterisks in Figure 3a). RT–PCR validation also confirmed that 10 genes in the MISR group were induced by serum in a Myc-independent manner, and that 9 out of 10 NRS genes showed no response, with only one (STAT3) showing a moderate induction (Supplementary Figure S3). Thus, RT–PCR confirmed the reliability of the regulatory patterns identified in our profiling experiments. We conclude that MDSR genes constitute the core transcriptional response built up by Myc during the G0→G1/S-phase progression.
Figure 3.
The MDSR genes are enriched for direct Myc targets. (a) Quantitative RT–PCR validation of 45 MDSR genes. The expression of each mRNA at the time point of maximal activation by serum is shown in Myc-expressing cells (−OHT, blue bars) alongside Myc-deleted cells (+OHT, red bars). The data are the average±s.d. of three independent experiments. The asterisks mark the three genes that were not validated according to our criteria (see text). (b) Binding of Myc to the corresponding promoters (TSS±1 kb), as assayed by qChIP (top) and ChIP-seq (bottom). Cells were either starved for 2 days (blue bars) or stimulated with serum for 8 h (red bars) before harvesting.
Analysis of Myc-repressed gene during serum stimulation
Myc has also been shown to repress transcription (Adhikary and Eilers, 2005). Upon serum stimulation, the mRNAs of Myc-repressed genes would be expected to show either of two patterns in Myc-deleted cells relative to controls: (i) loss of repression by serum (that is, Myc-dependent repression) or (ii) super-activation in the absence of Myc. In our microarray profiles, 130 genes apparently followed the first of these criteria (Supplementary Table S1 and Supplementary Figure S2B): however, when tested by quantitative RT–PCR, only 7/20 (35%) of the corresponding mRNAs confirmed this pattern, whereas nine (45%) were validated as repressed by serum but not as Myc-dependent (Supplementary Figure S4A). We also identified a group of 65 genes, which were super-induced by serum in the absence of Myc (Supplementary Table S1 and Supplementary Figure S2C). RT–PCR analysis validated the antagonizing role of Myc for 4/13 (30%) of these genes (Mmp9, Fdps, Abr and Cdkn1a, encoding p21Cip1) (Supplementary Figure S5). In both of the above groups, several other genes tested by RT–PCR showed an apparent Myc dependence, which however remained below statistical significance (Supplementary Figures S4A and S5).
We further compiled a list of 41 genes including (i) previously reported Myc-repressed genes, (ii) genes listed as serum-repressed and Myc-dependent in rat fibroblasts (Watson et al., 2002), and (iii) genes listed in the Myc database as repressed and Myc-bound in any species (Supplementary Table S2). However, these genes showed very disparate behaviors in our microarray profiles in terms of serum activation, repression or Myc-dependence (Supplementary Figure S2D). Only eight (20%) of these genes (Acp5, Gadd45, Rara, Tmsb4x, Vhl, Zfp36l1, Cdkn1a and Cdkn2b, encoding p15INK4b) showed a pattern consistent with their negative regulation by Myc (given our stringency criteria, only Cdkn1a had been included in the above ‘super-induced' group). RT–PCR analysis of Cdkn2b and Gadd45 validated their negative regulation by Myc (Supplementary Figure S4B); Gadd45a, in particular, is repressed by Myc overexpression (Marhin et al., 1997; Amundson et al., 1998; Tao and Umek, 1999) and, consistent with our data, was previously reported to be repressed by serum in a Myc-dependent manner (Bush et al., 1998). Altogether, and as concluded above for Myc-activated genes, only a fraction of the genes that are repressed by overexpressed Myc also show evidence for repression by endogenous Myc upon serum stimulation.
Our data are consistent with the concept that negative gene regulation by Myc represents an important facet of its activity in the mitogenic response. However, because Myc-dependent repression in our microarray profiles was validated to a lesser extent than Myc-dependent activation, we will concentrate here on the latter.
MDSR genes are direct Myc targets, but are a minor fraction of all the genes bound by Myc in the genome
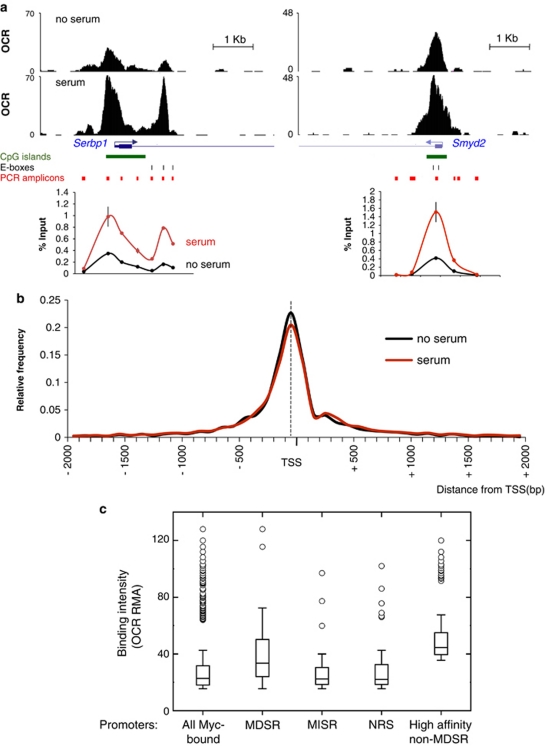
In parallel with gene expression, we used ChIP-seq to profile Myc binding in the genome of c-mycf/f 3T9 fibroblasts. Cells were fixed for ChIP analysis either before (0 h) or after serum stimulation (8 h), and chromatin was immunoprecipitated with a Myc-specific antibody. The recovered DNA was controlled by PCR amplification of known Myc-binding sites (data not shown), followed by ultra-high-throughput sequencing. In order to identify and quantify Myc-binding sites along the genome, we applied the following criteria. First, an overlap count value was determined for every base in the genome, indicating the number of sequence tags overlapping that position. Second, overlap count was normalized to sequencing depth in either library (0 and 8 h), yielding the ‘overlap count-resampled' (OCR) value, which represents a measure of binding intensity: to eliminate sequencing artifacts, we only retained sites showing over fivefold enrichment in OCR relative to input chromatin (Chen et al., 2008). The ChIP-seq profiles at two representative Myc-target loci are shown alongside their validation by quantitative ChIP (qChIP) with successive PCR amplicons (Figure 4a). qChIP validation of 76 peaks (45 at MDSR and 31 at randomly selected loci) showed a good correlation with relative ChIP-seq values, either before or after serum stimulation (Figure 3b and Supplementary Figure S6). As expected based on the induction of Myc by serum, Myc binding intensity was generally higher in serum-stimulated cells (Figure 3b and Supplementary Figure S1B).
Figure 4.
Distribution of Myc-binding peaks in the genome. (a) Examples of ChIP-seq profiles in serum-starved and stimulated cells are shown for two Myc-target loci, Serbp1 and Smyd2, aligned with CpG islands, E-boxes, the PCR amplicons used for validation and the corresponding qChIP profiles (bottom graphs). The qChIP data represent the average±s.d. from three independent experiments. (b) Relative frequency of peaks in the genomic window ±2 kb from all annotated TSSs, represented for cells without (black line) or with serum (red line). The vertical dotted line represents the mode of the distributions. (c) Box plots reporting the distribution of Myc-binding intensities (OCR-RMA) at peaks within ±1 kb from the TSS, shown separately for genes within each of the indicated regulatory patterns. Only genes bound above threshold were considered for each group (see text).
To facilitate the comparison of the two data sets, OCR values were further subjected to robust multi-array average (RMA) normalization (Irizarry et al., 2003). Whereas the OCR is a measure of Myc binding intensity at a given site, the RMA-normalized value (henceforth OCR-RMA) is proportional to the relative affinity of Myc for the corresponding site. Indeed, whereas OCR values were much higher in serum-stimulated relative to starved cells, reflecting differences in Myc levels (Supplementary Figure S1B), OCR-RMA values were generally closer between the two conditions (Supplementary Figure S1C). Interestingly, however, a subset of the sites targeted in serum-stimulated cells showed no significant binding in quiescent cells (Figure 3b and Supplementary Figure S1C), suggesting that a fraction of the Myc-binding sites are themselves induced by serum. The nature of this phenomenon remains to be addressed, but may conceivably be linked to changes in local chromatin configuration, which can modulate DNA recognition and Myc binding (Guccione et al., 2006).
