Abstract
Glycopeptides are extremely useful for basic research and clinical applications, but access to structurally-defined glycopeptides is limited by the difficulties in synthesizing this class of compounds. In this study, we demonstrate that many common peptide coupling conditions used to prepare O-linked glycopeptides result in substantial amounts of epimerization at the alpha position. In fact, epimerization resulted in up to 80% of the non-natural epimer, indicating that it can be the major product in some reactions. Through a series of mechanistic studies, we demonstrate that the enhanced epimerization relative to non-glycosylated amino acids is due to a combination of factors, including a faster rate of epimerization, an energetic preference for the unnatural epimer over the natural epimer, and a slower overall rate of peptide coupling. In addition, we demonstrate that use of 2,4,6-trimethylpyridine (TMP) as the base in peptide couplings produces glycopeptides with high efficiency and low epimerization. The information and improved reaction conditions will facilitate the preparation of glycopeptides as therapeutic compounds and vaccine antigens.
Keywords: epimerization, glyco-amino acid, solid-phase peptide synthesis, glycopeptides
Introduction
Glycopeptides are used extensively in basic research and a number of them are under investigation for various clinical applications.1 Glycopeptides are commonly used as structurally-defined models of complex glycoproteins for studying the roles of glycosylation, for understanding relationships between structure and function, and for modulating biological processes involving the parent glycoprotein.2 Glycopeptide fragments of glycoproteins also have significant potential as vaccine antigens.3-8 For example, glycopeptides bearing the Tn antigen (GalNAc alpha linked to a serine or threonine of a polypeptide chain), the TF antigen (Galβ1-3GalNAc alpha linked to a serine or threonine of a polypeptide chain), and the STn antigen (NeuAcα2-6GalNAc alpha linked to a serine or threonine of a polypeptide chain) have been investigated in a number of clinical trials as antigens for cancer vaccines.4, 5, 9, 10 Glycopeptides also have significant potential as therapeutic agents. Glycopeptides found in human urine from patients with interstitial cystitis have anti-proliferative activity, and analogs show promise as treatments for this debilitating disease.11, 12 Glycopeptides found in venoms, such as contalukin-G of Conus geographus13 and vespulakinins from yellow jacket venom,14 are of interest as analgesics.15 Furthermore, prokaryotes are known to produce a variety of anti-microbial glycopeptides, such as glycocin F16 and sublancin 168.17 For these reasons, glycopeptides have been the subject of intense research for many years.
Access to structurally-defined glycopeptides is crucial to study their biological properties as well as utilize them for clinical applications. Homogenous glycopeptides, however, are difficult to obtain from natural sources and are often only accessible through chemical synthesis.18 Synthesis typically involves solid phase peptide synthesis using amino acids that contain the protected glycan (glyco-amino acids) as building blocks. This approach allows precise control over the chemical structure as well as access to larger quantities of material. Moreover, one can obtain unnatural derivatives that are useful for studying structure-activity relationships (SAR).
While the synthesis of standard peptides has become routine and can be readily accomplished via automated methods, the synthesis of glycopeptides/glycoproteins can be significantly more challenging.1, 19 Couplings of glyco-amino acids to a growing peptide chain are often slow and inefficient. Moreover, the outcome can be highly variable from one glycopeptide to another. To complicate matters further, the building blocks themselves typically need to be synthesized or are expensive when commercially-available. Therefore, the use of a large excess of glyco-amino acid to increase the rate and drive the reaction to completion is impractical. As a result of these problems, scientists typically prepare the peptide precursor up to the glycosylation site on an automated solid phase peptide synthesizer and then carry out the coupling of the glyco-amino acid manually.1, 19 This allows one to follow the reaction carefully, extend the coupling time as needed, repeat the coupling if necessary, and examine alternative activating reagents and/or bases if necessary. Once the glyco-amino acid has been successfully incorporated, the remaining peptide can be completed via automated solid phase peptide synthesis. Although this approach can provide access to glycopeptides, it is not ideal. This process is especially disadvantageous when preparing large numbers of glycopeptides, such as in the construction of a glycopeptide library.
Glycan microarrays, arrays containing numerous carbohydrates immobilized on a solid support in a spatially-defined arrangement, have become valuable tools for high-throughput analysis of carbohydrate-macromolecule interactions.20-23 Given the importance of glycopeptides in biology, our group24-28 and others29-37 have been interested in obtaining large libraries of glycopeptides to enhance the diversity on glycan microarrays. To facilitate these efforts, we have focused on identifying efficient and general coupling conditions. In this study, we carried out a systematic comparison of published coupling conditions using four different Fmoc-protected glyco-amino acids as well as the non-glycosylated counterpart. Both efficiency and epimerization were evaluated using HPLC assays. Interestingly, high rates of epimerization were observed under many conditions, a fact that has important implications for solid phase synthesis of glycopeptides. Finally, we identify conditions that provide high yields and low epimerization for a range of glyco-amino acids.
Materials and Methods
General methods
Unless otherwise stated, reagents were purchased from commercial suppliers and used without purification. 2-Chlorotrityl chloride resin, Fmoc-Ser(Trt)-OH, Fmoc-d-Ser(Trt)-OH, Fmoc-OSu were purchased from AnaSpec, Inc. (San Jose, CA). Fmoc-6-aminohexanoic acid (Fmoc-Hex-OH), Fmoc-Gly-OH, and Fmoc-Pro-OPfp were purchased from Novabiochem (Darmstadt, Germany). 2-(7-Aza-1H-benzotriazol-1-yl)-N,N,N’,N’-tetramethyl uronium hexafluorophosphate (HATU), 1-hydroxy-7-azabenzotriazole (HOAt), 2-(1H-benzotriazol-1-yl)-N,N,N’,N’-tetramethyluronium hexafluorophosphate (HBTU), N-hydroxybenzotriazole (HOBt), N,N′-dicyclohexylcarbodiimide (DCC), benzotriazol-1-yl-oxy-tris-pyrrolidino-phosphonium hexafluorophosphate (PyBOP), dimethylformamide (DMF), N-methyl-2-pyrrolidone (NMP), N,N-diisopropylethylamine (DIEA), N-methylmorpholine (NMM), and 2,4,6-trimethylpyridine (TMP) were from Sigma (St. Louis, MO).
Solid-phase peptide coupling of glyco-amino acids and analysis of efficiency and epimerization
Synthesis, purification, and characterization of the Fmoc-Pro-Gly-Hex-resin starting material and the glycopeptide standards are described in the supporting information. Glycopeptides with the natural configuration (l) contain a number followed by an “a”, and those with the unnatural configuration (d) are followed by a “b”. Prior to coupling the glyco-amino acids, the Fmoc-Pro-Gly-Hex-resin (10.0mg, 0.0037 mmol) was swollen in DCM (2 mL) for 0.5 h at room temperature in a 3 mL Baker disposable filtration column (Phillipsburg, NJ). The resin was then treated with 20% piperidine in DMF (2× 0.5 mL) for 15 min to remove the Fmoc protecting group, followed by washing with DCM (3× 1.0 mL) and DMF (2× 1.0 mL).
Next, the appropriate amount of Fmoc-Ser(Trt)-OH or glyco-amino acid, activating reagents, and base were combined in solvent (150 μL total), mixed with a pipet 5 times, and then immediately added to the resin. In certain cases, Fmoc-Ser(Trt)-OH or the glyco-amino acids were pre-activated for a period of time and then added to the resin according to the specific conditions listed in Table 1. The reaction was allowed to mix at room temperature with an Eppendorf thermomixer 5436 (Hamburg, Germany) at a speed of 600 rpm, and then the resin was washed with DCM (3×2 mL) then DMF (2×1 mL). Next, any unreacted Pro-Gly-Hex-resin was capped by treatment with FmocOSu (12.3 mg, 0.037 mmol) and DIEA (6.4 μL, 0.037 mmol) in DMF (0.5 mL) for 2 h at 600 rpm at room temperature. The resin was washed with DMF (2×1 mL), DCM (3×2 mL), and the glycopeptide was cleaved off the resin by incubating with a mixture of TFA/DCM (90:10) (0.5 mL) at room temperature for 1.5 h at 500 rpm. After cleavage from the resin, the resulting peptides were dilute with DCM (2 mL), concentrated under reduced pressure with a rotary evaporator, dried under vacuum, and dissolved in acetonitrile (1 mL). Solutions were filtered and then product distributions were evaluated using RP-HPLC (for specific HPLC conditions for each substrate, see Supporting Information).
Table 1.
