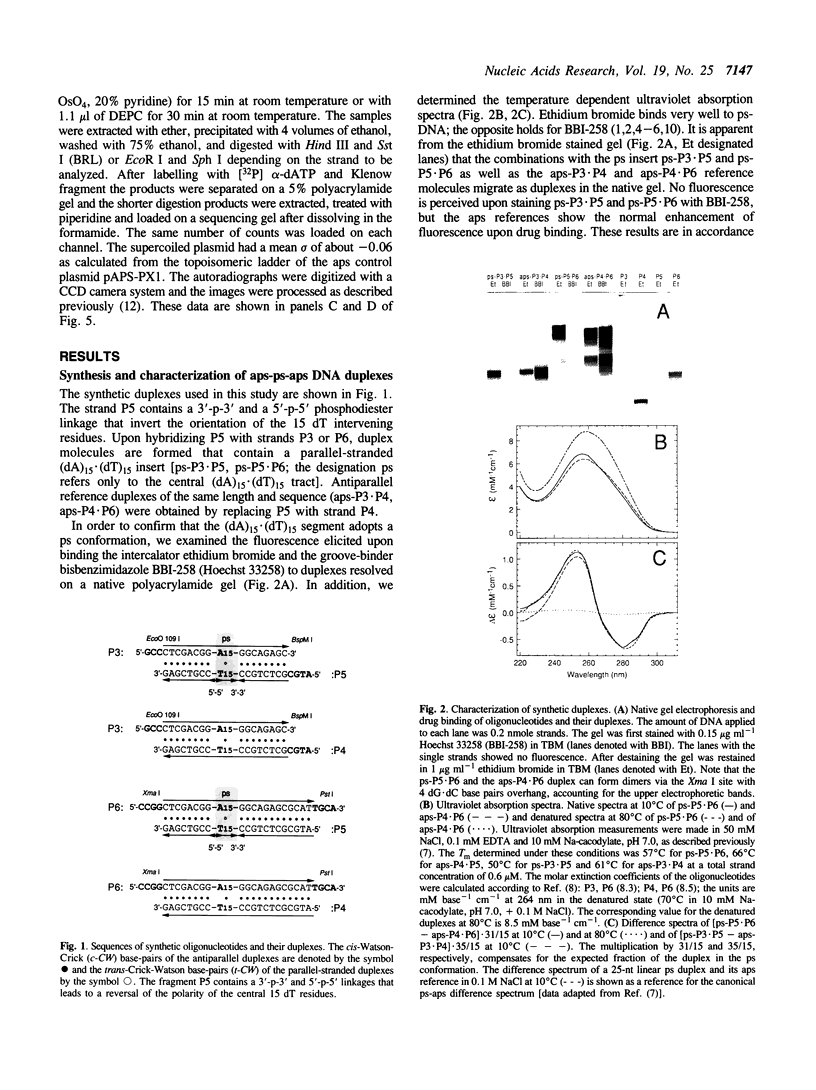
Abstract
DNA oligonucleotides with appropriate sequences can form a stable duplex in which the two strands are paired in a parallel orientation instead of as the conventional antiparallel double helix of B-DNA. In parallel-stranded DNA (ps-DNA) base pairing is noncanonical with the glycosidic bonds in a trans orientation. The two grooves are equivalent. We have synthesized DNA duplexes consisting of a central parallel-stranded (dA)15.(dT)15 tract flanked by normal antiparallel regions, and ligated them into the pUC18 plasmid. The effect of negative supercoiling on the covalently closed circular molecules was studied by two-dimensional agarose gel electrophoresis and by chemical modification with OsO4-pyridine (Os,py) and diethylpyrocarbonate (DEPC). The following results were obtained: (i) The ps insert, and by inference ps-DNA in general, adopts a right handed helical form. (ii) Upon increasing the negative superhelix density (-sigma) to greater than 0.03 the 15 bp ps insert undergoes a major transition leading to a relaxation corresponding to a reduction in twist of approximately 2.5 helical turns. The transition free surgery is approximately kcal/mol. (iii) The chemical modification pattern of the resulting structure suggests that the purine strand folds back and associates with the pyrimidine strand, forming a novel intramolecular triplex structure consisting of d(A.A.T) base triplets. A model for the triplex conformation is proposed and its thermodynamic properties are analyzed by statistical mechanics.
