Abstract
Covalent labeling along with mass spectrometry is a method that is increasingly used to study protein structure. Recently, it has been shown that diethylpyrocarbonate (DEPC) is a powerful labeling reagent because it can modify up to 30% of the residues in the average protein, including the N-terminus, His, Lys, Tyr, Ser, Thr, and Cys residues. We recently discovered, however, that Cys residues that form disulfide bonds appear to be modified by DEPC as well. In this work, we demonstrate that disulfide linked Cys residues are not actually reactive with DEPC, but instead once reduced free Cys residues can capture a carbethoxy group from other modified amino acids via a solution-phase reaction that can occur during the protein digestion step. This “scrambling” of carbethoxy groups decreases the amount of modification observed at other residues and can potentially provide incorrect protein structural information. Fortunately, label scrambling can be completely avoided by alkylating the free thiols after disulfide reduction.
Introduction
Mass spectrometry (MS) is increasingly being used as a means of studying the higher order structure of proteins and protein complexes. While other methods, such as NMR and X-ray crystallography, provide more detailed structural information, there are some limitations to these methods. For example, some proteins are difficult to study by these methods due to size, conformational flexibility, aggregation propensity, or limited sample amount. As such, MS combined with labeling techniques has been investigated to address some of these shortcomings. H/D exchange [1–6], chemical cross-linking [7–13], covalent labeling [14–23], and non-covalent labeling [24–26] are commonly used techniques. Each of these approaches results in the modification of multiple amino acids, and except for the non-covalent labeling approach protein structural information is obtained by identifying the protein sites that are modified by MS and/or MS/MS after proteolytic digestion. H/D exchange uses deuterium as a reagent to measure backbone amide hydrogen exchange rates. These rates are then used to provide information concerning protein structure, dynamics, and interactions. When combined with MS, H/D exchange is a very powerful tool because it gives information about virtually every backbone amide; however, it does not provide information about amino acid side chain positions and also suffers from back-exchange and scrambling if special care is not taken during the MS measurements. In chemical cross-linking experiments new intra- or intermolecular bonds are created that set distance constraints on the location of two amino acid side chains. The resulting set of distance constraints can then be used to deduce protein structure, especially for protein-protein and protein-ligand complexes. One challenge with cross-linking experiments is the identification and sequencing of the resulting cross-linked peptides.
Mass spectrometry has also being used in combination with covalent labeling reactions to study protein structure [21]. In comparison to H/D exchange, covalent labeling does not suffer much from label back-exchange or scrambling, and it also provides structural information about amino acid side chains, making it complementary to H/D exchange. Covalent labeling also has some similarities to chemical cross-linking in that it covalently modifies amino acid side chains. In covalent labeling experiments, however, only a single polypeptide chain is modified, making mass spectral and data analyses more straightforward. There are two general types of covalent labeling reagents – those that modify many different amino acids (e.g. hydroxyl radicals [14–20, 23, 27]) and those that react with specific amino acids (e.g. succinimides that react with lysine residues [21, 28, 29]). Hydroxyl radicals are commonly used and irreversibly modify proteins and other macromolecules. Amino acid specific labels also typically irreversibly modify amino acid side chains. Although, there are a few examples of labels, such as arginine-specific dicarbonyl compounds that label reversibly and therefore require additional reagents to stabilize them [21].
Our group has been investigating diethylpyrocarbonate (DEPC) as a multi-residue labeling reagent because we have found that it can probe up to 30% of the residues in the average protein, including the N-terminus, His, Lys, Tyr, Ser, Thr and Cys residues [22]. DEPC also generates only one product type, carbethoxylated residues, allowing for straightforward and sensitive analyses of the modified amino acids. We and others have shown that DEPC labeling can provide useful structural information for proteins and protein-protein interactions [22, 30–35]. Due to DEPC’s molecular size, proteins are typically only labeled at amino acids that are exposed to solvent on the protein surface. Amino acids that are buried inside the protein are usually not labeled. Thus, DEPC is particularly useful for studying protein-protein complexes.
In the course of our work with DEPC, we have noticed for some proteins that Cys residues that form disulfide bonds can be modified by DEPC. The thiol groups of free Cys residues are known to be very reactive with electrophilic compounds, but the disulfide formed by two Cys residues is typically much less reactive toward electrophiles. In the present work, we set out to understand this observed reactivity in order to establish whether or not disulfides indeed react with DEPC or whether something else explains the modification of disulfide bonded Cys residues. From a series of labeling experiments with proteins and Cys-containing peptides, we have discovered that unmodified free Cys residues can capture a carbethoxy group from other DEPC modified amino acids via a solution phase reaction. This “scrambling” of covalently modified residues is very atypical for covalent labeling/MS experiments and would seem to lessen the utility of DEPC as a labeling reagent. Fortunately, though, addition of an alkylating agent (e.g. iodoacetamide) during protein digestion prevents this scrambling and ensures the fidelity of the information obtained from the DEPC labeling experiments.
Experimental
Materials
Human β-2-microglobulin (β2m) was obtained from Fitzgerald Industries International, Inc (Concord, MA). Immobilized chymotrypsin and triethylamine acetate (pH 8.0) were obtained from Princeton Separations (Adelphia, NJ). Diethylpyrocarbonate (DEPC), imidazole, iodoacetamide, and tris(2-carboxyethyl)phosphine (TCEP) were purchased from Sigma-Aldrich (St. Louis, MO). Ammonium acetate, methanol, acetonitrile, and acetic acid were purchased from Fisher Scientific (Fair Lawn, NJ). Centricon molecular weight cutoff (MWCO) filters were obtained from Millipore (Burlington, MA). Deionized water was generated with a Millipore (Burlington, MA) Simplicity 185 water purification system. The peptides ANP (SLRRSSCFGGR), Angiotensin I (DRVYIHPFHL), and semastatin (SPWTKCSATCGGGHYMRTR) were from American Peptide Company (Sunnyvale, CA).