Summary of yields and epimerization rates for various peptide coupling conditions
| Fmoc-Ser(R)-OH, where R = | |||||||
|---|---|---|---|---|---|---|---|
| # | Coupling Conditionsa | Trt | Ac3GalNAcα | Ac4Galβ1-3Ac2GalNAcα | Ac3GlcNAcα | Ac3GlcNAcβ | |
| 1 | AAs: 2.5 eq | Yields (%)b | 75.1 | 69.1 | 83.9 | 100 | 69.0 |
| HATU/HOAt: 1.1/1.1 eq | Epimerization (D/(D+L) %)b | 0.8 | 5.1 | 4.3 | 7.7 | 8.1 | |
| NMM 2.2 eq | |||||||
| 0/3 h in DMF57 | |||||||
| 2 | AAs: 1.5 eq | Yields (%)b | 81.0 | 74.6 | 96.7 | 89.9 | 92.0 |
| HATU/HOAt: 1.2/1.2 eq | Epimerization (D/(D+L) %)b | 2.2 | 11.4 | 21.2 | 15.0 | 20.0 | |
| NMM: 2:4 eq | |||||||
| 0/8 h in DMF58 | |||||||
| 3 | AAs: 3.3 eq | Yields (%)b | 98.9 | 99.8 | 99.6 | 100 | 96.8 |
| HATU/HOAt: 3.65/3.7 eq | Epimerization (D/(D+L) %)b | 0.2 | 0.8 | 2.5 | 1.8 | 3.4 | |
| DIPEA: 7.22 eq | |||||||
| 0/12 h in DMF59 | |||||||
| 4 | AAs: 4.4 eq | Yields (%)b | 63.4 | 99.4 | 83.0 | 100 | 1.9 |
| HATU/HOAt: 4.4/0 eq | Epimerization (D/(D+L) %)b | 37.6 | 69.8 | 65.6 | 72.5 | 52.0 | |
| NMM: 8.8 eq | |||||||
| 3/12 h in NMP59 | |||||||
| 5 | AAs: 5 eq | Yields (%)b | 98.4 | 99.4 | 98.8 | 99.5 | 95.2 |
| HATU/HOAt: 6.25/6.25 eq | Epimerization (D/(D+L) %)b | 2.9 | 5.2 | 6.5 | 8.1 | 9.7 | |
| NMM: 12.5 eq | |||||||
| 0/6 h in NMP59 | |||||||
| 6 | AAs: 2 eq | Yields (%)b | 99.5 | 96.7 | 99.4 | 100 | 94.9 |
| HBTU/HOBt: 4.5/4.5 eq | Epimerization (D/(D+L) %)b | 0.4 | 7.9 | 7.4 | 11.5 | 7.3 | |
| DIPEA: 9 eq | |||||||
| 0/1 h in NMP60 | |||||||
| 7 | AAs: 3 eq: | Yields (%)b | 99.5 | 99.7 | 97.8 | 90.5 | 98.6 |
| DCC/HOBt: 18/18 eq | Epimerization (D/(D+L) %)b | 0.1 | 1.9 | 2.4 | 1.7 | 2.8 | |
| 1/64 h in NMP61 | |||||||
| 8 | AAs: 2 eq | Yields (%)b | 99.5 | 87.1 | 99.7 | 86.0 | 90.3 |
| PyBOP/HOBt: 2/2 eq | Epimerization (D/(D+L) %)b | 1.2 | 10.1 | 11.7 | 8.8 | 5.7 | |
| DIPEA: 4 eq | |||||||
| 0/24 h in DMF62 | |||||||
| 9 | AAs: 1.5 eq | Yields (%)b | 95.8 | 88.1 | 98.0 | 85.6 | 86.0 |
| HBTU/HOBt: 1.5/1.5 eq | Epimerization (D/(D+L) %)b | 0.7 | 2.1 | 3.7 | 4.4 | 3.9 | |
| DIPEA: 1.5 eq | |||||||
| 0/0.33 h in DMF63 | |||||||
| 10 | AAs: 5 eq | Yields (%)b | 99.6 | 99.9 | 99.5 | 100.0 | 96.7 |
| HBTU/HOBt: 5/7.5 eq | Epimerization (D/(D+L) %)b | 0.8 | 5.2 | 7.8 | 13.3 | 8.6 | |
| NMM: 10 eq | |||||||
| 0/5 h in DMF64 | |||||||
| 11 | AAs: 5 eq | Yields (%)b | 99.6 | 99.7 | 99.0 | 100 | 98.2 |
| DIC/HOBt: 10/10 | Epimerization (D/(D+L) %)b | 0.5 | 2.9 | 2.9 | 5.5 | 8.0 | |
| 0.5/0.5 h in DMF65 | |||||||
| 12 | AAs: 2.5 eq | Yields (%)b | 97.1 | 98.4 | 99.7 | 90.7 | 96.2 |
| BOP/HOBt: 2.5/2.5 eq | Epimerization (D/(D+L) %)b | 0.1 | 3.0 | 2.8 | 3.9 | 2.6 | |
| DIPEA: 2.5 eq | |||||||
| 0/4 h in DMF66 | |||||||
| 13 | AAs: 2.0 eq | Yields (%)b | 98.7 | 99.1 | 99.5 | 100 | 98.9 |
| HATU/HOAt: 2.0/2.0 eq | Epimerization (D/(D+L) %)b | 0.2 | 1.1 | 0.3 | 1.6 | 0.6 | |
| TMP: 2.0 eq | |||||||
| 0/2 h in DMF | |||||||
Glyco-amino acids were coupled to Pro-Gly-Hex resin. For the coupling times, the first number refers to the pre-incubation time and the second number refers to the reaction time (e.g. 3/12 h = 3 h pre-incubation followed by 12 h reaction time).
All yield and epimerization data have an error of less than 0.3%.
Solid-phase peptide coupling using condition 13
The appropriate glyco-amino acid (2 eq relative to the resin, 0.0073 mmol), HATU (2.78 mg, 0.0073 mmol), HOAt (1.0 mg, 0.0073 mmol), and TMP (0.98 μL, 0.0073 mmol) were mixed in DMF (150 μL) and immediately added to the resin (10 mg, 0.00366 mmol) in a 3 mL Baker disposable filtration column. The reaction was allowed to mix at a speed of 600 rpm at room temperature for 2 h, followed by washing with DCM (3×2 mL) then DMF (2×1 mL). Any unreacted amino groups were capped by adding FmocOSu (12.3 mg, 0.037 mmol) and DIEA (6.4 μL, 0.037 mmol) in DMF (0.5 mL) and shaking at room temperature for 2 h. The resin was washed with DMF (2×1 mL), DCM (3×2 mL), and the glycopeptide was cleaved off the resin by treating the resin with a mixture of TFA/DCM (90:10) (0.5 mL) at room temperature for 1.5 h.
Molecular Modeling
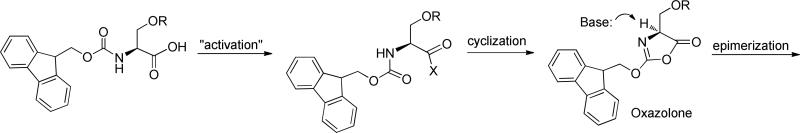
Calculation of conformational preferences of the putative intermediates in amino acid racemizations (oxazolone, see Figures 5 and 6) were performed using MacroModel within the Maestro user interface (Schrodinger. LLC, New York, NY). Structures were constructed within the Build facility in Maestro, and calculations were performed in the Conformational Search utility in MacroModel. Structures were first energy minimized using the OPLS-2005 force field for 2000 steps using the Polak-Ribieri Conjugate Gradient (PRCG) method with a convergence threshold of 0.001. Conformational searching was performed on a random minimized structure of each oxazolone with no restraints. No explicit or implicit solvent was used, but a distance dependent dielectric constant of 38 was used to simulate the dielectric value of DMF. Thirty structures were calculated around each rotatable bond and each conformation was minimized with either the PRCG or Truncated Newton Conjugate Gradient (TNCG) method to a threshold of 0.005 in the OPLS-2005 force field. Charges were read from the force field, and an extended bond cutoff of 8.0, 20.0 and 4.0 Å were used for Van der Waals, electrostatic and hydrogen bond distances, respectively. Mixed torsional/low-mode sampling was performed for the searches and structures were analyzed for energy values and dihedral angles about the Hα-Cα-Cβ-Oβ bond. Conformational clustering was performed using the conformer_cluster.py script in the Chemoinformatics utility in Maestro. Clusters were examined for energies, number-of-times-found for each conformation/cluster and appropriate bond angles.