Peaks with the lowest levels of enrichment in ChIP-seq may reflect non-sequence-specific interactions of Myc with chromatin, rather than binding to bona fide target sites. To discriminate among those modes of binding, we reasoned that peaks occurring only in quiescent cells and with the lowest intensities where the most likely to be nonspecific. Of all the peaks in the no-serum data set, 37% were not found in serum-stimulated cells: applying a cut-off at an OCR-RMA of 15 reduced this proportion to 16.3%. In addition, binding sites below that value in serum-stimulated cells were minimally enriched for the consensus E-box relative to unbound sites (see below; Table 1). We thus applied OCR-RMA ⩾15 as threshold of confidence for Myc binding in both data sets. Following this criterion, we identified 4749 and 8632 peaks in serum-starved and stimulated cells, respectively (Supplementary Table S4). As 82.2% of the peaks in the no-serum samples overlapped with those in serum-stimulated cells, we will concentrate mainly on the latter.
Table 1. For each of the five gene groups (i. to iv.), the table shows the percentage of TSSs located within a CpG island and the percentage of promoters containing a CACGTG motif within ±1 kb of the TSS.
| Group | Promoters | Total |
CpG islands |
CACGTG |
||
|---|---|---|---|---|---|---|
| % | P-value | % | P-value | |||
| i. | MDSR, Myc-bound | 234 | 86.8 | 0 | 62.4 | 0 |
| ii. | High-affinity non-MDSR promoters (RMA⩾35,5) | 718 | 87.0 | 0 | 59.1 | 0 |
| iii. | Low-affinity non-MDSR promoters (15<RMA<35,5) | 3355 | 83.5 | 0 | 41.3 | 0 |
| iv. | Myc peak below threshold (OCR-RMA ⩽15) | 4803 | 76.4 | 0 | 29.0 | 0.852 |
| v. | No peak | 10 672 | 23.4 | 1 | 22.3 | 1 |
Abbreviation: TSS, transcription start site.
The enrichment P-value for CpG islands was calculated by using binomial distribution assuming a frequency of TSS located in a CpG island of 0.473 (9731/20 570). The enrichment P-value for the CACGTG motif was calculated by using binomial distribution assuming a frequency of CACGTG motif within 1 kb of a TSS of 0.283325 (5828/20 570).
To determine the distribution of Myc relative to genes, we aligned our ChIP-seq peaks with the annotated RefSeq data set (Supplementary Table S4). In both data sets, Myc-binding sites were clearly concentrated around transcription start sites (TSSs) and decreased with distance from the TSS (Figure 4b and Supplementary Figure S7A). In serum-stimulated cells, 47.1% of the peaks occurred between −1000 and +1000 bp relative to the TSS, with maximal occurrence just before the TSS (Figure 4b, 535 peaks between 0 and −50). Based on this observation, we will define Myc-bound promoters as those with a peak within ±1 kb from the TSS. Another 22.4% of the peaks occurred further downstream inside genes (henceforth labeled ‘intragenic-only', that is, from >1000 bp from the TSS to the 3′ end) and 30.4% occurred further upstream or further downstream outside genes (‘intergenic' Supplementary Table S4; see Materials and methods). As expected, a majority (75.5%) of the TSS-proximal sites occurred within a CpG island. A total of 6.7% and 13.2% of the intragenic-only and intergenic sites, respectively, were also in CpG islands (Supplementary Table S4, statistics) and hence were most likely proximal to either an alternative TSS for the same gene, or the un-annotated TSS of an alternative locus. Indeed, 85% (2234/2628) of the intergenic sites occurred within ±1 kb from a CAGE tag (Supplementary Figure S7B), indicating their proximity to the 5′ end of an RNA transcript (Kawaji et al., 2009). Altogether, about half of all Myc-binding sites are proximal (within±1 kb) to a TSS, with most of these sites occurring within CpG islands.
We then asked which proportion of the MDSR genes were direct targets of Myc. Remarkably, 234 out of 298 MDSR genes (78.5%) were bound by Myc within ±1000 bp from the TSS at levels above the threshold of OCR-RMA ⩾15 (Supplementary Tables S5 and S6). One additional MDSR gene showed a Myc binding peak just outside the ±1-kb boundary and was thus reassigned as direct Myc-target (Tardbp, +1061 bp). Forty-five genes showed a peak within the same window but below the threshold, and 19 scored as devoid of any peak. Of the latter, 13 did not score as nearer neighbors of any peak in our ChIP-seq data set (and were thus absent from Supplementary Table S4), whereas six were nominally associated with a peak, but at much larger distances (between −11 and −73 kb, or +20 and +250 kb). The corresponding peaks were located either within a CpG island or near a more recently identified TSS (data not shown), indicating that they belonged to other loci, rather than to distal enhancers of these MDSR genes. We conclude that MDSR genes without a TSS-associated peak (19/298 or 6.4%) were indirectly regulated by Myc. Altogether Myc directly controls the activity of most MDSR genes, and does so through proximal promoter elements.
In our whole ChIP-seq data set, 4284 of 19157 annotated promoters (22.4%) scored as Myc-bound in serum-stimulated fibroblasts (with OCR-RMA ⩾15; Supplementary Table S6). MDSR loci represented a minor fraction of all Myc-bound promoters (234/4284 or 5.5%) but, most remarkably, also showed significantly higher Myc-binding signals (Figure 4c). Myc also associated with 47.8% (138/289) and 44.4% (280/631) of the MISR and NRS promoters, respectively (Supplementary Table S6), but unlike MDSR promoters these did not show elevated signals (Figure 4c). Most importantly, however, efficient binding by Myc was not a unique characteristic of MDSR promoters, as shown by the existence of 650 ‘high-affinity non-MDSR' peaks (Figure 4c). The latter were operationally defined by elevated Myc binding (OCR-RMA ⩾35.5 or the top 15% of all peaks in serum-stimulated cells) and their association with MISR promoters (23 peaks), NRS promoters (60 peaks) or promoters not listed in either category (567 peaks; Supplementary Table S4): accounting for bi-directional promoters, these sites were associated with a total of 718 genes (listed in Supplementary Table S6). Hence Myc-dependent regulation was not determined solely by Myc-binding affinity.
ChIP-seq reveals the hierarchical determinants of Myc–DNA interactions in vivo
CpG islands are preferred domains for deposition of the histone methylation mark H3K4me3 (Bernstein et al., 2006; Guenther et al., 2007) owing to their direct recognition by Cfp1, a subunit of the Setd1 methyl-transferase complex (Thomson et al., 2010). Consistent with this association, Myc-binding sites are preferentially located within CpG islands (Fernandez et al., 2003; Zeller et al., 2006) and are enriched for H3K4me3 (Guccione et al., 2006; Kim et al., 2008). We previously proposed that recognition of this active chromatin environment constitutes the limiting step for target recognition in vivo (Guccione et al., 2006). An untested corollary of this mechanism was that, even in the absence of a high-affinity binding site, Myc should be detectable at basal levels on promoters with a permissive chromatin environment. Taking CpG islands as surrogate markers for active chromatin allowed us to validate this prediction in our data set: first, relative to promoters devoid of any Myc-binding peak (Group-v.; Table 1), those with a Myc peak below threshold (OCR-RMA <15; Group-iv.) were already enriched for CpG islands, although not for the E-box consensus. Second, the E-box was enriched at intermediate levels in the promoters bound by Myc with low-to-medium affinity (OCR-RMA 15–35.5: Group-iii.) and at higher levels in both the MDSR and high-affinity non-MDSR groups (i./ii.). Another corollary was that, if Myc associates with transcriptionally active or poised promoters, loci with increasing Myc-binding efficiencies (as assessed following serum stimulation) should show correspondingly higher median mRNA levels prior to Myc activation (that is, before serum stimulation): analysis of our data confirmed this trend (Supplementary Figure S8). Altogether, these observations are fully consistent with the concept that Myc initially recognizes CpG island-associated chromatin in a non-sequence-specific manner, most likely involving interactions with other chromatin-associated proteins, followed by sequence specific DNA binding.
