Abstract
Toxoplasma gondii, like most apicomplexan parasites, possesses a nessential relict chloroplast, the apicoplast. Several apicoplast membrane proteins lack the bipartite targeting sequences of luminal proteins. Vesicles bearing these membrane proteins are detected during apicoplast enlargement, but the means of cargo selection remains obscure. We used a combination of deletion mutagenesis, point mutations, and protein chimeras to identify a short motif prior to the first transmembrane domain of the T. gondii apicoplast phosphate transporter 1 (APT1) that is necessary for apicoplast trafficking. Tyrosine 16 was essential for proper localization; any substitution resulted in misdirection of APT1 to the Golgi body. Glycine 17 was also important, with significant Golgi body accumulation in the alanine mutant. Separation of at least eight amino acids from the transmembrane domain was required for full motif function. Similarly placed YG motifs are present in apicomplexan APT1 orthologues and the corresponding N-terminal domain from Plasmodium vivax was able to route T. gondii APT1 to the apicoplast. Differential permeabilization demonstrated that both the N-and C- termini of APT1 are exposed to the cytosol. We propose that this YG motif facilitates APT1 trafficking via interactions that occur on the cytosolic face of nascent vesicles destined for the apicoplast.
Introduction
Most apicomplexan parasites, such as Toxoplasma gondii and Plasmodium spp., possess a degenerate non-photosynthetic plastid called the apicoplast, adopted from an ancient algal endosymbiont. The apicoplast, like the chloroplasts of related photosynthetic chromalveolates, is surrounded by four membranes. The outer membranes were presumably derived from the phagosomal membrane and the algal plasma membrane, and the inner two membranes from the outer and inner chloroplast membranes. The apicoplast is an essential organelle (1–3). Since it does not have a counterpart in the animal host, it is a promising target for therapeutic intervention in diseases such as toxoplasmosis or malaria.
Proteins targeted to the lumen of the apicoplast have a n N-terminal bipartite targeting sequence that is processed upon trafficking, and bioinformatic analysis has yielded apicoplast metabolomes for those apicomplexans with sequenced genomes including Toxoplasma, Plasmodium, Babesia, Theileria, Eimeria and Neospora. Among these, there is some variation in the metabolic pathways residing in the apicoplast (4). The apicoplasts of all species characterized to date contain an isoprenoid biosynthetic pathway and an iron/sulfur cluster assembly pathway (4). In Toxoplasma gondii and Plasmodium spp, the apicoplast also houses fatty acid, heme, and lipoic acid biosynthetic pathways. The carbon sources for apicoplast biosynthetic pathways, triose phosphate and phosphoenolpyruvate, are translocated across apicoplast membranes by an apicoplast phosphate transporter (APT) (5;6), which is a member of the plastid phosphate translocator family. Additionally APT1 functions in a triose phosphate/3-phosphoglycerate shuttle, allowing the capture of ATP and reducing equivalents within the apicoplast (6). T. gondii has one APT family member, APT1 (TGME49_061070), that may inhabit multiple membranes of the apicoplast (7), while Plasmodium has two APT proteins, PfoTPT in the outer membrane, and PfiTPT, likely in the innermost membrane (5;8). Other apicomplexans (excluding Cryptosporidium spp) are predicted to have APT1 orthologues, and some have additional APT paralogues as well. TgAPT1 is essential in T. gondii, since conditional knockdown abruptly stops parasite growth and kills the majority of cells within 3 days (6).
The Plasmodium falciparum transporter, PfiTPT, has the same type of bipartite targeting sequence at its N-terminus as those present on proteins targeted to the lumen of the apicoplast. These bipartite sequences, which contain an N-terminal signal sequence followed by a transit peptide resembling those that target proteins to chloroplasts, first direct nascent proteins to the ER and from there to the apicoplast. However, PfoTPT and T. gondii APT1 lack an identifiable bipartite targeting sequence as do two other proteins associated with membranes of the T. gondii apicoplast, the protease FtsH1 (9) and the thioredoxin ATrx1 (10) as well as some other recently identified proteins (11). Unlike many other apicoplast proteins, there is no evidence for processing of APT1 during trafficking.
We have shown that like apicoplast luminal proteins (12;13), both APT1 (7) and FtsH1 (9) likely traffic through the ER en route to the apicoplast. Unlike luminal proteins, these membrane proteins are readily detected on vesicles at the time of apicoplast enlargement prior to division (7;9). Like the apicoplast, these vesicles are decorated with phosphatidyl insoitol 3-phosphate. (14). By analogy with vesicular transport to other cellular destinations, cytosolically disposed regions of apicoplast membrane proteins could contain motifs that allow their packaging into vesicles. Here we show that a tyrosine-containing motif prior to the first transmembrane (TM) domain of APT1 is necessary for its proper trafficking. With the exception of a deletion mutant that did not escape the endoplasmic reticulum (ER), APT1 mutants that did not localize to the apicoplast were found in the Golgi body. We examine spacing of the motif from the TM domain and its conservation in Apicomplexa. Our experiments demonstrate that the N-terminus of APT1 faces the cytosol, suggesting that it may interact with proteins that collect membrane protein cargo for transit to the apicoplast.
Results
The APT1 N terminal domain is necessary for apicoplast targeting
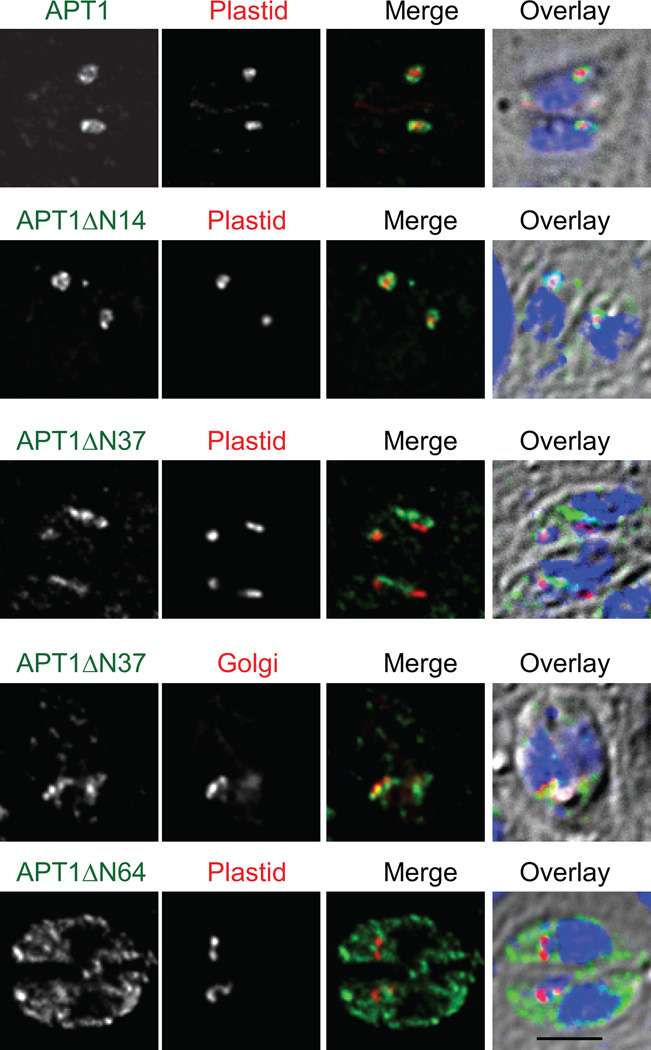
T, gondii APT1 has multiple TM domains, the first of which is preceded by 42 aa (or 41 aa, depending on the topology prediction algorithm). Given that the sequences mediating transport of many apicoplast proteins lie at the N-terminus, we tested whether N-terminal motifs might be required for localization of APT1. Three N-terminal deletion constructs of APT1 C-terminally tagged with HA epitopes were prepared (all APT1 constructs used the APT1 promoter unless otherwise noted). Plasmids were transfected into T. gondii cell lines expressing either an apicoplast-targeted red fluorescent protein marker or the Golgi marker GRASP55 fused to YFP (15) and the parasites subjected to immunofluorescence analysis (IFA) (Figure 1). APT1 lacking aa 2–14 (APT1ΔN14-HA) showed the characteristic circumplastid staining pattern seen with wild type APT1 (7). In contrast, the deletion protein which begins at aa 38 (APT1ΔN37-HA), did not show circumplastid localization, but localized primarily to the Golgi body as shown by colocalization with GRASP55-YFP. Deleting through the first TM domain to aa 64 (APT1ΔN64-HA) yielded a protein that was retained in the ER. Since loss of a TM domain is likely to impact proper folding and assembly, this deletion may indirectly abrogate trafficking. Thus, a motif necessary to direct APT1 to the apicoplast appeared to be encoded in the N-terminal region prior to the first TM domain, between aa 14 and aa 38.
Figure 1. N-terminal deletions of APT1 alter its targeting.