Full text
PDF

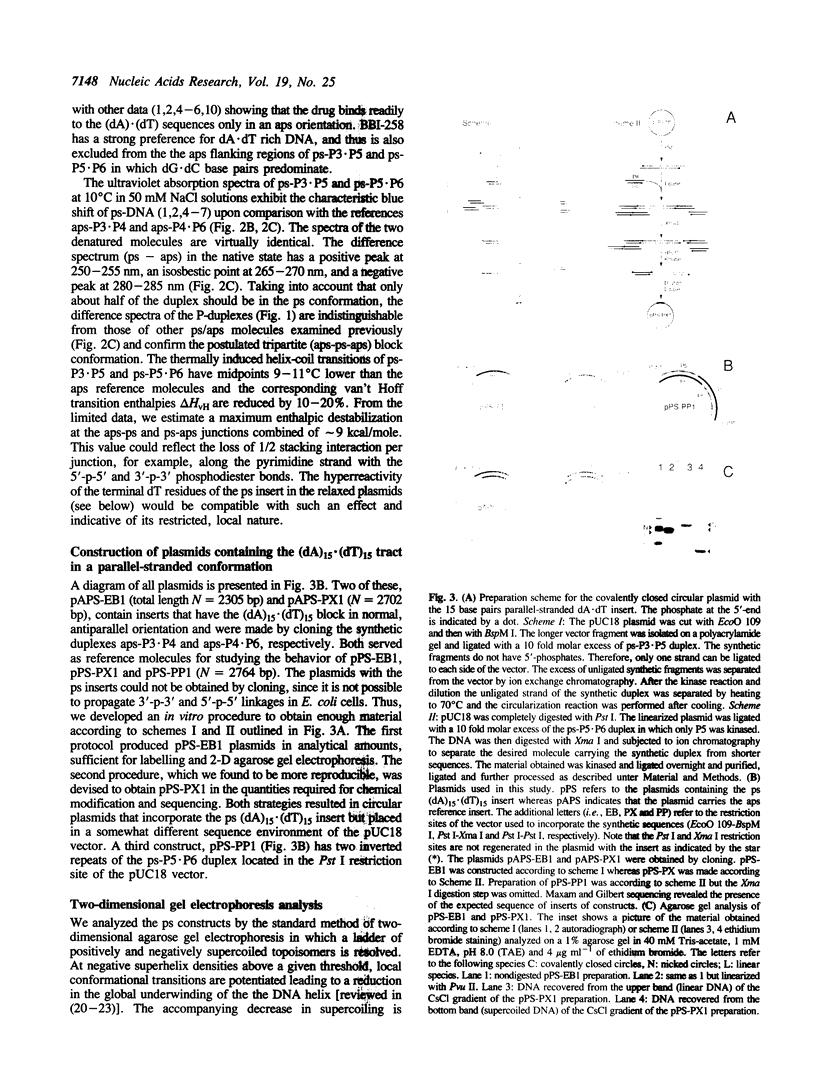

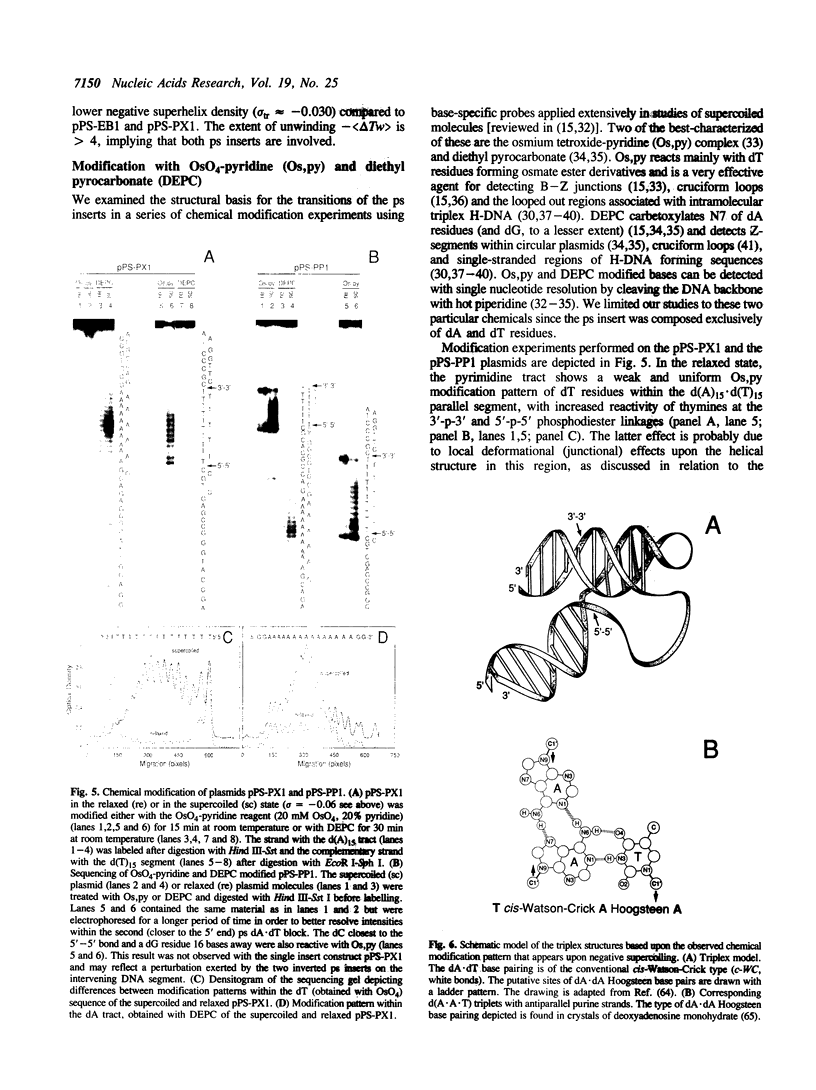




Images in this article
Selected References
These references are in PubMed. This may not be the complete list of references from this article.
- Anshelevich V. V., Vologodskii A. V., Frank-Kamenetskii M. D. A theoretical study of formation of DNA noncanonical structures under negative superhelical stress. J Biomol Struct Dyn. 1988 Oct;6(2):247–259. doi: 10.1080/07391102.1988.10507711. [DOI] [PubMed] [Google Scholar]
- Beal P. A., Dervan P. B. Second structural motif for recognition of DNA by oligonucleotide-directed triple-helix formation. Science. 1991 Mar 15;251(4999):1360–1363. doi: 10.1126/science.2003222. [DOI] [PubMed] [Google Scholar]
- Bernués J., Beltrán R., Casasnovas J. M., Azorín F. DNA-sequence and metal-ion specificity of the formation of *H-DNA. Nucleic Acids Res. 1990 Jul 25;18(14):4067–4073. doi: 10.1093/nar/18.14.4067. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bernués J., Beltrán R., Casasnovas J. M., Azorín F. Structural polymorphism of homopurine--homopyrimidine sequences: the secondary DNA structure adopted by a d(GA.CT)22 sequence in the presence of zinc ions. EMBO J. 1989 Jul;8(7):2087–2094. doi: 10.1002/j.1460-2075.1989.tb03617.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Blake R. D., Delcourt S. G. Electrostatic forces at helix-coil boundaries in DNA. Biopolymers. 1990 Feb 5;29(2):393–405. doi: 10.1002/bip.360290211. [DOI] [PubMed] [Google Scholar]
- Blake R. D., Delcourt S. G. Loop energy in DNA. Biopolymers. 1987 Dec;26(12):2009–2026. doi: 10.1002/bip.360261204. [DOI] [PubMed] [Google Scholar]
- Broitman S. L., Im D. D., Fresco J. R. Formation of the triple-stranded polynucleotide helix, poly(A.A.U). Proc Natl Acad Sci U S A. 1987 Aug;84(15):5120–5124. doi: 10.1073/pnas.84.15.5120. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Courey A. J., Wang J. C. Cruciform formation in a negatively supercoiled DNA may be kinetically forbidden under physiological conditions. Cell. 1983 Jul;33(3):817–829. doi: 10.1016/0092-8674(83)90024-7. [DOI] [PubMed] [Google Scholar]
- Fox K. R. Long (dA)n.(dT)n tracts can form intramolecular triplexes under superhelical stress. Nucleic Acids Res. 1990 Sep 25;18(18):5387–5391. doi: 10.1093/nar/18.18.5387. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Furlong J. C., Lilley D. M. Highly selective chemical modification of cruciform loops by diethyl pyrocarbonate. Nucleic Acids Res. 1986 May 27;14(10):3995–4007. doi: 10.1093/nar/14.10.3995. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Galazka G., Palecek E., Wells R. D., Klysik J. Site-specific OsO4 modification of the B-Z junctions formed at the (dA-dC)32 region in supercoiled DNA. J Biol Chem. 1986 May 25;261(15):7093–7098. [PubMed] [Google Scholar]