Figure 5.
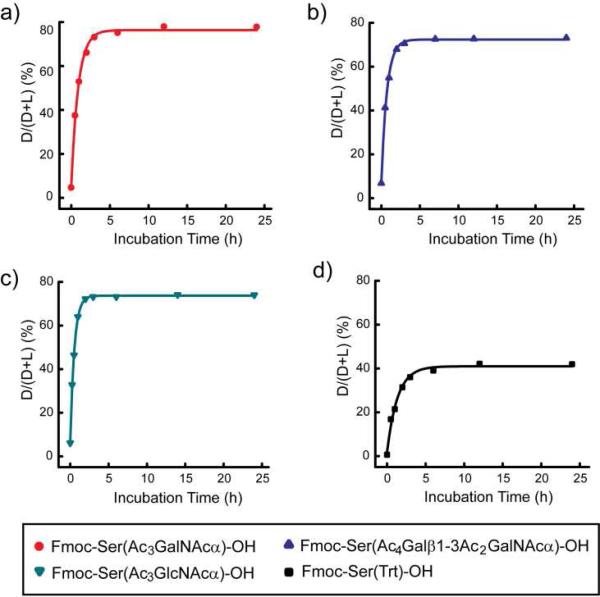
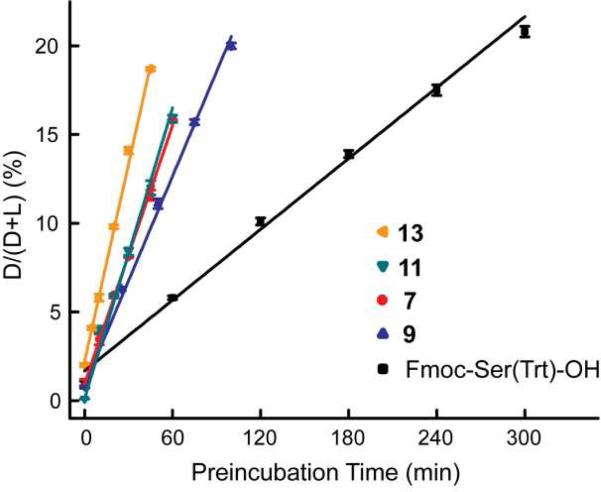
Epimerization as a function of pre-activation time. Fmoc-Ser(Trt)-OH and glyco-amino acids were coupled using the following coupling reagents and ratios: AAs/HATU/NMM=1:1:2. Each amino acid was pre-activated for varying times and then the extent of epimerization was measured as described in the Materials and Methods: a) Fmoc-Ser(Ac3GalNAcα)-OH (7); b) Fmoc-Ser(Ac4Galβ1-3Ac2GalNAcα)-OH (9); c) Fmoc-Ser(Ac3GlcNAcα)-OH (11); and d) Fmoc-Ser(Trt)-OH.
Figure 6.
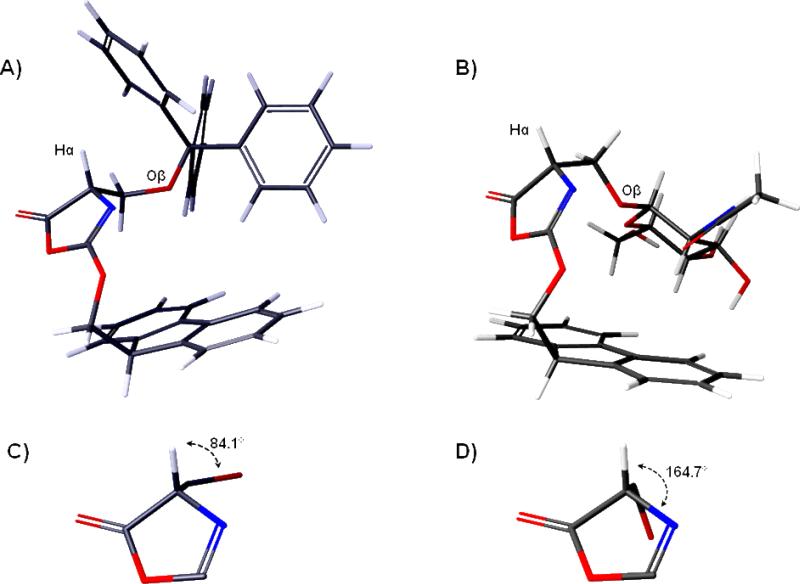
Low energy models of the oxazolone intermediates derived from A) Fmoc-Ser(Trt)-OH and B) Fmoc-Ser(Ac3GlcNAcβ)-OH (13) generated from the Conformational Search utility in Macromodel. 2011. Panels (C) and (D) are expansions of the oxazolone ring with the Hα-Cα-Cβ-Oβ dihedral angle value labeled.
Results and Discussion
Approach and Design
Many factors, such as coupling reagents, reaction time, pre-incubation time, structure of the glyco-amino acids, and the structure of the accepting peptides on the solid support could affect the yields and epimerization rates of glycopeptides during solid phase synthesis. To find the optimal coupling conditions for glyco-amino acids, we investigated a variety of published conditions that had previously been used for the solid phase synthesis of glycopeptides to determine which conditions would provide the highest and most consistent yields and the lowest levels of epimerization. Based on a survey of the literature, we initially selected 12 conditions (see Table 1) that encompassed the most widely used activating agents (HATU, HBTU, DCC, PyBOP), bases (DIEA, NMM), and solvents (DMF, NMP). In addition, the conditions were selected to include some variety in terms of reaction times, equivalents of base, and equivalents of reagents. A thirteenth condition was also evaluated, based on a rationale that is discussed below.
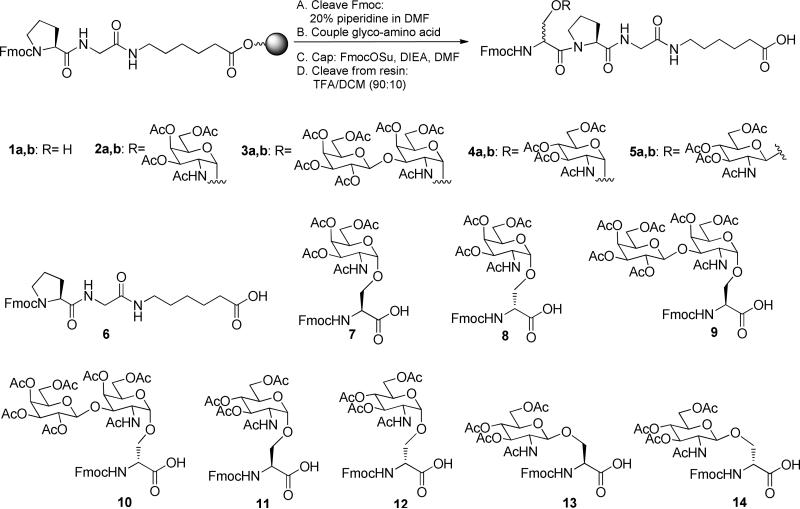
Since the outcome of the coupling could be dependent on the structure of the glyco-amino acid, we opted to evaluate four different glyco-amino acids (see Figure 1) that are commonly found in O-linked glycans. Two of the glyco-amino acids, Fmoc-Ser(Ac3GalNAcα)-OH (7) and Fmoc-Ser(Ac4Galβ1-3Ac2GalNAcα)-OH (9), are protected derivatives of the tumor-associated carbohydrate antigens, Tn and TF. Synthesis of Tn and TF-containing glycopeptides is highly relevant to cancer research and the development of Tn and TF-based cancer vaccines.5, 38, 39 We also looked at the coupling of other glyco-amino acids, Fmoc-Ser(Ac3GlcNAcα)-OH (11) and its beta linked analog, Fmoc-Ser(Ac3GlcNAcβ)-OH (13). Attachment of an O-β-GlcNAc to specific serine and threonine residues of many cytosolic proteins is important for regulating their biological activity40, 41, and the alpha linked analog is found in prokaryotes.42 While not a complete evaluation of every potential glyco-amino acid, this set would allow us to compare effects of certain features, such as alpha vs beta linkages, GalNAc vs GlcNAc configuration, and mono- vs di-saccharides, for a biologically important group of glyco-amino acids. In each case, the carbohydrate fragment was attached to a serine residue. For comparison, we evaluated the coupling of Fmoc-Ser(Trt)-OH, a protected form of serine that has been commonly used in solid phase peptide synthesis.
Figure 1.
Model coupling reaction and key synthetic materials. The “a” and “b” designations in the numbering refer to the l and d isomers of the glycopeptides (or non-glycosylated peptides), respectively.