APT1 or its mutants lacking aa 2–14 (ΔN14), 2–37 (ΔN37), or 2–64 (ΔN64) were C-terminally tagged with 4 HA epitopes and co-expressed with a plastid marker (S+T(ACP)-HcRed) or a Golgi marker (GRASP55-YFP) in T. gondii parasites, as indicated. Stable transfectants were processed for IFA. In this and other figures, APT1 in parasites bearing the plastid marker was revealed with anti-HA mAb coupled to FITC whereas in those bearing the Golgi marker it was revealed with anti-HA mAb coupled to Alexa 594. Nonetheless, in all color images, anti-HA is shown in green, the organelle markers in red and DAPI in blue. The three color merge is overlaid onto a DIC image. Bar = 2 µM.
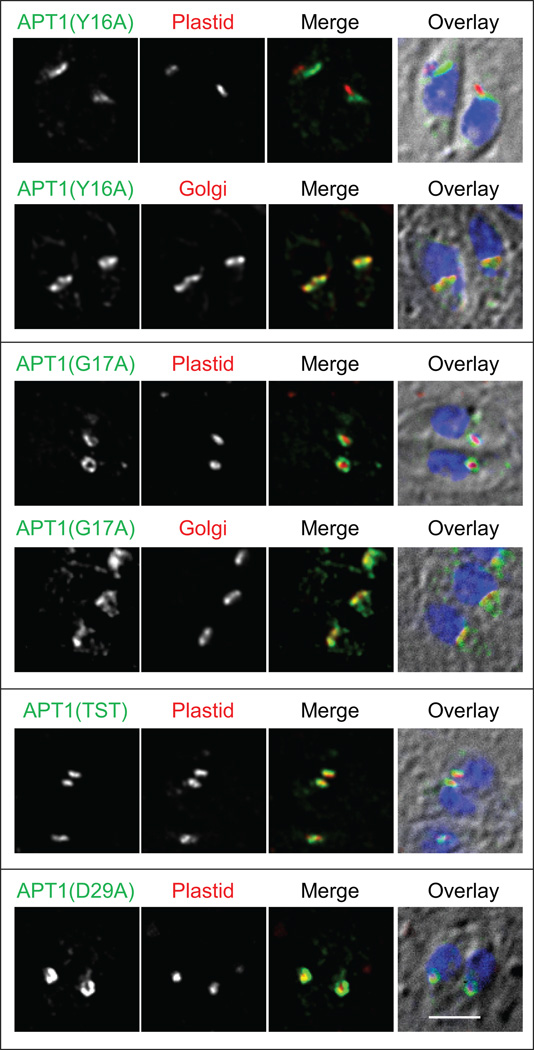
To identify amino acids in this region of the protein that are important for targeting, non-alanine residues between aa 15 and 37 were mutated individually or in combination to alanine (Table 1). Changing either Y16 or G17 to alanine redirected all (Y16) or most (G17) of the detected protein from the apicoplast to the Golgi in transient tranfectants, but alanine substitution at other residues retained apicoplast localization (Figure 2 and Table 1). Although vesicles bearing wild type APT1 are commonly observed during apicoplast elongation ((7) see example in Figure S1), no vesicles bearing APT1(Y16A) were detected. Parasites expressing N-terminally tagged APT1 and APT1(Y16A) recapitulated the findings seen with C-terminally tagged protein (not shown).
Table 1.
Alanine scanning mutants of residues 15–36 and their subcellular location following transient transfection
| Mutationa | Location |
|---|---|
| Q15A | apicoplast |
| Y16A | Golgi |
| G17A | Golgi+apicoplast |
| T18A | apicoplast |
| V19A | apicoplast |
| S20A | apicoplast |
| T21A | apicoplast |
| T18A S20A T21A | apicoplast |
| G22A G23A | apicoplast |
| R25A P26A K28A | apicoplast |
| D29A | apicoplast |
| L30A E31A S32A | apicoplast |
| Q33A S35A P36A | apicoplast |
Residues 24, 27, and 34 are alanine in wild type APT1.
Figure 2. Tyrosine 16 and glycine 17 are important for apicoplast targeting of APT1.
Amino acids 15–36 were changed to alanine either individually or in combination and the mutant proteins were expressed in T. gondii bearing either the apicoplast marker or the Golgi marker as indicated. IFA was conducted as in Figure 1. Labels above the grayscale panels indicate the color shown in the merged images. IFAs of APT1(G17A)-HA with GRASP55 and the triple mutant APT1(T18A,S20A,T21A)-HA were performed with stable transfectants; the remainder are from transient expression. Bar = 2 µM.
Tyrosine-based motifs involved in sorting of proteins to vesicles follow the consensus YXXϕ, where ϕ is a bulky hydrophobic aa (16–19). Mutating V19 (three residues after Y16) to alanine (Figure S1) or to serine (not shown) did not abrogate targeting. The hydroxylated residues following the targeting sequence were also dispensable. Di-acidic motifs, typically DXE or DXD (where X can be any amino acid), can function as a “ticket” for protein exit from the ER via interaction with Sec24 (20) and have been shown to be functional in P. falciparum (21). However, disrupting APT1’s sole di-acidic motif by mutation of either D29 or E31 to alanine did not alter apicoplast targeting (Figure 2). The mutant APTs which retained strong apicoplast targeting showed not only circumplastid targeting in parasites with a compact apicoplast but also vesicle-type staining in parasites with an elongated apicoplast (Figure S1 shows the example of V19A). Thus a YG motif near the N-terminus of APT1 is necessary for its inclusion into vesicles and targeting to the apicoplast, whereas alanine is tolerated at other residues.
A similar deletion strategy to examine the C-terminus was not productive since no expression of proteins lacking the C-terminal 5, 128, 226 or 250 aa was seen following transient transfection or in stable transfectants. The presence of the deletion construct was confirmed by sequencing PCR products amplified from genomic DNA from the latter three clones. Interestingly, APT1 terminates five residues after the last predicted TM domain in a YG dipeptide. However concurrent mutation of these two residues to alanine did not affect trafficking to the apicoplast membrane (not shown).
APT1-nucleotide sugar transporter chimeras suggest that the N-terminal motif is not sufficient for proper trafficking
To assess whether the N-terminus of APT1 was sufficient for targeting to the apicoplast, we made chimeras between APT1 and a related transporter. Since T. gondii possesses only one transporter in the plastid phosphate translocator family, we turned to the next most closely related family, the nucleotide sugar transporters (22). We searched for such transporters in ToxoDB version 3 and identified a gene (TgME49_067380) that encoded a protein with strong similarity to human UDP-N-acetyl glucosamine transporter (E-value = 2 × 10−102). This gene, provisionally dubbed nucleotide sugar transporter 1 (NST1), was expressed in T. gondii along with the Golgi marker GRASP55-YFP. As shown in Figure S2A, the protein colocalized with GRASP55 to the bar-like Golgi structure. Carbonate extraction (Figure S2B) demonstrated that NST1 was, as predicted, an integral membrane protein. As NST1 was readily detected and localized to the next compartment of the default pathway after the ER, it was chosen as the chimera partner.
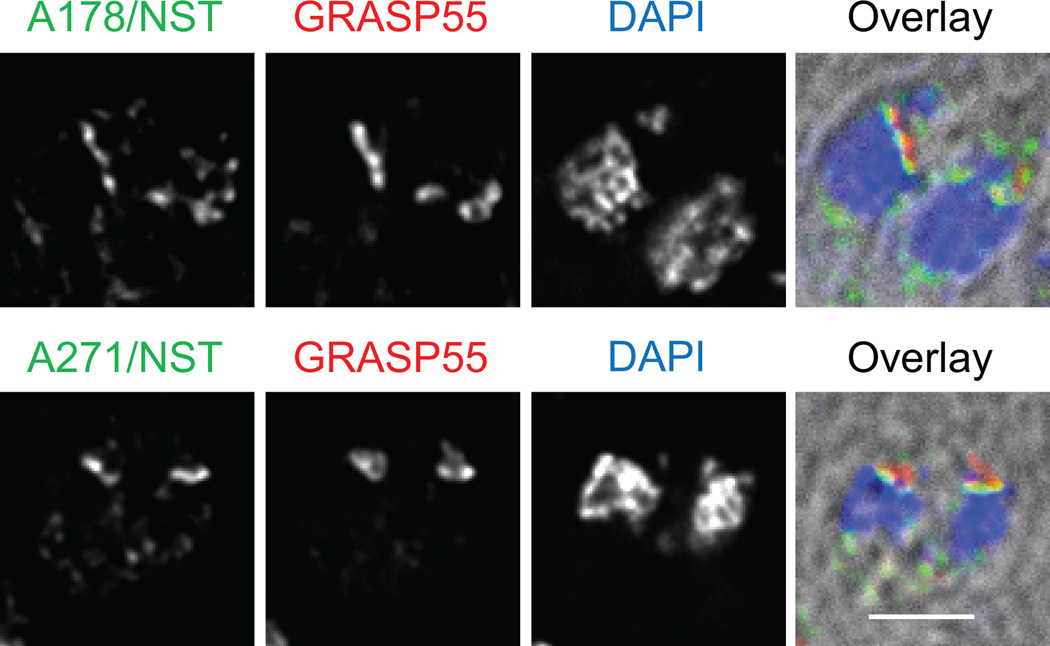
NST1 and APT1 have low primary sequence homology, so they were aligned based on a combination of alignment between plant sugar phosphate transporters and nucleotide sugar transporters (23), similarities in hydropathy plots, and local amino acid alignments. Chimeric proteins bearing the first 178 or 271 aa of APT1 fused to aa 188 or 253 of NST1 respectively were observed to colocalize with the Golgi apparatus (Figure 3). Thus the N-terminus of APT1 is not sufficient to achieve apicoplast targeting in this context. Since ER to Golgi trafficking in most organisms requires proper protein folding and assembly, the localization of these proteins to the Golgi body indicates that the proteins achieved a transport competent structure. However, transfectants stably expressing the chimeric proteins could not be isolated. Reversed chimeras of those above did not yield detectable expression, although the first 347 aa of NST1 followed by the C-terminal tail of APT1 (5 aa) was expressed and Golgi-localized.