- Germann M. W., Kalisch B. W., van de Sande J. H. Relative stability of parallel- and antiparallel-stranded duplex DNA. Biochemistry. 1988 Nov 1;27(22):8302–8306. doi: 10.1021/bi00422a002. [DOI] [PubMed] [Google Scholar]
- Germann M. W., Vogel H. J., Pon R. T., van de Sande J. H. Characterization of a parallel-stranded DNA hairpin. Biochemistry. 1989 Jul 25;28(15):6220–6228. doi: 10.1021/bi00441a013. [DOI] [PubMed] [Google Scholar]
- Hanvey J. C., Klysik J., Wells R. D. Influence of DNA sequence on the formation of non-B right-handed helices in oligopurine.oligopyrimidine inserts in plasmids. J Biol Chem. 1988 May 25;263(15):7386–7396. [PubMed] [Google Scholar]
- Herr W. Diethyl pyrocarbonate: a chemical probe for secondary structure in negatively supercoiled DNA. Proc Natl Acad Sci U S A. 1985 Dec;82(23):8009–8013. doi: 10.1073/pnas.82.23.8009. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Horowitz D. S., Wang J. C. Torsional rigidity of DNA and length dependence of the free energy of DNA supercoiling. J Mol Biol. 1984 Feb 15;173(1):75–91. doi: 10.1016/0022-2836(84)90404-2. [DOI] [PubMed] [Google Scholar]
- Htun H., Dahlberg J. E. Single strands, triple strands, and kinks in H-DNA. Science. 1988 Sep 30;241(4874):1791–1796. doi: 10.1126/science.3175620. [DOI] [PubMed] [Google Scholar]
- Johnston B. H., Rich A. Chemical probes of DNA conformation: detection of Z-DNA at nucleotide resolution. Cell. 1985 Oct;42(3):713–724. doi: 10.1016/0092-8674(85)90268-5. [DOI] [PubMed] [Google Scholar]
- Johnston B. H. The S1-sensitive form of d(C-T)n.d(A-G)n: chemical evidence for a three-stranded structure in plasmids. Science. 1988 Sep 30;241(4874):1800–1804. doi: 10.1126/science.2845572. [DOI] [PubMed] [Google Scholar]
- Kelleher R. J., 3rd, Ellison M. J., Ho P. S., Rich A. Competitive behavior of multiple, discrete B-Z transitions in supercoiled DNA. Proc Natl Acad Sci U S A. 1986 Sep;83(17):6342–6346. doi: 10.1073/pnas.83.17.6342. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Klysik J., Rippe K., Jovin T. M. Reactivity of parallel-stranded DNA to chemical modification reagents. Biochemistry. 1990 Oct 23;29(42):9831–9839. doi: 10.1021/bi00494a012. [DOI] [PubMed] [Google Scholar]