Effect of core E-box variants and flanking nucleotides on DNA binding in vivo
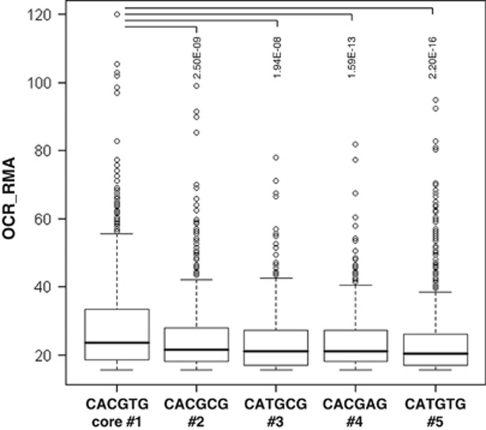
We exploited our ChIP-seq data set to further address the sequence specificity of Myc–DNA interactions in vivo. First, we addressed the enrichment of all possible hexamers in promoters bound by Myc with low-to-intermediate (OCR-RMA 15–35.5) or high affinity (OCR-RMA >35.5) (Supplementary Table S7A). The consensus CACGTG (henceforth numbered #1) was the most significantly enriched motif in both groups. Four core variants, CACGCG, CATGCG, CACGAG and CATGTG (henceforth #2–5), were also enriched significantly, albeit at lower levels, most of the high-affinity binding by Myc being associated with the presence of the canonical core #1 (Figure 5). Remarkably, the same variant motifs were identified previously as Myc/Max-binding sites based on oligonucleotide selection in vitro (Blackwell et al., 1993) and sequencing of 27 sites recovered by ChIP (Grandori et al., 1996). Other significantly enriched hexamers (Supplementary Table S7A) contained partial matches to the above core motifs (in particular half-sites), and in a few cases the Ets motif (see below), but none of the 13 remaining CANNTG E-box variants were enriched.
Figure 5.
Relative Myc-binding affinity for the different E-box core motifs. The DNA sequence in the window ±250 bp from each Myc-binding peak in serum-treated cells was analyzed. Peaks with OCR-RMA <15 were not included. Only sequences containing exactly one type of core E-box variant (#1–5) were considered. The distribution of OCR-RMA values associated with each peak is shown as box plots, along with t-test P-values relative to core #1.
We then addressed the effect of the nucleotides flanking either side of the core motifs #1–5: For this analysis, each one of the positions at ±1, ±2 or ±3 was considered independently from the others, thus reflecting the tendency of each core to enrich for a given base at each of those positions (Supplementary Table S7B–D; summarized in Table 2). Core #1 was the most tolerant at the proximal positions (±1), with a strong preference for C or G, and to a lower extent for A on the 5′ side (and their complement on the 3′), in various combinations. Sites with T before or A after the core were not enriched, consistent with experiments using in vitro oligonucleotide selection with Myc/Max dimers (Solomon et al., 1993) or yeast as a heterologous in vivo system (Fisher et al., 1993). Although with different levels of significance, Myc binding to core #1 enriched for any combination of bases at positions ±2 and ±3, allowing a simple consensus with this canonical E-box (Table 2). The core variants #2–5 allowed fewer combinations of flanking nucleotides at either position (Table 2 and Supplementary Table S7): at position ±1 for example, the preferred combination was C-core-G for core #4 and G-core-C/T for core #3. In two instances (position ±2 with core #2 and position ±3 with core #5), the base on one side influenced that on the other, resulting in different combinations of preferred bases (Table 2). Most remarkably, several of the motifs that were not enriched among high-affinity targets were still enriched among the low-affinity targets (for example, TCACGTGG or GGACGCGC; Supplementary Table S7B). Altogether, our data indicate that variations in the core allow efficient binding, provided the presence of a restricted set of flanking bases. This accounts for the fact that the canonical CACGTG consensus is present in only 40–60% of all Myc-binding sites in vivo (Table 1).
Table 2. Core E-box motifs with flanking nucleotides at each of the indicated positions were analyzed for enrichment in high-affinity peaks (OCR-RMA ⩾35.5) as described in Supplementary Table S7.
| Core: | CACGTG (#1) | CACGCG (#2) | CATGCG (#3) | CACGAG (#4) | CATGTG (#5) |
|---|---|---|---|---|---|
| Positions ±1 | VCACGTGB | SCACGCGG | GCATGCGY | CCACGAGG | CCATGTGC |
| Positions ±2 | N-CACGTG-N | C-CACGCG-M A-CACGCG-C T-CACGCG-T | C-CATGCG-A | A-CACGAG-C | W-CATGTG-T |
| Positions ±3 | N--CACGTG--N | A--CACGCG--C | G--CATGCG--R | G--CACGAG--C | T--CATGTG--C G--CATGTG--T |
| consensus | NNVCACGTGBNN | ACSCACGCGGMC AASCACGCGGCC ATSCACGCGGTC | GCGCATGCGYAR | GACCACGAGGCC | TWCCATGTGCTC GWCCATGTGCTT |
Abbreviations: OCR, overlap count-resampled; RMA, robust multi-array average.
Motifs with an enrichment P-value ⩽0.001 were considered as significantly enriched and are shown using IUPAC codes.
Effect of DNA-binding sites on gene regulation
We then addressed whether the Myc-dependent serum response—as opposed to DNA binding per se—might be determined by distinct sequence elements in target promoters. The Myc-bound MDSR promoters and the high-affinity non-MDSR promoters showed very similar percentages of either the consensus E-box (CACGTG) or CpG islands (groups i. and ii.; Table 1). Both of these features were significantly enriched relative to the promoters with no Myc-binding peak in ChIP-seq (Group-v.), fully consistent with their roles in determining Myc–DNA interactions in vivo (see below). Most importantly here, our data also demonstrate that these features are insufficient to determine Myc-dependent regulation. In addition, the binding sites (defined as Position Weight Matrices) that were enriched among the Myc-bound MDSR promoters were also present in the high-affinity non-MDSR group (Supplementary Table S8, see legend). Thus, at this level of resolution the differential regulation of MDSR genes could be attributed neither to preferential DNA binding by Myc, nor to the presence of secondary transcription factor-binding sites.
It is noteworthy that other motifs, in particular ETS consensus sites, were also enriched at Myc-binding sites (Supplementary Tables S7A and S8), suggesting that Myc may positively interact with other factor(s) for DNA binding. The molecular basis for this association remains to be addressed.
Finally, the binding of Myc to negatively regulated genes deserves a note here: the Myc-bound fraction for these genes is lower than in the other groups, being 23% (15/65) and 22% (29/130) for the super-induced and serum repressed Myc-dependent promoters, respectively (Supplementary Table S6). Motif analysis for the genes super-induced without Myc failed to find a significant enrichment for Myc-binding sites, whereas it revealed two motifs in common with the list of the high-affinity non-MDSR group (Klf7 and Sp4) and three additional motifs, NF-Y, TBP and SRF (Supplementary Table S8). These observations point to possible roles of the latter factors in negative gene regulation by Myc—consistent with previous observations for NF-Y (Izumi et al., 2001)—but given the lower rate of RT–PCR validation for the negatively regulated groups in our experiments (see above), their relevance remains to formally be addressed.
MDSR genes are enriched for specific regulatory pathways
Gene Ontology (GO) analysis with the NIH-DAVID suite (http://david.abcc.ncifcrf.gov/home.jsp) (Dennis et al., 2003) and with Ingenuity Pathway Analysis 8.6 (http://www.ingenuity.com) consistently indicated that the MDSR group was enriched for genes involved in metabolic processes, nucleotide/DNA/RNA metabolism, ribosome biogenesis and translation, RNA/rRNA/tRNA processing, gene expression, DNA replication and cell-cycle control (Supplementary Table S9). In line with previous observations, MDSR genes included key enzymes in purine and pyrimidine biosynthetic pathways, including Prps1, Ppat, Gart, Pfas, Impdh2, Umps and Rrm2 (Liu et al., 2008), as well as genes involved in one-carbon pool and methionine metabolism (Nikiforov et al., 2002). Other MDSR genes encoded proteins directly involved in DNA replication, including four subunits of the Mcm helicase complex (Blow and Dutta, 2005; Blow and Tanaka, 2005) as well as Dbf4, the regulatory subunit of the Cdc7 kinase, a regulator of Mcm activity (Masai et al., 2005). Thus, in this experimental setting, Myc regulated a substantial group of genes involved in cell-cycle progression, cell growth, RNA, DNA and nucleotide metabolism, and DNA replication.
MDSR gene products also included three subunits of the exosome complex, which has a central role in mRNA decay/surveillance and in the processing of rRNA, snoRNA and snRNA molecules (Vanacova and Stefl, 2007; Schmid and Jensen, 2008). The mammalian exosome has a ring-shaped part, composed of six different polypeptides, and a top cap composed of three proteins, required for the stability and function of the whole structure (Liu et al., 2006; Shen and Kiledjian, 2006). Exosc8 (or Rrp43) in the ring-shaped moiety, and Exosc2 (or Rrp4) and Exosc3 (or Rrp40) in the cap, were encoded by MDSR genes suggesting that Myc activity may be limiting for exosome function.
