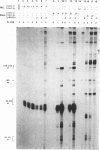
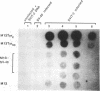
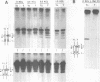
Abstract
We have analyzed proteins bound to active and to inactive tRNA(tyr) genes imbedded in the late coding region of SV40 minichromosomal DNA. Bal-31 nuclease resection from the 5' and 3' sides of the active tRNA(tyr) gene reveals proteins bound to the 5' flank, to the promoter 'A' block, to an intragenic sequence, to the promoter 'B' block and to a 3' downstream terminator/pause sequence. The proteins bound near the promoter 'B' block and the downstream terminator/pause sequence are reduced or eliminated by an inactivating deletion in the tRNA(tyr) 'B block'. That proteins are detected in the 5' flank and over the promoter 'A block' of the inactive gene contrasts with current notions regarding the requirement for a functional 'B' block for binding of transcription factors.
Full text
PDF

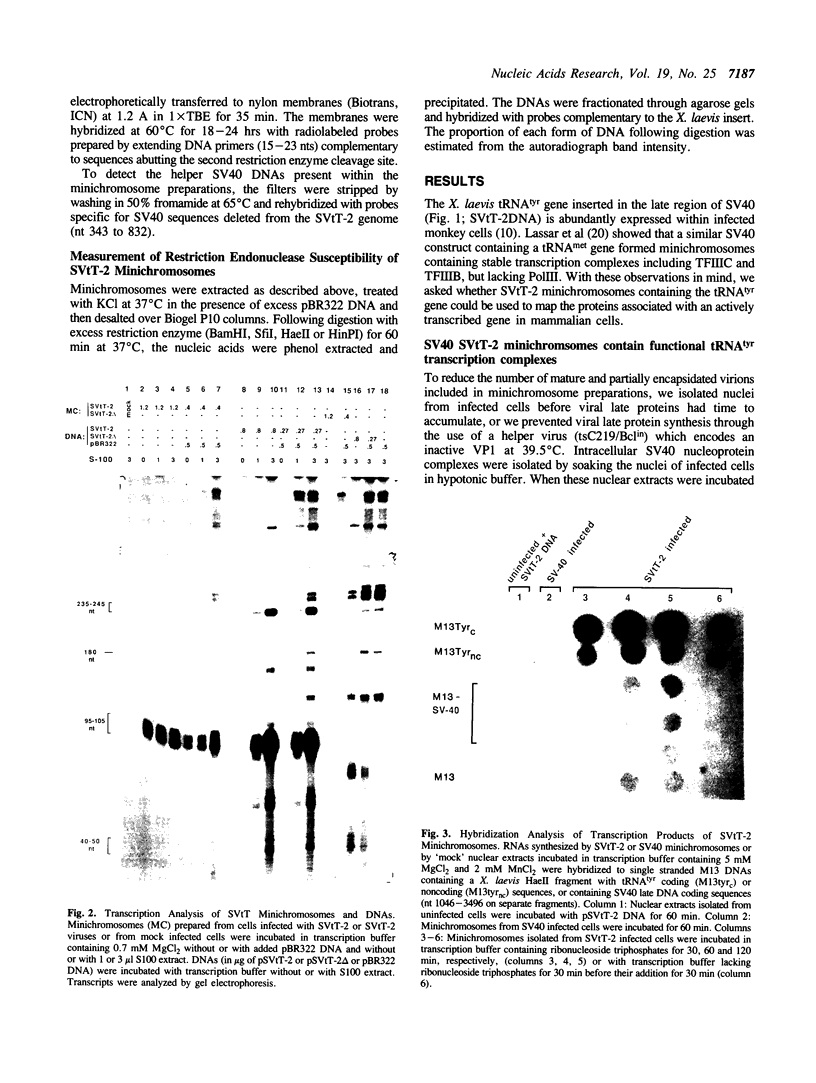




Images in this article
Selected References
These references are in PubMed. This may not be the complete list of references from this article.