- Klysik J., Zacharias W., Galazka G., Kwinkowski M., Uznanski B., Okruszek A. Structural interconversion of alternating purine-pyrimidine inverted repeats cloned in supercoiled plasmids. Nucleic Acids Res. 1988 Jul 25;16(14B):6915–6933. doi: 10.1093/nar/16.14.6915. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kohwi Y., Kohwi-Shigematsu T. Magnesium ion-dependent triple-helix structure formed by homopurine-homopyrimidine sequences in supercoiled plasmid DNA. Proc Natl Acad Sci U S A. 1988 Jun;85(11):3781–3785. doi: 10.1073/pnas.85.11.3781. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Letai A. G., Palladino M. A., Fromm E., Rizzo V., Fresco J. R. Specificity in formation of triple-stranded nucleic acid helical complexes: studies with agarose-linked polyribonucleotide affinity columns. Biochemistry. 1988 Dec 27;27(26):9108–9112. doi: 10.1021/bi00426a007. [DOI] [PubMed] [Google Scholar]
- Lilley D. M., Hallam L. R. Thermodynamics of the ColE1 cruciform. Comparisons between probing and topological experiments using single topoisomers. J Mol Biol. 1984 Nov 25;180(1):179–200. doi: 10.1016/0022-2836(84)90436-4. [DOI] [PubMed] [Google Scholar]
- Lilley D. M., Palecek E. The supercoil-stabilised cruciform of ColE1 is hyper-reactive to osmium tetroxide. EMBO J. 1984 May;3(5):1187–1192. doi: 10.1002/j.1460-2075.1984.tb01949.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lyamichev V. I., Mirkin S. M., Frank-Kamenetskii M. D. Structure of (dG)n.(dC)n under superhelical stress and acid pH. J Biomol Struct Dyn. 1987 Oct;5(2):275–282. doi: 10.1080/07391102.1987.10506393. [DOI] [PubMed] [Google Scholar]
- Lyamichev V. I., Mirkin S. M., Frank-Kamenetskii M. D. Structures of homopurine-homopyrimidine tract in superhelical DNA. J Biomol Struct Dyn. 1986 Feb;3(4):667–669. doi: 10.1080/07391102.1986.10508454. [DOI] [PubMed] [Google Scholar]
- Lyamichev V. I., Mirkin S. M., Kumarev V. P., Baranova L. V., Vologodskii A. V., Frank-Kamenetskii M. D. Energetics of the B-H transition in supercoiled DNA carrying d(CT)x.d(AG)x and d(C)n.d(G)n inserts. Nucleic Acids Res. 1989 Nov 25;17(22):9417–9423. doi: 10.1093/nar/17.22.9417. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lyamichev V. I., Panyutin I. G., Frank-Kamenetskii M. D. Evidence of cruciform structures in superhelical DNA provided by two-dimensional gel electrophoresis. FEBS Lett. 1983 Mar 21;153(2):298–302. doi: 10.1016/0014-5793(83)80628-0. [DOI] [PubMed] [Google Scholar]
- Lyamichev V. I., Voloshin O. N., Frank-Kamenetskii M. D., Soyfer V. N. Photofootprinting of DNA triplexes. Nucleic Acids Res. 1991 Apr 11;19(7):1633–1638. doi: 10.1093/nar/19.7.1633. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Maxam A. M., Gilbert W. Sequencing end-labeled DNA with base-specific chemical cleavages. Methods Enzymol. 1980;65(1):499–560. doi: 10.1016/s0076-6879(80)65059-9. [DOI] [PubMed] [Google Scholar]
