Abstract
Seed size is important to crop domestication and natural selection and is affected by the balance of maternal and paternal genomes in endosperm. Endosperm, like placenta in mammals, provides reserves to the developing embryo. Interploidy crosses disrupt the genome balance in endosperm and alter seed size. Specifically, paternal-excess crosses (2 × 4) delay endosperm cellularization (EC) and produce larger seeds, whereas maternal-excess crosses (4 × 2) promote precocious EC and produce smaller seeds. The mechanisms for responding to the parental genome dosage imbalance and for gene expression changes in endosperm are unknown. In plants, RNA polymerase IV (PolIV or p4) encoded by NRPD1a is required for biogenesis of a major class of 24-nt small interfering RNAs (also known as p4-siRNAs), which are predominately expressed in developing endosperm. Here we show that p4-siRNA accumulation depends on the maternal genome dosage, and maternal p4-siRNAs target transposable elements (TEs) and TE-associated genes (TAGs) in seeds. The p4-siRNAs correlate negatively with expression levels of AGAMOUS-LIKE (AGL) genes in endosperm of interploidy crosses. Moreover, disruption of maternal NRPD1a expression is associated with p4-siRNA reduction and AGL up-regulation in endosperm of reciprocal crosses. This is unique genetic evidence for maternal siRNAs in response to parental genome imbalance and in control of transposons and gene expression during endosperm development.
Keywords: epigenetics, imprinting, polyploidy, reproduction, RNA interference
Crop seeds provide nearly 70–80% of calories and 60–70% of all proteins consumed by the human population (1). Endosperm is the direct or indirect source for most of the nutritional content of the seed, and it is similar to the placenta in mammals (2), which is the source of nutrition for embryo development (3).
In angiosperms the endosperm is formed after pollination of the egg by a male gamete (pollen) that contains two sperm nuclei. One sperm fertilizes the egg to form a zygote with a 1:1 maternal-to-paternal genome ratio (1m:1p), whereas the other fertilizes two central cell nuclei to form an endosperm cell with a 2:1 maternal-to-paternal genome ratio (2m:1p). In Arabidopsis thaliana, increasing the paternal genome ratio (2m:2p) in endosperm by pollinating a diploid “mother” with a tetraploid “father” (2 × 4) delays endosperm cellularization (EC) and produces larger seeds. In contrast, increasing the maternal genome ratio (4m:1p) in endosperm by pollinating a tetraploid mother with a diploid father (4 × 2) leads to precocious EC and smaller seeds (4, 5).
Transcription factors including AGAMOUS-LIKE proteins (AGLs) affect endosperm development (6–9). AGLs are members of the plant type I MADS domain subfamily (6), and it is likely that they have a role in reproductive development because they are expressed in female gametophyte or developing seeds (7). Mutations in AGL62 lead to precocious EC and arrest of embryo growth, suggesting a direct effect of AGL62 in endosperm development (8). AGL36 is maternally imprinted and has a potential role in endosperm development, although no obvious phenotype is found in the agl36 mutant probably because of redundancy in this subfamily (9). Moreover, up-regulation of AGL62 and AGL90 is related to the postzygotic barrier between A. thaliana and Arabidopsis arenosa, which is associated with endosperm overproliferation and delayed development (10), similar to that in paternal-excess interploidy crosses.
Mechanisms for responding to parental genome dosage and for regulating AGL expression in endosperm are largely unknown. The model of parental genome balance to explain this effect requires a parent-of-origin–specific factor and a mechanism for balancing the level of this factor relative to the other parental genome. In principle, this parent-of-origin–specific factor could involve imprinted genes, including MEA in A. thaliana (11) and PEG1 and FIE101 in maize (12). However, expression patterns of many imprinted genes are contradictory to the predictions in interploidy crosses (5, 13). For example, maternally expressed genes including MEA, FWA, and FIS2 are up-regulated in the paternal-excess endosperm, similar to that of paternally expressed genes such as PHE1. It is therefore unlikely that they are the parent-of-origin–specific factor.
An alternative mechanism could involve 24-nt small interfering (si)RNAs that are dependent on NRPD1a. The NRPD1a protein is the largest subunit of RNA polymerase IV (PolIV or p4), a homolog of DNA-dependent RNA polymerase II (14–16), and p4-siRNAs in developing seed of A. thaliana are predominately expressed from the maternal genome in endosperm (14). Some 24-nt siRNAs are associated with target genes in leaves of A. thaliana hybrids (17). Here we test the possibility that maternally expressed p4-siRNAs are the factors sensitive to the parental genome dosage and regulate AGL expression levels in seeds through a mechanism in which, as in leaves of A. thaliana hybrids (17), they silence gene expression.
Results
Parent-of-Origin Effects on Endosperm Size and siRNA Production in Reciprocal Triploids.