Proteolytic Digestion
Before the addition of enzyme, DEPC-reacted β2m was purified using a 10,000 MWCO filter to reduce the concentration of the reagent used to quench the DEPC reaction (see DEPC modification procedure below). Before filtering, the volume of the β2m sample was 100 µL. The volume was then reduced to approximately 10 µL during the filtering step, and then the sample was reconstituted with deionized water to a volume of about 40 µL, resulting in a β2m concentration of about 250 µM. β2m was then incubated in a buffer solution (100 mM triethylamine at pH 8.0) with 10% (v/v) acetonitrile at 50 °C for 45 min prior to digestion. β2m samples were then reacted with TCEP (protein:TCEP=1:40 molar ratio) to reduce the disulfide bond, while for proteins in which the disulfide bond was not reduced, TCEP was not added. In some cases, iodoacetamide was also added (~80 fold molar excess over the protein) at the same time as TCEP to alkylate the free Cys residues. Under these reaction conditions, the cysteine residues were 100% alkylated as determined by LC/MS analysis. Stock solutions of iodoacetamide were prepared in triethylamine acetate at pH 8.0, and alkylation reactions were carried out in the dark for 30 min. Immobilized chymostrypsin was added to yield an enzyme/substrate ratio of 1:10, and the protein/enzyme mixture was incubated at 37 °C. After 2 h of digestion, the enzyme was separated from the protein by centrifuging the reaction mixture for 2 min at 1000 RPM. In all cases, the modified proteins were analyzed right after digestion.
DEPC Modification
The modification reactions of peptides and proteins were performed for 1 min at 37 °C and were initiated by adding DEPC in acetonitrile in a molar excess of 4. The total amount of acetonitrile added was 1 µL and the total reaction volume was 100 µL. After 1 min, 10 mM imidazole was added to quench the reaction [22].
Instrumentation
Mass spectra were acquired on a Bruker AmaZon (Billerica, MA) quadrupole ion trap mass spectrometer equipped with an electrospray ionization source. Typically, the electrospray needle voltage was kept at ~4 kV, and the capillary temperature was set to 250 °C. Either CID or ETD was used to obtain tandem mass spectra. In the CID experiments, an activation time of 40 ms was chosen, and the amplitude of the resonance excitation voltage was optimized to ensure efficient ion dissociation. For the ETD experiments, fluoranthene radical anions, which were generated via negative chemical ionization with methane reagent gas, were used with an accumulation time of 10 ms. The ETD parameters that were used are the following: low m/z cutoff (LMCO) values ranging from 50 to 180, reaction times ranging from 50 ms to 180 ms. The values were chosen to give the most efficient dissociation. For direct injection experiments, samples were delivered at 2 µl/min with a syringe pump. An HP1100 HPLC system (Agilent, Wilmington, DE) with a Discovery C18 column (15 cm × 2.1 mm, 5 µm particle size, Supelco, St. Louis, MO) was used for HPLC analysis. Peptide mixtures or peptide fragments from the proteolytic digests were eluted using a linear gradient of methanol increased from 10 to 100% methanol for 30 min. The mobile phase contained 0.1% acetic acid and had a flow rate of 0.25 mL/min.
Results and Discussion
Modification of cysteines in a buried disulfide bond
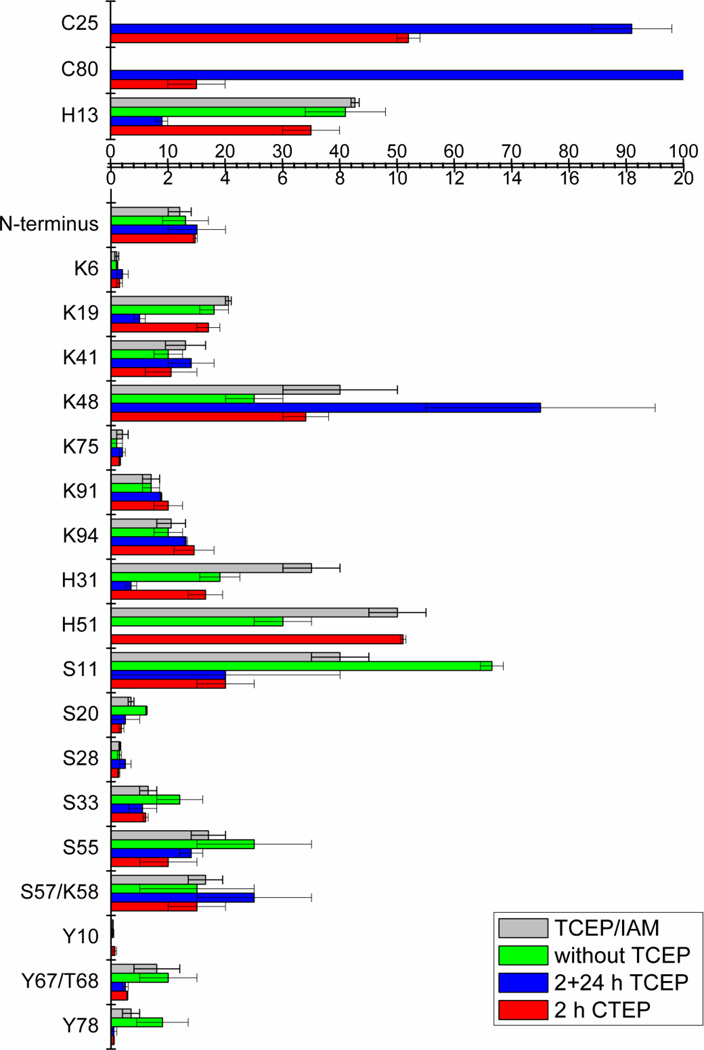
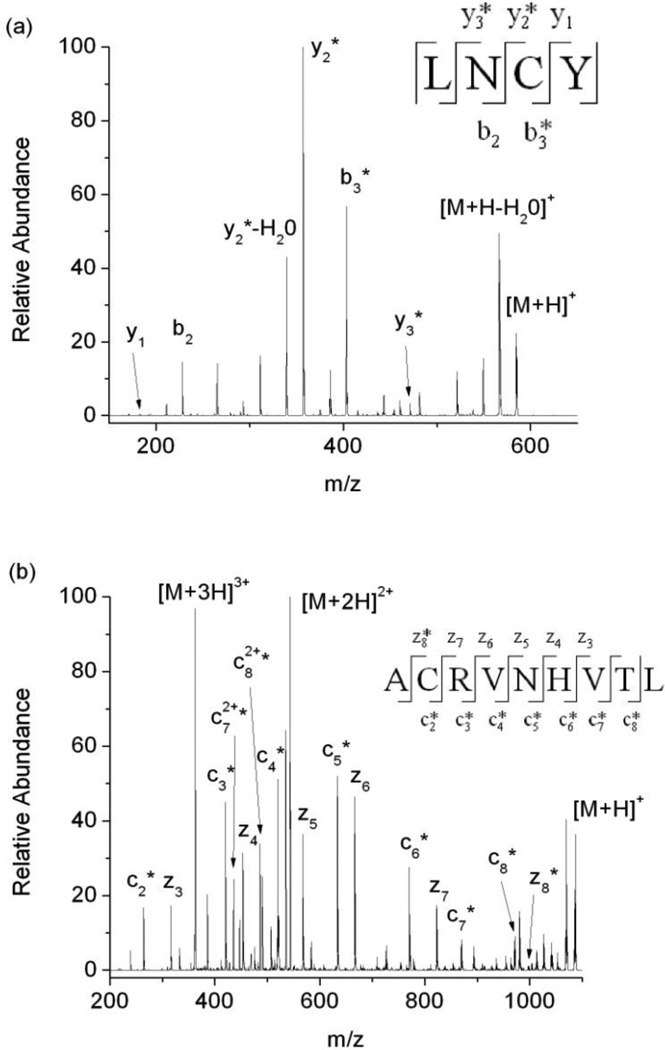
