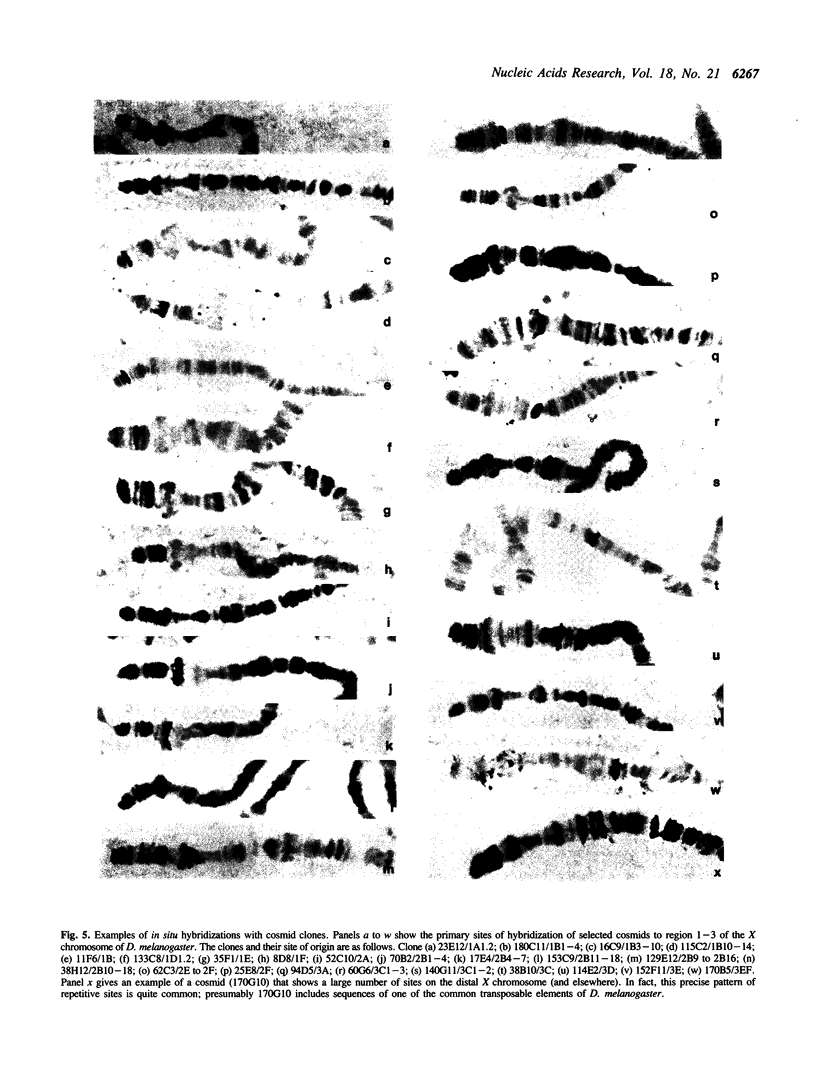
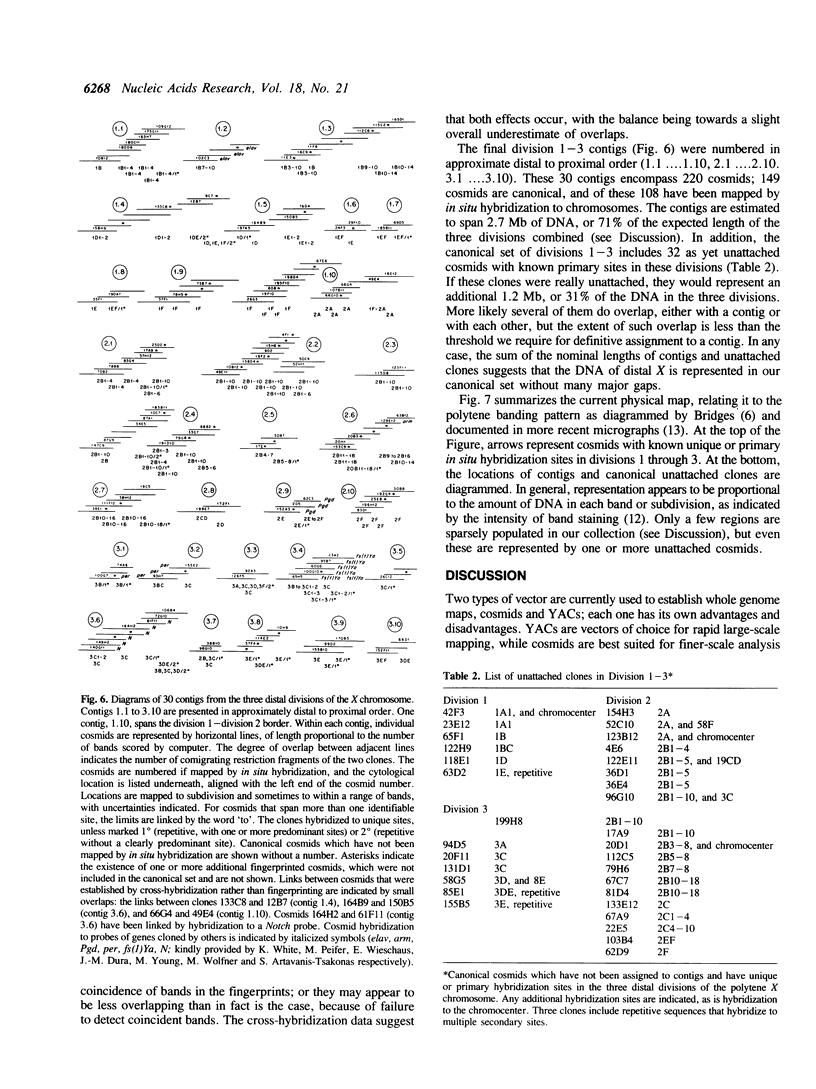
Abstract
A physical map of the D. melanogaster genome is being constructed, in the form of overlapping cosmid clones that are assigned to specific polytene chromosome sites. A master library of ca. 20,000 cosmids is screened with probes that correspond to numbered chromosomal divisions (ca. 1% of the genome); these probes are prepared by microdissection and PCR-amplification of individual chromosomes. The 120 to 250 cosmids selected by each probe are fingerprinted by Hinfl digestion and gel electrophoresis, and overlaps are detected by computer analysis of the fingerprints, permitting us to assemble sets of contiguous clones (contigs). Selected cosmids, both from contigs and unattached, are then localized by in situ hybridization to polytene chromosomes. Crosshybridization analysis using end probes links some contigs, and hybridization to previously cloned genes relates the physical to the genetic map. This approach has been used to construct a physical map of the 3.8 megabase DNA in the three distal divisions of the x chromosome. The map is represented by 181 canonical cosmids, of which 108 clones in contigs and 32 unattached clones have been mapped individually by in situ hybridization to chromosomes. Our current database of in situ hybridization results also includes the beginning of a physical map for the rest of the genome: 162 cosmids have been assigned by in situ hybridization to 129 chromosomal subdivisions elsewhere in the genome, representing 5 to 6 megabases of additional mapped DNA.
Full text
PDF







Images in this article
Selected References
These references are in PubMed. This may not be the complete list of references from this article.
- Artavanis-Tsakonas S., Schedl P., Tschudi C., Pirrotta V., Steward R., Gehring W. J. The 5S genes of Drosophila melanogaster. Cell. 1977 Dec;12(4):1057–1067. doi: 10.1016/0092-8674(77)90169-6. [DOI] [PubMed] [Google Scholar]
- Bender W., Spierer P., Hogness D. S. Chromosomal walking and jumping to isolate DNA from the Ace and rosy loci and the bithorax complex in Drosophila melanogaster. J Mol Biol. 1983 Jul 25;168(1):17–33. doi: 10.1016/s0022-2836(83)80320-9. [DOI] [PubMed] [Google Scholar]
- Bingham P. M., Levis R., Rubin G. M. Cloning of DNA sequences from the white locus of D. melanogaster by a novel and general method. Cell. 1981 Sep;25(3):693–704. doi: 10.1016/0092-8674(81)90176-8. [DOI] [PubMed] [Google Scholar]
- Burke D. T., Carle G. F., Olson M. V. Cloning of large segments of exogenous DNA into yeast by means of artificial chromosome vectors. Science. 1987 May 15;236(4803):806–812. doi: 10.1126/science.3033825. [DOI] [PubMed] [Google Scholar]