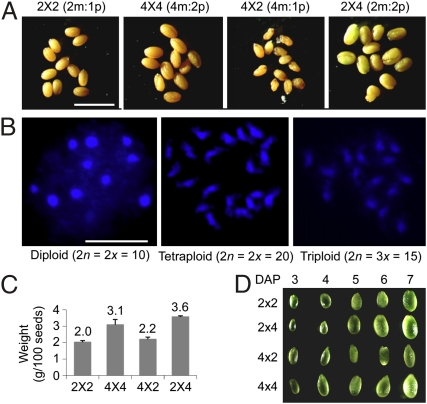
In A. thaliana, reciprocal interploidy crosses between diploid (2×) and tetraploid (4×) plants produce variable seed sizes in Col-0 or C24 ecotypes (Fig. 1A and Fig. S1A). These plants contain the expected ploidy number of chromosomes (Fig. 1B). As reported previously (4), the paternal genome excess (2 × 4) results in larger seeds, whereas the maternal genome excess (4 × 2) leads to smaller seeds (Fig. 1C and Fig. S1 B and C). The response to paternal genome excess in 2 × 4 crosses was dependent on genotypes. Larger and normal seeds were produced in Ler and C24, whereas in Col the seeds were aborted during seed coat development because the expression of TTG2 and other genes was disrupted (18). However, during early seed development the response to altered parental genome dosage is consistent among all ecotypes tested. The endosperm size, reflected in the seed size (4), was noticeably different in the paternal- and maternal-excess seeds 5–6 d after pollination (DAP) in Col-0 (Fig. 1D).
Fig. 1.
Seed morphology and chromosome counts in interploidy crosses. (A) Seed size and morphology in diploids, tetraploids, and triploids (2 × 4 or 4 × 2) in A. thaliana C24. m, maternal; p, paternal. By convention, the maternal parent is listed first in a genetic cross. (B) Chromosome counts in diploid, triploid, and tetraploid flowers in A. thaliana Col-0. (C) Seed weight in interploidy crosses. (D) Developing seeds dissected at 3–7 d after pollination (DAP) in Col-0.
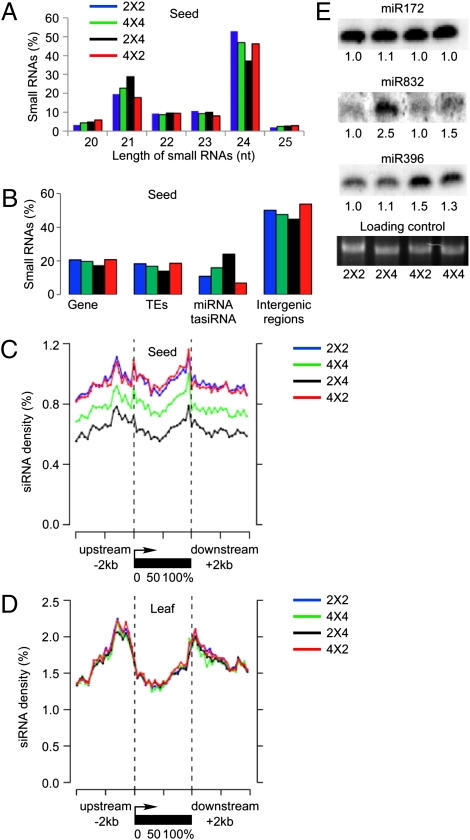
We manually dissected hundreds of seeds (containing endosperm and embryo) out of siliques at 6 DAP of reciprocal crosses (2 × 4 and 4 × 2) and their Col-0 parents, diploids (2 × 2) and tetraploids (4 × 4). At this stage, expression of p4-siRNA is most abundant in endosperm (14), and the seed size was obviously different between reciprocal crosses (Fig. 1D). Eight small RNA libraries were made from these immature seeds and from rosette leaves in reciprocal crosses and their parents. A total of ∼80 million small RNA reads were generated by Illumina sequencing, and ∼64 million reads (∼80%) were mapped (Tables S1 and S2). To reduce ambiguity, only the reads that perfectly matched sequences of the annotated genome (TAIR9) were normalized to reads per 10 million for further analysis. In seeds, the most abundant small RNAs were 21- and 24-nt long, representing 20–29% and 37–53% of total small RNAs (Fig. 2A). The proportions of 21-nt (18–23%) and 24-nt (46–50%) small RNAs were similar in endosperm of diploids (2 × 2) and tetraploids (4 × 4). However, the 24-nt siRNA population was ∼9% lower in 2 × 4 (37%) than in 4 × 2 (46%) seeds, but not in leaves (Fig. S1D). The 24-nt siRNA densities in a 10-kp sliding window were significantly lower in 2 × 4 than in 4 × 2 seeds (Wilcoxon paired ranks sum test, P = 0), whereas the 24-nt siRNA density difference between 2 × 4 and 4 × 2 crosses in leaves was insignificant (P = 0.2).
Fig. 2.
Small RNA distribution in interploidy crosses. (A) Size distribution of 20–25 nt small RNA reads in Col-0 seeds of 2 × 2 (blue), 4 × 4 (green), 2 × 4 (cyan), and 4 × 2 (magenta). (B) Distribution of 20–25 nt small RNAs in genes, TEs, miRNA, and ta-siRNA targets, and intergenic regions. (C and D) 24-nt small RNA densities (100-bp sliding window) in 5′ upstream (2 kb), transcribed, and 3′ downstream (2 kb) regions of TE genes in seeds (C) and leaves (D). (E) Small RNA blot analysis of miR172, miR832, and miR396 in diploids, triploids, and tetrpaloids at 6 DAP.
Ploidy-Dependent siRNAs Are Derived from Transposable Elements (TEs) and TE-Associated Genes (TAGs) in Endosperm.
Average distributions of small RNAs were 16.9% in TEs, 19.5% in genes, 49.1% in intergenic regions (IGRs), and 14.4% in microRNAs (miRNAs) and transacting siRNA (ta-siRNAs) (Fig. 2B). The 24-nt siRNAs were enriched in TEs and intergenic regions, whereas 21-nt siRNAs were derived from miRNA and ta-siRNA loci. Consistent with the reduction of 24-nt siRNAs in 2 × 4 seeds, there was a lower proportion of small RNA reads in TEs (14%), genes (17%), and IGRs (45%) in 2 × 4 than in 4 × 2 seeds (19/21/54%).