Upon DEPC labeling of the protein β2m, 70% (19/27) of the modifiable solvent exposed amino acids are modified (Figure 1 and Table S1, column 5 in the Supporting Information), which is consistent with previous work [22, 36]. Residues with ≥ 30% solvent accessibility are typically considered to be solvent exposed, and residues with 10–30% solvent accessibility are usually recognized as partially exposed [37]. The DEPC reaction appears to be fairly selective for labeling solvent exposed residues as only 4 partially exposed residues and 3 “buried” residues are labeled by DEPC. The modification percentages for partially exposed residues, such as Ser11, Ser28, Lys41 and Tyr67/Thr68 are relatively low, ranging from ~0.3 to 4%, which is consistent with these residues being only partially exposed. The buried residues that are modified are Cys25, Cys80, and Tyr78. The modification percentage for Tyr78 is very low, only ~0.1%. This low level of modification may reflect protein dynamics that partially expose this residue to solvent. Indeed, in the NMR structures used to determine solvent accessibilities for β2m, a few of the structures have this residue partially exposed [38]. The relatively high modification percentages for Cys25 and Cys80 (Figure 1, red bars), however, are surprising. The residues are not only completely buried in all the NMR structures, but they also form a disulfide bond. As such, they should not react with DEPC at all. Nonetheless, after reacting the protein as described in the experimental section and then reducing and digesting the protein, we find two peptides, 23LNCY26 and 79ACRVNHVTL87, that contain DEPC-modified cysteine residues (Figure 2). In addition, these two Cys residues are modified to significant extents.
Figure 1.
DEPC modification percentages for residues in β-2-microglobulin under different conditions as explained in the text and experimental section. All experiments were repeated three times, and the means and standard deviations are reported. Table S1 in the Supporting Information provides a list of the solvent accessible surface areas for each of these labeled residues and the numerical modification percentages for each residue. TCEP corresponds to tris(2-carboxyethyl)phosphine, and IAM corresponds to iodoacetamide.
Figure 2.
(a) Tandem mass spectrum acquired after CID of the [M+H]+ ion of the Leu23-Tyr26 proteolytic fragment of β2m. Unmodified b2, y1 and modified b3, y2, y3 ions confirm that Cys25 is the site of modification. (b) Tandem mass spectrum acquired after ETD of the [M+3H]3+ ion of the Ala79-Leu87 fragment of β2m. A series of modified c2 to c8 ions and a series unmodified z3 to z7 with modified z8 confirm that Cys80 is the site of modification. The product ions with an asterisk are the product ions that contain the DEPC modification.
One possible explanation for the modification of Cys25 and Cys80 is that DEPC is not fully removed during the quenching step with imidazole such that this reagent can react with the free cysteine groups that are made available during the reduction and digestion steps. To test whether there is still DEPC left in the sample, the peptide angiotensin I (DRVYIHPFHL) was added into the sample after the quenching and filtering steps. If DEPC remained in the solution, we would expect this peptide to be modified by DEPC at one or more of its His residues, but no modification of angiotensin I is observed (data not shown), indicating that the DEPC is fully quenched by imidazole.