- McCarthy J. G., Williams L. D., Rich A. Chemical reactivity of potassium permanganate and diethyl pyrocarbonate with B DNA: specific reactivity with short A-tracts. Biochemistry. 1990 Jun 26;29(25):6071–6081. doi: 10.1021/bi00477a027. [DOI] [PubMed] [Google Scholar]
- Mirkin S. M., Lyamichev V. I., Kumarev V. P., Kobzev V. F., Nosikov V. V., Vologodskii A. V. The energetics of the B-Z transition in DNA. J Biomol Struct Dyn. 1987 Aug;5(1):79–88. doi: 10.1080/07391102.1987.10506376. [DOI] [PubMed] [Google Scholar]
- Murchie A. I., Clegg R. M., von Kitzing E., Duckett D. R., Diekmann S., Lilley D. M. Fluorescence energy transfer shows that the four-way DNA junction is a right-handed cross of antiparallel molecules. Nature. 1989 Oct 26;341(6244):763–766. doi: 10.1038/341763a0. [DOI] [PubMed] [Google Scholar]
- Otto C., Thomas G. A., Rippe K., Jovin T. M., Peticolas W. L. The hydrogen-bonding structure in parallel-stranded duplex DNA is reverse Watson-Crick. Biochemistry. 1991 Mar 26;30(12):3062–3069. doi: 10.1021/bi00226a012. [DOI] [PubMed] [Google Scholar]
- Palecek E. Local supercoil-stabilized DNA structures. Crit Rev Biochem Mol Biol. 1991;26(2):151–226. doi: 10.3109/10409239109081126. [DOI] [PubMed] [Google Scholar]
- Pattabiraman N. Can the double helix be parallel? Biopolymers. 1986 Sep;25(9):1603–1606. doi: 10.1002/bip.360250903. [DOI] [PubMed] [Google Scholar]
- Peck L. J., Wang J. C. Energetics of B-to-Z transition in DNA. Proc Natl Acad Sci U S A. 1983 Oct;80(20):6206–6210. doi: 10.1073/pnas.80.20.6206. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pilch D. S., Levenson C., Shafer R. H. Structure, stability, and thermodynamics of a short intermolecular purine-purine-pyrimidine triple helix. Biochemistry. 1991 Jun 25;30(25):6081–6088. doi: 10.1021/bi00239a001. [DOI] [PubMed] [Google Scholar]
- Ramsing N. B., Jovin T. M. Parallel stranded duplex DNA. Nucleic Acids Res. 1988 Jul 25;16(14A):6659–6676. doi: 10.1093/nar/16.14.6659. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rippe K., Jovin T. M. Substrate properties of 25-nt parallel-stranded linear DNA duplexes. Biochemistry. 1989 Nov 28;28(24):9542–9549. doi: 10.1021/bi00450a044. [DOI] [PubMed] [Google Scholar]
- Rippe K., Ramsing N. B., Jovin T. M. Spectroscopic properties and helical stabilities of 25-nt parallel-stranded linear DNA duplexes. Biochemistry. 1989 Nov 28;28(24):9536–9541. doi: 10.1021/bi00450a043. [DOI] [PubMed] [Google Scholar]
- Rippe K., Ramsing N. B., Klement R., Jovin T. M. A parallel stranded linear DNA duplex incorporating dG.dC base pairs. J Biomol Struct Dyn. 1990 Jun;7(6):1199–1209. doi: 10.1080/07391102.1990.10508559. [DOI] [PubMed] [Google Scholar]