Since our goal was to identify conditions that would be suitable for many different couplings, including difficult linkages, we initially evaluated couplings of all glyco-amino acids to a short peptide with an N-terminal proline residue, which we anticipated would be challenging. In addition, peptides with several other N-terminal residues were coupled with Fmoc-Ser(Ac3GalNAcα)-OH (7) to test how the terminal residue of peptides affected yields and epimerization. In each case, standards of the truncated peptide, and peptides containing the S and R epimers of the glyco-amino acids and serine were synthesized, and HPLC assays were used to measure yields and epimerization (for example, see Figure 2).
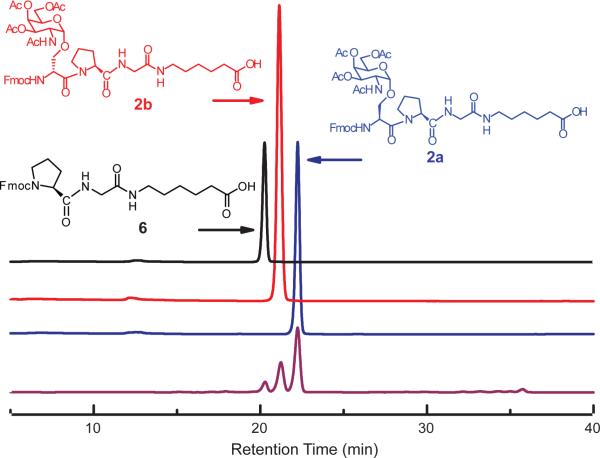
Figure 2.
Representative RP-HPLC analysis of coupling reactions. Traces of three potential products in the coupling reaction with Fmoc-Ser(Ac3GalNAcα)-OH: Fmoc-Pro-Gly-Hex-OH (6) (starting material, black); Fmoc-d-Ser(Ac3GalNAcα)-Pro-Gly-Hex-OH (2b, non-natural epimer, red); Fmoc-l-Ser(Ac3GalNAcα)-Pro-Gly-Hex-OH (2a, desired product, blue); mixture of three components at a ratio of 1:2:4 (purple).
Serine
Serine has previously been found to have a higher rate of racemization than most other natural amino acids.43 Nevertheless, many existing coupling conditions produce high efficiency couplings with low racemization. Of the thirteen conditions that we tested, ten produced the desired product with greater than 95% yield, and all but one condition produced the peptide with less than 3% racemization. Condition 4, which involved pre-activation of Fmoc-Ser(Trt)-OH with HATU/HOAt/NMM/NMP for three hours, led to both suboptimal yield (63.4%) and substantial racemization (37.6%). Higher rates of racemization were not entirely unexpected for this condition, since long pre-activation times are known to increase racemization.44
Tn
Next, we evaluated the coupling efficiency of an Fmoc- protected Tn-derivative, Fmoc-Ser(Ac3GalNAcα)-OH (7). Compound 7 was synthesized following reported methods.45 The unnatural glyco-amino acid, Fmoc-d-Ser(Ac3GalNAcα)-OH (8), was synthesized with similar procedures, and the details can be found in the Supporting Information.
Results for the protected Tn antigen highlight the difficulties of coupling glyco-amino acids. Although couplings of glyco-amino acids can be less efficient than the non-glycosylated counterparts, efficiency was not a significant problem for the conditions we tested. The yields obtained for compound 7 were similar to that of serine, where nine of thirteen conditions provided over 95% yields. Epimerization, however, was a much greater problem for Fmoc-Ser(Ac3GalNAcα)-OH (7) than for non-glycosylated Fmoc-Ser(Trt)-OH. Eight of thirteen conditions led to an epimerization of over 3%, and three of those conditions led to over 10% epimerization. Moreover, condition 4 produced the unnatural epimer as the major product in the reaction (70%). It is important to note that unlike Fmoc-Ser(Trt)-OH, epimerization of Fmoc-Ser(Ac3GalNAcα)-OH at the alpha position of the amino acid produces a new diastereomer, rather than an enantiomer. The production of 70% of the unnatural epimer suggests that this diastereomer is energetically favored, or that L-proline selectively reacts with the unnatural epimer. In either case, the major product of the reaction is the unnatural epimer, demonstrating that one cannot assume that the unnatural epimer will only be a minor contaminant.
The epimerization process is thought to occur through abstraction of the alpha hydrogen of an oxazolone intermediate by the base (see Figure 3).44 Therefore, one would expect that lower amounts of base and use of a weaker base would lead to less epimerization. Through a systematic comparison of bases, Carpino and colleagues demonstrated that 2,4,6-trimethylpyridine (TMP) is an excellent base for peptide couplings, providing high yields with low epimerization.46 However, use of TMP as a base for coupling glyco-amino acids has, to our knowledge, only been used once previously, and the conditions employed microwave irradiation to accelerate coupling.47 Therefore, it was not clear if TMP would be suitable for coupling glyco-amino acids under non-microwave conditions. To test this, we examined a thirteenth condition with 2 equivalents of glyco-amino acid, 2 equivalents of HATU, 2 equivalents of HOAt, and 2 equivalents of TMP as the base. The coupling proceeded smoothly and produced very little epimerization (see Table 1, entry 13).
Figure 3.
General pathway for formation of the oxazolone and the mechanism of epimerization.
TF
Glyco-amino acids with larger glycan chains could potentially be more difficult to attach to a growing peptide during solid phase peptide synthesis. To address this issue, we prepared Fmoc-Ser(Ac4Galβ1-3Ac2GalNAcα)-OH (9) and Fmoc-d-Ser(Ac4Galβ1-3Ac2GalNAcα)-OH (10), the natural and unnatural epimers of a protected TF antigen that contains a disaccharide residue on the side chain of the amino acid. The corresponding peptides were also prepared, and an HPLC assay was developed to evaluate potential products from the peptide coupling reactions. The detailed descriptions of the syntheses of the glyco-amino acids and glycopeptides are in the Supporting Information.
Like the protected Tn amino acid, Fmoc-Ser(Ac4Galβ1-3Ac2GalNAcα)-OH produced substantially higher levels of epimerization than the non-glycosylated Fmoc-Ser(Trt)-OH. The larger glycan fragment, however, did not significantly affect the yield or epimerization relative to the mono-glycosylated amino acid, indicating that the second monosaccharide residue did not have a major effect on the outcome. As with the Fmoc-Ser(Ac3GalNAcα)-OH, coupling with TMP as a base (condition 13) produced excellent results.
Alpha-GlcNAc
Next, we evaluated the coupling of Fmoc-Ser(Ac3GlcNAcα)-OH. Fmoc-Ser(Ac3GlcNAcα)-OH (11) and Fmoc-d-Ser(Ac3GlcNAcα)-OH (12) (see Figure 1) were each synthesized from per-acetylated 2-azido-2-deoxy-d-glucopyranosyl bromide according to a reported method (see Supporting Information for details).45 The yields for peptide couplings with Fmoc-Ser(Ac3GlcNAcα)-OH under the various published conditions were similar to those of serine, Tn, and TF. The extent of epimerization, however, was even higher for this glyco-amino acid. For example, the percentage of epimerized product observed under conditions 1 and 10 were 10 and 17 times higher for compound 11 than that of Fmoc-Ser(Trt)-OH. Moreover, all but three conditions produced over 3% epimerization. As previously observed for Tn and TF, condition 4 provided the highest epimerization (72.5 %), and conditions 3, 7 and 13 provided the lowest epimerization (1.8%, 1.7 % and 1.6%, respectively).
Beta-GlcNAc
To study the effects of the stereochemistry of the glycosidic-linkage on efficiency and epimerization, we evaluated the coupling of Fmoc-Ser(Ac3GlcNAcβ)-OH (13). The synthesis of Fmoc-Ser(Ac3GlcNAcβ)-OH (13) and Fmoc-D-Ser(Ac3GlcNAcβ)-OH (14)were each carried out in one step according to a reported method48 starting from D-glucosamine pentaacetate (see Supporting Information). As summarized in Table 1, nine of the thirteen conditions provided good yields (≥ 95%), and three of the conditions ranged from 69-92% yield. Condition 4, however, provided an unexpectedly low yield (2%). Repeating condition 4 without pre-activation and with 1 h pre-activation provided 74% and 32% yield, with 8% and 45% epimerization, respectively. This demonstrated that extensive decomposition and epimerization occurs during the pre-activation step for condition 4. As previously described for the other glyco-amino acids, the epimerization for Fmoc-Ser(Ac3GlcNAcβ)-OH was much higher than the protected serine analog. For example, the percentage of epimerized product observed under conditions 3 and 11 were 17 and 16 times higher for Fmoc-Ser(Ac3GlcNAcβ)-OH than that of Fmoc-Ser(Trt)-OH. In the case of Fmoc-Ser(Ac3GlcNAcβ)-OH, only conditions 7, 12 and 13 had under 3% epimerization.