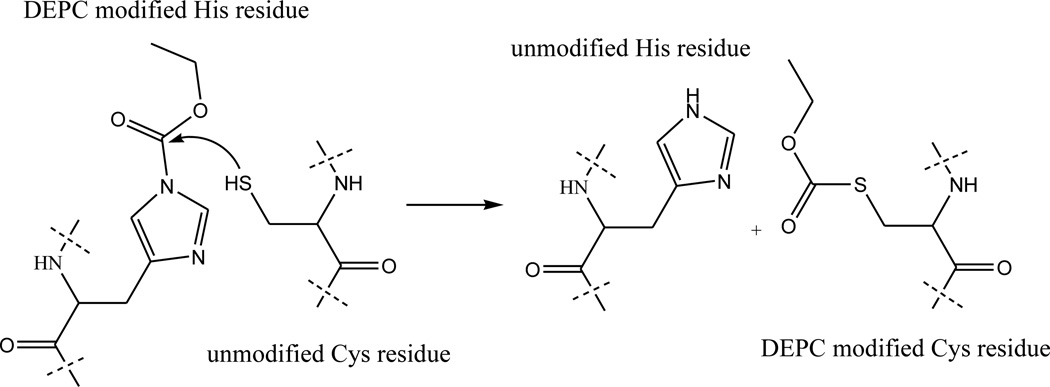
Another possible explanation for the modification of Cys25 and Cys80 is that the free thiols of these residues are able to capture a carbethoxy group from another modified amino acid via an intra- or intermolecular reaction (Scheme 1) after the disulfide bond is reduced. Several experiments were carried out to test this hypothesis. First, the modified protein was enzymatically digested without adding TCEP to reduce the disulfide bond. LC/MS experiments reveal the presence of the disulfide-linked peptide, 23LNCY26/79ACRVNHVTL87, without any measurable modification. This result indicates that the disulfide linked Cys residues do not react with DEPC in the intact protein or in peptide fragments. Moreover, the modification levels for many of the other residues, especially His, Ser, and Tyr residues, are found to increase (Figure 1, compare red and green bars and Table S1, compare columns 5 and 7), which is consistent with the idea that Cys can no longer capture the carbethoxy group from these other residues. In a second set of experiments, the modified protein with all residual DEPC completely removed (as confirmed by the failure to modify angiotensin I [data not shown]), was reduced by TCEP, digested by immobilized chymotrypsin, and then the peptide fragments were allowed to remain in solution for 24 h after removing chymotrypsin. After remaining in solution for 24 h, the modification extent for Cys25 increased from 52 ± 2 to 91 ± 7%. Similarly, the modification extent for Cys80 increased from 15 ± 5 to 100% (Figure 1, blue bars and Table S1, column 6). It is important to note that the modification extents for some of the other residues, especially His, Ser, and Tyr residues, decrease over time, which is consistent with the Cys residues capturing the carbethoxy group; although, the modification losses of some of these residues are partially due to hydrolysis [21,22, 39, 40]. As a whole, these data are consistent with the idea that the free thiols of Cys can capture a carbethoxy group from other DEPC-modified amino acids in solution.
Scheme 1.
Hypothesized mechanism of cysteine capture of carbethoxy-modified histidine.
Carbethoxy group capture by peptides in solution
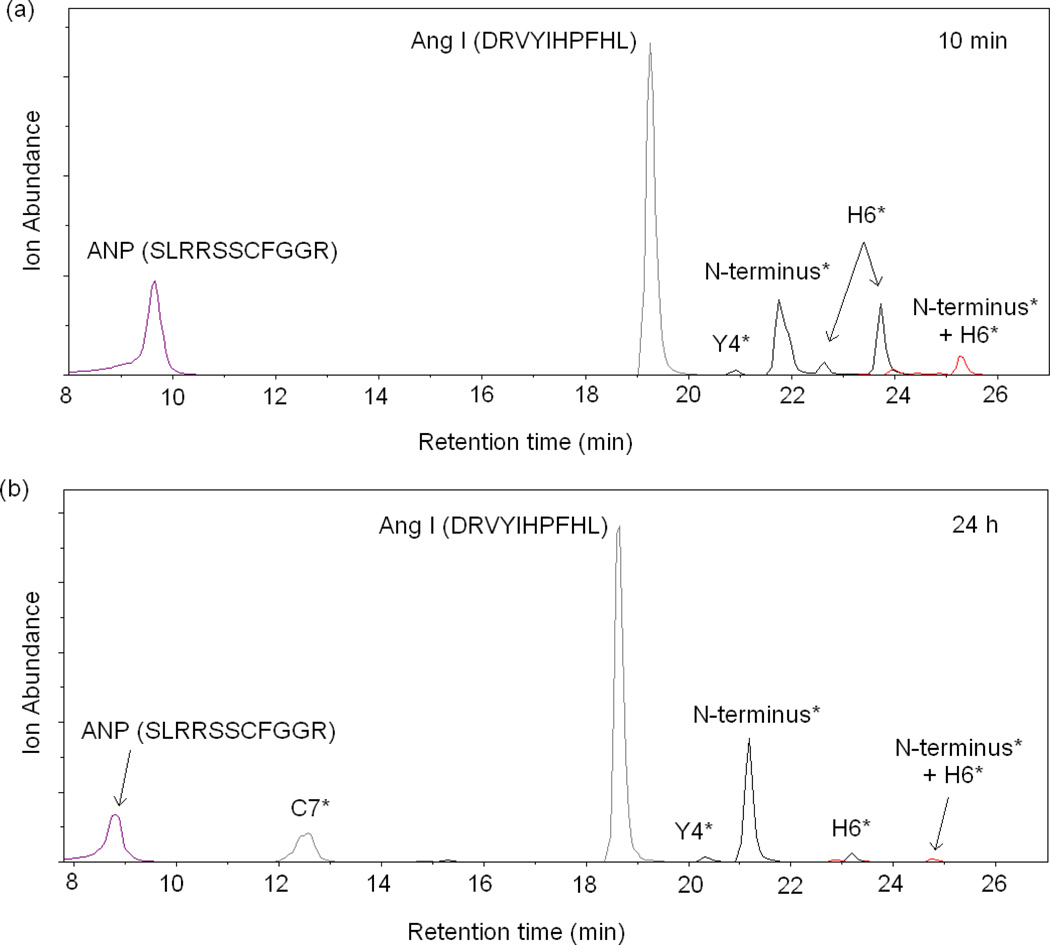
To further test this idea, we examined the reactivity of unmodified cysteine-containing peptides with other DEPC-modified peptides. Angiotensin I was reacted with DEPC, and the reaction was quenched by imidazole. Then, the cysteine-containing peptide ANP (SLRRSSCFGGR) was added to the solution containing DEPC-modified angiotensin, and TCEP was also added to minimize intermolecular disulfide formation between two ANP molecules. The sample was then analyzed by LC/MS as soon as possible (Figure 3a) and after 24 h (Figure 3b). Figure 3 indicates that the cysteine-containing peptide ANP is able to capture a carbethoxy group from angiotensin I after reacting with the peptide for 24 h. The chromatogram in Figure 3a shows that no modification of ANP is observed when the sample is analyzed within 10 min; this is because no free DEPC is present in the solution and the capture reaction has not occurred yet. After incubating the two peptides together for 24 h, however, 37 ± 2% of ANP is found modified at Cys7 (Figure 3b, peak at retention time of 12.5 min). Tandem MS of the peptide corresponding to this peak at 12.5 min confirms that the modification site on ANP is Cys7 with no modification observed at the N-terminus (Figure S1 in the Supporting Information). Importantly, if ANP is reacted directly with DEPC for 1 min, both the N-terminus and Cys7 are found modified (Figure S2 in the Supporting Information). In addition to modification of ANP at Cys7, the ion abundances of the peaks corresponding to DEPC-modified angiotensin I decrease. For example, angiotensin I peaks corresponding to modifications at His6 decrease in abundance. These decreases are likely due to cysteine capture, but they also could be due to hydrolysis of the carbethoxylated residues as indicated above [22, 33, 39, 40]. One might argue that the hydrolysis product, ethyl hydrogen carbonate, could react with the thiol group of Cys, but this product is not only unreactive with nucleophiles but is well known to be highly unstable in water, like all alkyl hydrogen carbonates [41]. These experiments with peptides further suggest that cysteine can capture a carbethoxy group from other DEPC-modified amino acids.