Figure 3. Chimeras of the N-terminus of APT1 and the C-terminus of NST1 traffic to the Golgi body.
T. gondii stably expressing GRASP55-YFP were transiently transfected with constructs encoding the Chimeric proteins APT1(1–178)/NST1(188–347)-HA or APT1(1–271)/NST1(254–347)-HA. Parasites were stained with DAPI and with anti-HA mAb. Bar = 2 µM.
Targeting motif analysis
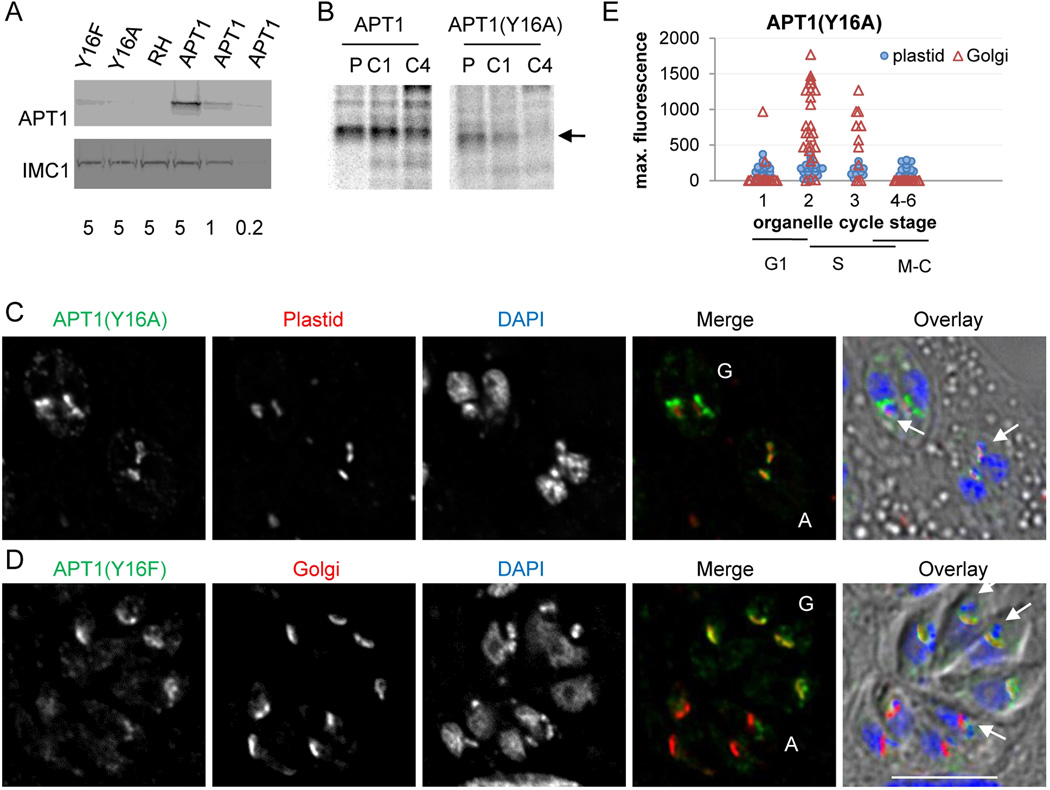
To test whether any other amino acid could functionally substitute for Y16, we systematically mutated it to all other residues and transfected the constructs into T. gondii (Table S1). Parasites were analyzed by IFA following transient transfection. Interestingly, even the most conservative mutations at this position did not permit significant targeting to the apicoplast, but showed Golgi body localization similar to the Y16A mutant. These include mutations to Thr (hydroxylated), Phe (aromatic) and Glu (potential substitute for phosphotyrosine). The expression level of the tagged mutant proteins in clonal lines of stable transfectants expressing APT1(Y16A) or APT1(Y16F) was lower than the properly targeted, tagged wild type protein as shown by western blot analysis (Figure 4A), indicating that the localization to the Golgi body was not the result of overexpression. Pulse chase immunoprecipitation analysis of the APT1 wild type and Y16A mutant cell lines demonstrated that the mutant protein is less stable than wild type (Figure 4B).
Figure 4. APT1 Y16 mutants are not overexpressed in clonal lines but show variable localization coordinated with the cell cycle.
A) Western blot of protein extracted from the indicated clonal lines of stable tranfectants probed with anti-HA mAb (upper panel) and anti-IMC1 antibodies as a loading control (lower panel). Numbers below the lanes indicate cell equivalents per lane (in millions). B) Pulse-chase analysis. Cultures of fibroblasts infected with T. gondii expressing either APT1-HA or APT1(Y16A)-HA were labeled with 35S cysteine and methionine for 1 hour (p) or labeled then chased for 1 hr (c1) or 4 hr (c4). Following immunoprecipitation with anti-HA mAb, samples were separated by SDSPAGE and the dried gels were exposed to a phosphor screen. The arrow marks full-length APT1. C) IFA was performed as in Figure 1. APT1(Y16A)-HA co-expressed with the plastid marker. D) APT1(Y16F)-HA co-expressed with the Golgi marker. For panels C and D, vacuoles where APT1 is predominantly Golgi-localized are marked “G”, while those where APT1 is predominantly in the apicoplast are marked “A” in the two color images. Arrows indicate examples of colocalization with the apicoplast in the overlay images. Bar = 5 µM E) Cell-cycle associated variation in staining pattern of APT1(Y16A). A clonal cell line co-expressing APT1(Y16A)-HA and the plastid marker S+T(ACP)-HcRed was stained with FITC-conjugated anti-HA, rabbit anti-IMC1, and DAPI. Vacuoles were analyzed to assess their stage in the organelle division cycle (stages 1–6) / cell cycle (G1, S, and mitosis / cytokinesis (M-C)) and APT1(Y16A) location and maximum staining intensity in the area over the apicoplast and in the Golgi body. Maximum staining in the Golgi body and apicoplast for each vacuole is plotted against their stage in the organelle division cycle. In vacuoles where no Golgi staining was detected, background fluorescence for that image is plotted.
IFA showed that in each of several clonal lines expressing APT1 Y16 mutant proteins (Y16A, Y16F, Y16T, and Y16P) Golgi staining was present in parasites in some vacuoles but absent in others. Additionally, faint apicoplast staining that was not apparent in transient transfectants was seen in some parasites within clonal lines stably expressing Y16A, Y16F, and Y16T (but not Y16P) (Figure 4 C, D; Y16T not shown). Parasites within a vacuole transit the cell cycle together suggesting the above variation in subcellular localization could be linked to the cell cycle and organellar division. Vacuoles bearing parasites from the APT1(Y16A)-HA stable clonal line co-expressing the plastid marker S+T(ACP)-HcRed were staged as described (24) by assessing nucleus division, apicoplast shape and location, and formation of the inner membrane complex (Figure 4E). The signal from APT1(Y16A) at the apicoplast and Golgi body was assessed by measuring the peak intensity of signal at the HcRed stained plastid and the Golgi body (when detected). Overall, in this experiment, approximately 25% of parasites showed no staining, 20% had plastid staining only (at least 2-fold above background), 25% had Golgi body staining only, and 30% had signal at both organelles. The signal at the plastid varied from undetectable to about 3 fold above background regardless of the parasite stage. The low level of plastid-associated signal may be why we did not detect it in transient transfections, where there was less time for protein accumulation. Golgi body staining was absent in cells undergoing mitosis or cytokinesis (plastid division stages 4–6) and rare in early G1 (stage 1). In contrast, the percentage of cells with localization to the Golgi body as well as the intensity of staining increased later in G1 and S (stages 2–3), although with significant inter-vacuole variation in staining intensity. APT1 mutant proteins were never seen in the plasma membrane or secretory organelles of the parasites. Taken together our findings suggest that APT1 mutants with residues other than Y at position 16 are predominantly misdirected to the Golgi, and are degraded shortly after Golgi body division.