Relationship between the MDSR signature and other Myc-associated profiles
To address whether the gene lists identified here were significantly represented in previous data sets, we selected 85 relevant publications and used meta-analysis to determine significant overlaps (Supplementary Table S10). The list of MDSR genes showed statistically significant overlaps with a variety of Myc-associated expression profiles in either cultured cells or transgenic mice, but very poor overlap with others, consistent with the concept that Myc-regulated transcription is largely context-dependent. The same is true for the list of genes super-induced without Myc, which overlaps with statistical significance with some lists of Myc-repressed genes but not with others.
Of particular relevance to the present study, the response to serum in human fibroblasts was functionally related to a wound-healing response (Iyer et al., 1999), a serum/wound-like profile was enriched in a variety of tumors (Chang et al., 2004) and overexpressed Myc and CSN5 were identified as coordinate inducers of this profile (Adler et al., 2006). Of the 234 Myc-bound MDSR genes identified here, 17 where also induced by serum in human fibroblasts in one study (Chang et al., 2004) (Supplementary Table S10, list 14737219_1), representing a moderate but statistically significant enrichment (of note, the significance of this overlap increased if we considered all our serum-induced genes, that is, MDSR, MISR and super-induced without Myc). However, only four of these genes were part of the Myc/CSN5-induced wound-healing profile (list 16518402_1), pointing to significant differences in the programs driven by endogenous and overexpressed Myc in those settings. Another serum-response profile in human cells (Iyer et al., 1999) showed no significant overlap with our MDSR group.
Myc was identified as a driver of a cancer-associated, ESC-like signature in either human or mouse (Wong et al., 2008): the mouse ESC-like module (Supplementary Table S10, list 18397753_1) as well as the human/mouse module (list 18397753_4) were enriched among our MDSR genes, in either the bound or unbound subgroups. Thus, the ‘ESC-like' module appears highly related to the function of Myc during serum stimulation. It has been shown, however, that this original ‘ESC-like' module is unrelated to the program regulated by pluripotency factors in ES cells, but rather corresponds to a Myc program common to ES and cancer cells (Kim et al., 2010). Indeed, our MDSR genes showed significant overlap with the Myc module, but not with the core pluripotency model of the latter study (Supplementary Table S10, lists 20946988_1 and 3).
Myc is also required for the maintenance of pluripotency in ES cells (Cartwright et al., 2005) and when co-expressed with the ES-specific factors Oct3/4, Sox2 and Klf4, can contribute to reprogram fibroblasts to an ‘induced pluripotent state' (iPS) analogous to that of ES cells (Takahashi and Yamanaka, 2006). Myc binding in our work (whether total or the MDSR subset) showed very significant overlaps with the Myc-binding profiles in ES cells (lists 19030024_1, 18555785_1, 18358816_9, 19167336_3, 18555785_2 and 19079543_1), as well as in fibroblasts either fully or partially reprogrammed to the induced pluripotent state (lists 19167336_7 and 19167336_11) (Sridharan et al., 2009). These overlaps in binding profiles are summarized in Supplementary Table S11. However, we also note that a set of genes reported to be induced by Myc activation in mouse embryonic fibroblasts (Sridharan et al., 2009) showed overlap neither with our MDSR genes (Supplementary Table S10, list 19167336_20) nor with our own Myc activation profile in mouse embryonic fibroblasts (unpublished data), calling for a re-evaluation of this issue in future work. In particular, it remains unclear whether Myc contributes a true ‘reprogramming' activity or, most likely, endows cells with a growth state ideal for reprogramming by the other factors, which constitutes the common denominator of its activities in ES cells, iPS reprogramming and cancer cells.
Another interesting observation is the overlap of MDSR genes with clusters of genes regulated by serum in mouse embryonic fibroblasts in different cell-cycle phases (Ishida et al., 2001). Myc-bound MDSR genes specifically overlapped with the ‘G1 growth' group of genes that were induced in G1 and remained constant through the following phases, as well as with the ‘G1/S growth' and ‘G1/S cycle' groups, which were both induced at the G1/S transition and either remained constant later, or declined and were re-induced at the following G1/S transition (Supplementary Table S10, lists 11416145_4, 11416145_5 and 11416145_6). The ‘G1/S growth' and ‘G1/S cycle' clusters, like the MDSR group, were enriched for genes involved in transcription, RNA metabolism and DNA replication. Instead, we did not see a significant overlap between MDSR genes and the subset of E2F-regulated ‘G1/S cycle' genes described in the same work (list 11416145_8). Consistent with this finding, several E2F-bound or regulated lists of genes considered in our meta-analysis showed no significant overlap with our MDSR genes, and we observed no enrichment of the E2F consensus-binding site at Myc-bound promoters (data not shown). Thus, although cooperativity on a restricted subset of target genes remains fully plausible (Leung et al., 2008), our data do not support a general cross-talk between Myc and E2F-family proteins. Instead our group of genes super-induced without Myc shows a significant overlap with the ‘Early G1 cluster' and ‘G1 cycle cluster' groups, confirming the relevance of negative gene regulation by Myc during cell-cycle re-entry.
Functional screening of MDSR genes
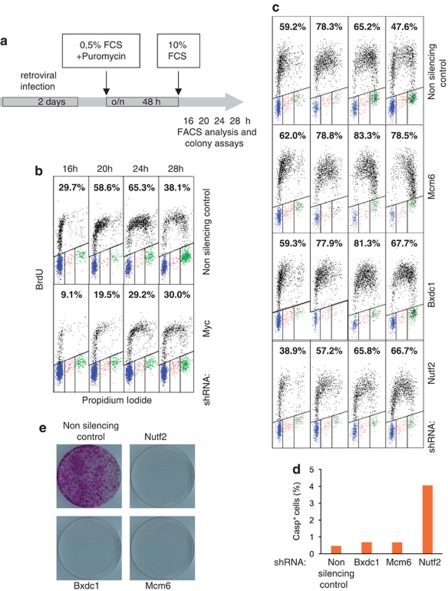
Conditional deletion experiments show that c-myc is required for cells to enter the cell cycle and reach S-phase following mitogenic stimulation (Figure 1d) (Trumpp et al., 2001). Therefore, MDSR genes—or a subset of them—are likely to be important for G0→S-phase progression. To address this question, near-confluent c-mycf/f 3T9 cells (the same cells used above, but without CreER) were infected with a recombinant lentivirus or retrovirus expressing a specific small-hairpin RNA (shRNA), after which the cells were concomitantly selected in puromycin and serum-starved for 48 h (Figure 6a). In this manner, RNA interference was induced while the cells were becoming quiescent (through starvation and contact inhibition), circumventing prior antiproliferative defects. The cells were then re-seeded in serum-containing medium and harvested after 16, 20, 24 and 28 h: just before harvesting, the cells were exposed to a 20-min pulse of 5-bromodeoxyuridine (BrdU) incorporation for flow-cytometric analysis of DNA synthesis. In parallel, plates were seeded at lower densities for colony formation assays. A pilot experiment with an shRNA insert targeting c-myc (which achieved 88% knockdown of the mRNA; data not shown) showed reduced and delayed progression into the S and G2/M phases relative to cells infected with a control vector (Figure 6b).
Figure 6.
Some MDSR genes are required for entry/progression into S-phase following serum stimulation. (a) A schematic representation of the experimental approach: cells infected with shRNA vectors for the genes indicated were starved for 2 days and then stimulated with serum. (b, c) Two-dimensional fluorescence-activated cell sorter profiles of cells stimulated for the indicated time period are shown, along with the percent of BrdU-positive cells measured at each time point. (d) Percentages of apoptotic cells, as assayed by cleaved caspase-3 staining and flow cytometry, following knockdown of the indicated genes and 24 h of serum stimulation. (e) Cells were infected with the indicated shRNA retroviruses or with a non-silencing control shRNA. Puromycin-selected cells were trypsinized and plated in duplicate for colony formation assay. After 10–12 days of culture, cells were fixed and stained with crystal violet. A representative experiment is shown.