In contrast, the fraction of miRNA and ta-siRNA reads was higher in 2 × 4 (24%) than in 4 × 2 seeds (7%). Among the up-regulated miRNAs and ta-siRNAs in 2 × 4 seeds, many were from a few loci with abundant reads in seed (Fig. S2 A–E and Dataset S1). Correspondingly we demonstrated that miR832 but not miR172 and miR396 accumulated to higher levels in 2 × 4 seeds than in the other samples (Fig. 2E). However, there was no correlation between expression levels of miRNAs and ta-siRNAs and their targets in 2 × 4 and 4 × 2 seeds (Fig. S2 F–I). A role for these RNAs in seed development is not clear.
Interestingly, certain siRNA loci were markedly over- or underrepresented in these samples. Among 3,901 annotated TE genes, 24-nt siRNA densities in a 100-bp sliding window were 42% lower in 2 × 4 than in 4 × 2 seeds (Fig. 2C and Fig. S1 E and F) (Wilcoxon paired ranks sum test, P = 0). The 24-nt siRNA densities were 20% lower in tetraploids than in diploids (P = 0), suggesting a dosage compensation mechanism for siRNA expression in seeds that are larger in tetraploids than in diploids. Among 418 TE families each generating 100 or more unique 24-nt siRNA reads, most siRNAs were expressed at significantly lower levels in 2 × 4 than in 4 × 2 seeds (P = 0, Wilcoxon paired rank sum test, Dataset S2). In contrast, 24-nt siRNA densities in leaves did not show a significant difference between 2 × 4 and 4 × 2 crosses (Fig. 2D) (P = 0.7). The data suggest parent-of-origin effects of siRNAs on TE genes in developing seeds.
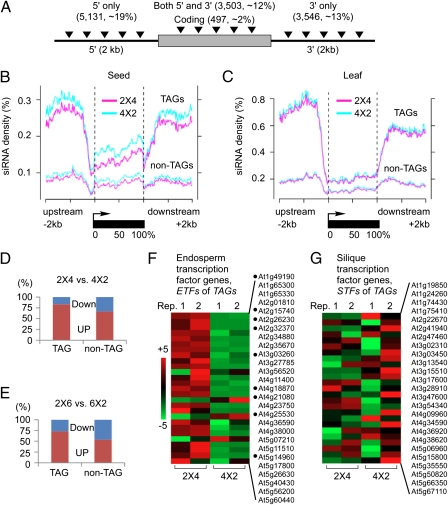
Corresponding to siRNA accumulation in TE genes, there was also an effect of siRNAs on TAGs. We mapped genomic coordinates of 31,189 TEs and TE fragments onto transcribed regions and 2-kb regions upstream and downstream of 27,379 protein-coding genes (TAIR9). We found that 12,676 protein-coding genes (∼46%) contained at least one TE or TE fragment (Fig. 3A and Dataset S3). Among them, ∼19% contained TEs in the 5′ upstream (∼2 kb), ∼13% in the 3′ downstream (∼2 kb), ∼12% in both upstream and downstream, and ∼2% in the transcribed regions. Interestingly, siRNA densities in 5′ and 3′ regions of TAGs were significantly lower in 2 × 4 than in 4 × 2 seeds (Fig. 3B, P = 2.20E-16, Wilcoxon paired rank sum test), but not in leaves (Fig. 3C, P = 0.04).
Fig. 3.
Distribution of TE-associated genes (TAGs) and parent-of-origin effects of siRNAs on TAG expression in reciprocal interploidy crosses. (A) Proportions of TAGs and locations of TEs (triangles) in coding sequences (gray box) and within 2 kb up or downstream of 5′ and 3′ regions (extended lines) in A. thaliana Col-0. (B and C) Small RNA densities (100-bp sliding window) in 5′ upstream (2 kb), transcribed, and 3′ downstream (2 kb) regions of TAGs and non-TAGs in seeds (B) and leaves (C). (D and E) Percentage of up-regulated (red) and down-regulated (blue) TAGs or non-TAGs in 2 × 4 vs. 4 × 2 crosses (D) and in 2 × 6 vs. 6 × 2 crosses (E). (F and G) Heat maps of gene expression changes in endosperm transcription factor (EFT) genes (n = 27) (F) and silique transcription factor (STF) genes (n = 25) (G) in two replicated experiments (Reps. 1 and 2); color bar indicates up (red) and down (green) regulation; black dots indicate up-regulated genes at statistically significant levels between 2 × 4 and 4 × 2 endosperm.
Does the reduction of siRNAs in 2 × 4 relative to 4 × 2 seeds affect gene expression in endosperm? To address this question we first used published microarray data in reciprocal 2 × 4 and 4 × 2 or 2 × 6 and 6 × 2 crosses in young siliques (5), and we identified 9,742 TAGs and 13,029 non-TAGs on the array data. There were 151 TAGs that were differentially expressed between 4 × 2 and 2 × 4 siliques and, of those, 83% were more abundant in 2 × 4 than in 4 × 2 crosses. In contrast, of 90 non-TAGs, only 67% were more abundant in the 2 × 4 cross (χ2 = 19.13, P = 1.2 × 10−5) (Fig. 3D). The same trend was observed in reciprocal tetraploids (2 × 6 and 6 × 2) between diploid and hexaploid lines (χ2 = 17.37, P = 3.1 × 10−5) (Fig. 3E). These data indicated a tendency of increased expression of TAGs in siliques of paternal-excess crosses. These TAGs could be expressed in endosperm together with the maternally expressed p4-siRNAs (14), and their up-regulation in the paternal-excess crosses could be due to the reduced level of these siRNAs.