A stable clonal line expressing APT1 mutated at G17 to glutamic acid also showed protein mistargeted to the Golgi body in some vacuoles (like the Y16 mutants, others had no Golgi staining,). However, all of the parasites showed significant plastid accumulation, often reaching maximal levels similar to those seen in the Golgi. Thus the APT1(G17E) mutant, like APT1(G17A), is partially functional in localization. In contrast, when G17 was mutated to proline, the detected protein in transient transfectants was exclusively Golgi-localized. The order of the residues (YG) is important since inverting the dipeptide to GY mis-directed the protein to the Golgi (not shown).
Conservation of the targeting motif
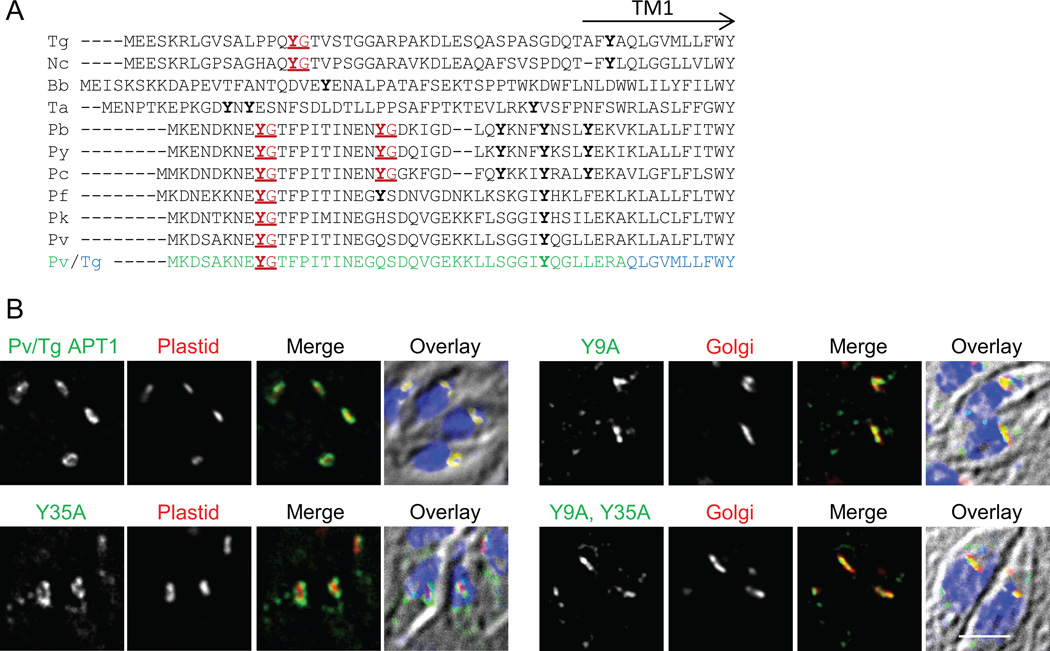
We reasoned that a targeting motif might be conserved among APT1 orthologues in other apicomplexans. Sequence comparisons between the N-terminus of T. gondii APT1 and its orthologues revealed a YG dipeptide in the closely related parasite Neospora caninum as well as in multiple Plasmodium species (Figure 5A). Although in T. gondii, N. caninum and Plasmodium species the motif is preceded by an amino acid with a side chain containing a carbonyl (glutamine or glutamic acid) and is followed by a threonine, changing these positions to alanine did not interrupt trafficking to the T. gondii apicoplast (see Figure 2 and Table S1). Theileria and Eimeria also had a tyrosine a similar distance from the first TM domain but it was followed by glutamic acid instead of glycine, suggesting that YE, which is partially functional in T. gondii APT1 (as described above), might be a functional motif in these species.
Figure 5. N-terminal extensions of apicomplexan APT1 homologues have similar YG motifs.
A) Alignment of the N-terminal extension of various apicomplexan APT1 predicted proteins. Y residues are bold, YG motifs are highlighted red and underlined. The predicted location of the first TM domain of T. gondii APT1 is indicated (TM1). The species and systematic gene identifiers (from ToxoDB, PiroplasmaDB and PlasmoDB) are: Nc, Neospora caninum (NCLIV_026210); Bb, Babesia bovis (I004950); Ta, Theileria annulata (TA05100); Pb, Plasmodium berghei (PBANKA_110790); Pv, P. vivax (pvx_097975); Pc, P. chabaudi (PCHAS_11076); Pf, P. falciparum (PFE0410w), Pk, P. knowlesi (PKH_102460); and Py, P. yoelii (PY00389). Pv/Tg depicts the sequence of the chimeric protein commencing with the N-terminal region of the P. vivax orthologue (green), followed by the T. gondii sequence (blue).
B) The YG motif of the P. vivax N-terminal domain can function in apicoplast targeting of APT1. Transiently transfected parasites expressing the Pv/Tg APT1 fusion protein depicted in panel A) or tyrosine to alanine mutants thereof were subjected to IFA as described in Figure 1. Parasites also expressed a marker of the indicated organelles. Bar = 2 µM.
If the tyrosine motif at the N-terminus of APT1 is a functional feature required for proper targeting, then the N-terminus of APT1 orthologues should allow APT1 targeting and that targeting should be disrupted by mutation of the YG targeting motif. To test these predictions, a chimera was constructed fusing aa 1–42 of the APT1 orthologue of P. vivax, which contains a YG and a YQ dipeptide (Figure 5A), to aa 47 of T. gondii APT1. The junction region of this chimera lies just before or within the first TM domain in a region of strong similarity (the start of first TM domain of the P. vivax protein is predicted to be at aa 38, 41, or 44, depending on the topology prediction algorithm). Like TgAPT1, this chimera targeted to the apicoplast periphery (Figure 5B). The tyrosines corresponding to the YG at 9–10 and YQ at 35–36 were mutated to alanine individually as well as together, and the trafficking of the proteins was assessed (Figure 5B). In transient and stable tranfectants, mutation of Y9, as well as concurrent mutation of both tyrosines, abrogated apicoplast localization. In contrast mutation of Y35 alone had no discernable effect on trafficking. Therefore the YG motif at residue 9–10 is essential for trafficking of the chimeric protein while the YQ at 35–36 is not. Thus the N-terminal region of APT1 from another apicomplexan also contains a YG motif that is functional in apicoplast targeting.
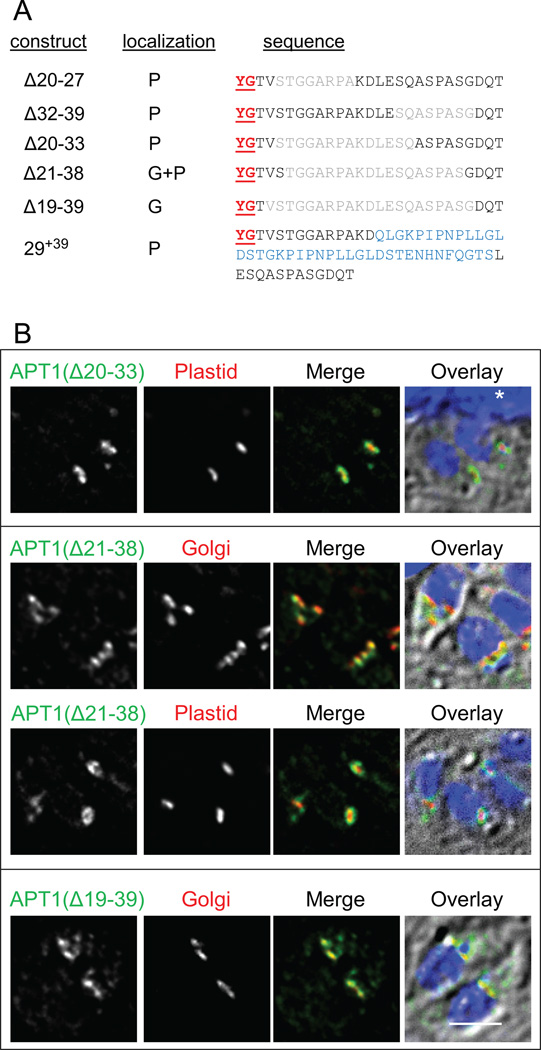
Spacing of the YG motif from the membrane is important
Previous studies have indicated that the C-terminal YG motifs of LAMP1 and HLA-DM to be at least 6 aa from the TM domain to function in targeting to the lysosome (25;26). To test whether spacing between the YG targeting motif and the TM domain of APT1 affects trafficking we constructed a series of deletion and insertion mutants with modifications between the YG motif and the first TM domain (Figure 6A). The YG motif at aa 16–17 is ~25 residues from the predicted start of the first TM domain. We therefore analyzed the spacing constraints of the APT1 YG motif with respect to the TM domain. We generated overlapping constructs deleting as few as 8 and up to 21 residues between the YG motif and the TM domain (Figure 6B). In transient transfectants, deletion of up to 14 aa had no detectable effect on localization, whereas deletion of 18 aa (Δ21–38) caused predominantly but not exclusively Golgi body localization. Deleting 21 aa (Δ19–39), such that the YG motif is separated from the TM domain by only ~4 aa, yielded only Golgi body localization. This is unlikely to be due to a specific sequence requirement since each residue had either been mutated or deleted in other constructs without abrogating targeting. In contrast to requirement for a minimal separation from the TM domain, addition of 39 aa between residues 29 and 30 did not interfere with trafficking. Thus for full function the YG motif must be more than 8 residues from the predicted TM domain but may be as much as 64 residues distant.