- Scheffler I. E., Elson E. L., Baldwin R. L. Helix formation by d(TA) oligomers. II. Analysis of the helix-coli transitions of linear and circular oligomers. J Mol Biol. 1970 Feb 28;48(1):145–171. doi: 10.1016/0022-2836(70)90225-1. [DOI] [PubMed] [Google Scholar]
- Shimizu M., Hanvey J. C., Wells R. D. Intramolecular DNA triplexes in supercoiled plasmids. I. Effect of loop size on formation and stability. J Biol Chem. 1989 Apr 5;264(10):5944–5949. [PubMed] [Google Scholar]
- Singleton C. K., Wells R. D. The facile generation of covalently closed, circular DNAs with defined negative superhelical densities. Anal Biochem. 1982 May 15;122(2):253–257. doi: 10.1016/0003-2697(82)90277-9. [DOI] [PubMed] [Google Scholar]
- Sklenár V., Feigon J. Formation of a stable triplex from a single DNA strand. Nature. 1990 Jun 28;345(6278):836–838. doi: 10.1038/345836a0. [DOI] [PubMed] [Google Scholar]
- Stokrová J., Vojtisková M., Palecek E. Electron microscopy of supercoiled pEJ4 DNA containing homopurine.homopyrimidine sequences. J Biomol Struct Dyn. 1989 Apr;6(5):891–898. doi: 10.1080/07391102.1989.10506520. [DOI] [PubMed] [Google Scholar]
- Vojtísková M., Mirkin S., Lyamichev V., Voloshin O., Frank-Kamenetskii M., Palecek E. Chemical probing of the homopurine.homopyrimidine tract in supercoiled DNA at single-nucleotide resolution. FEBS Lett. 1988 Jul 18;234(2):295–299. doi: 10.1016/0014-5793(88)80102-9. [DOI] [PubMed] [Google Scholar]
- Voloshin O. N., Mirkin S. M., Lyamichev V. I., Belotserkovskii B. P., Frank-Kamenetskii M. D. Chemical probing of homopurine-homopyrimidine mirror repeats in supercoiled DNA. Nature. 1988 Jun 2;333(6172):475–476. doi: 10.1038/333475a0. [DOI] [PubMed] [Google Scholar]
- Watson D. G., Sutor D. J., Tollin P. The crystal structure of deoxyadenosine monohydrate. Acta Crystallogr. 1965 Jul 10;19(1):111–124. doi: 10.1107/s0365110x65002852. [DOI] [PubMed] [Google Scholar]
- Yagil G. Paranemic structures of DNA and their role in DNA unwinding. Crit Rev Biochem Mol Biol. 1991;26(5-6):475–559. doi: 10.3109/10409239109086791. [DOI] [PubMed] [Google Scholar]
- Zheng G. X., Sinden R. R. Effect of base composition at the center of inverted repeated DNA sequences on cruciform transitions in DNA. J Biol Chem. 1988 Apr 15;263(11):5356–5361. [PubMed] [Google Scholar]
- van de Sande J. H., Ramsing N. B., Germann M. W., Elhorst W., Kalisch B. W., von Kitzing E., Pon R. T., Clegg R. C., Jovin T. M. Parallel stranded DNA. Science. 1988 Jul 29;241(4865):551–557. doi: 10.1126/science.3399890. [DOI] [PubMed] [Google Scholar]