- Caccone A., Amato G. D., Powell J. R. Rates and patterns of scnDNA and mtDNA divergence within the Drosophila melanogaster subgroup. Genetics. 1988 Apr;118(4):671–683. doi: 10.1093/genetics/118.4.671. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cooley L., Kelley R., Spradling A. Insertional mutagenesis of the Drosophila genome with single P elements. Science. 1988 Mar 4;239(4844):1121–1128. doi: 10.1126/science.2830671. [DOI] [PubMed] [Google Scholar]
- Coulson A., Sulston J., Brenner S., Karn J. Toward a physical map of the genome of the nematode Caenorhabditis elegans. Proc Natl Acad Sci U S A. 1986 Oct;83(20):7821–7825. doi: 10.1073/pnas.83.20.7821. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dowsett A. P. Closely related species of Drosophila can contain different libraries of middle repetitive DNA sequences. Chromosoma. 1983;88(2):104–108. doi: 10.1007/BF00327329. [DOI] [PubMed] [Google Scholar]
- Dowsett A. P., Young M. W. Differing levels of dispersed repetitive DNA among closely related species of Drosophila. Proc Natl Acad Sci U S A. 1982 Aug;79(15):4570–4574. doi: 10.1073/pnas.79.15.4570. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Feinberg A. P., Vogelstein B. "A technique for radiolabeling DNA restriction endonuclease fragments to high specific activity". Addendum. Anal Biochem. 1984 Feb;137(1):266–267. doi: 10.1016/0003-2697(84)90381-6. [DOI] [PubMed] [Google Scholar]
- Garza D., Ajioka J. W., Burke D. T., Hartl D. L. Mapping the Drosophila genome with yeast artificial chromosomes. Science. 1989 Nov 3;246(4930):641–646. doi: 10.1126/science.2510296. [DOI] [PubMed] [Google Scholar]
- Gibson T. J., Rosenthal A., Waterston R. H. Lorist6, a cosmid vector with BamHI, NotI, ScaI and HindIII cloning sites and altered neomycin phosphotransferase gene expression. Gene. 1987;53(2-3):283–286. doi: 10.1016/0378-1119(87)90017-5. [DOI] [PubMed] [Google Scholar]
- Glover D. M., White R. L., Finnegan D. J., Hogness D. S. Characterization of six cloned DNAs from Drosophila melanogaster, including one that contains the genes for rRNA. Cell. 1975 Jun;5(2):149–157. doi: 10.1016/0092-8674(75)90023-9. [DOI] [PubMed] [Google Scholar]
- Grunstein M., Hogness D. S. Colony hybridization: a method for the isolation of cloned DNAs that contain a specific gene. Proc Natl Acad Sci U S A. 1975 Oct;72(10):3961–3965. doi: 10.1073/pnas.72.10.3961. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hinton T. A Comparative Study of Certain Heterochromatic Regions in the Mitotic and Salivary Gland Chromosomes of Drosophila Melanogaster. Genetics. 1942 Jan;27(1):119–127. doi: 10.1093/genetics/27.1.119. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Johnson D. H. Molecular cloning of DNA from specific chromosomal regions by microdissection and sequence-independent amplification of DNA. Genomics. 1990 Feb;6(2):243–251. doi: 10.1016/0888-7543(90)90563-a. [DOI] [PubMed] [Google Scholar]