Figure 6. Space between the targeting motif and the membrane is important for targeting.
Insertions and small deletions do not affect targeting, while large deletions re-direct the protein to the Golgi body. A) APT1 deletion and insertion constructs used in this study. Dominant targeting to the plastid (P) and Golgi body (G) are indicated. Deleted amino acids are colored gray, those inserted are blue. B) Localization of deletion mutants. APT1 with the indicated deletions was co-expressed with either the plastid or the Golgi marker and IFA was conducted as described in Figure 1. The asterisk in the Δ20–33 overlay marks a portion of the host cell nucleus. Bar = 2 µM.
The N-terminus of APT1 is cytosolic in a Golgi localized mutant
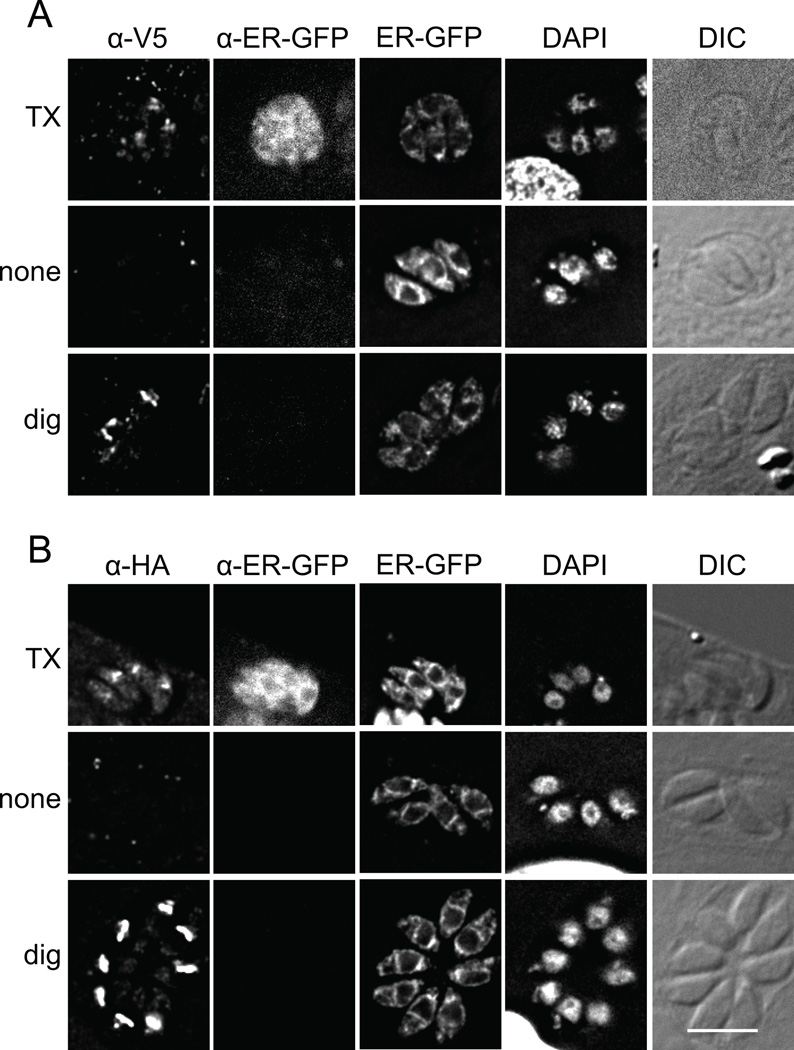
Membrane proteins which exit the ER for the Golgi typically are packaged into vesicles by virtue of motifs within cytosolic domains. In silico predictions using TopPred (27) suggested the N-terminal domain was cytosolic and had seven TM domains, TMHMM (28) predicted with 68% likelihood the N-terminus was cytosolic and had six TM domains, and TMPred (29) stated the orientation was ambiguous and the protein had eight TM domains. Earlier studies of PfoTPT, suggested that the N-and C-termini of the protein face the cytosol (8), although controls for these experiments could be complicated by the multiple membranes of the apicoplast, and their possible fragility during lysis. To experimentally assess whether the YG targeting motif is exposed to the cytosol or the ER lumen upon synthesis, we performed IFA on differentially permeabilized cells. These experiments were simplified by using the Golgi-localized mutant Y16A, since it is not known which membrane(s) of the apicoplast harbor APT1, and because markers available for the various apicoplast subcompartments are lacking. Because topology of proteins is stable once inserted into the ER membrane, analysis of the Golgi-localized mutant APT1 would reveal the original topology in the ER and avoid the complication of multiple membranes. A stable T. gondii line was prepared expressing APT1(Y16A) tagged with V5 and HA epitopes at the N-terminus (between aa 3 and 4) and C-terminus respectively. To assist in monitoring permeablization of the endomembrane system, the parasites also expressed a soluble GFP protein localized to the ER lumen, signal(P30)-GFP-HDEL (30).
Transfectants were used to infect fibroblasts on coverslips. Prior to addition of antibodies, the samples were treated with either Triton X-100 to permeabilize all membranes, digitonin to permeablize the plasma membrane of host cell and parasite, or buffer alone. ER-luminal GFP and the N-terminal and C-terminal epitope tags on APT1(Y16A) were detected following Triton X-100 permeabilization while none of these were detected in the absence of detergent (Figure 7). Upon digitonin treatment, both N-terminal and C-terminal tags on APT1(Y16A) were accessed by antibodies, but ER-luminal GFP was not. Thus under conditions in which the ER membrane remains intact, both termini of APT1 were available to antibodies, indicating that they are located within the cytosol. Hence the T. gondii protein has an even number of TM domains.
Figure 7. The N-terminus of APT1 faces the cytosol during trafficking.
T. gondii expressing V5t-APT1(Y16A)-HA and an ER-luminal GFP marker were fixed then permeabilized with Triton X-100, digitonin, or no detergent as indicated. Samples were then probed with anti-V5 mAb (A) or anti-HA mAb (B) as well as rabbit anti-GFP antibodies. The antibodies were premixed with Fab anti-mouse Ig coupled to Alexa 568 and Fab anti-rabbit Ig coupled to Alexa 680 respectively. Intrinsic fluorescence of ER-localized GFP and DAPI staining are also shown, along with DIC images. To facilitate proper scaling of the fluorescence signal, wild type parasites were included on each coverslips as a negative control (not shown). In some DAPI images, part of the host nucleus is visible. Bar = 5 µM.
Discussion
Like a few other known apicoplast membrane proteins, APT1 lacks a canonical bipartite apicoplast targeting motif—raising the question of which motifs or structures route it to the organelle. The studies reported here show that the region of the protein prior to the first TM domain is essential for proper localization. This section of the protein contains sequences that resemble the tyrosine-based sorting motifs and di-acidic motifs that function within the secretory system of other eukaryotes. The di-acidic motif was shown to be dispensable for proper targeting while the YG motif was essential. Interestingly, in contrast to the decreased efficiency of ER exit seen after mutation of tyrosine-based motifs of some membrane proteins (16–19), probably by reducing cargo assembly into COPII vesicles (31), the YG mutants of APT1 left the ER and localized to the Golgi body. No residues were able to substitute for Y16, although replacement of G17 with glutamic acid retained some plastid targeting function. This YG motif is followed by threonine and valine and thus closely resembles a tyrosine-based motif (YXXϕ). However, unlike canonical tyrosine-based motifs, mutation of the hydrophobic residue (valine) to alanine or serine did not abrogate trafficking. The use of multiple secondary structure prediction algorithms suggested that the N-terminus of APT1 is not folded into α helices or β strands, reminiscent of the recent finding that transit peptides of proteins destined for the apicoplast lumen are unstructured (32).
Like the C-terminal tyrosine-based motifs of LAMP1 and HLA-DM, the N-terminal APT1 YG motif was not functional if placed within a few amino acids of the TM domain. These findings may explain why the C-terminal YG motif of T. gondii does not appear to be relevant for targeting. In contrast with the tyrosine motif of HLA-DM which functioned optimally with a 7–13 aa spacing from the TM domain (26), the APT1 YG motif tolerated a 64 aa separation.