- Ambrose C., Blasquez V., Bina M. A block in initiation of simian virus 40 assembly results in the accumulation of minichromosomes containing an exposed regulatory region. Proc Natl Acad Sci U S A. 1986 May;83(10):3287–3291. doi: 10.1073/pnas.83.10.3287. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ambrose C., Lowman H., Rajadhyaksha A., Blasquez V., Bina M. Location of nucleosomes in simian virus 40 chromatin. J Mol Biol. 1990 Aug 20;214(4):875–884. doi: 10.1016/0022-2836(90)90342-J. [DOI] [PubMed] [Google Scholar]
- Bartholomew B., Kassavetis G. A., Braun B. R., Geiduschek E. P. The subunit structure of Saccharomyces cerevisiae transcription factor IIIC probed with a novel photocrosslinking reagent. EMBO J. 1990 Jul;9(7):2197–2205. doi: 10.1002/j.1460-2075.1990.tb07389.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bina M., Blasquez V., Ng S. C., Beecher S. SV40 morphogenesis. Cold Spring Harb Symp Quant Biol. 1983;47(Pt 1):565–569. doi: 10.1101/sqb.1983.047.01.066. [DOI] [PubMed] [Google Scholar]
- Boulanger P. A., Yoshinaga S. K., Berk A. J. DNA-binding properties and characterization of human transcription factor TFIIIC2. J Biol Chem. 1987 Nov 5;262(31):15098–15105. [PubMed] [Google Scholar]
- Carey M. F., Gerrard S. P., Cozzarelli N. R. Analysis of RNA polymerase III transcription complexes by gel filtration. J Biol Chem. 1986 Mar 25;261(9):4309–4317. [PubMed] [Google Scholar]
- Clarkson S. G., Kurer V., Smith H. O. Sequence organization of a cloned tDNA met fragment from Xenopus laevis. Cell. 1978 Jul;14(3):713–724. doi: 10.1016/0092-8674(78)90253-2. [DOI] [PubMed] [Google Scholar]
- Fernandez-Munoz R., Coca-Prados M., Hsu M. T. Intracellular forms of simian virus 40 nucleoprotein complexes. I. Methods of isolation and characterization in CV-1 cells. J Virol. 1979 Feb;29(2):612–623. doi: 10.1128/jvi.29.2.612-623.1979. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Geiduschek E. P., Tocchini-Valentini G. P. Transcription by RNA polymerase III. Annu Rev Biochem. 1988;57:873–914. doi: 10.1146/annurev.bi.57.070188.004301. [DOI] [PubMed] [Google Scholar]
- Germond J. E., Bellard M., Oudet P., Chambon P. Stability of nucleosomes in native and reconstituted chromatins. Nucleic Acids Res. 1976 Nov;3(11):3173–3192. doi: 10.1093/nar/3.11.3173. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gottlieb E., Steitz J. A. The RNA binding protein La influences both the accuracy and the efficiency of RNA polymerase III transcription in vitro. EMBO J. 1989 Mar;8(3):841–850. doi: 10.1002/j.1460-2075.1989.tb03445.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hart R. P., Folk W. R. Structure and organization of a mammalian 5 S gene cluster. J Biol Chem. 1982 Oct 10;257(19):11706–11711. [PubMed] [Google Scholar]
- Huibregtse J. M., Engelke D. R. Genomic footprinting of a yeast tRNA gene reveals stable complexes over the 5'-flanking region. Mol Cell Biol. 1989 Aug;9(8):3244–3252. doi: 10.1128/mcb.9.8.3244. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Huibregtse J. M., Evans C. F., Engelke D. R. Comparison of tRNA gene transcription complexes formed in vitro and in nuclei. Mol Cell Biol. 1987 Sep;7(9):3212–3220. doi: 10.1128/mcb.7.9.3212. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jahn D., Wingender E., Seifart K. H. Transcription complexes for various class III genes differ in parameters of formation and stability towards salt. J Mol Biol. 1987 Jan 20;193(2):303–313. doi: 10.1016/0022-2836(87)90221-x. [DOI] [PubMed] [Google Scholar]
- Kasher M. S., Pintel D., Ward D. C. Rapid enrichment of HeLa transcription factors IIIB and IIIC by using affinity chromatography based on avidin-biotin interactions. Mol Cell Biol. 1986 Sep;6(9):3117–3127. doi: 10.1128/mcb.6.9.3117. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kassavetis G. A., Braun B. R., Nguyen L. H., Geiduschek E. P. S. cerevisiae TFIIIB is the transcription initiation factor proper of RNA polymerase III, while TFIIIA and TFIIIC are assembly factors. Cell. 1990 Jan 26;60(2):235–245. doi: 10.1016/0092-8674(90)90739-2. [DOI] [PubMed] [Google Scholar]
- Laski F. A., Alzner-DeWeerd B., RajBhandary U. L., Sharp P. A. Expression of a X. laevis tRNATyr gene in mammalian cells. Nucleic Acids Res. 1982 Aug 11;10(15):4609–4626. doi: 10.1093/nar/10.15.4609. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Laski F. A., Fire A. Z., RajBhandary U. L., Sharp P. A. Characterization of tRNA precursor splicing in mammalian extracts. J Biol Chem. 1983 Oct 10;258(19):11974–11980. [PubMed] [Google Scholar]