In our second approach to address the possibility that maternal p4-siRNAs are mediators of silencing in the endosperm, we exploited previous analysis that had identified endosperm-preferred early seed stage (EP-ESS, also known as (aka) endosperm transcription factor, ETF) genes and silique tissue-preferred early seed stage (OST-ESS, aka silique transcription factor, STF) genes (5, 19, 20). The ETF and STF genes included 779 and 448 genes, respectively. Of the ETF genes, 60 were up-regulated, and 2 were repressed in 2 × 4 seeds relative to 4 × 2 seeds (Fig. S3 A and B). In contrast, only 4 STF genes were up-regulated and 1 was repressed in 2 × 4 seeds. Similar results were obtained from interploidy crosses between 2 × 6 and 6 × 2. Overall, the differentially expressed genes in interploidy crosses were enriched in gene ontology groups of hydrolase, receptor binding, and transcription factor activities (Fig. S3C).
These data indicate that up-regulation of ETF genes is correlated positively with increased paternal genome dosage and negatively with increased maternal genome dosage (hypergeometric test, P = 0). Moreover, among 27 ETF genes that are validated as regulators of endosperm development (19), the majority were up-regulated in 2 × 4 seeds in two biological replicates (Fig. 3F and Dataset S4). No obvious trend was found among 25 STF genes (Fig. 3G). Remarkably, 20 of 27 (∼74%) ETF genes are TAGs, and they generated siRNAs. In comparison, of all other genes, only ∼46% are TAGs that generate siRNA (P = 0.0001). Up-regulation of these ETF genes may play a role in overproliferation of endosperm in the paternal-excess triploids (4, 5).
Maternal siRNAs Regulate AGAMOUS-LIKE (AGL) Gene Expression.
The ETFs include a large family of AGLs that are members of the plant type I MADS domain subfamily (6). Many AGLs are expressed in female gametophyte or developing seeds and play a role in reproductive development (7). Indeed, most AGLs were highly induced in seeds 3–8 d after pollination (Fig. S4).
We next tested whether p4-siRNAs are associated with expression of AGLs. Thirteen of 61 (∼21%) type I genes and 5 of 45 (∼11%) type II genes, respectively, contained 10 or more normalized p4-siRNA reads in upstream, transcribed, and downstream regions (Table S3). All type I genes that generated siRNAs belong to Mα, Mγ, and Mδ subgroups (Fig. S5) that are expressed predominately in developing endosperm. The Mβ-type genes are mostly expressed in female gametophyte (7), and none of them was found to be associated with p4-siRNAs. Type II genes such as AGL42 are expressed in vegetative tissues and during floral transition (21).
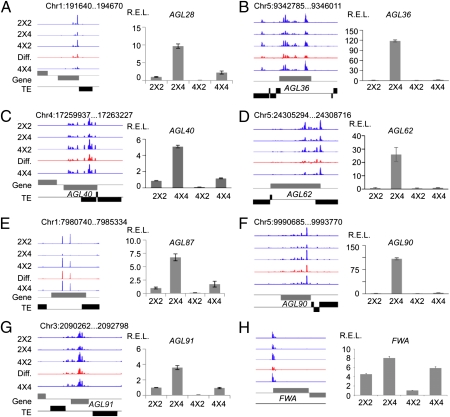
Interestingly, the expression levels of all 13 siRNA-containing AGLs were higher in 2 × 4 than in 4 × 2 seeds (Fig. 4 A–G and Fig. S6), as observed in siliques (5). The transcript levels of these AGLs were inversely correlated with siRNA levels that were lower in 2 × 4 than in 4 × 2 seeds (R2 = 0.67, P = 0.025) (Fig. S3D).
Fig. 4.
Maternal siRNAs are associated with expression of AGL genes and FWA. (A–H) siRNA hotspots (Left) and qRT-PCR analysis (Right) of AGL28 (A), AGL36 (B), AGL40 (C), AGL62 (D), AGL87 (E), AGL90 (F), AGL91 (G), and FWA (H) in 2 × 4 and 4 × 2 triploids and their parents (2 × 2 and 4 × 4). Diff., siRNA differences between 4 × 2 and 2 × 4; positive, Above the line; negative, Below the line; gray box, gene; black box, transposon. Genomic coordinates are shown Above each diagram, and SEs were calculated from three biological replicates.
We also tested the expression of additional ETF genes including six known imprinted genes and four candidate imprinted genes (Dataset S4), many of which overlapped with TEs that generated 24-nt siRNAs in 5′ or 3′ regions. Some without obvious TEs generated 24-nt siRNAs from their 5′ upstream regions, suggesting presence of TE fragments or repeats that have not been annotated. The expression levels of these genes were higher in 2 × 4 than in 4 × 2 seeds but not obviously correlated with siRNA densities, with one exception. FWA is an imprinted gene in endosperm (22). The maternally expressed FWA was up-regulated in paternal-excess seeds (2 × 4) (Fig. 4H), which correlated with lower siRNA densities in endosperm in 2 × 4 than in 4 × 2.