Tyrosine offers multiple opportunities for interactions with other residues, for example, the μ2 subunit of AP2 adaptor protein binds tyrosine-based motifs and employs a combination of hydrophobic interactions with the tyrosine’s aromatic ring and hydrogen bonding with its hydroxyl moiety, providing specificity and binding affinity (33). FtsH1, the other apicoplast TM protein lacking a bipartite targeting sequence, does not bear a YG motif close to its N-terminus, but it does have multiple tyrosines prior to its sole TM domain. Further experimentation will be required to determine whether one of these tyrosines could potentially fulfill the same targeting function. Tyrosine-based targeting motifs have been proposed as targeting sequences for other proteins in T. gondii. While initial studies suggested that T. gondii ROP2 was targeted to rhoptries via AP1 binding a cytoplasmically facing tyrosine-based motif (34), those studies have been complicated by additional work which questioned the topology of this family of proteins (35) and demonstrated that the tyrosine-based motif of the related protein ROP4 was dispensable for localization (36).
To address whether sequences in addition to the N-terminal extension were required for APT1 trafficking we tested chimeras with a distantly related Golgi-localized TM protein. When the C-terminal one-third of the APT1 (downstream of aa 271) was replaced by the homologous region of this protein, the chimera was able to exit the ER (indicating successful folding), but localized to the Golgi body. This suggests that other residues of APT1, perhaps acting in concert with the N-terminal YG motif, are required for localization to the apicoplast. An alternative explanation is that the NST1 region of the chimera bears a motif that strongly enhances packaging into COPII vesicles destined for the Golgi body, behaving dominantly over the YG apicoplast-targeting motif. In fact this region of NST1 region bears a DXD motif. It also bears two tyrosine-based motifs, but they are either within or immediately adjacent to a TM domain and therefore are unlikely to be functional in this respect.
Three routes have been proposed for trafficking proteins from the ER to the apicoplast: 1) vesicular trafficking from the ER to the Golgi body and then to the apicoplast; 2) vesicle trafficking directly from the ER to the apicoplast; and 3) transient contact between the ER and the apicoplast, enabling vesicle-independent trafficking (37). The pathway of trafficking of membrane proteins to the apicoplast has not been dissected, but earlier studies showed that luminal proteins are not routed through the Golgi body (38;39). Furthermore, we have demonstrated the presence of vesicles bearing apicoplast membrane proteins (7;9;10) and in preliminary studies have observed their presence when Golgi trafficking is inhibited by brefeldin A. Previous immunoelectron microscopy analysis of APT1-HA did not reveal any APT1 in the Golgi body (7). These data suggest that model 2 is likely correct, at least for some membrane proteins. However, the data presented here are compatible with any of the three models, with the difference being location in the cell where the targeting motif functions. In model 2 the interaction would occur while the protein is in the ER, allowing packing into vesicles destined for the apicoplast. Movement of proteins from the ER to the Golgi is considered to be a default pathway that requires proper protein folding, although its efficiency can be enhanced by specific motifs. Thus, an apicoplast membrane protein mutated to lack a recognizable apicoplast targeting signal would likely be routed to the Golgi by bulk flow, as we observed. Based on these results as well as other data described above we propose that mistargeting of the APT1 mutants described above is the result of altering a motif required for specific packaging of TM cargo into ER-derived vesicles destined for the apicoplast. In some cases (e.g., Y16P and Δ19–39) the alterations completely blocked apicoplast targeting, whereas in others (e.g., Y16F and Δ21–38) a small amount of apicoplast localization was observed. Why the APT1 mutants are present in the Golgi body but absent from later secretory system destinations (e.g., plasma membrane) remains unclear. It is possible that the mutant APT1 is retained in the Golgi body by virtue of fortuitous characteristics, such as the lengths of the TM domains as has been seen to play a role in localization of single-pass membrane proteins within the secretory system of other organisms (40;41). Another possibility is that the mutant proteins are not retained, but rather are unstable if routed to more distal compartments.
The proteins required for generating the APT1-bearing vesicles remain unknown, but by analogy with other vesicles one would expect that membrane proteins would be recruited by virtue of cytosolic domains that interact with a component of the vesicle. We propose the existence of a cargo binding protein which is related to one of the other cargo binding proteins of the secretory system. Examples of such proteins are Sec24 in the COPII coat and the mu subunits of adaptor complexes mentioned above (17;42;43), which facilitate tyrosine-based sequestration of cargo during endocytic vesicle formation. T. gondii has a full complement of adaptor complexes (although their specific roles are not clear), but some apicomplexans appear to be missing specific subunits (43). In addition to Sec23 and Sec24, T. gondii has an additional Sec24-related protein of unknown function (TgGT1_081660), that is also present in P. falciparum (21) as well as other apicomplexans. The outermost membrane of the apicoplast is thought to derive from the endocytic vacuole of the progenitor apicomplexan that engulfed an alga bearing the gift of a chloroplast. The required trafficking machinery could have been generated by gene duplication and divergence, by repurposing existing trafficking machinery no longer required, or by utilizing machinery derived from the algal endosymbiont. Indeed three paralogues of components of the ER-associated degradation (ERAD) machinery, Der1, CDC48 and Ufd, are encoded on the residual algal nucleus of T. gondii’s chromalveolate cousin Guillardia theta (44) and are thought to be involved in protein import into its chloroplast. These three proteins have homologues targeted to the apicoplast in Plasmodium (45) and T. gondii (46) and one of these, Der1, is essential for importing proteins into the apicoplast of T. gondii (46). Thus there is a precedent for repurposing existing proteins to facilitate protein targeting to the apicoplast.
Materials and Methods
Parasite culture and transfection
T. gondii was grown in human foreskin fibroblasts in DMEM plus NuSerum as previously described (7). Plasmids were transfected as previously described into RHΔHXGPRT, or RHΔHXGPRT expressing the apicoplast marker S + T(ACP)-HcRed or the Golgi marker GRASP55-YFP, both driven by the TubA promoter.
Plasmid construction and mutagenesis
Primers for all plasmid constructions are listed in Table S2. APT1 and chimera expression plasmids were derived from the plasmid pHXGPRT apt1-APT1-HA (7), which encodes APT1 with a C-terminal 4 HA tag driven by the APT1 promoter. Expression of three of the C-terminal deletion constructs (APT1Δ128, Δ226 and Δ250) was driven by the DHFR promoter. Point mutants were constructed using oligonucleotide-based mutagenesis. APT1 was N-terminally tagged using site-directed mutagenesis to add Mun1 and Spe1 restriction sites between codons 3 and 4, and then inserting linkers (with Mun1 and Spe1 compatible ends) that encoded two V5 epitopes (and a TEV protease site) between the two restriction sites to yield V5t-APT1-HA. V5t-APT1(Y16A)-HA was generated by site directed mutagenesis of this plasmid. Similarly, Mun1 and Spe1 sites were introduced into APT1 at residues 29–30 and the same coding sequence (39 aa) was inserted to yield APT1-(29+39)-HA. In some constructs, the C-terminal HA tag was removed leaving just the N-terminal V5 tag. To replace the N-terminus of T. gondii with that of P. vivax, a Mun1 restriction site was introduced via a silent mutation at codons 47–48. Linkers encoding aa 1–42 (followed by a Mun1 half site) of the P. vivax orthologue were introduced into the Mun1 site. NST1 was amplified from RH strain cDNA, cloned into pGEM t-easy. To construct chimeric proteins, the desired regions of APT1 and NST1 were separately amplified using primers designed such that the products would contain the junction region. The two fragments were then stitched together by PCR to yield the gene fusion. NST1 was subcloned into pHXGPRT with a 4 HA tag, with expression driven from the DHFR promoter.
Immunofluorescence analysis and differential permeabilization
Fibroblasts growing on 12 mm round coverslips were inoculated with T. gondii and allowed to grow for 16–24 hr. Samples were fixed for 30 min at 4°C in 4% formaldehyde plus 0.0075% glutaraldehyde in PBS, and then blocked and permeablilized in PBS containing 2% BSA, 5% goat serum, and 0.1% NP-40 except as noted. HA-tagged proteins were detected with anti-HA monoclonal antibody (mAb) 3F10 coupled to FITC (Roche) or anti-HA mAb16B12 coupled to Alexa 594 (Invitrogen). IMC1 was detected with anti-IMC1 antiserum (47)(gift of Dr. Con Beckers) followed by goat anti-rabbit Ig Alexa 680 (Invitrogen). Samples were stained with DAPI and Texas red coupled streptavidin (where indicated). Samples were viewed on a DeltaVision RT imaging system (Applied Precision) using a 100 × 1.35 na objective; image stacks were deconvolved using the conservative ratio method, and a single image plane is presented. Images used for quantitation in Figure 4E were all acquired with the same exposure settings. Signal intensity was measured using the SoftWorx data inspector tool. Mean signal intensity of the six brightest contiguous pixels is presented for the region that co-localized with the apicoplast marker or the Golgi body (when detected) or listed as background for the Golgi body when staining was not detected.