Figure 3.
Extracted ion chromatograms (EIC) of the products of the reaction of DEPC-modified angiotensin I and the peptide ANP with TCEP added to prevent disulfide formation. Shown are the products (a) after a 10 min reaction and (b) after a 24 h reaction. Chromatographic peaks notated with a residue(s) having an asterisk indicate that the corresponding chromatographic peak is the peptide labeled at only the indicated residue(s).
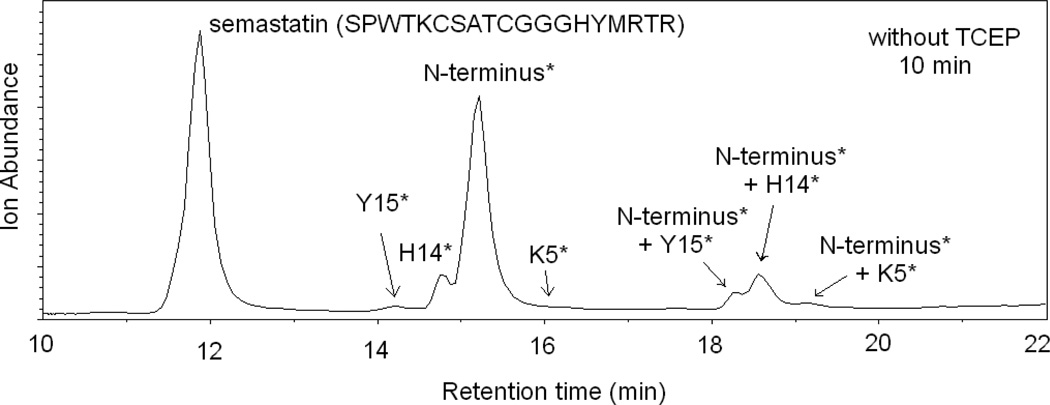
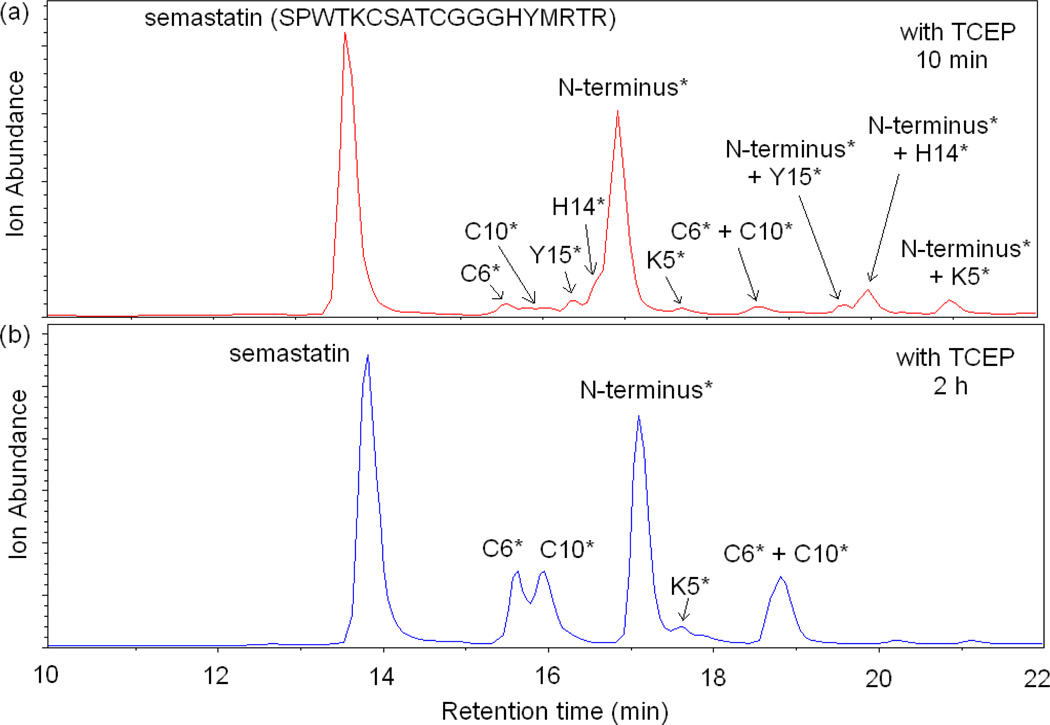
We also investigated the possibility of capture by Cys in a peptide that forms a disulfide bond. For this purpose we used the peptide semastatin (SPWTKCSATCGGGHYMRTR). Figure 4 shows the total ion chromatogram (TIC) of semastatin after reacting with DEPC at a 1:4 peptide:DEPC ratio. The reaction was quenched by adding imidazole, and no TCEP was added to reduce the disulfide bond. Four different modification sites are identified by LC/MS/MS, including Tyr15, His14, the N-terminus and Lys5. Peaks corresponding to peptides with two DEPC modifications on the same peptide are also observed around 18 min as indicated in Figure 4. No modification of the two Cys residues is observed because the two Cys residues form a disulfide bond and are therefore unreactive with DEPC. To test whether free Cys residues in semastatin could capture the modification from another residue, TCEP was added after quenching the DEPC reaction in order to reduce the disulfide bond before analysis. Figure 5 shows the resulting LC/MS data after letting the disulfide reduced peptide sit in solution for 10 min (Figure 5a) and 2 h (Figure 5b). As shown in the figure, there is some minor modification at Cys6 and Cys10 even after 10 min, indicating that the capture reaction is fast for this peptide; however, the data also shows that Tyr15, His14, the N-terminus, and Lys5 are still modified. In addition, a peak is observed that corresponds to a peptide with two DEPC modifications on the same peptide; MS/MS confirms that both Cys6 and Cys10 are modified in the ion corresponding to this peak. After sitting in solution for 2 h, the modification levels at Cys6 and Cys10 have increased quite noticeably, while the modifications at Tyr15 and His14 are no longer measurable. Also the intensity of the peak that corresponds to a peptide with two DEPC modifications at Cys6 and Cys10 also increased. The decrease in the modification extents for Tyr15 and His14 could be due to the hydrolysis of the carbethoxylated residues [22, 36, 39, 40] rather than capture of the carbethoxy group by the Cys residues. A control experiment, however, shows that hydrolysis is not the primary cause (see Figure S3). Overall, these data indicate that Cys residues can capture a modification from another residue but only when the Cys residues are free thiols and not when they are part of a disulfide bond.
Figure 4.
Total ion chromatogram (TIC) of semastatin modified by DEPC at a peptide: DEPC ratio of 1:4 without TCEP added after the reaction. Chromatographic peaks notated with a residue(s) having an asterisk indicate that the corresponding chromatographic peak is the peptide labeled at only the indicated residue(s).
Figure 5.
Total ion chromatograms (TIC) of semastatin modified by DEPC at a peptide:DEPC ratio of 1:4 with TCEP added after the reaction with DEPC (a) Products after allowing the peptide to remain in solution for 10 min after TCEP addition and (b) 2 h after adding TCEP. Chromatographic peaks notated with a residue(s) having an asterisk indicate that the corresponding chromatographic peak is the peptide labeled at only the indicated residue(s).
Avoiding carbethoxy transfer to cysteine