Effects of N-terminal Residue
The nature of the N-terminal coupling partner can have a significant effect on the rate of a peptide bond formation. We used a peptide with an N-terminal proline residue for all of our initial studies. Glycosylated serine residues in nature are often found next to various other amino acids.49 To test the effects of N-terminal amino acid structure on efficiency and epimerization, we evaluated coupling of Fmoc-Ser(Ac3GalNAcα)-OH (7) to peptides with a terminal serine and leucine, two amino acid residues that are often found adjacent to the Tn antigen on mucins. In general, the yields were high, but epimerization was substantial for many of the coupling conditions (see Table 2). The production of about 70% of the unnatural epimer under condition 4 demonstrates that the energetic preference for this diastereomer is not dependent on an N-terminal proline.
Table 2.
Effects of the N-terminal amino acid residue on efficiency and epimerization
| Coupling of Fmoc-Ser(Ac3GalNAcα)OH to X-Gly-Hex-Resin, where X = |
||||||||
|---|---|---|---|---|---|---|---|---|
| Pro | Ser(Trt) | Leu | Ser(Ac3GalNAcα) | |||||
| #a | Yields (%)b | D/(D+L) (%)b | Yields (%)b | D/(D+L) (%)b | Yields (%)b | D/(D+L) (%)b | Yields (%)b | D/(D+L) (%)b |
| 1 | 69.1 | 5.1 | 99.0 | 2.6 | 97.7 | 3.8 | 92.5 | 3.9 |
| 2 | 74.6 | 11.4 | 99.6 | 6.7 | 98.0 | 7.1 | 91.5 | 6.6 |
| 3 | 99.8 | 0.8 | 99.6 | 1.3 | 98.3 | 2.3 | 99.1 | 1.9 |
| 4 | 99.4 | 69.8 | 99.9 | 71.7 | 98.5 | 69.3 | 99.2 | 70.8 |
| 5 | 99.4 | 5.2 | 99.4 | 3.6 | 98.3 | 4.4 | 96.1 | 3.9 |
| 6 | 99.3 | 9.7 | 99.8 | 4.3 | 97.4 | 2.9 | 97.8 | 3.0 |
| 7 | 99.7 | 1.9 | 99.1 | 1.0 | 98.4 | 1.7 | 95.4 | 3.8 |
| 8 | 87.1 | 10.1 | 99.8 | 3.0 | 98.2 | 4.2 | 96.9 | 3.8 |
| 9 | 88.1 | 2.1 | 99.7 | 2.1 | 98.1 | 2.6 | 96.2 | 5.7 |
| 10 | 99.9 | 5.2 | 99.6 | 1.4 | 98.1 | 2.7 | 96.9 | 4.9 |
| 11 | 99.7 | 2.9 | 99.8 | 1.0 | 98.2 | 2.1 | 97.2 | 3.3 |
| 12 | 98.4 | 3.0 | 99.8 | 0.8 | 98.5 | 1.6 | 96.9 | 3.2 |
| 13 | 99.1 | 1.1 | 98.3 | 0.2 | 99.3 | 1.5 | 96.8 | 0.5 |
Entry number corresponds to the coupling conditions listed in Table 1.
All yield and epimerization data have an error of less than 0.3%.
Synthesis of Clustered Tn Peptides
Mucins and other glycoproteins often contain 2 or more glycosylated serine and/or threonine residues linked consecutively on a peptide chain. When these residues are all glycosylated, these arrangements are referred to as “clustered” glycans. Clustered Tn and STn antigens are more tumor-specific than single antigens, and clustered Tn and STn glycopeptides are of high interest as vaccine antigens.45, 50, 51 We anticipated that coupling of Fmoc-Ser(Ac3GalNAcα)-OH (7) to a peptide chain containing an N-terminal Ser(Ac3GalNAcα)-OH residue could be even more difficult than coupling to a proline, serine, or leucine. As before, we synthesized the appropriate standards and developed an HPLC assay to detect the potential products. While the yields were above 90% for all the conditions tested, only two conditions, 3 and 13, had an epimerization of less than 3%. It is important to note that the preparation of clustered Tn peptides requires at least two peptide couplings using glyco-amino acids. Since the attachment of successive glyco-amino acids residues is more difficult than coupling of the first glyco-amino acid and the effects of epimerization are cumulative, the preparation of clustered glycopeptides can be especially difficult. Therefore, it is crucial to use optimal peptide coupling conditions.
Mechanistic Studies
The differences in the levels of epimerization observed for glyco-amino acids versus serine could be due to a variety of reasons, such as differential rates of epimerization, different rates of peptide coupling, different energetic preferences for the natural versus unnatural epimer, and enhanced coupling of the unnatural D-configured activated esters relative to their L-counterparts.
To better understand the origin of the differences, we first measured the initial rates of epimerization. To accurately measure the initial rates, we needed conditions wherein the reactants would not be consumed to an appreciable degree during the reaction. In addition, we needed conditions where the epimerization would be slow enough to conveniently measure using our assay. Since the base is consumed in the reaction (through protonation), we needed to use a large excess of base such that the concentration would not change significantly during the reaction; however, a large excess of DIEA or NMM would result in much faster rates of epimerization, as well as undesired side reactions, making analysis difficult. For these reasons, we chose to couple using 12 equivalents of TMP as the base.
As shown in Figure 4, epimerization rates of the four glyco-amino acids were significantly faster than the epimerization rate of Fmoc-Ser(Trt)-OH. Fmoc-Ser(Ac3GlcNAcβ)-OH (13) displayed the fastest initial rate of epimerization and was about 6 times faster than Fmoc-Ser(Trt)-OH. Taken together, these results show that a faster rate of epimerization is one factor that contributes to the higher levels of epimerization for glyco-amino acids.
Figure 4.
Initial rates of epimerization. Initial rates were measured using the following coupling conditions: AAs/HATU/TMP=1:1:12. The initial rates were as follows: Fmoc-Ser(Ac3GlcNAcβ)-OH (13), k = 0.37±0.01%/min; Fmoc-Ser(Ac3GlcNAcα)-OH (11), k = 0.27±0.01%/min; Fmoc-Ser(Ac3GalNAcα)-OH (7), k = 0.241±0.002%/min; Fmoc-Ser(Ac4Galβ1-3Ac2GalNAcα)-OH (9), k = 0.197±0.005%/min; Fmoc-Ser(Trt)-OH, k = 0.066±0.003%/min.
Next, the extent of epimerization of glyco-amino acids was measured. Condition 4, which included a 3 h pre-activation step, produced high levels of epimerization, and in some cases, produced the unnatural epimer as the major product. We hypothesized that the pre-incubation step allows for equilibration of the natural and unnatural epimer at the alpha position. If so, extending the pre-incubation time would allow us to measure the equilibrium ratios of natural and unnatural epimers. To test this, we repeated this reaction using a variety of pre-incubation times ranging from 0 to 24 h (see Figure 5). Since the activated esters of the d and l glyco-amino acids are diastereomers and could potentially have different coupling rates to an N-terminal proline on the resin, an excess of resin was used to assure that all d and l isomers would be captured on the resin and detected in the assay. For each of the glyco-amino acids tested, much less epimerization was observed when no pre-incubation was used, and the amount of epimerization increased as the pre-incubation time increased, indicating that the epimerization occurs during the pre-incubation period. The extent of epimerization of Fmoc-Ser(Ac3GalNAcα)-OH (7) (protected Tn, Figure 5a) reached a maximum of approximately 80%, indicating that the equilibrium composition is 4:1. Similar results were obtained with Fmoc-Ser(Ac4Galβ1-3Ac2GalNAcα)-OH (9, Figure 5b) and Fmoc-Ser(Ac3GlcNAcα)-OH (11, Figure 5c). The extent of epimerization could not be accurately determined for Fmoc-Ser(Ac3GlcNAcβ)-OH due to degradation of the starting material during the pre-incubation period. These results indicate that the unnatural epimer of these glyco-amino acids are favored over the natural epimer and that this energetic preference is another factor that contributes to the higher observed levels of epimerization for glyco-amino acids.
