Abstract
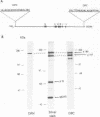
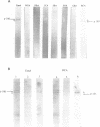
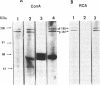
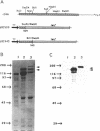
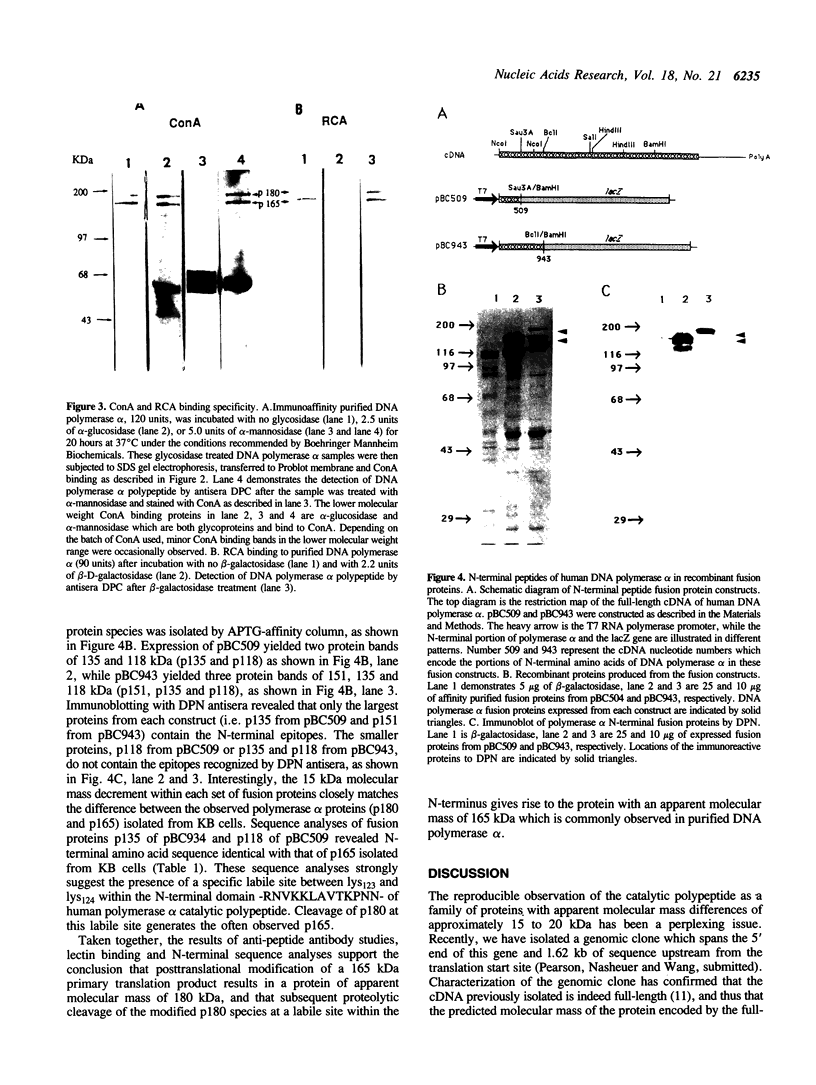
The catalytic polypeptide of DNA polymerase alpha is often observed in vitro as a family of phosphopolypeptides predominantly of 180 and 165 kDa derived from a single primary structure. The estimated Mr of this polypeptide deduced from the full-length cDNA is 165 kDa. Immunoblot analysis with polyclonal antibodies against peptides of the N- and C-termini of the deduced primary sequence indicates that the observed family of polypeptides from 180 kDa to lower molecular weight results from proteolytic cleavage from the N-terminus. Antibodies against the N-terminal peptide detect only the 180 kDa species suggesting that this higher molecular weight polypeptide may be the result of posttranslational modification of the 165 kDa primary translation product. The catalytic polypeptide is not only phosphorylated but is also found to react with lectins ConA and RCA. N-terminal sequencing of the isolated catalytic polypeptide from human cells and of the recombinant fusion proteins indicates that the often observed 165 kDa polypeptide is the in vitro proteolytic cleavage product of the modified 180 kDa protein at the specific site between lys123 and lys124 within the sequence -RNVKKLAVTKPNN-.
Full text
PDF





Images in this article
Selected References
These references are in PubMed. This may not be the complete list of references from this article.