- Knott V., Rees D. J., Cheng Z., Brownlee G. G. Randomly picked cosmid clones overlap the pyrB and oriC gap in the physical map of the E. coli chromosome. Nucleic Acids Res. 1988 Mar 25;16(6):2601–2612. doi: 10.1093/nar/16.6.2601. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kohara Y., Akiyama K., Isono K. The physical map of the whole E. coli chromosome: application of a new strategy for rapid analysis and sorting of a large genomic library. Cell. 1987 Jul 31;50(3):495–508. doi: 10.1016/0092-8674(87)90503-4. [DOI] [PubMed] [Google Scholar]
- Lander E. S., Waterman M. S. Genomic mapping by fingerprinting random clones: a mathematical analysis. Genomics. 1988 Apr;2(3):231–239. doi: 10.1016/0888-7543(88)90007-9. [DOI] [PubMed] [Google Scholar]
- Lemeunier F., Ashburner M. A. Relationships within the melanogaster species subgroup of the genus Drosophila (Sophophora). II. Phylogenetic relationships between six species based upon polytene chromosome banding sequences. Proc R Soc Lond B Biol Sci. 1976 May 18;193(1112):275–294. doi: 10.1098/rspb.1976.0046. [DOI] [PubMed] [Google Scholar]
- Lis J. T., Prestidge L., Hogness D. S. A novel arrangement of tandemly repeated genes at a major heat shock site in D. melanogaster. Cell. 1978 Aug;14(4):901–919. doi: 10.1016/0092-8674(78)90345-8. [DOI] [PubMed] [Google Scholar]
- Lüdecke H. J., Senger G., Claussen U., Horsthemke B. Cloning defined regions of the human genome by microdissection of banded chromosomes and enzymatic amplification. Nature. 1989 Mar 23;338(6213):348–350. doi: 10.1038/338348a0. [DOI] [PubMed] [Google Scholar]
- Martin G., Wiernasz D., Schedl P. Evolution of Drosophila repetitive-dispersed DNA. J Mol Evol. 1983;19(3-4):203–213. doi: 10.1007/BF02099967. [DOI] [PubMed] [Google Scholar]
- O'Kane C. J., Gehring W. J. Detection in situ of genomic regulatory elements in Drosophila. Proc Natl Acad Sci U S A. 1987 Dec;84(24):9123–9127. doi: 10.1073/pnas.84.24.9123. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Olson M. V., Dutchik J. E., Graham M. Y., Brodeur G. M., Helms C., Frank M., MacCollin M., Scheinman R., Frank T. Random-clone strategy for genomic restriction mapping in yeast. Proc Natl Acad Sci U S A. 1986 Oct;83(20):7826–7830. doi: 10.1073/pnas.83.20.7826. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Olson M., Hood L., Cantor C., Botstein D. A common language for physical mapping of the human genome. Science. 1989 Sep 29;245(4925):1434–1435. doi: 10.1126/science.2781285. [DOI] [PubMed] [Google Scholar]