Differential permeabilization was similar to (48) with modifications to make the procedure amenable to T. gondii. Fibroblasts growing on cover slips were co-infected with 104 RH cells plus 2×104 cells expressing V5t-APT1(Y16A)-HA plus signalP30-GFP-HDEL and allowed to grow for 16 – 24 hours. Cover slips were fixed as above and washed in PIPES buffer (200 mM sucrose, 10 mM PIPES pH 6.8, 100 mM KCl, 2.5 mM MgCl2, 1 mM EDTA) (all washes were 5 min). After a 5 min treatment with PIPES buffer alone or buffer containing 0.1% Triton X-100 or 40 µg/ml digitonin, coverslips were washed twice in PIPES buffer and then blocked for 30 minutes in 2% BSA in PIPES buffer. Anti-V5 mAb (Invitrogen) was labeled with Zenon goat anti-mouse Ig Alexa 594 (Invitrogen), and anti-GFP (a gift from Dr. Jim Cregg) was labeled with Zenon goat anti-rabbit Ig Alexa 680 (Invitrogen) as described by the manufacturer. Samples were incubated with labeled antibodies for 90 min, then washed for in PIPES buffer, followed by a wash in PBS 0.1% Triton X-100. After two additional washes in PBS, samples were again fixed in 4% paraformaldehyde in PBS at room temperature for 5 min to stabilize the Zenon reagent. They were then washed in PBS, DAPI stained and viewed as described above using a 60×, 1.4 NA objective.
Immunoblot analysis and immunoprecipitation
Immunoblots utilized previously described procedures (7) in which samples were lysed in Laemmli sample buffer and heat denatured at 55°C for 15 minutes. After SDS-PAGE and transfer to nitrocellulose, membranes were probed with mouse anti-HA.11 mAb (16B2, Covance) and rabbit-anti-IMC1 or anti-Mic5 (49) (a gift of Dr. Vern Carruthers) which were detected by anti-mouse IgG coupled to IRdye 800 and anti-rabbit IgG coupled to IRdye 680. Samples were scanned on an Odyssey imaging system (LiCor). Images were scaled with a gamma setting of 1.0.
In vivo labeling and immunoprecipitation were performed as described in (38), except that cells were lysed directly in the lysis buffer and precipitated with mono HA.11 and protein A coupled to Dynabeads (Invitrogen). Samples were separated by SDS-PAGE, and transferred to nitrocellulose followed by phosphorimaging.
Supplementary Material
Acknowledgements
We would like to thank Dr. Jeffrey Shaver for assistance in the laboratory, Dr. Con Beckers for anti-Mic5 antiserum, Dr. Vern Carruthers for anti-IMC1 antiserum, and Dr. Jim Cregg for anti-GFP antiserum. This work was supported by NIAID R01 AI50506 and R01 AI050506-08S1; the authors are solely responsible for its content.
Reference List
- 1.Fichera ME, Roos DS. A plastid organelle as a drug target in apicomplexan parasites. Nature. 1997;390:407–409. doi: 10.1038/37132. [DOI] [PubMed] [Google Scholar]
- 2.Goodman CD, Su V, McFadden GI. The effects of anti-bacterials on the malaria parasite Plasmodium falciparum. Mol Biochem Parasitol. 2007;152:181–191. doi: 10.1016/j.molbiopara.2007.01.005. [DOI] [PubMed] [Google Scholar]
- 3.Dahl EL, Shock JL, Shenai BR, Gut J, DeRisi JL, Rosenthal PJ. Tetracyclines specifically target the apicoplast of the malaria parasite Plasmodium falciparum. Antimicrob Agents Chemother. 2006;50:3124–3131. doi: 10.1128/AAC.00394-06. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Fleige T, Limenitakis J, Soldati-Favre D. Apicoplast: keep it or leave it. Microbes Infect. 2010;12:253–262. doi: 10.1016/j.micinf.2009.12.010. [DOI] [PubMed] [Google Scholar]
- 5.Lim L, McFadden GI. The evolution, metabolism and functions of the apicoplast. Philos Trans R Soc Lond B Biol Sci. 2010;365:749–763. doi: 10.1098/rstb.2009.0273. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Brooks CF, Johnsen H, van Dooren GG, Muthalagi M, Lin SS, Bohne W, Fischer K, Striepen B. The Toxoplasma apicoplast phosphate translocator links cytosolic and apicoplast metabolism and is essential for parasite survival. Cell Host Microbe. 2010;7:62–73. doi: 10.1016/j.chom.2009.12.002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Karnataki A, DeRocher A, Coppens I, Nash C, Feagin JE, Parsons M. Cell cycle-regulated vesicular trafficking of Toxoplasma APT1, a protein localized to multiple apicoplast membranes. Mol Microbiol. 2007;63:1653–1668. doi: 10.1111/j.1365-2958.2007.05619.x. [DOI] [PubMed] [Google Scholar]
- 8.Mullin KA, Lim L, Ralph SA, Spurck TP, Handman E, McFadden GI. Membrane transporters in the relict plastid of malaria parasites. Proc Natl Acad Sci USA. 2006;103:9572–9577. doi: 10.1073/pnas.0602293103. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Karnataki A, Derocher AE, Coppens I, Feagin JE, Parsons M. A membrane protease is targeted to the relict plastid of Toxoplasma via an internal signal sequence. Traffic. 2007;8:1543–1553. doi: 10.1111/j.1600-0854.2007.00637.x. [DOI] [PubMed] [Google Scholar]
- 10.Derocher AE, Coppens I, Karnataki A, Gilbert LA, Rome ME, Feagin JE, Bradley PJ, Parsons M. A thioredoxin family protein of the apicoplast periphery identifies abundant candidate transport vesicles in Toxoplasma gondii. Eukaryot Cell. 2008;7:1518–1529. doi: 10.1128/EC.00081-08. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Sheiner L, Demerly JL, Poulsen N, Beatty WL, Lucas O, Behnke MS, White MW, Striepen B. A systematic screen to discover and analyze apicoplast proteins identifies a conserved and essential protein import factor. PLoS Pathog. 2011;7 doi: 10.1371/journal.ppat.1002392. e1002392. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.DeRocher A, Hagen CB, Froehlich JE, Feagin JE, Parsons M. Analysis of targeting sequences demonstrates that trafficking to the Toxoplasma gondii plastid branches off the secretory system. J Cell Sci. 2000;113:3969–3977. doi: 10.1242/jcs.113.22.3969. [DOI] [PubMed] [Google Scholar]
- 13.Waller RF, Reed MB, Cowman AF, McFadden GI. Protein trafficking to the plastid of Plasmodium falciparum is via the secretory pathway. EMBO J. 2000;19:1794–1802. doi: 10.1093/emboj/19.8.1794. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Tawk L, Dubremetz JF, Montcourrier P, Chicanne G, Merezegue F, Richard V, Payrastre B, Meissner M, Vial HJ, Roy C, Wengelnik K, Lebrun M. Phosphatidylinositol 3-monophosphate is involved in toxoplasma apicoplast biogenesis. PLoS Pathog. 2011;7 doi: 10.1371/journal.ppat.1001286. e1001286. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Pelletier L, Stern CA, Pypaert M, Sheff D, Ngo HM, Roper N, He CY, Hu K, Toomre D, Coppens I, Roos DS, Joiner KA, Warren G. Golgi biogenesis in Toxoplasma gondii. Nature. 2002;418:548–552. doi: 10.1038/nature00946. [DOI] [PubMed] [Google Scholar]
- 16.Heineman TC, Connolly P, Hall SL, Assefa D. Conserved cytoplasmic domain sequences mediate the ER export of VZV HSV-1, and HCMV gB. Virology. 2004;328:131–141. doi: 10.1016/j.virol.2004.07.011. [DOI] [PubMed] [Google Scholar]
- 17.Renard HF, Demaegd D, Guerriat B, Morsomme P. Efficient ER exit and vacuole targeting of yeast Sna2p require two tyrosine-based sorting motifs. Traffic. 2010;11:931–946. doi: 10.1111/j.1600-0854.2010.01070.x. [DOI] [PubMed] [Google Scholar]
- 18.Sato K, Nakano A. Emp47p and its close homolog Emp46p have a tyrosine-containing endoplasmic reticulum exit signal and function in glycoprotein secretion in Saccharomyces cerevisiae. Mol Biol Cell. 2002;13:2518–2532. doi: 10.1091/mbc.E02-01-0027. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Bonifacino JS, Traub LM. Signals for sorting of transmembrane proteins to endosomes and lysosomes. Annu Rev Biochem. 2003;72:395–447. doi: 10.1146/annurev.biochem.72.121801.161800. [DOI] [PubMed] [Google Scholar]