- Bensch K. G., Tanaka S., Hu S. Z., Wang T. S., Korn D. Intracellular localization of human DNA polymerase alpha with monoclonal antibodies. J Biol Chem. 1982 Jul 25;257(14):8391–8396. [PubMed] [Google Scholar]
- Bernatowicz M. S., Matsueda G. R. Preparation of peptide-protein immunogens using N-succinimidyl bromoacetate as a heterobifunctional crosslinking reagent. Anal Biochem. 1986 May 15;155(1):95–102. doi: 10.1016/0003-2697(86)90231-9. [DOI] [PubMed] [Google Scholar]
- Bhattacharya P., Basu S. Probable involvement of a glycoconjugate in IMR-32 DNA synthesis: decrease of DNA polymerase alpha 2 activity after tunicamycin treatment. Proc Natl Acad Sci U S A. 1982 Mar;79(5):1488–1491. doi: 10.1073/pnas.79.5.1488. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Casadaban M. J., Chou J., Cohen S. N. In vitro gene fusions that join an enzymatically active beta-galactosidase segment to amino-terminal fragments of exogenous proteins: Escherichia coli plasmid vectors for the detection and cloning of translational initiation signals. J Bacteriol. 1980 Aug;143(2):971–980. doi: 10.1128/jb.143.2.971-980.1980. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Davis L. I., Blobel G. Identification and characterization of a nuclear pore complex protein. Cell. 1986 Jun 6;45(5):699–709. doi: 10.1016/0092-8674(86)90784-1. [DOI] [PubMed] [Google Scholar]
- Douglass J., Civelli O., Herbert E. Polyprotein gene expression: generation of diversity of neuroendocrine peptides. Annu Rev Biochem. 1984;53:665–715. doi: 10.1146/annurev.bi.53.070184.003313. [DOI] [PubMed] [Google Scholar]
- Elbein A. D. Inhibitors of the biosynthesis and processing of N-linked oligosaccharide chains. Annu Rev Biochem. 1987;56:497–534. doi: 10.1146/annurev.bi.56.070187.002433. [DOI] [PubMed] [Google Scholar]
- Fisher P. A., Korn D. DNA polymerase-alpha. Purification and structural characterization of the near homogeneous enzyme from human KB cells. J Biol Chem. 1977 Sep 25;252(18):6528–6535. [PubMed] [Google Scholar]
- Gallagher P., Henneberry J., Wilson I., Sambrook J., Gething M. J. Addition of carbohydrate side chains at novel sites on influenza virus hemagglutinin can modulate the folding, transport, and activity of the molecule. J Cell Biol. 1988 Dec;107(6 Pt 1):2059–2073. doi: 10.1083/jcb.107.6.2059. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Goldstein I. J., Hayes C. E. The lectins: carbohydrate-binding proteins of plants and animals. Adv Carbohydr Chem Biochem. 1978;35:127–340. doi: 10.1016/s0065-2318(08)60220-6. [DOI] [PubMed] [Google Scholar]
- Hart G. W., Haltiwanger R. S., Holt G. D., Kelly W. G. Glycosylation in the nucleus and cytoplasm. Annu Rev Biochem. 1989;58:841–874. doi: 10.1146/annurev.bi.58.070189.004205. [DOI] [PubMed] [Google Scholar]
- Holt G. D., Hart G. W. The subcellular distribution of terminal N-acetylglucosamine moieties. Localization of a novel protein-saccharide linkage, O-linked GlcNAc. J Biol Chem. 1986 Jun 15;261(17):8049–8057. [PubMed] [Google Scholar]
- Holt G. D., Snow C. M., Senior A., Haltiwanger R. S., Gerace L., Hart G. W. Nuclear pore complex glycoproteins contain cytoplasmically disposed O-linked N-acetylglucosamine. J Cell Biol. 1987 May;104(5):1157–1164. doi: 10.1083/jcb.104.5.1157. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hunkapiller M. W., Hewick R. M., Dreyer W. J., Hood L. E. High-sensitivity sequencing with a gas-phase sequenator. Methods Enzymol. 1983;91:399–413. doi: 10.1016/s0076-6879(83)91038-8. [DOI] [PubMed] [Google Scholar]
- Jackson S. P., Tjian R. O-glycosylation of eukaryotic transcription factors: implications for mechanisms of transcriptional regulation. Cell. 1988 Oct 7;55(1):125–133. doi: 10.1016/0092-8674(88)90015-3. [DOI] [PubMed] [Google Scholar]
- Johnson L. M., Snyder M., Chang L. M., Davis R. W., Campbell J. L. Isolation of the gene encoding yeast DNA polymerase I. Cell. 1985 Nov;43(1):369–377. doi: 10.1016/0092-8674(85)90042-x. [DOI] [PubMed] [Google Scholar]
- Kaguni L. S., Rossignol J. M., Conaway R. C., Banks G. R., Lehman I. R. Association of DNA primase with the beta/gamma subunits of DNA polymerase alpha from Drosophila melanogaster embryos. J Biol Chem. 1983 Aug 10;258(15):9037–9039. [PubMed] [Google Scholar]