- Painter T S. A New Method for the Study of Chromosome Aberrations and the Plotting of Chromosome Maps in Drosophila Melanogaster. Genetics. 1934 May;19(3):175–188. doi: 10.1093/genetics/19.3.175. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Painter T. S. A NEW METHOD FOR THE STUDY OF CHROMOSOME REARRANGEMENTS AND THE PLOTTING OF CHROMOSOME MAPS. Science. 1933 Dec 22;78(2034):585–586. doi: 10.1126/science.78.2034.585. [DOI] [PubMed] [Google Scholar]
- Pardu M. L., Gerbi S. A., Eckhardt R. A., Gall J. G. Cytological localization of DNA complementary to ribosomal RNA in polytene chromosomes of Diptera. Chromosoma. 1970;29(3):268–290. doi: 10.1007/BF00325943. [DOI] [PubMed] [Google Scholar]
- Rasch E. M., Barr H. J., Rasch R. W. The DNA content of sperm of Drosophila melanogaster. Chromosoma. 1971;33(1):1–18. doi: 10.1007/BF00326379. [DOI] [PubMed] [Google Scholar]
- Saunders R. D., Glover D. M., Ashburner M., Siden-Kiamos I., Louis C., Monastirioti M., Savakis C., Kafatos F. PCR amplification of DNA microdissected from a single polytene chromosome band: a comparison with conventional microcloning. Nucleic Acids Res. 1989 Nov 25;17(22):9027–9037. doi: 10.1093/nar/17.22.9027. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sulston J., Mallett F., Durbin R., Horsnell T. Image analysis of restriction enzyme fingerprint autoradiograms. Comput Appl Biosci. 1989 Apr;5(2):101–106. doi: 10.1093/bioinformatics/5.2.101. [DOI] [PubMed] [Google Scholar]
- Sulston J., Mallett F., Staden R., Durbin R., Horsnell T., Coulson A. Software for genome mapping by fingerprinting techniques. Comput Appl Biosci. 1988 Mar;4(1):125–132. doi: 10.1093/bioinformatics/4.1.125. [DOI] [PubMed] [Google Scholar]
- Wensink P. C., Finnegan D. J., Donelson J. E., Hogness D. S. A system for mapping DNA sequences in the chromosomes of Drosophila melanogaster. Cell. 1974 Dec;3(4):315–325. doi: 10.1016/0092-8674(74)90045-2. [DOI] [PubMed] [Google Scholar]
- Werman S. D., Davidson E. H., Britten R. J. Rapid evolution in a fraction of the Drosophila nuclear genome. J Mol Evol. 1990 Mar;30(3):281–289. doi: 10.1007/BF02099998. [DOI] [PubMed] [Google Scholar]
- Wesley C. S., Ben M., Kreitman M., Hagag N., Eanes W. F. Cloning regions of the Drosophila genome by microdissection of polytene chromosome DNA and PCR with nonspecific primer. Nucleic Acids Res. 1990 Feb 11;18(3):599–603. doi: 10.1093/nar/18.3.599. [DOI] [PMC free article] [PubMed] [Google Scholar]