In order to target individual MDSR genes, we used shRNA-mir inserts, which mimic the structure of natural primary microRNA transcripts (Silva et al., 2005). A series of inserts targeting 29 of our MDSR genes was initially obtained in the second-generation lentiviral vector pGIPZ (Thermo Fisher Scientific, Huntsville, AL, USA): however, this caused a suboptimal knockdown of all target mRNAs in 3T9 fibroblasts. Upon subcloning in the retroviral vector MLP (Dickins et al., 2005), the same shRNA-mir inserts effectively knocked down their target mRNAs (Supplementary Table S12). Knockdown of three MDSR genes, Mcm6, Bxdc1 and Nutf2, caused defects in entry/progression into S-phase following serum stimulation (Figure 6c). Closer examination of fluorescence-activated cell sorter profiles showed that Mcm6 knockdown prevented neither entry, nor early progression (until 20 h), but prevented the completion of DNA replication. Knockdown of Bxdc1 also did not impair entry, but caused a delay of ∼4 h in S-phase progression, whereas that of Nutf2 delayed both S-phase entry and progression, suggesting a defect in the G0–G1 and/or G1–S transitions. Cleaved caspase-3 staining profiles determined 28 h following serum stimulation revealed enhanced apoptosis in Nutf2-targeted cells, but not in the others (Figure 6d). Knockdown of the three genes also heavily impaired colony outgrowth (Figure 6e). The above data were reproduced with two independent shRNA-mir hairpins for either Mcm6 or Bxdc1 (data not shown). Of three additional Myc-target genes tested, Cad also showed a requirement for S-phase entry but Odc1 and Cul1 did not (Supplementary Table S11), contrasting previous conclusions on Cul1 (O'Hagan et al., 2000). Finally, targeting 11 MDSR genes (Serbp1, Cct7, Prmt3, Sfrs7, Mybbp1a, Hspd1, Gart, Thop1, Gpatch4, Mcm5 and Exosc2) caused no delay in the first S-phase, but impaired colony formation (data not shown; Supplementary Table S11), implying a defect in proliferation and/or survival at later stages, whereas 15 other MDSR genes showed no obvious requirement in our experiments.
Discussion
We have combined mRNA profiling and genome-wide mapping of Myc–DNA interactions to identify MDSR genes in mouse fibroblasts. For a gene to be included in the MDSR group, its mRNA had to be induced by at least 1.5-fold either 4 or 8 h following serum stimulation, and its response had to be reduced by 1.5-fold or more in the absence of Myc, as determined in our microarray data set (see Materials and methods). Validation by quantitative RT–PCR showed that MDSR genes were identified with high confidence by these criteria. Conceivably, subtler regulation of a larger number of genes may also be a relevant aspect of Myc's activity, but is below the threshold of confidence for microarray analysis. As such, our list of 298 MDSR genes can be considered as the core mRNA program controlled by Myc upon mitogenic stimulation in mouse fibroblasts.
Consistent with the fact that Myc can also repress transcription (Adhikary and Eilers, 2005), we confirmed the existence of genes that are either repressed in a Myc-dependent manner, or super-induced in the absence of Myc. As the rates of RT–PCR validation for these gene groups in our data set were lower than for the Myc-activated MDSR genes, we concentrated here on the latter. Another aspect that remains to be addressed in the context of mitogenic stimulation is the role of Myc in regulating microRNAs, a phenomenon that is central to Myc's transforming activity (Klapproth and Wirth, 2010).
Our ChIP-seq analysis revealed that about half of all Myc-binding sites in the genome clustered in the interval between −1000 and +1000 bp from a TSS. As many as 78.5% (234/298) of the MDSR promoters—against 22.4% (4284/19157) of all the promoters identified in our data set—were bound by Myc within this interval, implying that most MDSR genes were directly regulated by Myc. Those genes represented a minor part (234/4284 or 5.5%) of all the genes targeted by Myc in the genome, but on average were bound with much higher efficiency, raising the possibility that Myc-binding levels determined Myc-dependent regulation. This was ruled out, however, as we were able to identify 718 non-MDSR genes that were bound by Myc at 650 different sites as efficiently—if not better—as most MDSR genes. Thus, efficient DNA binding by Myc did not automatically lead to Myc-dependent transcriptional activation.
All of the promoters with high affinity for Myc (whether MDSR or not) showed frequent occurrences of CpG islands and E-boxes, indicating that those sequence elements determine promoter recognition, as expected (Fernandez et al., 2003; Zeller et al., 2006), but not gene regulation per se. Our data are also consistent with the concept that promoter recognition by Myc initially occurs through non-sequence specific interactions with active chromatin domains (Guccione et al., 2006), followed in a second step by direct DNA binding. Relative to promoters with no trace of Myc binding (that is, no peak at all in ChIP-seq), those with low levels of Myc interaction (OCR-RMA ⩽15) already showed significant enrichment for CpG islands, most likely reflecting the permissive chromatin configuration generally present at those domains. The E-box, on the other hand, was most significantly enriched in the promoters with the highest Myc levels, reflecting the sequence-specific interaction of Myc with those sites.
Our ChIP-seq data also indicated that several variants of the core E-box and flanking nucleotides are tolerated in vivo (Table 2), largely confirming predictions made based on DNA binding in vitro or in yeast (Blackwell et al., 1993; Grandori et al., 1996). Remarkably, the core hexamer influenced the preferred flanking nucleotides (analyzed here independently for positions ±1, ±2 and ±3). In the presence of a canonical CACGTG element (core #1), T and A were strongly counter-selected at the −1 and +1 positions, respectively (Fisher et al., 1993; Solomon et al., 1993) but other bases were all tolerated, and positions ±2 and ±3 had no significant influence on Myc binding. In the presence of variant cores, the consensus at all flanking positions was more restrictive, with fewer variants being allowed. In two instances (position ±2 with core #2 and position ±3 with core #5) distinct combinations were preferred, the base on one side influencing that on the other (Table 2). It is noteworthy that the sub-optimal variant TCACGTGA is the preferred consensus for TFE3, a bHLH-LZ factor that regulates lysosomal genes (Sardiello et al., 2009), indicating that flanking nucleotides are important in determining the selectivity of bHLH-family proteins that bind to the same E-box toward distinct groups of genes.
Our data directly illustrate the fact that Myc binds to a large number of genes, but is critical for the regulation of only a subset of those genes in any given physiological context. We speculate that differences in environment, signaling events and/or cell type will determine the regulation of alternative subsets of target genes, possibly with relatively unchanged DNA-binding profiles. An example is provided by Snai1, a gene activated by transforming growth factor-β in a Myc-dependent manner in mammary epithelial cells: Snai1 was bound by Myc also in fibroblasts, but showed a Myc-independent serum response (MISR; Supplementary Table S4 and Supplementary Figure S3) (Smith et al., 2009). Five genes that were co-regulated by Myc and the estrogen receptor in human mammary epithelial cells (GABPB2, HK2, RCC2, SAMHD1 and SYVN1) (Cheng et al., 2006) were also bound by Myc in mouse fibroblasts, but were not found in the MDSR group. In addition, target genes showing no Myc-dependent regulation in a given physiological setting may respond to the oncogenic activation of Myc in the same cell type: for example, more than 200 of the Myc-bound non-MDSR genes identified here were induced upon ectopic Myc activation in mouse embryonic fibroblasts (unpublished data).
The MDSR group was enriched for genes involved in metabolic and biosynthetic processes, with particular representation of genes favoring ribosome biogenesis, protein biosynthesis, nucleotide biosynthesis and DNA replication (see section Results), consistent with previous lists of Myc-regulated genes and with the critical roles of Myc in these processes (Schlosser et al., 2003; Barna et al., 2008; Liu et al., 2008; Dang et al., 2009; Herold et al., 2009; van Riggelen et al., 2010), and in particular in the regulation of cell growth (Mateyak et al., 1997; Iritani and Eisenman, 1999; Schuhmacher et al., 1999).
In order to address the function of MDSR genes, we knocked down 29 individual mRNAs in quiescent cells: three of these MDSR gene products were required for normal S-phase entry or progression following serum stimulation, 11 were needed to sustain colony formation, whereas 15 others showed no obvious requirement in our experiments (Supplementary Table S12). Of particular interest, several subunits of the Mcm complex were included in our MDSR list: of these, we targeted Mcm5 and Mcm6 by RNA interference. Neither of these proteins was required for S-phase entry and early progression following serum stimulation. This is in line with the notion that the Mcm complex (Mcm2–7), which is required for origin licensing, is pre-loaded on replication origins in the precedent mitosis (Blow and Dutta, 2005; Blow and Tanaka, 2005). Therefore, after G0 exit, Mcm proteins do not need to be synthesized de novo for origin firing in the first S-phase. The Mcm complex is also required for the progression of replication forks (Blow and Dutta, 2005; Blow and Tanaka, 2005). In our hands, Mcm6 but not Mcm5 knockdown prevented the completion of S-phase, suggesting that Mcm6 may turn over more rapidly, and that its synthesis becomes limiting already during the first cell cycle. As expected, both Mcm5 and Mcm6 were essential for subsequent proliferation, as judged by the colony formation assays.