Expression of p4-siRNAs and AGLs in Interploidy Crosses Is Dependent on PolIV.
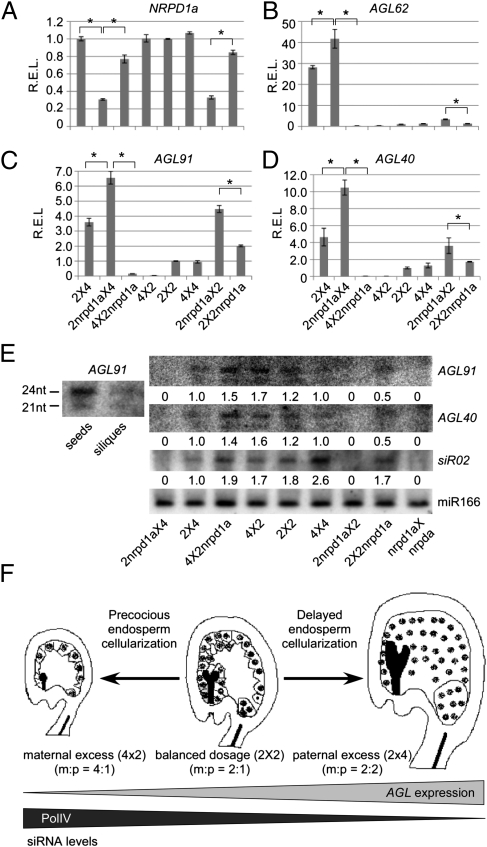
Biogenesis of p4-siRNAs is, by definition, dependent on RNA polymerase IV (PolIV), a homolog of RNA polymerase II (14–16). NRPD1a encodes the largest subunit of PolIV. Absence of maternal NRPD1a reduces or eliminates 24-nt siRNA expression in endosperm (14). To test the effects of NRPD1a on AGL expression, we crossed a diploid nrpd1a (2nrpd1a) mutant (14) with diploid (2×) and tetraploid (4×) wild-type plants in Col-0. NRPD1a expression was lower in 2nrpd1aX4 and 2nrpd1aX2 seeds than in corresponding reciprocal hybrids (Fig. 5A). A previously reported p4-siRNA, siR02 (14), was undetectable in 2nrpd1aX4 and 2nrpdl1aX2 seeds (Fig. 5E).
Fig. 5.
siRNA production and AGL expression are dependent on RNA polymerase IV (NRPD1A) in endosperm and a model for endosperm development in interploidy crosses. (A) qRT-PCR analysis (relative expression levels, REL) of NRPD1a expression in endosperm of interploidy crosses. (B–D) qRT-PCR analyses of AGL62 (B), AGL91 (C), and AGL40 (D) expression (n = 3 biological replicates). (E) Small RNA blot analysis of p4-siRNA (siR02), AGL40-siRNA, and AGL91-siRNA in endosperm of interploidy crosses (n = 2 biological replicates). (Left) AGL91-siRNAs were present in seeds but not in siliques. miR166 was used as a control. (F) Model for the role of maternal siRNA-mediated AGL expression in endosperm and seed development (see text for explanation). Multiple dots and an elongated black rod in each diagram represent the endosperm and embryo cells, respectively.
Absence of maternal NRPD1a transcripts in these mutant crosses was associated with up-regulation of 11 AGLs including AGL40, AGL62, and AGL91, which were two- to threefold higher in 2nrpd1ax4 than in 2 × 4, as well as in 2nrpd1aX2 seeds than in 2 × 2nrpdl1a (Fig. 5 B–D and Fig. S7 A–C). Up-regulation of AGL91 and AGL40 in 2nrpd1aX4 and 2nrpdl1aX2 seeds correlated with down-regulation of p4-siRNAs (Fig. 5E, Right and Fig. S7D). The p4-siRNAs associated with AGL91 were present in seeds but not in siliques from which the seeds were removed (Fig. 5E, Left). A lower amount of 21-nt siRNAs was also associated with AGL91. The data indicate a link of maternal nrpd1a repression with reduction of maternal siRNAs and up-regulation of AGLs.
Discussion
Our data collectively suggest a unique model that explains the role for PolIV-dependent maternal siRNAs in AGL expression and endosperm development (Fig. 5F). Proper seed development requires an endosperm balance number of 2m:1p in diploids (2 × 2) (2, 4). In the maternal-excess endosperm (4 × 2, 4m:1p), maternal p4-siRNA expression levels increase, and p4-siRNA-associated AGLs are repressed, causing precocious cellularization of endosperm and development of smaller seeds. In contrast, in the paternal-excess endosperm (2 × 4, 2m:2p) a low abundance of maternal and p4-siRNAs leads to up-regulation of AGLs, promoting endosperm nuclear proliferation and enlarging seeds.
Genome-wide demethylation in endosperm (23, 24) is predicted to produce p4-siRNAs that are dependent on maternal genome dosage. During endosperm development, these maternal p4-siRNAs may directly silence AGL targets and TEs, as observed in this study or indirectly through a mechanism of RNA-directed DNA methylation (25, 26). It is unclear whether these maternal siRNAs present in endosperm affect TEs and gene expression in embryos. Movement of 21-nt siRNAs is predicted in vegetative and sperm nuclei of pollen (27). Alternatively, some of these maternal siRNAs, most likely 21-nt siRNAs (Table S3), may participate in the posttranscriptional silencing pathway and trigger the secondary siRNA cascades through a ta-siRNA–like mechanism (28, 29). Indeed, AGL91, AGL40, and AGL36 had a significantly high probability (P < 0.01) of generating 21-nt phased siRNAs (Table S3) (28).