- Lehman I. R., Kaguni L. S. DNA polymerase alpha. J Biol Chem. 1989 Mar 15;264(8):4265–4268. [PubMed] [Google Scholar]
- Nasheuer H. P., Grosse F. DNA polymerase alpha-primase from calf thymus. Determination of the polypeptide responsible for primase activity. J Biol Chem. 1988 Jun 25;263(18):8981–8988. [PubMed] [Google Scholar]
- Nyame K., Cummings R. D., Damian R. T. Schistosoma mansoni synthesizes glycoproteins containing terminal O-linked N-acetylglucosamine residues. J Biol Chem. 1987 Jun 15;262(17):7990–7995. [PubMed] [Google Scholar]
- Pizzagalli A., Valsasnini P., Plevani P., Lucchini G. DNA polymerase I gene of Saccharomyces cerevisiae: nucleotide sequence, mapping of a temperature-sensitive mutation, and protein homology with other DNA polymerases. Proc Natl Acad Sci U S A. 1988 Jun;85(11):3772–3776. doi: 10.1073/pnas.85.11.3772. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Plevani P., Foiani M., Valsasnini P., Badaracco G., Cheriathundam E., Chang L. M. Polypeptide structure of DNA primase from a yeast DNA polymerase-primase complex. J Biol Chem. 1985 Jun 10;260(11):7102–7107. [PubMed] [Google Scholar]
- Schindler M., Hogan M., Miller R., DeGaetano D. A nuclear specific glycoprotein representative of a unique pattern of glycosylation. J Biol Chem. 1987 Jan 25;262(3):1254–1260. [PubMed] [Google Scholar]
- Tabor S., Richardson C. C. A bacteriophage T7 RNA polymerase/promoter system for controlled exclusive expression of specific genes. Proc Natl Acad Sci U S A. 1985 Feb;82(4):1074–1078. doi: 10.1073/pnas.82.4.1074. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tanaka S., Hu S. Z., Wang T. S., Korn D. Preparation and preliminary characterization of monoclonal antibodies against human DNA polymerase alpha. J Biol Chem. 1982 Jul 25;257(14):8386–8390. [PubMed] [Google Scholar]
- Torres C. R., Hart G. W. Topography and polypeptide distribution of terminal N-acetylglucosamine residues on the surfaces of intact lymphocytes. Evidence for O-linked GlcNAc. J Biol Chem. 1984 Mar 10;259(5):3308–3317. [PubMed] [Google Scholar]
- Towbin H., Staehelin T., Gordon J. Electrophoretic transfer of proteins from polyacrylamide gels to nitrocellulose sheets: procedure and some applications. Proc Natl Acad Sci U S A. 1979 Sep;76(9):4350–4354. doi: 10.1073/pnas.76.9.4350. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tseng B. Y., Ahlem C. N. A DNA primase from mouse cells. Purification and partial characterization. J Biol Chem. 1983 Aug 25;258(16):9845–9849. [PubMed] [Google Scholar]
- Ullmann A. One-step purification of hybrid proteins which have beta-galactosidase activity. Gene. 1984 Jul-Aug;29(1-2):27–31. doi: 10.1016/0378-1119(84)90162-8. [DOI] [PubMed] [Google Scholar]
- Walter G. Production and use of antibodies against synthetic peptides. J Immunol Methods. 1986 Apr 17;88(2):149–161. doi: 10.1016/0022-1759(86)90001-3. [DOI] [PubMed] [Google Scholar]
- Wang T. S., Hu S. Z., Korn D. DNA primase from KB cells. Characterization of a primase activity tightly associated with immunoaffinity-purified DNA polymerase-alpha. J Biol Chem. 1984 Feb 10;259(3):1854–1865. [PubMed] [Google Scholar]
- Wang T. S., Wong S. W., Korn D. Human DNA polymerase alpha: predicted functional domains and relationships with viral DNA polymerases. FASEB J. 1989 Jan;3(1):14–21. doi: 10.1096/fasebj.3.1.2642867. [DOI] [PubMed] [Google Scholar]
- Wong S. W., Paborsky L. R., Fisher P. A., Wang T. S., Korn D. Structural and enzymological characterization of immunoaffinity-purified DNA polymerase alpha.DNA primase complex from KB cells. J Biol Chem. 1986 Jun 15;261(17):7958–7968. [PubMed] [Google Scholar]
- Wong S. W., Wahl A. F., Yuan P. M., Arai N., Pearson B. E., Arai K., Korn D., Hunkapiller M. W., Wang T. S. Human DNA polymerase alpha gene expression is cell proliferation dependent and its primary structure is similar to both prokaryotic and eukaryotic replicative DNA polymerases. EMBO J. 1988 Jan;7(1):37–47. doi: 10.1002/j.1460-2075.1988.tb02781.x. [DOI] [PMC free article] [PubMed] [Google Scholar]