Finally, we measured the relative rates of coupling to the resin using a competition assay. The epimerization process is generally thought to occur via the activated intermediate(s) (see Figure 3); once the glyco-amino acid has been coupled to the growing peptide chain, the resulting amide would no longer epimerize under the coupling conditions. A slower overall coupling rate could allow more time for epimerization. We used a competition assay to measure the relative coupling rates wherein glyco-amino acids were mixed with an equivalent amount of Fmoc-Ser(Trt)-OH, activated, and then captured with resin as before. Using an HPLC assay, we measured relative amounts of the glyco-peptides and serine containing peptides, which provide a measure of the relative rates of coupling. Under standard conditions, the initial coupling rates were too fast to measure in this assay. Therefore, the reaction solution was diluted ten-fold and the coupling reaction was stopped at 5 minutes to ensure the yield of each peptide was below 20% (total yield below 40%). Two equivalents of each amino acid were used, which ensured that the consumption of each amino acid was below 10%.
As summarized in Table 3, most glyco-amino acids coupled more slowly than Fmoc-Ser(Trt)-OH. The slowest glyco-amino acid, 9, showed an approximately 50% decrease in relative coupling rate. The Fmoc-d-Ser(Trt)-OH gave the same coupling rate as its enantiomer. Interestingly, the unnatural d isomer of Fmoc-Ser(Ac3GlcNAcα)-OH had a noticeably higher coupling rate than the natural isomer.
Table 3.
Relative overall rates of peptide couplinga
| Amino Acid | Relative reaction rate GAA/Ser |
|---|---|
| Fmoc-Ser(Trt)-OH | 1 |
| Fmoc-D-Ser(Trt)-OH | 1.06±0.05 |
| Fmoc-Ser(Ac3GalNAcα)-OH | 0.69±0.02 |
| Fmoc-D-Ser(Ac3GalNAcα)-OH | 0.84±0.03 |
| Fmoc-Ser(Ac4Galβ1-3Ac2GalNAcα)-OH | 0.46±0.01 |
| Fmoc-Ser(Ac3GlcNAcα)-OH | 0.64±0.02 |
| Fmoc-D-Ser(Ac3GlcNAcα)-OH | 1.19±0.03 |
| Fmoc-Ser(Ac3GlcNAcβ)-OH | 0.99±0.02 |
| Fmoc-D-Ser(Ac3GlcNAcβ)-OH | 1.20±0.09 |
All reactions were carried out by mixing Fmoc-Ser(Trt)-OH (2 eq.) and the listed glyco-amino acid (GAA, 2 eq.) with HATU (4 eq.) and TMP (4 eq.) in DMF. The total reaction time was 5 m and no pre-activation was used.
Steric, Electronic, and Conformational Factors Affecting Epimerization
Although the experiments described above demonstrated that initial rates of epimerization, the amounts of D-isomer formed and the coupling times of glyco-amino acids are all quite distinct from their non-glycosylated counterparts, a clearer understanding of the structural features that may contribute to the higher epimerization rates was desired. Therefore, we considered several factors that could influence the relative rates of epimerization.
One factor that could potentially contribute to enhanced epimerization of glyco-amino acids relative to non-glycosylated amino acids are inductive effects. Monosaccharide rings are expected to exert a stronger electron withdrawing inductive effect than a trityl group. This could lead to enhance acidity of the alpha hydrogen. To evaluate this possibility, we examined the proton NMR chemical shifts of the alpha hydrogens. Interestingly, we observed no differences in chemical shifts of the alpha hydrogens for any of the serine analogs used in this study. In contrast, the beta hydrogens of glycosylated serine residues were shifted downfield approximately 0.5 ppm relative to the beta hydrogens of trityl protected serine. While this difference could be due to inductive and/or anisotropic effects, we note that the chemical shifts of the alpha and beta hydrogens of tert-butyl protected serine were nearly identical with those of the trityl-protected serine, indicating that anisotropic effects of the trityl group did not have a significant effect on the chemical shifts of the alpha and beta hydrogens. Therefore, the NMR data supports a stronger electron withdrawing effect exerted by the sugar ring; however, this effect does not extend to the alpha position of the amino acids and does not appear to contribute to the increase in epimerization.
A second factor that could contribute to the enhanced epimerization of glyco amino acids are conformational effects; however, defining the preferred conformations of small, flexible glyco-amino acids by NMR is difficult due to the limited number of NMR-derived distance and angle restraints. Moreover, peptide coupling and epimerization reactions can proceed through intermediates that may have distinct conformational preferences from the starting amino acids, but these intermediates are not easily characterized due to their instability. Therefore, we undertook a modeling study to evaluate conformational preferences. Epimerization/racemization is generally thought to proceed through an intermediate oxazolone (see Figure 3).52, 53 As a side note, β-elimination could also occur through a similar mechanism, but we did not observe β-elimination in any of our peptide coupling reactions. We reasoned that structural differences between the various oxazolones could potentially contribute to differences in epimerization. Therefore, we compared conformational preferences of the putative oxazolone intermediates of Fmoc-Ser(Trt)-OH and the glyco-amino acid that is most prone to epimerization, viz. Fmoc-Ser(Ac3GlcNAcβ)-OH. The oxazolones were constructed with the Build facility in Maestro (Schrodinger, Inc.) and the program MacroModel 11.1 was used to perform searches and analyze data. A series of structures was calculated based on various criteria as the input to the Conformational Search utility in Macromodel. To mimic our reaction solvent, we used a distance-dependent dielectric constant equal to the value for DMF (38). Full descriptions of the search criteria (force fields, minimization settings, search parameters) can be found in the Materials and Methods section.
The preferred conformations for the two oxazolones were significantly different. Figure 6 shows the lowest energy conformers of the two oxazolones modeled (Figures 6A and 6B) along with the values of the Hα-Cα-Cβ-Oβ dihedral angles in each (Figures 6C and 6D). For the glycosylated oxazolone, the Hα and Oβ atoms are nearly anti to each other (~165°), whereas these atoms adopt an angle of about 85° for the trityl-protected oxazolone. The difference in preferred conformations has two key effects. First, the alpha hydrogen of Fmoc-Ser(Trt)-OH is more sterically shielded than the alpha hydrogen of Fmoc-Ser(Ac3GlcNAcβ)-OH. The steric hindrance could make it more difficult for the base to abstract the hydrogen from Fmoc-Ser(Trt)-OH. Second, the anti configuration between the Hα and Oβ atoms of Fmoc-Ser(Ac3GlcNAcβ)-OH maximizes orbital overlap between the C-H sigma bond and the C-O sigma* anti-bonding orbital, which would facilitate abstraction of the alpha hydrogen from Fmoc-Ser(Ac3GlcNAcβ)-OH. Although additional evidence will be needed to verify this hypothesis, our initial analysis indicates that a combination of steric and stereoelectronic effects contribute to the enhanced rate of epimerization of glycosylated amino acids relative to the non-glycosylated counterparts.
Conclusion
In this study, we demonstrate that glycosylated amino acids are much more prone to epimerization than their corresponding non-glycosylated counterparts. The enhanced epimerization during solid phase synthesis is due to a faster rate of epimerization, an energetic preference for the unnatural epimer, a slower overall rate of peptide coupling, and a conformational preference in the putative oxazolone intermediate that can be potentially stabilizing toward α-hydrogen abstraction. Of the thirteen peptide coupling conditions that we systematically evaluated, conditions 3 (HATU/HOAt/DIPEA/glyco-amino acid at a ratio of about 1:1:2:1), 7 (DCC/HOBt/ glyco-amino acid at a ratio of 6:6:1) and 13 (HATU/HOAt/TMP/glyco-amino acid at a ratio of about 1:1:1:1) provided the best overall yields (>95%) and the lowest levels of epimerization (≤3%). These conditions are compatible with automated solid phase glycopeptide synthesis, but condition 7 is less desirable due to the long reaction time and large excess of reagents used in the reaction. Therefore, conditions 3 and 13 represent the best current options for coupling glycosylated amino acids to a growing peptide chain. It is important to note, however, that additional studies are needed to more fully evaluate the scope of the epimerization problem and the performance of these peptide coupling conditions. For example, other factors, such as the protecting groups on the glyco-amino acid and the nature of the core amino acid (serine vs. threonine), could also affect the outcome. Moreover, longer sequences and/or more aggregation prone sequences may be more difficult to synthesize and more susceptible to side reactions. Nevertheless, this study provides an important step toward general and efficient peptide coupling conditions for synthesizing glycopeptides.