The endosperm and seed size is critical to the fitness of plants. Alteration in seed size is a manifestation of parental genome conflict in plants (2). Our model is consistent with that feature of parental genome conflict because the p4-siRNAs and their AGL transcription factor targets are all expressed in the endosperm. Endosperm is a triploid that contains two maternal (central cell) and one paternal (sperm) genomes. During the evolution of angiosperms, these maternal p4-siRNAs are responsive to parental genome dosage and regulate expression of genes such as AGLs, which are important to metabolism and nourishing function of maternal tissues (2, 3). We predict that these maternal p4-siRNAs also serve as the factor for balancing and recognizing heterologous maternal and parental genomes in hybrids (30). The p4-siRNAs would allow the differentiation between paternal and maternal alleles, which could relieve the repressive maternal effects on the hybrids resulting from the parental genome imbalance and conflict in gametogenesis, fertilization, and early zygotic development (30, 31). The imbalance between the maternal siRNAs and their target genes in endosperm could lead to endosperm failure, a common syndrome observed in many interspecific hybrids, but the embryos are viable and can be rescued to regenerate plants under tissue culture conditions (32). These predicted effects could be readily tested in the hybrids within and between species.
Materials and Methods
Plant Materials and Growth Conditions.
Diploids (2n = 2x = 10) and tetraploids (2n = 4x = 20) of A. thaliana Col-0, C24, and Ler ecotypes were grown under 16-h light at 22 °C and 8-h darkness at 20 °C. Reciprocal interploidy crosses were made by pollinating diploid flowers with tetraploid pollens (2 × 4) or tetraploid flowers with diploid pollens (4 × 2) 24 h after manual emasculation. Diploid and tetraploid flowers were manually self-pollinated to serve as balanced dosage controls. Seeds were manually dissected from the siliques at 3, 4, 5, 6, and 7 DAP to eliminate maternal tissue contamination. Small RNA library construction and gene expression assays were performed using the seeds dissected at 6 DAP. Rosette leaves of F1 and their parents were collected before bolting for small RNA and gene expression studies.
Chromosome Counts.
A published protocol was adopted (33). In brief, young floral buds were fixed in Carnoy's fixative (ethanol:glacial acetic acid, 3:1) and digested with pectolytic enzyme mixture (0.3% (wt/vol) cellulase, 0.3% (wt/vol) pectolyase, and 0.3% (wt/vol) cytohelicase (all from Sigma in citrate buffer) at 37 °C for 5 h. Flower buds were then homogenized and spread on a glass slide by repeatedly adding 60% acetic acid and Carnoy's fixative. The chromosome spread was stained with 4′,6-diamidino-2-phenylindole (Sigma) and examined under a wide-field florescent microscope (Axiovert 200 M; Carl Zeiss). Three flower buds were examined per plant.
Small RNA Library Construction.
Total RNA was extracted from seeds and leaves using Plant RNA reagent (Invitrogen) and subjected to electrophoresis in a 15% urea-polyacrylamide gel. The small RNA fraction (18–30 nt) was recovered from the gel. The small RNAs were ligated to 5′ and 3′ RNA oligo adapters (Table S4) and reverse transcribed to produce first strand cDNAs, which were amplified by PCR and sequenced by Illumina Genome Analyzer II. Small RNA data are deposited in short read archives (http://www.ncbi.nlm.nih.gov/sra) (GSE25280).
Bioinformatic Analysis.
Short reads (40 nt) were parsed to remove 3′ adapters and mapped to A. thaliana genome (TAIR9, June 2009 release) using CASHX (http://asrp.cgrb.oregonstate.edu/db/download.html) (34). To reduce ambiguity, only the perfectly matched reads were used for further analysis. The sequences from chloroplast and mitochondrial and structural noncoding RNAs including ribosomal RNAs, transfer RNAs, snoRNAs, and snRNAs were excluded from the analysis. Small RNA reads were normalized by library size and number of hits to the genome using the same weight for each matched locus.
Protein coding genes with adjacent TEs were identified by overlapping genomic coordinates of TEs and TE fragments with those of protein-coding genes using a Python script. Natural antisense gene pairs were defined as two loci overlapping with each other in the opposite direction. miRNA and ta-siRNA targets were downloaded from The Arabidopsis Small RNA Project (ASRP) database (http://carringtonlab.org/resources) (35). GOSlim terms were downloaded from http://www.arabidopsis.org/tools/bulk/go/index.jsp. The significance of enrichment was tested using hypergeometric test and Bonferroni multiple-testing correction (36) (P = 0.01).
Microarray Analysis.
The design and analysis of microarray datasets on interploidy crosses were described (5). Normalized data for 2 × 4, 4 × 2, 2 × 6, and 6 × 2 five-DAP siliques (two replicates; Affymetrix) were downloaded from Gene Expression Omnibus (GSE20007). All statistic analyses were performed using R (http://www.r-project.org). Gene expression changes were estimated on the basis of a t test between reciprocal crosses 2 × 4 vs. 4 × 2 and 2 × 6 vs. 6 × 2, respectively. Probe sets were called differentially expressed when P ≤ 0.01 and log-twofold change ≥2.
Quantitative RT-PCR Analysis.