Avoiding the modification of disulfide linked or buried Cys residues is important for maintaining accurate protein structural information from DEPC labeling experiments. As was shown above for β2m, this can be done by not reducing the disulfide bond, but this, of course, would lead to low digestion efficiency and a diminished ability to find modified amino acids. So, we investigated the ability of iodoacetamide to prevent carbethoxy transfer to the free cysteine residues that are produced after disulfide reduction. Iodoacetamide is commonly used in proteomics to alkylate free thiols and prevent peptide fragments from forming disulfides after digestion. So, we reasoned that this reagent might rapidly alkylate the free thiols, thereby preventing transfer of the carbethoxy groups to the newly reduced Cys residues. The gray bars in Figure 1 and the last column in Table S1 show that when iodoacetamide is added during the β2m disulfide bond reduction step, no DEPC modification is observed at the two Cys residues. Moreover, the modification percentages of all the other residues, as compared to the case when no TCEP is added (Figure 1, compare green and gray bars and Table S1, compare columns 7 and 8), are mostly unaffected, indicating iodoacetamide has no effect on the DEPC labeled peptides.
Additional experiments were also carried out to ensure that alkylation by iodoacetamide does not displace DEPC labels from Cys residues that are properly labeled by DEPC. To test this possible scenario, the peptide ANP was reacted with DEPC for 1 min at a 1:4 peptide:DEPC ratio, and LC/MS/MS data indicate modifications at the N-terminus and Cys7. The percentage of peptides with a modification at the N-terminus was found to be 5 ± 1%, and the percentage of modified Cys7 residues was found to be 3.8 ± 0.6%. To the same sample TCEP and iodoacetamide were added and allowed to react for 30 min. After this 30 min reaction, the remaining free thiols were modified 100% by iodoacetamide. The DEPC modification percentages of the N-terminus and Cys7 were found to be almost identical to the percentages measured before alkylation, with the N-terminus being modified 6 ± 1% and Cys7 modified 2.9 ± 0.4%. These results indicate that, even with a vast excess of iodoacetamide, alkylation does not displace a DEPC label from cysteine or other modified amino acids. Therefore, blocking cysteine groups by iodoacetamide can effectively prevent label scrambling without influencing the modification levels of solvent accessible free cysteine residues.
Conclusions
From the data presented here, we can conclude that disulfide linked cysteine residues do not react with DEPC, but when a disulfide bond is reduced to form free thiols, these reduced cysteines can capture carbethoxy groups from other modified amino acids. This process can accelerate the loss of DEPC modification from certain amino acids, especially His, Ser, Thr, and Tyr residues, and lead to less structural information. In addition, the modification of buried cysteines can also lead to incorrect structural information about a protein. Hence, it is important to avoid the transfer of carbethoxy groups to cysteine. Digesting the protein without adding a reducing agent is one possible solution, but this decreases digestion efficiencies. A better solution is to add iodoacetamide during the disulfide reduction step to alkylate free thiols, thereby preventing transfer of carbethoxy groups from modified residues to cysteines. While the described results are specific for DEPC labeling experiments, we predict that other electrophilic labeling reagents could conceivably exhibit similar scrambling behavior.