The two other MDSR genes showing a requirement in the first S-phase were Bxdc1 and Nutf2. The Bxdc1 product is a Brix-domain protein that localizes to the nucleolus in an RNA-dependent manner and participates in rRNA maturation (Hirano et al., 2009). Nutf2, on the other hand, mediates the nuclear import of the Ran GTPase (Ribbeck et al., 1998; Smith et al., 1998), a key regulator of nucleocytoplasmic trafficking (Stewart, 2007). Nutf2 is essential for viability both in Caenorhabditis elegans and Saccharomyces cerevisiae, and in the latter its interaction with Ran is required for cell-cycle progression (Corbett and Silver, 1996; Quimby et al., 2000a, 2000b).
In summary, we have identified a group of 298 MDSR genes, which constitutes the core mRNA expression program driven by Myc in this physiological setting. This group is highly consistent with the role of Myc in cell-cycle entry, cell growth, protein biosynthesis and DNA replication. We surmise that concerted upregulation of these genes is critical for the optimal response of cells to mitogenic stimuli, as well as for sustained cell growth and proliferation. The binding of Myc to a much larger number of loci, and the concept that different subsets of target genes are regulated by Myc under different physiological conditions, permit rationalizing the involvement of this transcription factor in diverse signaling pathways and biological responses. This complexity may also underlie the involvement of Myc as a driving oncogene in a variety of neoplasias.
Materials and methods
Details on Materials and methods are provided in full detail in the Supplementary information.
Acknowledgments
We thank Gioacchino Natoli for critical discussion; Lucilla Luzi for help with data analysis; Ivan Muradore and Simona Ronzoni for help with imaging; Simone Minardi and Davide Cittaro for help with microarrays; Loris Bernard, Valentina Dall' Olio and Laura Tizzoni for help with real-time PCR; and Atif Shahab and CheeSeng Chan for technical guidance on processing and mapping sequencing data. We also acknowledge the Genome Technology and Biology Group at the Genome Institute of Singapore, in particular the GIS sequencing team, for technical support. Part of this work was supported by grants from the Agency for Science, Technology and Research (A*STAR) of Singapore and NIH ENCODE to C-LW, and from the Swiss National Science Foundation to AT. Work in the Amati lab was supported by grants from the EU-FP7 Program EuroSyStem, the Association for International Cancer Research (AICR), the Italian health ministry and the Italian Association for Cancer Research (AIRC).
The authors declare no conflict of interest.
Footnotes
Supplementary Information accompanies the paper on the Oncogene website (http://www.nature.com/onc)
Supplementary Material
References
- Adhikary S, Eilers M. Transcriptional regulation and transformation by Myc proteins. Nat Rev Mol Cell Biol. 2005;6:635–645. doi: 10.1038/nrm1703. [DOI] [PubMed] [Google Scholar]
- Adler AS, Lin MF, Horlings H, Nuyten DS, van de Vijver MJ, Chang HY. Genetic regulators of large-scale transcriptional signatures in cancer. Nat Genet. 2006;38:421–430. doi: 10.1038/ng1752. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Amundson SA, Zhan Q, Penn LZ, Fornace AJ., Jr Myc suppresses induction of the growth arrest genes gadd34, gadd45, and gadd153 by DNA-damaging agents. Oncogene. 1998;17:2149–2154. doi: 10.1038/sj.onc.1202136. [DOI] [PubMed] [Google Scholar]
- Barna M, Pusic A, Zollo O, Costa M, Kondrashov N, Rego E, et al. Suppression of Myc oncogenic activity by ribosomal protein haploinsufficiency. Nature. 2008;456:971–975. doi: 10.1038/nature07449. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bernstein BE, Mikkelsen TS, Xie X, Kamal M, Huebert DJ, Cuff J, et al. A bivalent chromatin structure marks key developmental genes in embryonic stem cells. Cell. 2006;125:315–326. doi: 10.1016/j.cell.2006.02.041. [DOI] [PubMed] [Google Scholar]
- Blackwell TK, Huang J, Ma A, Kretzner L, Alt FW, Eisenman RN, et al. Binding of myc proteins to canonical and noncanonical DNA sequences. Mol Cell Biol. 1993;13:5216–5224. doi: 10.1128/mcb.13.9.5216. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Blow JJ, Dutta A. Preventing re-replication of chromosomal DNA. Nat Rev Mol Cell Biol. 2005;6:476–486. doi: 10.1038/nrm1663. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Blow JJ, Tanaka TU. The chromosome cycle: coordinating replication and segregation. Second in the cycles review series. EMBO Rep. 2005;6:1028–1034. doi: 10.1038/sj.embor.7400557. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bouchard C, Dittrich O, Kiermaier A, Dohmann K, Menkel A, Eilers M, et al. Regulation of cyclin D2 gene expression by the Myc/Max/Mad network: Myc-dependent TRRAP recruitment and histone acetylation at the cyclin D2 promoter. Genes Dev. 2001;15:2042–2047. doi: 10.1101/gad.907901. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bouchard C, Marquardt J, Bras A, Medema RH, Eilers M. Myc-induced proliferation and transformation require Akt-mediated phosphorylation of FoxO proteins. EMBO J. 2004;23:2830–2840. doi: 10.1038/sj.emboj.7600279. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bush A, Mateyak M, Dugan K, Obaya A, Adachi S, Sedivy J, et al. c-myc null cells misregulate cad and gadd45 but not other proposed c-Myc targets. Genes Dev. 1998;12:3797–3802. doi: 10.1101/gad.12.24.3797. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cartwright P, McLean C, Sheppard A, Rivett D, Jones K, Dalton S. LIF/STAT3 controls ES cell self-renewal and pluripotency by a Myc-dependent mechanism. Development. 2005;132:885–896. doi: 10.1242/dev.01670. [DOI] [PubMed] [Google Scholar]
- Cawley S, Bekiranov S, Ng HH, Kapranov P, Sekinger EA, Kampa D, et al. Unbiased mapping of transcription factor binding sites along human chromosomes 21 and 22 points to widespread regulation of noncoding RNAs. Cell. 2004;116:499–509. doi: 10.1016/s0092-8674(04)00127-8. [DOI] [PubMed] [Google Scholar]
- Chang HY, Sneddon JB, Alizadeh AA, Sood R, West RB, Montgomery K, et al. Gene expression signature of fibroblast serum response predicts human cancer progression: similarities between tumors and wounds. PLoS Biol. 2004;2:E7. doi: 10.1371/journal.pbio.0020007. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chen X, Xu H, Yuan P, Fang F, Huss M, Vega VB, et al. Integration of external signaling pathways with the core transcriptional network in embryonic stem cells. Cell. 2008;133:1106–1117. doi: 10.1016/j.cell.2008.04.043. [DOI] [PubMed] [Google Scholar]
- Cheng AS, Jin VX, Fan M, Smith LT, Liyanarachchi S, Yan PS, et al. Combinatorial analysis of transcription factor partners reveals recruitment of c-MYC to estrogen receptor-alpha responsive promoters. Mol Cell. 2006;21:393–404. doi: 10.1016/j.molcel.2005.12.016. [DOI] [PubMed] [Google Scholar]
- Corbett AH, Silver PA. The NTF2 gene encodes an essential, highly conserved protein that functions in nuclear transport in vivo. J Biol Chem. 1996;271:18477–18484. doi: 10.1074/jbc.271.31.18477. [DOI] [PubMed] [Google Scholar]