Total RNA was extracted from leaves and dissected seeds using Plant RNA reagent (Invitrogen) and treated with DNaseI (Promega). First-strand cDNA was synthesized using SuperScriptIII reverse transcriptase (Invitrogen). Primer sequences are listed in Table S5. Actin 2 (ACT2) was used as the internal control. qRT-PCR is performed using Applied Biosystems 7500 Real-Time PCR Systems (ABI).
Small RNA Blot Analysis.
Small RNA (<200 nt) was enriched from 50 μg total RNA using mirVana miRNA purification kit (Ambion). Small RNA was separated on a 15% denaturing 19:1 acrylamide:bisacrylamide gel with 1X TBE and 7 M urea and transferred to HybondN+ membrane (GE/Amersham). Oligonucleotides were end labeled with [γ-32P]ATP and T4 polynucleotide kinase (New England Biolabs) and hybridized with the membrane in Church's buffer (37) at 37 °C overnight. A mixture of oligonucleotides corresponding to the most abundant siRNAs from each AGL locus in the sequencing libraries was used as the probe to detect AGL-related siRNAs. The blots were washed twice in 2× SSC, 0.1% SDS at 50 °C before exposure to phosphor storage screens. Probe sequences are listed in Table S6.
Supplementary Material
Acknowledgments
We thank Mary Gehring for sharing the data of differential methylation regions in embryo and endosperm and Rod J. Scott for sharing microarray data in interploidy crosses. We also thank Luca Comai, Charles Nicolet, and Wang Kit Danny Ng for coordinating Illumina sequencing; Craig Dupree for administrating a high-performance computer cluster system; and Attila Molnar and other members in the D.C.B. and Z.J.C. laboratories for insightful discussions to improve the manuscript. The work is supported by the National Science Foundation Grant MCB1110957 and a Fulbright United States–United Kingdom Scholar Award (to Z.J.C.). David Baulcombe is a Royal Society Research Professor.
Footnotes
The authors declare no conflict of interest.
Data deposition: The sequence reported in this paper has been deposited in the Sequence Read Archive, http://www.ncbi.nlm.nih.gov/sra (accession no. GSE25280).
This article contains supporting information online at www.pnas.org/lookup/suppl/doi:10.1073/pnas.1203094109/-/DCSupplemental.
References
- 1.Borlaug NE. Civilization's Future: A call for international granaries. Science and Public Affairs. 1973;24:7–15. [Google Scholar]
- 2.Moore T, Haig D. Genomic imprinting in mammalian development: A parental tug-of-war. Trends Genet. 1991;7:45–49. doi: 10.1016/0168-9525(91)90230-N. [DOI] [PubMed] [Google Scholar]
- 3.Stebbins GL. Seeds, seedlings, and the origin of angiosperms. In: Beck CB, editor. Origin and Early Evolution of Angiosperms. New York: Columbia Univ Press; 1976. pp. 300–311. [Google Scholar]
- 4.Scott RJ, Spielman M, Bailey J, Dickinson HG. Parent-of-origin effects on seed development in Arabidopsis thaliana. Development. 1998;125:3329–3341. doi: 10.1242/dev.125.17.3329. [DOI] [PubMed] [Google Scholar]
- 5.Tiwari S, et al. Transcriptional profiles underlying parent-of-origin effects in seeds of Arabidopsis thaliana. BMC Plant Biol. 2010;10:72. doi: 10.1186/1471-2229-10-72. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Parenicová L, et al. Molecular and phylogenetic analyses of the complete MADS-box transcription factor family in Arabidopsis: New openings to the MADS world. Plant Cell. 2003;15:1538–1551. doi: 10.1105/tpc.011544. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Bemer M, Heijmans K, Airoldi C, Davies B, Angenent GC. An atlas of type I MADS box gene expression during female gametophyte and seed development in Arabidopsis. Plant Physiol. 2010;154:287–300. doi: 10.1104/pp.110.160770. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Kang IH, Steffen JG, Portereiko MF, Lloyd A, Drews GN. The AGL62 MADS domain protein regulates cellularization during endosperm development in Arabidopsis. Plant Cell. 2008;20:635–647. doi: 10.1105/tpc.107.055137. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Shirzadi R, et al. Genome-wide transcript profiling of endosperm without paternal contribution identifies parent-of-origin-dependent regulation of AGAMOUS-LIKE36. PLoS Genet. 2011;7:e1001303. doi: 10.1371/journal.pgen.1001303. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Josefsson C, Dilkes B, Comai L. Parent-dependent loss of gene silencing during interspecies hybridization. Curr Biol. 2006;16:1322–1328. doi: 10.1016/j.cub.2006.05.045. [DOI] [PubMed] [Google Scholar]
- 11.Erilova A, et al. Imprinting of the polycomb group gene MEDEA serves as a ploidy sensor in Arabidopsis. PLoS Genet. 2009;5:e1000663. doi: 10.1371/journal.pgen.1000663. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Gutierrez-Marcos JF, Pennington PD, Costa LM, Dickinson HG. Imprinting in the endosperm: A possible role in preventing wide hybridization. Philos Trans R Soc Lond B Biol Sci. 2003;358:1105–1111. doi: 10.1098/rstb.2003.1292. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Jullien PE, Berger F. Parental genome dosage imbalance deregulates imprinting in Arabidopsis. PLoS Genet. 2010;6:e1000885. doi: 10.1371/journal.pgen.1000885. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Mosher RA, et al. Uniparental expression of PolIV-dependent siRNAs in developing endosperm of Arabidopsis. Nature. 2009;460:283–286. doi: 10.1038/nature08084. [DOI] [PubMed] [Google Scholar]