- 20.Barlowe C. Signals for COPII-dependent export from the ER: what's the ticket out? Trends Cell Biol. 2003;13:295–300. doi: 10.1016/s0962-8924(03)00082-5. [DOI] [PubMed] [Google Scholar]
- 21.Lee MC, Moura PA, Miller EA, Fidock DA. Plasmodium falciparum Sec24 marks transitional ER that exports a model cargo via a diacidic motif. Mol Microbiol. 2008;68:1535–1546. doi: 10.1111/j.1365-2958.2008.06250.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Weber AP, Linka M, Bhattacharya D. Single, ancient origin of a plastid metabolite translocator family in plantae from an endomembrane-derived ancestor. Eukaryot Cell. 2006;5:609–612. doi: 10.1128/EC.5.3.609-612.2006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Knappe S, Flugge UI, Fischer K. Analysis of the plastidic phosphate translocator gene family in Arabidopsis and identification of new phosphate translocator-homologous transporters, classified by their putative substrate-binding site. Plant Physiol. 2003;131:1178–1190. doi: 10.1104/pp.016519. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Striepen B, Crawford MJ, Shaw MK, Tilney LG, Seeber F, Roos DS. The plastid of Toxoplasma gondii is divided by association with the centrosomes. J Cell Biol. 2000;151:1423–1434. doi: 10.1083/jcb.151.7.1423. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Rohrer J, Schweizer A, Russell D, Kornfeld S. The targeting of Lamp1 to lysosomes is dependent on the spacing of its cytoplasmic tail tyrosine sorting motif relative to the membrane. J Cell Biol. 1996;132:565–576. doi: 10.1083/jcb.132.4.565. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Potter PK, Copier J, Sacks SH, Calafat J, Janssen H, Neefjes JJ, Kelly AP. Accurate intracellular localization of HLA-DM requires correct spacing of a cytoplasmic YTPL targeting motif relative to the transmembrane domain. Eur J Immunol. 1999;29:3936–3944. doi: 10.1002/(SICI)1521-4141(199912)29:12<3936::AID-IMMU3936>3.0.CO;2-K. [DOI] [PubMed] [Google Scholar]
- 27.von Heijne G. Membrane protein structure prediction. Hydrophobicity analysis and the positive-inside rule. J Mol Biol. 1992;225:487–494. doi: 10.1016/0022-2836(92)90934-c. [DOI] [PubMed] [Google Scholar]
- 28.Krogh A, Larsson B, von Heijne G, Sonnhammer EL. Predicting transmembrane protein topology with a hidden Markov model: application to complete genomes. J Mol Biol. 2001;305:567–580. doi: 10.1006/jmbi.2000.4315. [DOI] [PubMed] [Google Scholar]
- 29.Hoffman K, Stoffel W. TMbase - A database of membrane spanning proteins segments. Biol. Chem. Hoppe-Seyler. 1993;374:166. [Google Scholar]
- 30.Hager KM, Striepen B, Tilney LG, Roos DS. The nuclear envelope serves as an intermediary between the ER and Golgi complex in the intracellular parasite Toxoplasma gondii. J Cell Sci. 1999;112:2631–2638. doi: 10.1242/jcs.112.16.2631. [DOI] [PubMed] [Google Scholar]
- 31.Miller EA, Beilharz TH, Malkus PN, Lee MC, Hamamoto S, Orci L, Schekman R. Multiple cargo binding sites on the COPII subunit Sec24p ensure capture of diverse membrane proteins into transport vesicles. Cell. 2003;114:497–509. doi: 10.1016/s0092-8674(03)00609-3. [DOI] [PubMed] [Google Scholar]
- 32.Gallagher JR, Matthews KA, Prigge ST. Plasmodium falciparum apicoplast transit peptides are unstructured in vitro and during apicoplast import. Traffic. 2011;12:1124–1138. doi: 10.1111/j.1600-0854.2011.01232.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Owen DJ, Evans PR. A structural explanation for the recognition of tyrosine-based endocytotic signals. Science. 1998;282:1327–1332. doi: 10.1126/science.282.5392.1327. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Hoppe HC, Ngo HM, Yang M, Joiner KA. Targeting to rhoptry organelles of Toxoplasma gondii involves evolutionarily conserved mechanisms. Nat Cell Biol. 2000;2:449–456. doi: 10.1038/35017090. [DOI] [PubMed] [Google Scholar]
- 35.El HH, Lebrun M, Fourmaux MN, Vial H, Dubremetz JF. Inverted topology of the Toxoplasma gondii ROP5 rhoptry protein provides new insights into the association of the ROP2 protein family with the parasitophorous vacuole membrane. Cell Microbiol. 2006;9:54–64. doi: 10.1111/j.1462-5822.2006.00767.x. [DOI] [PubMed] [Google Scholar]
- 36.Bradley PJ, Li N, Boothroyd JC. A GFP-based motif-trap reveals a novel mechanism of targeting for the Toxoplasma ROP4 protein. Mol Biochem Parasitol. 2004;137:111–120. doi: 10.1016/j.molbiopara.2004.05.003. [DOI] [PubMed] [Google Scholar]
- 37.Tonkin CJ, Kalanon M, McFadden GI. Protein targeting to the malaria parasite plastid. Traffic. 2008;9:166–175. doi: 10.1111/j.1600-0854.2007.00660.x. [DOI] [PubMed] [Google Scholar]
- 38.DeRocher A, Gilbert B, Feagin JE, Parsons M. Dissection of brefeldin A-sensitive and - insensitive steps in apicoplast protein targeting. J Cell Sci. 2005;118:565–574. doi: 10.1242/jcs.01627. [DOI] [PubMed] [Google Scholar]
- 39.Tonkin CJ, Struck NS, Mullin KA, Stimmler LM, McFadden GI. Evidence for Golgi-independent transport from the early secretory pathway to the plastid in malaria parasites. Mol Microbiol. 2006;61:614–630. doi: 10.1111/j.1365-2958.2006.05244.x. [DOI] [PubMed] [Google Scholar]
- 40.Ronchi P, Colombo S, Francolini M, Borgese N. Transmembrane domain-dependent partitioning of membrane proteins within the endoplasmic reticulum. J Cell Biol. 2008;181:105–118. doi: 10.1083/jcb.200710093. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Bulbarelli A, Sprocati T, Barberi M, Pedrazzini E, Borgese N. Trafficking of tail-anchored proteins: transport from the endoplasmic reticulum to the plasma membrane and sorting between surface domains in polarised epithelial cells. J Cell Sci. 2002;115:1689–1702. doi: 10.1242/jcs.115.8.1689. [DOI] [PubMed] [Google Scholar]
- 42.Kelly BT, Owen DJ. Endocytic sorting of transmembrane protein cargo. Curr Opin Cell Biol. 2011;23:404–412. doi: 10.1016/j.ceb.2011.03.004. [DOI] [PubMed] [Google Scholar]
- 43.Nevin WD, Dacks JB. Repeated secondary loss of adaptin complex genes in the Apicomplexa. Parasitol Int. 2009;58:86–94. doi: 10.1016/j.parint.2008.12.002. [DOI] [PubMed] [Google Scholar]
- 44.Sommer MS, Gould SB, Lehmann P, Gruber A, Przyborski JM, Maier UG. Der1-mediated pre-protein import into the periplastid compartment of chromalveolates? Mol Biol Evol. 2007;24:918–928. doi: 10.1093/molbev/msm008. [DOI] [PubMed] [Google Scholar]
- 45.Spork S, Hiss JA, Mandel K, Sommer M, Kooij TW, Chu T, Schneider G, Maier UG, Przyborski JM. An unusual ERAD-like complex is targeted to the apicoplast of Plasmodium falciparum. Eukaryot Cell. 2009;8:1134–1145. doi: 10.1128/EC.00083-09. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Agrawal S, van Dooren GG, Beatty WL, Striepen B. Genetic evidence that an endosymbiont-derived ERAD system functions in import of apicoplast proteins. J Biol Chem. 2009;284:33683–33691. doi: 10.1074/jbc.M109.044024. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Mann T, Beckers C. Characterization of the subpellicular network, a filamentous membrane skeletal component in the parasite Toxoplasma gondii. Mol Biochem Parasitol. 2001;115:257–268. doi: 10.1016/s0166-6851(01)00289-4. [DOI] [PubMed] [Google Scholar]
- 48.Lin S, Cheng D, Liu MS, Chen J, Chang TY. Human acyl-CoA:cholesterol acyltransferase-1 in the endoplasmic reticulum contains seven transmembrane domains. J Biol Chem. 1999;274:23276–23285. doi: 10.1074/jbc.274.33.23276. [DOI] [PubMed] [Google Scholar]
- 49.Brydges SD, Sherman GD, Nockemann S, Loyens A, Daubener W, Dubremetz JF, Carruthers VB. Molecular characterization of TgMIC5, a proteolytically processed antigen secreted from the micronemes of Toxoplasma gondii. Mol Biochem Parasitol. 2000;111:51–66. doi: 10.1016/s0166-6851(00)00296-6. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.