While we observed substantial amounts of epimerization in many reactions, epimerization has been noted only infrequently in previous publications. There are several factors that could account for this discrepancy. First, glycopeptides containing an unnatural epimer are often extremely difficult to separate from the corresponding glycopeptides containing the natural epimer. As the peptide sequence increases in length, the separation becomes even more challenging. In many previous studies, the glycopeptides of interest were over 10 amino acids long. In our study, we used short sequences containing 3 amino acid residues, which facilitated the separation of natural and unnatural epimers. In addition, we chemically synthesized the unnatural epimers as defined standards, which enabled the development of suitable conditions for separating the natural and unnatural epimers by HPLC. It should be noted, however, that even for our short glycopeptides, separation was not trivial. Second, it can be extremely difficult to detect the presence of the unnatural epimer by standard characterization methods, such as mass spectroscopy, elemental analysis, and NMR. Therefore, these impurities could go undetected. Finally, certain peptide linkages may be easier to form, leading to less epimerization.
Epimerization of glycopeptides could have a major impact in a number of situations. Many glycopeptides are in clinical development as vaccine antigens. Previous studies have shown that the immune system can mount a much stronger response to peptides containing an unnatural epimer than to natural peptides.54, 55 Thus, even if the unnatural epimer is a minor contaminant, it may be the dominant antigen recognized by the immune system. Therefore, the presence or absence of the unnatural epimer could have a major impact on the clinical efficacy of a glycopeptide vaccine. For biologically active glycopeptides, compounds containing an unnatural epimer can have completely different activities. For example, the anti-proliferative factor (APF) present in urine of patients with interstitial cystitis potently blocks proliferation of bladder epithelial cells.11, 56 In contrast, the corresponding glycopeptide containing a D-proline at position 3 potently antagonizes the activity of APF.11 Although the unnatural epimer is not at a glycosylated position, this example demonstrates that variation of stereochemistry at a single position can lead to a complete switch in bioactivity. Our results demonstrate that formation of the unnatural epimer can be a major problem and that judicious choice of coupling conditions can minimize this undesired side product.
Supplementary Material
Acknowledgment
This research was supported by the Intramural Research Program of the NIH, NCI. We gratefully acknowledge James A. Kelley (NIH/NCI) for obtaining mass spectrometry data for all the compounds.
Footnotes
Supporting Information Available. Experimental details for the synthesis of glyco-amino acids and glycopeptides, characterization data, and supplemental HPLC analyses of glycopeptides are available free of charge via the internet at http://pubs.acs.org.
References
- 1.Buskas T, Ingale S, Boons GJ. Glycobiology. 2006;16:113R–136R. doi: 10.1093/glycob/cwj125. [DOI] [PubMed] [Google Scholar]
- 2.Grogan MJ, Pratt MR, Marcaurelle LA, Bertozzi CR. Annu. Rev. Biochem. 2002;71:593–634. doi: 10.1146/annurev.biochem.71.110601.135334. [DOI] [PubMed] [Google Scholar]
- 3.Andersson IE, Dzhambazov B, Holmdahl R, Linusson A, Kihlberg J. J. Med. Chem. 2007;50:5627–5643. doi: 10.1021/jm0705410. [DOI] [PubMed] [Google Scholar]
- 4.Buskas T, Thompson P, Boons G-J. Chem. Commun. 2009:5335–5349. doi: 10.1039/b908664c. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Zhu J-L, Warren JD, Danishefsky SJ. Expert Rev. Vaccines. 2009;8:1399–1413. doi: 10.1586/erv.09.95. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Batsalova T, Dzhambazov B, Klaczkowska D, Holmdahl R. J. Immunol. 2010;185:2701–2709. doi: 10.4049/jimmunol.1000385. [DOI] [PubMed] [Google Scholar]
- 7.Andersson IE, Andersson CD, Batsalova T, Dzhambazov B, Holmdahl R, Kihlberg J, Linusson A. PLoS One. 2011;6:e17881. doi: 10.1371/journal.pone.0017881. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Hoffmann-Roeder A, Schoenhentz J, Wagner S, Schmitt E. Chem. Commun. 2011;47:382–384. doi: 10.1039/c0cc02250k. [DOI] [PubMed] [Google Scholar]
- 9.Springer GF. Science. 1984;224:1198–1206. doi: 10.1126/science.6729450. [DOI] [PubMed] [Google Scholar]
- 10.Holmberg LA, Sandmaier BM. Expert Rev. Vaccines. 2004;3:655–663. doi: 10.1586/14760584.3.6.655. [DOI] [PubMed] [Google Scholar]
- 11.Kaczmarek P, Keay SK, Tocci GM, Koch KR, Zhang C-O, Barchi JJ, Jr., Grkovic D, Guo L, Michejda CJ. J. Med. Chem. 2008;51:5974–5983. doi: 10.1021/jm8002763. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Keay S, Kaczmarek P, Zhang C-O, Koch K, Szekely Z, Barchi JJ, Jr., Michejda C. Chem. Biol. Drug Des. 2011;77:421–430. doi: 10.1111/j.1747-0285.2011.01108.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Kang J, Low W, Norberg T, Meisenhelder J, Hansson K, Stenflo J, Zhou G-P, Imperial J, Olivera Baldomero M, Rigby Alan C, Craig AG. Eur. J. Biochem. 2004;271:4939–49. doi: 10.1111/j.1432-1033.2004.04464.x. [DOI] [PubMed] [Google Scholar]
- 14.Yoshida H, Geller RG, Pisano JJ. Biochemistry. 1976;15:61–4. doi: 10.1021/bi00646a010. [DOI] [PubMed] [Google Scholar]
- 15.Dhanasekaran M, Polt R. Curr. Drug Delivery. 2005;2:59–73. doi: 10.2174/1567201052772843. [DOI] [PubMed] [Google Scholar]
- 16.Venugopal H, Edwards PJB, Schwalbe M, Claridge JK, Libich DS, Stepper J, Loo T, Patchett ML, Norris GE, Pascal SM. Biochemistry. 2011;50:2748–2755. doi: 10.1021/bi200217u. [DOI] [PubMed] [Google Scholar]
- 17.Paik SH, Chakicherla A, Hansen JN. J. Biol. Chem. 1998;273:23134–42. doi: 10.1074/jbc.273.36.23134. [DOI] [PubMed] [Google Scholar]
- 18.Wu C-Y, Wong C-H. Chem. Commun. 2011;47:6201–6207. doi: 10.1039/c0cc04359a. [DOI] [PubMed] [Google Scholar]
- 19.Hojo H, Nakahara Y. Biopolymers. 2007;88:308–324. doi: 10.1002/bip.20699. [DOI] [PubMed] [Google Scholar]
- 20.Wu CY, Liang PH, Wong CH. Org. Biomol. Chem. 2009;7:2247–2254. doi: 10.1039/b902510n. [DOI] [PubMed] [Google Scholar]
- 21.Oyelaran O, Gildersleeve JC. Curr. Opin. Chem. Biol. 2009;13:406–13. doi: 10.1016/j.cbpa.2009.06.021. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Lonardi E, Balog CI, Deelder AM, Wuhrer M. Expert Rev. Proteomics. 2010;7:761–774. doi: 10.1586/epr.10.41. [DOI] [PubMed] [Google Scholar]
- 23.Rillahan CD, Paulson JC. Annu. Rev. Biochem. 2011;80:797–823. doi: 10.1146/annurev-biochem-061809-152236. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Manimala JC, Li Z, Jain A, VedBrat S, Gildersleeve JC. ChemBioChem. 2005;6:2229–2241. doi: 10.1002/cbic.200500165. [DOI] [PubMed] [Google Scholar]