- Lassar A. B., Hamer D. H., Roeder R. G. Stable transcription complex on a class III gene in a minichromosome. Mol Cell Biol. 1985 Jan;5(1):40–45. doi: 10.1128/mcb.5.1.40. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lassar A. B., Martin P. L., Roeder R. G. Transcription of class III genes: formation of preinitiation complexes. Science. 1983 Nov 18;222(4625):740–748. doi: 10.1126/science.6356356. [DOI] [PubMed] [Google Scholar]
- Marschalek R., Dingermann T. Identification of a protein factor binding to the 5'-flanking region of a tRNA gene and being involved in modulation of tRNA gene transcription in vivo in Saccharomyces cerevisiae. Nucleic Acids Res. 1988 Jul 25;16(14B):6737–6752. doi: 10.1093/nar/16.14.6737. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mertz J. E. A detailed genetic analysis of the late complementation groups of simian virus 40. Virology. 1984 Jan 15;132(1):173–185. doi: 10.1016/0042-6822(84)90101-6. [DOI] [PubMed] [Google Scholar]
- Morse R. H. Nucleosomes inhibit both transcriptional initiation and elongation by RNA polymerase III in vitro. EMBO J. 1989 Aug;8(8):2343–2351. doi: 10.1002/j.1460-2075.1989.tb08362.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nedospasov S. A., Georgiev G. P. Non-random cleavage of SV40 DNA in the compact minichromosome and free in solution by micrococcal nuclease. Biochem Biophys Res Commun. 1980 Jan 29;92(2):532–539. doi: 10.1016/0006-291x(80)90366-6. [DOI] [PubMed] [Google Scholar]
- Oei S. L., Pieler T. A transcription stimulatory factor binds to the upstream region of Xenopus 5 S RNA and tRNA genes. J Biol Chem. 1990 May 5;265(13):7485–7491. [PubMed] [Google Scholar]
- Ottonello S., Rivier D. H., Doolittle G. M., Young L. S., Sprague K. U. The properties of a new polymerase III transcription factor reveal that transcription complexes can assemble by more than one pathway. EMBO J. 1987 Jul;6(7):1921–1927. doi: 10.1002/j.1460-2075.1987.tb02452.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Palmer J. M., Folk W. R. Unraveling the complexities of transcription by RNA polymerase III. Trends Biochem Sci. 1990 Aug;15(8):300–304. doi: 10.1016/0968-0004(90)90018-7. [DOI] [PubMed] [Google Scholar]
- Rhodes D. Structural analysis of a triple complex between the histone octamer, a Xenopus gene for 5S RNA and transcription factor IIIA. EMBO J. 1985 Dec 16;4(13A):3473–3482. doi: 10.1002/j.1460-2075.1985.tb04106.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Scott W. A., Walter C. F., Cryer B. L. Barriers to nuclease Bal31 digestion across specific sites in simian virus 40 chromatin. Mol Cell Biol. 1984 Apr;4(4):604–610. doi: 10.1128/mcb.4.4.604. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Setzer D. R., Brown D. D. Formation and stability of the 5 S RNA transcription complex. J Biol Chem. 1985 Feb 25;260(4):2483–2492. [PubMed] [Google Scholar]
- Wingender E., Jahn D., Seifart K. H. Association of RNA polymerase III with transcription factors in the absence of DNA. J Biol Chem. 1986 Jan 25;261(3):1409–1413. [PubMed] [Google Scholar]
- Workman J. L., Taylor I. C., Kingston R. E. Activation domains of stably bound GAL4 derivatives alleviate repression of promoters by nucleosomes. Cell. 1991 Feb 8;64(3):533–544. doi: 10.1016/0092-8674(91)90237-s. [DOI] [PubMed] [Google Scholar]
- Wu C. The 5' ends of Drosophila heat shock genes in chromatin are hypersensitive to DNase I. Nature. 1980 Aug 28;286(5776):854–860. doi: 10.1038/286854a0. [DOI] [PubMed] [Google Scholar]
- Yoshinaga S. K., Boulanger P. A., Berk A. J. Resolution of human transcription factor TFIIIC into two functional components. Proc Natl Acad Sci U S A. 1987 Jun;84(11):3585–3589. doi: 10.1073/pnas.84.11.3585. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yoshinaga S., Dean N., Han M., Berk A. J. Adenovirus stimulation of transcription by RNA polymerase III: evidence for an E1A-dependent increase in transcription factor IIIC concentration. EMBO J. 1986 Feb;5(2):343–354. doi: 10.1002/j.1460-2075.1986.tb04218.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Young L. S., Dunstan H. M., Witte P. R., Smith T. P., Ottonello S., Sprague K. U. A class III transcription factor composed of RNA. Science. 1991 Apr 26;252(5005):542–546. doi: 10.1126/science.1708526. [DOI] [PubMed] [Google Scholar]