- Cotterman R, Jin VX, Krig SR, Lemen JM, Wey A, Farnham PJ, et al. N-Myc regulates a widespread euchromatic program in the human genome partially independent of its role as a classical transcription factor. Cancer Res. 2008;68:9654–9662. doi: 10.1158/0008-5472.CAN-08-1961. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dang CV, Le A, Gao P. MYC-induced cancer cell energy metabolism and therapeutic opportunities. Clin Cancer Res. 2009;15:6479–6483. doi: 10.1158/1078-0432.CCR-09-0889. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dennis G, Jr, Sherman BT, Hosack DA, Yang J, Gao W, Lane HC, et al. DAVID: Database for Annotation, Visualization, and Integrated Discovery. Genome Biol. 2003;4:P3. [PubMed] [Google Scholar]
- Dickins RA, Hemann MT, Zilfou JT, Simpson DR, Ibarra I, Hannon GJ, et al. Probing tumor phenotypes using stable and regulated synthetic microRNA precursors. Nat Genet. 2005;37:1289–1295. doi: 10.1038/ng1651. [DOI] [PubMed] [Google Scholar]
- Eilers M, Eisenman RN. Myc's broad reach. Genes Dev. 2008;22:2755–2766. doi: 10.1101/gad.1712408. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Eisen MB, Spellman PT, Brown PO, Botstein D. Cluster analysis and display of genome-wide expression patterns. Proc Natl Acad Sci U S A. 1998;95:14863–14868. doi: 10.1073/pnas.95.25.14863. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ellwood-Yen K, Graeber TG, Wongvipat J, Iruela-Arispe ML, Zhang J, Matusik R, et al. Myc-driven murine prostate cancer shares molecular features with human prostate tumors. Cancer Cell. 2003;4:223–238. doi: 10.1016/s1535-6108(03)00197-1. [DOI] [PubMed] [Google Scholar]
- Fernandez P, Frank SR, Wang L, Schroeder M, Liu S, Greene J, et al. Genomic targets of the human c-Myc protein. Genes Dev. 2003;17:1115–1129. doi: 10.1101/gad.1067003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fisher F, Crouch DH, Jayaraman PS, Clark W, Gillespie DAF, Goding CR. Transcription activation by Myc and Max: flanking sequences target activation to a subset of CACGTG motifs in vivo. EMBO J. 1993;12:5075–7082. doi: 10.1002/j.1460-2075.1993.tb06201.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Frank SR, Schroeder M, Fernandez P, Taubert S, Amati B. Binding of c-Myc to chromatin mediates mitogen-induced acetylation of histone H4 and gene activation. Genes Dev. 2001;15:2069–2082. doi: 10.1101/gad.906601. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Grandori C, Mac J, Siebelt F, Ayer DE, Eisenman RN. Myc–Max heterodimers activate a DEAD box gene and interact with multiple E box-related sites in vivo. EMBO J. 1996;15:4344–4357. [PMC free article] [PubMed] [Google Scholar]
- Guccione E, Martinato F, Finocchiaro G, Luzi L, Tizzoni L, Dall' Olio V, et al. Myc-binding-site recognition in the human genome is determined by chromatin context. Nat Cell Biol. 2006;8:764–770. doi: 10.1038/ncb1434. [DOI] [PubMed] [Google Scholar]
- Guenther MG, Levine SS, Boyer LA, Jaenisch R, Young RA. A chromatin landmark and transcription initiation at most promoters in human cells. Cell. 2007;130:77–88. doi: 10.1016/j.cell.2007.05.042. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Herold S, Herkert B, Eilers M. Facilitating replication under stress: an oncogenic function of MYC. Nat Rev Cancer. 2009;9:441–444. doi: 10.1038/nrc2640. [DOI] [PubMed] [Google Scholar]
- Hirano Y, Ishii K, Kumeta M, Furukawa K, Takeyasu K, Horigome T. Proteomic and targeted analytical identification of BXDC1 and EBNA1BP2 as dynamic scaffold proteins in the nucleolus. Genes Cells. 2009;14:155–166. doi: 10.1111/j.1365-2443.2008.01262.x. [DOI] [PubMed] [Google Scholar]
- Iritani BM, Eisenman RN. c-Myc enhances protein synthesis and cell size during B lymphocyte development. Proc Natl Acad Sci U S A. 1999;96:13180–13185. doi: 10.1073/pnas.96.23.13180. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Irizarry RA, Hobbs B, Collin F, Beazer-Barclay YD, Antonellis KJ, Scherf U, et al. Exploration, normalization, and summaries of high density oligonucleotide array probe level data. Biostatistics. 2003;4:249–264. doi: 10.1093/biostatistics/4.2.249. [DOI] [PubMed] [Google Scholar]
- Ishida S, Huang E, Zuzan H, Spang R, Leone G, West M, et al. Role for E2F in control of both DNA replication and mitotic functions as revealed from DNA microarray analysis. Mol Cell Biol. 2001;21:4684–4699. doi: 10.1128/MCB.21.14.4684-4699.2001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Iyer VR, Eisen MB, Ross DT, Schuler G, Moore T, Lee JC, et al. The transcriptional program in the response of human fibroblasts to serum. Science. 1999;283:83–87. doi: 10.1126/science.283.5398.83. [DOI] [PubMed] [Google Scholar]
- Izumi H, Molander C, Penn LZ, Ishisaki A, Kohno K, Funa K. Mechanism for the transcriptional repression by c-Myc on PDGF beta-receptor. J Cell Sci. 2001;114:1533–1544. doi: 10.1242/jcs.114.8.1533. [DOI] [PubMed] [Google Scholar]
- Kawaji H, Severin J, Lizio M, Waterhouse A, Katayama S, Irvine KM, et al. The FANTOM web resource: from mammalian transcriptional landscape to its dynamic regulation. Genome Biol. 2009;10:R40. doi: 10.1186/gb-2009-10-4-r40. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kidder BL, Yang J, Palmer S. Stat3 and c-Myc genome-wide promoter occupancy in embryonic stem cells. PLoS ONE. 2008;3:e3932. doi: 10.1371/journal.pone.0003932. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kim J, Chu J, Shen X, Wang J, Orkin SH. An extended transcriptional network for pluripotency of embryonic stem cells. Cell. 2008;132:1049–1061. doi: 10.1016/j.cell.2008.02.039. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kim J, Woo AJ, Chu J, Snow JW, Fujiwara Y, Kim CG, et al. A Myc network accounts for similarities between embryonic stem and cancer cell transcription programs. Cell. 2010;143:313–324. doi: 10.1016/j.cell.2010.09.010. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Klapproth K, Wirth T. Advances in the understanding of MYC-induced lymphomagenesis. Br J Haematol. 2010;149:484–497. doi: 10.1111/j.1365-2141.2010.08159.x. [DOI] [PubMed] [Google Scholar]
- Knoepfler PS, Zhang XY, Cheng PF, Gafken PR, McMahon SB, Eisenman RN. Myc influences global chromatin structure. EMBO J. 2006;25:2723–2734. doi: 10.1038/sj.emboj.7601152. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Leung JY, Ehmann GL, Giangrande PH, Nevins JR. A role for Myc in facilitating transcription activation by E2F1. Oncogene. 2008;27:4172–4179. doi: 10.1038/onc.2008.55. [DOI] [PubMed] [Google Scholar]
- Li Z, Van Calcar S, Qu C, Cavenee WK, Zhang MQ, Ren B. A global transcriptional regulatory role for c-Myc in Burkitt's lymphoma cells. Proc Natl Acad Sci U S A. 2003;100:8164–8169. doi: 10.1073/pnas.1332764100. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Liu Q, Greimann JC, Lima CD. Reconstitution, activities, and structure of the eukaryotic RNA exosome. Cell. 2006;127:1223–1237. doi: 10.1016/j.cell.2006.10.037. [DOI] [PubMed] [Google Scholar]
- Liu YC, Li F, Handler J, Huang CR, Xiang Y, Neretti N, et al. Global regulation of nucleotide biosynthetic genes by c-Myc. PLoS ONE. 2008;3:e2722. doi: 10.1371/journal.pone.0002722. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mao DY, Watson JD, Yan PS, Barsyte-Lovejoy D, Khosravi F, Wong WW, et al. Analysis of Myc bound loci identified by CpG island arrays shows that Max is essential for Myc-dependent repression. Curr Biol. 2003;13:882–886. doi: 10.1016/s0960-9822(03)00297-5. [DOI] [PubMed] [Google Scholar]
- Marhin WW, Chen S, Facchini LM, Fornace AJ, Jr, Penn LZ. Myc represses the growth arrest gene gadd45. Oncogene. 1997;14:2825–2834. doi: 10.1038/sj.onc.1201138. [DOI] [PubMed] [Google Scholar]