- 15.Onodera Y, et al. Plant nuclear RNA polymerase IV mediates siRNA and DNA methylation-dependent heterochromatin formation. Cell. 2005;120:613–622. doi: 10.1016/j.cell.2005.02.007. [DOI] [PubMed] [Google Scholar]
- 16.Herr AJ, Jensen MB, Dalmay T, Baulcombe DC. RNA polymerase IV directs silencing of endogenous DNA. Science. 2005;308:118–120. doi: 10.1126/science.1106910. [DOI] [PubMed] [Google Scholar]
- 17.Groszmann M, et al. Changes in 24-nt siRNA levels in Arabidopsis hybrids suggest an epigenetic contribution to hybrid vigor. Proc Natl Acad Sci USA. 2011;108:2617–2622. doi: 10.1073/pnas.1019217108. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Dilkes BP, et al. The maternally expressed WRKY transcription factor TTG2 controls lethality in interploidy crosses of Arabidopsis. PLoS Biol. 2008;6:2707–2720. doi: 10.1371/journal.pbio.0060308. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Day RC, Herridge RP, Ambrose BA, Macknight RC. Transcriptome analysis of proliferating Arabidopsis endosperm reveals biological implications for the control of syncytial division, cytokinin signaling, and gene expression regulation. Plant Physiol. 2008;148:1964–1984. doi: 10.1104/pp.108.128108. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Le BH, et al. Global analysis of gene activity during Arabidopsis seed development and identification of seed-specific transcription factors. Proc Natl Acad Sci USA. 2010;107:8063–8070. doi: 10.1073/pnas.1003530107. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Dorca-Fornell C, et al. The Arabidopsis SOC1-like genes AGL42, AGL71 and AGL72 promote flowering in the shoot apical and axillary meristems. Plant J. 2011;67:1006–1017. doi: 10.1111/j.1365-313X.2011.04653.x. [DOI] [PubMed] [Google Scholar]
- 22.Kinoshita T, et al. One-way control of FWA imprinting in Arabidopsis endosperm by DNA methylation. Science. 2004;303:521–523. doi: 10.1126/science.1089835. [DOI] [PubMed] [Google Scholar]
- 23.Gehring M, Bubb KL, Henikoff S. Extensive demethylation of repetitive elements during seed development underlies gene imprinting. Science. 2009;324:1447–1451. doi: 10.1126/science.1171609. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Hsieh TF, et al. Genome-wide demethylation of Arabidopsis endosperm. Science. 2009;324:1451–1454. doi: 10.1126/science.1172417. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Matzke M, Kanno T, Huettel B, Daxinger L, Matzke AJ. Targets of RNA-directed DNA methylation. Curr Opin Plant Biol. 2007;10:512–519. doi: 10.1016/j.pbi.2007.06.007. [DOI] [PubMed] [Google Scholar]
- 26.Law JA, Jacobsen SE. Establishing, maintaining and modifying DNA methylation patterns in plants and animals. Nat Rev Genet. 2010;11:204–220. doi: 10.1038/nrg2719. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Slotkin RK, et al. Epigenetic reprogramming and small RNA silencing of transposable elements in pollen. Cell. 2009;136:461–472. doi: 10.1016/j.cell.2008.12.038. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Chen HM, Li YH, Wu SH. Bioinformatic prediction and experimental validation of a microRNA-directed tandem trans-acting siRNA cascade in Arabidopsis. Proc Natl Acad Sci USA. 2007;104:3318–3323. doi: 10.1073/pnas.0611119104. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Montgomery TA, et al. Specificity of ARGONAUTE7-miR390 interaction and dual functionality in TAS3 trans-acting siRNA formation. Cell. 2008;133:128–141. doi: 10.1016/j.cell.2008.02.033. [DOI] [PubMed] [Google Scholar]
- 30.Ng DW, Lu J, Chen ZJ. Big roles for small RNAs in polyploidy, hybrid vigor, and hybrid incompatibility. Curr Opin Plant Biol. 2012 doi: 10.1016/j.pbi.2012.01.007. [DOI] [PubMed] [Google Scholar]
- 31.Bourc'his D, Voinnet O. A small-RNA perspective on gametogenesis, fertilization, and early zygotic development. Science. 2010;330:617–622. doi: 10.1126/science.1194776. [DOI] [PubMed] [Google Scholar]
- 32.Sharma DR, Kaur R, Kumar K. Embryo rescue in plants: A review. Euphytica. 1996;89:325–337. [Google Scholar]
- 33.Lysak M, Fransz P, Schubert I. Cytogenetic analyses of Arabidopsis. Methods Mol Biol. 2006;323:173–186. doi: 10.1385/1-59745-003-0:173. [DOI] [PubMed] [Google Scholar]
- 34.Fahlgren N, et al. Computational and analytical framework for small RNA profiling by high-throughput sequencing. RNA. 2009;15:992–1002. doi: 10.1261/rna.1473809. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Gustafson AM, et al. ASRP: The Arabidopsis Small RNA Project Database. Nucleic Acids Res. 2005;33:D637–D640. doi: 10.1093/nar/gki127. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Holm S. A simple sequentially rejective multiple test procedure. Scand J Stat. 1979;6:65–70. [Google Scholar]
- 37.Church GM, Gilbert W. Genomic sequencing. Proc Natl Acad Sci USA. 1984;81:1991–1995. doi: 10.1073/pnas.81.7.1991. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.