Supplementary Material
Acknowledgements
This work was supported by a grant from the National Institutes of Health (R01 GM075092).
References
- 1.Wales TE, Engen JR. Hydrogen exchange mass spectrometry for the analysis of protein dynamics. Mass Spectrom. Rev. 2006;25:158–170. doi: 10.1002/mas.20064. [DOI] [PubMed] [Google Scholar]
- 2.Englander SW. Hydrogen Exchange and Mass Spectrometry: A Historical Perspective. J. Am. Soc. Mass Spectrom. 2006;17:1481–1489. doi: 10.1016/j.jasms.2006.06.006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Tsutsui Y, Wintrode PL. Hydrogen/deuterium exchange-mass spectrometry: a powerful tool for probing protein structure, dynamics and interactions. Curr. Med. Chem. 2007;14:2344–2358. doi: 10.2174/092986707781745596. [DOI] [PubMed] [Google Scholar]
- 4.Engen JR. Analysis of Protein Conformation and Dynamics by Hydrogen/Deuterium Exchange MS. Anal. Chem. 2009;81:7870–7875. doi: 10.1021/ac901154s. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Marcsisin SR, Engen JR. Hydrogen exchange mass spectrometry: what is it and what can it tell us? Analyt. Bioanalyt. Chem. 2010;397:967–972. doi: 10.1007/s00216-010-3556-4. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Pan J, Han J, Borchers CH, Konermann L. Conformer-Specific Hydrogen Exchange Analysis of Aβ(1–42) Oligomers by Top-Down Electron Capture Dissociation Mass Spectrometry. Anal. Chem. 2011;83:5386–5393. doi: 10.1021/ac200906v. [DOI] [PubMed] [Google Scholar]
- 7.Steen H, Jensen ON. Analysis of protein-nucleic acid interactions by photochemical cross-linking and mass spectrometry. Mass Spectrom. Rev. 2002;21:163–182. doi: 10.1002/mas.10024. [DOI] [PubMed] [Google Scholar]
- 8.Sinz A. Chemical cross-linking and mass spectrometry to map three-dimensional protein structures and protein-protein interactions. Mass Spectrom. Rev. 2006;25:663–682. doi: 10.1002/mas.20082. [DOI] [PubMed] [Google Scholar]
- 9.Chakravarti B, Lewis SJ, Chakravarti DN, Raval A. Three dimensional structures of proteins and protein complexes from chemical cross-linking and mass spectrometry: a biochemical and computational overview. Curr. Proteomics. 2006;3:1–21. [Google Scholar]
- 10.Petrotchenko EV, Borchers CH. Crosslinking combined with mass spectrometry for structural proteomics. Mass Spectrom. Rev. 2010;29:862–876. doi: 10.1002/mas.20293. [DOI] [PubMed] [Google Scholar]
- 11.Sinz A. Investigation of protein-protein interactions in living cells by chemical crosslinking and mass spectrometry. Analyt. Bioanalyt. Chem. 2010;397:3433–3440. doi: 10.1007/s00216-009-3405-5. [DOI] [PubMed] [Google Scholar]
- 12.Leitner A, Walzthoeni T, Kahraman A, Herzog F, Rinner O, Beck M, Aebersold R. Probing native protein structures by chemical cross-linking, mass spectrometry and bioinformatics. Mol. Cell. Proteomics. 2010;9:1634–1649. doi: 10.1074/mcp.R000001-MCP201. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Singh P, Panchaud A, Goodlett DR. Chemical Cross-Linking and Mass Spectrometry As a Low-Resolution Protein Structure Determination Technique. Anal. Chem. 2010;82:2636–2642. doi: 10.1021/ac1000724. [DOI] [PubMed] [Google Scholar]
- 14.Xu G, Chance MR. Hydroxyl Radical-Mediated Modification of Proteins as Probes for Structural Proteomics. Chem. Rev. 2007;107:3514–3543. doi: 10.1021/cr0682047. [DOI] [PubMed] [Google Scholar]
- 15.Kiselar JG, Chance MR. Future directions of structural mass spectrometry using hydroxyl radical footprinting. J. Mass Spectrom. 2010;45:1373–1382. doi: 10.1002/jms.1808. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Wang L-W, Chance MR. Structural Mass Spectrometry of Proteins Using Hydroxyl Radical Based Protein Footprinting. Anal. Chem. 2011;83:7234–7241. doi: 10.1021/ac200567u. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Hambly DM, Gross ML. Laser Flash Photolysis of Hydrogen Peroxide to Oxidize Protein Solvent-Accessible Residues on the Microsecond Timescale. J. Am. Soc Mass Spectrom. 2005;16:2057–2063. doi: 10.1016/j.jasms.2005.09.008. [DOI] [PubMed] [Google Scholar]
- 18.Hambly DM, Gross ML. Laser Flash Photochemical Oxidation to Locate Heme Binding and Conformational Changes in Myoglobin. Int. J. Mass Spectrom. 2007;259:124–129. [Google Scholar]
- 19.Gau BC, Sharp JS, Rempel DL, Gross ML. Fast Photochemical Oxidation of Protein Footprints Faster than Protein Unfolding. Anal. Chem. 2009;81:6563–6571. doi: 10.1021/ac901054w. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Chen J, Rempel DL, Gross ML. T-jump and Fast Photochemical Oxidation Probe Sun Millisecond Protein Folding. J. Am. Chem. Soc. 2010;132:15502–15504. doi: 10.1021/ja106518d. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Mendoza VL, Vachet RW. Probing protein structure by amino acid-specific covalent labeling and mass spectrometry. Mass Spectrom. Rev. 2009;28:785–815. doi: 10.1002/mas.20203. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Mendoza VL, Vachet RW. Protein Surface Mapping Using Diethylpyrocarbonate with Mass Spectrometric Detection. Anal. Chem. 2008;80:2895–2904. doi: 10.1021/ac701999b. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Konermann L, Stocks BB, Pan Y, Tong X. Mass spectrometry combined with oxidative labeling for exploring protein structure and folding. Mass Spectrom. Rev. 2010;29:651–667. doi: 10.1002/mas.20256. [DOI] [PubMed] [Google Scholar]