- 25.Li Q, Anver MR, Butcher DO, Gildersleeve JC. Mol. Cancer Ther. 2009;8:971–979. doi: 10.1158/1535-7163.MCT-08-0934. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Zhang Y, Li Q, Rodriguez LG, Gildersleeve JC. J. Am. Chem. Soc. 2010;132:9653–9662. doi: 10.1021/ja100608w. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Li Q, Anver MR, Li Z, Butcher DO, Gildersleeve JC. Int. J. Cancer. 2010;126:459–68. doi: 10.1002/ijc.24716. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Li Q, Rodriguez LG, Farnsworth DF, Gildersleeve JC. ChemBioChem. 2010;11:1686–91. doi: 10.1002/cbic.201000235. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Laurent N, Voglmeir J, Wright A, Blackburn J, Pham NT, Wong SCC, Gaskell SJ, Flitsch SL. ChemBioChem. 2008;9:883–887. doi: 10.1002/cbic.200700692. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Westerlind U, Schroeder H, Hobel A, Gaidzik N, Kaiser A, Niemeyer CM, Schmitt E, Waldmann H, Kunz H. Angew. Chem., Int. Ed. 2009;48:8263–8267. doi: 10.1002/anie.200902963. [DOI] [PubMed] [Google Scholar]
- 31.Ohyabu N, Hinou H, Matsushita T, Izumi R, Shimizu H, Kawamoto K, Numata Y, Togame H, Takemoto H, Kondo H, Nishimura S-I. J. Am. Chem. Soc. 2009;131:17102–17109. doi: 10.1021/ja903361f. [DOI] [PubMed] [Google Scholar]
- 32.Song X, Lasanajak Y, Rivera-Marrero C, Luyai A, Willard M, Smith DF, Cummings RD. Anal. Biochem. 2009;395:151–160. doi: 10.1016/j.ab.2009.08.024. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Kracun SK, Clo E, Clausen H, Levery SB, Jensen KJ, Blixt O. J. Proteome Res. 2010;9:6705–6714. doi: 10.1021/pr1008477. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Pedersen JW, Blixt O, Bennett EP, Tarp MA, Dar I, Mandel U, Poulsen SS, Pedersen AE, Rasmussen S, Jess P, Clausen H, Wandall HH. Int. J. Cancer. 2010;128:1860–1871. doi: 10.1002/ijc.25778. [DOI] [PubMed] [Google Scholar]
- 35.Steentoft C, Schjoldager KT, Clo E, Mandel U, Levery SB, Pedersen JW, Jensen K, Blixt O, Clausen H. Glycoconjugate J. 2010;27:571–582. doi: 10.1007/s10719-010-9301-6. [DOI] [PubMed] [Google Scholar]
- 36.Wandall HH, Blixt O, Tarp MA, Pedersen JW, Bennett EP, Mandel U, Ragupathi G, Livingston PO, Hollingsworth MA, Taylor-Papadimitriou J, Burchell J, Clausen H. Cancer Res. 2010;70:1306–1313. doi: 10.1158/0008-5472.CAN-09-2893. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Weinrich D, Koehn M, Jonkheijm P, Westerlind U, Dehmelt L, Engelkamp H, Christianen PCM, Kuhlmann J, Maan JC, Nuesse D, Schroeder H, Wacker R, Voges E, Breinbauer R, Kunz H, Niemeyer CM, Waldmann H. ChemBioChem. 2010;11:235–247. doi: 10.1002/cbic.200900559. [DOI] [PubMed] [Google Scholar]
- 38.Dalgleish AG. Expert Rev. Vaccines. 2004;3:665–668. doi: 10.1586/14760584.3.6.665. [DOI] [PubMed] [Google Scholar]
- 39.Ouerfelli O, Warren JD, Wilson RM, Danishefsky SJ. Expert Rev. Vaccines. 2005;4:677–685. doi: 10.1586/14760584.4.5.677. [DOI] [PubMed] [Google Scholar]
- 40.Vosseller K, Sakabe K, Wells L, Hart GW. Curr. Opin. Chem. Biol. 2002;6:851–857. doi: 10.1016/s1367-5931(02)00384-8. [DOI] [PubMed] [Google Scholar]
- 41.Wells L, Hart GW. FEBS Lett. 2003;546:154–158. doi: 10.1016/s0014-5793(03)00641-0. [DOI] [PubMed] [Google Scholar]
- 42.Olszewski NE, West CM, Sassi SO, Hartweck LM. Biochim. Biophys. Acta, Gen. Subj. 2010;1800:49–56. doi: 10.1016/j.bbagen.2009.11.016. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Di Fenza A, Tancredi M, Galoppini C, Rovero P. Tetrahedron Lett. 1998;39:8529–8532. [Google Scholar]
- 44.Albericio F, Carpino LA. Methods Enzymol. 1997;289:104–126. doi: 10.1016/s0076-6879(97)89046-5. [DOI] [PubMed] [Google Scholar]
- 45.Kuduk SD, Schwarz JB, Chen X-T, Glunz PW, Sames D, Ragupathi G, Livingston PO, Danishefsky SJ. J. Am. Chem. Soc. 1998;120:12474–12485. [Google Scholar]
- 46.Carpino LA, Ionescu D, El-Faham A. J. Org. Chem. 1996;61:2460–5. [Google Scholar]
- 47.Lee DJ, Harris PWR, Kowalczyk R, Dunbar PR, Brimble MA. Synthesis. 2010:763–769. [Google Scholar]
- 48.Arsequell G, Krippner L, Dwek RA, Wong SYC. J. Chem. Soc., Chem. Commun. 1994:2383–4. [Google Scholar]
- 49.Kim YS, Gum J, Jr., Brockhausen I. Glycoconjugate J. 1996;13:693–707. doi: 10.1007/BF00702333. [DOI] [PubMed] [Google Scholar]
- 50.Coltart DM, Royyuru AK, Williams LJ, Glunz PW, Sames D, Kuduk SD, Schwarz JB, Chen X-T, Danishefsky SJ, Live DH. J. Am. Chem. Soc. 2002;124:9833–9844. doi: 10.1021/ja020208f. [DOI] [PubMed] [Google Scholar]
- 51.Sakai K, Yuasa N, Tsukamoto K, Takasaki-Matsumoto A, Yajima Y, Sato R, Kawakami H, Mizuno M, Takayanagi A, Shimizu N, Nakata M, Fujita-Yamaguchi Y. J. Biochem. 2010;147:809–817. doi: 10.1093/jb/mvq014. [DOI] [PubMed] [Google Scholar]
- 52.Goodman M, McGahren WJ. Tetrahedron. 1967;23:2031–50. doi: 10.1016/0040-4020(67)80037-1. [DOI] [PubMed] [Google Scholar]
- 53.Young AR, Barcham GJ, Kemp JM, Dunphy JL, Nash A, Meeusen EN. Glycoconjugate J. 2009;26:423–432. doi: 10.1007/s10719-008-9190-0. [DOI] [PubMed] [Google Scholar]
- 54.Stupp Y, Sela M. Biochim. Biophys. Acta, Protein Struct. 1967;140:349–59. doi: 10.1016/0005-2795(67)90475-8. [DOI] [PubMed] [Google Scholar]
- 55.Pollack SJ, Hsiun P, Schultz PG. J. Am. Chem. Soc. 1989;111:5961–2. [Google Scholar]
- 56.Kim J, Keay SK, Dimitrakov JD, Freeman MR. FEBS Lett. 2007;581:3795–3799. doi: 10.1016/j.febslet.2007.06.058. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57.Brocke C, Kunz H. Synthesis. 2004:525–542. doi: 10.1002/chem.200400228. [DOI] [PubMed] [Google Scholar]
- 58.Karch F, Hoffmann-Roeder A. Beilstein. J. Org. Chem. 2010;6:47. doi: 10.3762/bjoc.6.47. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59.Becker T, Kaiser A, Kunz H. Synthesis. 2009:1113–1122. [Google Scholar]
- 60.Hojo H, Matsumoto Y, Nakahara Y, Ito E, Suzuki Y, Suzuki M, Suzuki A, Nakahara Y. J. Am. Chem. Soc. 2005;127:13720–13725. doi: 10.1021/ja053711b. [DOI] [PubMed] [Google Scholar]
- 61.Nakahara Y, Nakahara Y, Ogawa T. Carbohydr. Res. 1996;292:71–81. doi: 10.1016/s0008-6215(96)91027-7. [DOI] [PubMed] [Google Scholar]
- 62.Campo VL, Carvalho I, Allman S, Davis BG, Field RA. Org. Biomol. Chem. 2007;5:2645–2657. doi: 10.1039/b707772f. [DOI] [PubMed] [Google Scholar]
- 63.Matsushita T, Hinou H, Kurogochi M, Shimizu H, Nishimura S. Org. Lett. 2005;7:877–880. doi: 10.1021/ol0474352. [DOI] [PubMed] [Google Scholar]
- 64.Seitz O, Wong C-H. J. Am. Chem. Soc. 1997;119:8766–8776. [Google Scholar]
- 65.Shin Y, Winans KA, Backes BJ, Kent SBH, Ellman JA, Bertozzi CR. J. Am. Chem. Soc. 1999;121:11684–11689. [Google Scholar]
- 66.Chen L, Jensen KJ, Tejbrant J, Taylor JE, Morgan BA, Barany G. J. Pept. Res. 2000;55:81–91. doi: 10.1034/j.1399-3011.2000.00154.x. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.