- Martinato F, Cesaroni M, Amati B, Guccione E. Analysis of Myc-induced histone modifications on target chromatin. PLoS ONE. 2008;3:e3650. doi: 10.1371/journal.pone.0003650. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Masai H, You Z, Arai K. Control of DNA replication: regulation and activation of eukaryotic replicative helicase, MCM. IUBMB Life. 2005;57:323–335. doi: 10.1080/15216540500092419. [DOI] [PubMed] [Google Scholar]
- Mateyak MK, Obaya AJ, Adachi S, Sedivy JM. Phenotypes of c-Myc-deficient rat fibroblasts isolated by targeted homologous recombination. Cell Growth Differ. 1997;8:1039–1048. [PubMed] [Google Scholar]
- Nikiforov MA, Chandriani S, O'Connell B, Petrenko O, Kotenko I, Beavis A, et al. A functional screen for Myc-responsive genes reveals serine hydroxymethyltransferase, a major source of the one-carbon unit for cell metabolism. Mol Cell Biol. 2002;22:5793–5800. doi: 10.1128/MCB.22.16.5793-5800.2002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- O'Hagan RC, Ohh M, David G, de Alboran IM, Alt FW, Kaelin WG, Jr, et al. Myc-enhanced expression of Cul1 promotes ubiquitin-dependent proteolysis and cell cycle progression. Genes Dev. 2000;14:2185–2191. doi: 10.1101/gad.827200. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Quimby BB, Lamitina T, L'Hernault SW, Corbett AH. The mechanism of ran import into the nucleus by nuclear transport factor 2. J Biol Chem. 2000a;275:28575–28582. doi: 10.1074/jbc.M005055200. [DOI] [PubMed] [Google Scholar]
- Quimby BB, Wilson CA, Corbett AH. The interaction between Ran and NTF2 is required for cell cycle progression. Mol Biol Cell. 2000b;11:2617–2629. doi: 10.1091/mbc.11.8.2617. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rahl PB, Lin CY, Seila AC, Flynn RA, McCuine S, Burge CB, et al. c-Myc regulates transcriptional pause release. Cell. 2010;141:432–445. doi: 10.1016/j.cell.2010.03.030. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ribbeck K, Lipowsky G, Kent HM, Stewart M, Gorlich D. NTF2 mediates nuclear import of Ran. EMBO J. 1998;17:6587–6598. doi: 10.1093/emboj/17.22.6587. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sardiello M, Palmieri M, di Ronza A, Medina DL, Valenza M, Gennarino VA, et al. A gene network regulating lysosomal biogenesis and function. Science. 2009;325:473–477. doi: 10.1126/science.1174447. [DOI] [PubMed] [Google Scholar]
- Schlosser I, Holzel M, Murnseer M, Burtscher H, Weidle UH, Eick D. A role for c-Myc in the regulation of ribosomal RNA processing. Nucleic Acids Res. 2003;31:6148–6156. doi: 10.1093/nar/gkg794. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Schmid M, Jensen TH. The exosome: a multipurpose RNA-decay machine. Trends Biochem Sci. 2008;33:501–510. doi: 10.1016/j.tibs.2008.07.003. [DOI] [PubMed] [Google Scholar]
- Schuhmacher M, Staege MS, Pajic A, Polack A, Weidle UH, Bornkamm GW, et al. Control of cell growth by c-Myc in the absence of cell division. Curr Biol. 1999;9:1255–1258. doi: 10.1016/s0960-9822(99)80507-7. [DOI] [PubMed] [Google Scholar]
- Seoane J, Le HV, Massague J. Myc suppression of the p21(Cip1) Cdk inhibitor influences the outcome of the p53 response to DNA damage. Nature. 2002;419:729–734. doi: 10.1038/nature01119. [DOI] [PubMed] [Google Scholar]
- Shen V, Kiledjian M. A view to a kill: structure of the RNA exosome. Cell. 2006;127:1093–1095. doi: 10.1016/j.cell.2006.11.035. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Silva JM, Li MZ, Chang K, Ge W, Golding MC, Rickles RJ, et al. Second-generation shRNA libraries covering the mouse and human genomes. Nat Genet. 2005;37:1281–1288. doi: 10.1038/ng1650. [DOI] [PubMed] [Google Scholar]
- Smith A, Brownawell A, Macara IG. Nuclear import of Ran is mediated by the transport factor NTF2. Curr Biol. 1998;8:1403–1406. doi: 10.1016/s0960-9822(98)00023-2. [DOI] [PubMed] [Google Scholar]
- Smith AP, Verrecchia A, Faga G, Doni M, Perna D, Martinato F, et al. A positive role for Myc in TGFbeta-induced Snail transcription and epithelial-to-mesenchymal transition. Oncogene. 2009;28:422–430. doi: 10.1038/onc.2008.395. [DOI] [PubMed] [Google Scholar]
- Solomon DLC, Amati B, Land H. Distinct DNA binding preferences for the c-Myc/Max and Max/Max dimers. Nucleic Acids Res. 1993;21:5372–5376. doi: 10.1093/nar/21.23.5372. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sridharan R, Tchieu J, Mason MJ, Yachechko R, Kuoy E, Horvath S, et al. Role of the murine reprogramming factors in the induction of pluripotency. Cell. 2009;136:364–377. doi: 10.1016/j.cell.2009.01.001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Staller P, Peukert K, Kiermaier A, Seoane J, Lukas J, Karsunky H, et al. Repression of p15INK4b expression by Myc through association with Miz-1. Nat Cell Biol. 2001;3:392–399. doi: 10.1038/35070076. [DOI] [PubMed] [Google Scholar]
- Stewart M. Molecular mechanism of the nuclear protein import cycle. Nat Rev Mol Cell Biol. 2007;8:195–208. doi: 10.1038/nrm2114. [DOI] [PubMed] [Google Scholar]
- Takahashi K, Yamanaka S. Induction of pluripotent stem cells from mouse embryonic and adult fibroblast cultures by defined factors. Cell. 2006;126:663–676. doi: 10.1016/j.cell.2006.07.024. [DOI] [PubMed] [Google Scholar]
- Tao H, Umek RM. Reciprocal regulation of gadd45 by C/EBP alpha and c-Myc. DNA Cell Biol. 1999;18:75–84. doi: 10.1089/104454999315646. [DOI] [PubMed] [Google Scholar]
- Thomson JP, Skene PJ, Selfridge J, Clouaire T, Guy J, Webb S, et al. CpG islands influence chromatin structure via the CpG-binding protein Cfp1. Nature. 2010;464:1082–1086. doi: 10.1038/nature08924. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Trumpp A, Refaeli Y, Oskarsson T, Gasser S, Murphy M, Martin GR, et al. c-Myc regulates mammalian body size by controlling cell number but not cell size. Nature. 2001;414:768–773. doi: 10.1038/414768a. [DOI] [PubMed] [Google Scholar]
- van Riggelen J, Yetil A, Felsher DW. MYC as a regulator of ribosome biogenesis and protein synthesis. Nat Rev Cancer. 2010;10:301–309. doi: 10.1038/nrc2819. [DOI] [PubMed] [Google Scholar]
- Vanacova S, Stefl R. The exosome and RNA quality control in the nucleus. EMBO Rep. 2007;8:651–657. doi: 10.1038/sj.embor.7401005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Watson JD, Oster SK, Shago M, Khosravi F, Penn LZ. Identifying genes regulated in a Myc-dependent manner. J Biol Chem. 2002;277:36921–36930. doi: 10.1074/jbc.M201493200. [DOI] [PubMed] [Google Scholar]
- Winkles JA. Serum- and polypeptide growth factor-inducible gene expression in mouse fibroblasts. Prog Nucleic Acid Res Mol Biol. 1998;58:41–78. doi: 10.1016/s0079-6603(08)60033-1. [DOI] [PubMed] [Google Scholar]
- Wong DJ, Liu H, Ridky TW, Cassarino D, Segal E, Chang HY. Module map of stem cell genes guides creation of epithelial cancer stem cells. Cell Stem Cell. 2008;2:333–344. doi: 10.1016/j.stem.2008.02.009. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zeller KI, Jegga AG, Aronow BJ, O'Donnell KA, Dang CV. An integrated database of genes responsive to the Myc oncogenic transcription factor: identification of direct genomic targets. Genome Biol. 2003;4:R69. doi: 10.1186/gb-2003-4-10-r69. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zeller KI, Zhao X, Lee CW, Chiu KP, Yao F, Yustein JT, et al. Global mapping of c-Myc binding sites and target gene networks in human B cells. Proc Natl Acad Sci U S A. 2006;103:17834–17839. doi: 10.1073/pnas.0604129103. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.