- 24.Liu ZJ, Cheng SJ, Gailie DR, Julian RR. Exploring the Mechanism of Selective Noncovalent Adduct Protein Probing Mass Spectrometry Utilizing Site-Directed Mutagenesis To Examine Ubiquitin. Anal. Chem. 2008;80:3846–3852. doi: 10.1021/ac800176u. [DOI] [PubMed] [Google Scholar]
- 25.Ly T, Julian RR. Protein–Metal Interactions of Calmodulin and α-Synuclein Monitored by Selective Noncovalent Adduct Protein Probing Mass Spectrometry. J. Am. Soc. Mass Spectrom. 2008;19:1663–1672. doi: 10.1016/j.jasms.2008.07.006. [DOI] [PubMed] [Google Scholar]
- 26.Ly T, Julian RR. Using ESI-MS to Probe Protein Structure by Site-Specific Noncovalent Attachment of 18-Crown-6. J. Am. Soc. Mass Spectrom. 2006;17:1209–1215. doi: 10.1016/j.jasms.2006.05.007. [DOI] [PubMed] [Google Scholar]
- 27.Whitelegge J. Protein Mass Spectrometry. 1st edn. Amsterdam: Elsevier Science; 2009. [Google Scholar]
- 28.Knock SL, Miller BT, Blankenship JE, Nagle GT, Smith JS, Kurosky A. N-acylation of aplysia egg-laying hormone with biotin. J. Biol. Chem. 1991;266:24413–24419. [PubMed] [Google Scholar]
- 29.Glocker MO, Borchers C, Fiedler W, Suckau D, Przybylski M. Molecular characterization of surface topology in protein tertiary structures by amino-acylation and mass spectrometric peptide mapping. Bioconj. Chem. 1994;5:583–590. doi: 10.1021/bc00030a014. [DOI] [PubMed] [Google Scholar]
- 30.Glocker MO, Kalkum M, Yamamoto R, Schreurs J. Selective Biochemical Modification of Functional Residues in Recombinant Human Macrophage Colony-Stimulating Factor β (rhM-CSF β): Identification by Mass Spectrometry. Biochem. 1996;35:14625–14633. doi: 10.1021/bi961199o. [DOI] [PubMed] [Google Scholar]
- 31.Dage JL, Sun H, Halsall HB. Determination of diethylpyrocarbonate-modified amino acid residues in α1-acid glycoprotein by high performance liquid chromatography electrospray ionization-mass spectrometry and matrix-assisted laser desorption/ionization time-of-flight-mass spectrometry. Analyt. Biochem. 1998;257:176–185. doi: 10.1006/abio.1997.2552. [DOI] [PubMed] [Google Scholar]
- 32.Kalkum M, Przybylski M, Glocker MO. Structure Characterization of Functional Histidine Residues and Carbethoxylated Derivatives in Peptides and Proteins by Mass Spectrometry. Bioconj. Chem. 1998;9:226–235. doi: 10.1021/bc970162t. [DOI] [PubMed] [Google Scholar]
- 33.Tsubaki M, Kobayashi K, Ichise T, Takeuchi F, Tagawa S. Diethyl Pyrocarbonate Modification Abolishes Fast Electron Accepting Ability of Cytochrome b561 from Ascorbate but Does Not Influence Electron Donation to Monodehydroascorbate Radical: Identification of the Modification Sites by Mass Spectrometric Analysis. Biochem. 2000;39:3276–3284. doi: 10.1021/bi991883d. [DOI] [PubMed] [Google Scholar]
- 34.Jin XR, Abe Y, Li CY, Hamasaki N. Histidine-834 of Human Erythrocyte Band 3 Has an Essential Role in the Conformational Changes That Occur during the Band 3-Mediated Anion Exchange. Biochem. 2003;42:12927–12932. doi: 10.1021/bi0350809. [DOI] [PubMed] [Google Scholar]
- 35.Qin K, Yang Y, Mastrangelo P, Westaway D. Mapping Cu(II) binding sites in prion proteins by diethyl pyrocarbonate modification and matrix-assisted laser desorption ionization-time of flight (MALDI-TOF) mass spectrometric footprinting. J. Biol. Chem. 2002;277:1981–1990. doi: 10.1074/jbc.M108744200. [DOI] [PubMed] [Google Scholar]
- 36.Mendoza VL, Antwi K, Baron-Rodriguez MA, Blanco C, Vachet RW. Structure of the Preamyloid Dimer of β-2-Microglobulin from Covalent Labeling and Mass Spectrometry. Biochem. 2010;49:1522–1532. doi: 10.1021/bi901748h. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Fraczkiewicz R, Braun W. Exact and efficient analytical calculation of the accessible surface areas and their gradients for macromolecules. J. Comput. Chem. 1998;19:319–333. [Google Scholar]
- 38.Verdone G, Corazza A, Viglino P, Pettirossi F, Giorgetti S, Mangione P, Andreola A, Stoppini M, Bellotti V, Esposito G. The solution structure of human β2-microglobulin reveals the prodromes of its amyloid transition. Prot. Sci. 2002;11:487–499. doi: 10.1110/ps.29002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Srikanth R, Mendoza VL, Bridgewater JD, Zhang G, Vachet RW. Copper Binding to β-2-Microglobulin and Its Pre-Amyloid Oligomers. Biochem. 2009;48:9871–9881. doi: 10.1021/bi901172y. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Mendoza VL, Baron-Rodriguez MA, Blanco C, Vachet RW. Structural Insights into the Pre-Amyloid Tetramer of β-2-Microglobulin from Covalent Labeling and Mass Spectrometry. Biochem. 2011;50:6711–6722. doi: 10.1021/bi2004894. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Solomons TWG, Fryhle C. Organic Chemistry. 9th edn. New York: John wiley & Sons; 1998. [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.