Polymeric vesicles, a.k.a. “polymersomes”, are enclosed membranes formed by the self-assembly of amphiphilic block copolymers in water.[1] In recent years polymersomes have attracted much attention due to their unique features, such as improved mechanical properties, high stability, and long circulation half-lives in the body compared to liposomes, as well as their ability to incorporate both hydrophilic compounds in the aqueous core and hydrophobic compounds in the membrane.[1a] Furthermore, amphiphilic block copolymers can be designed to be noncytotoxic, efficiently internalized by cells, etc.[2] Polymersomes can be decorated with proteins and/or antibodies, either by chemically attaching the active moieties to the hydrophilic brushes[3] or by inserting membrane proteins across the hydrophobic membrane.[4] Recently, fine control over polymersome surface topology and its consequences on cell internalization kinetics has been demonstrated by the authors.[5] Thus the ability to examine the surface properties of polymersomes, as well as other water-borne nanoparticles, in situ on the nanoscale is becoming increasingly important in several fields. Imaging of wet nanoparticles is normally performed by transmission or scanning electron microscopy (EM). However, the high vacuum conditions necessary for such imaging require either dried or frozen (e.g., cryogenic EM) samples, potentially causing artefacts. Wet imaging can be performed by optical microscopy and recently the problem of diffraction-limited spatial resolution has been overcome by new fluorescence-based microscopy, such as scanning near-field optical microscopy (SNOM), photo-activated localization microscopy (PALM), stimulated emission depletion microscopy (STED), and structured illumination microscopy (SIM).[6] Atomic force microscopy (AFM) is another valuable analytical tool that has been used for both imaging and also for assessing mechanical, electrical, and surface properties.[7]
However, AFM requires particles to be immobilized when imaging in liquid. Immobilization of particles and polymersomes in particular has been achieved by both covalent attachment[8] and noncovalent bonding, e.g., by exploiting biotin/streptavidin complexation[9] or Coulombic interactions.[10]
Here we present two strategies for the facile functionalization of polymersomes with biotin and their subsequent immobilization on streptavidin-coated surfaces. This generic approach enables force spectroscopy mapping (FSM)[11] to image neutral polymersomes in an aqueous environment. As illustrated in Figure 1A, the first approach employs supramolecular functionalization by the addition of commercially available Biotin-1,2-distearoyl-sn-glycero-3-phosphoethanolamine- N -[biotinyl(polyethylene glycol)-2000] (Biotin–PEG–DSPE) to an amphiphilic block copolymer mixture. The second approach is the chemical functionalization of the block copolymer with biotin. In both cases, a poly((2-methacryloyloxy)ethyl phosphorylcholine)- block -poly(2-(diisopropylamino)ethyl methacrylate) (PMPC25 –PDPA70) diblock copolymers were used. PMPC–PDPA polymersomes are pH-sensitive and have been reported to efficiently enter cells and deliver their cargo within their cytosol with no toxic effects.[5a,12]
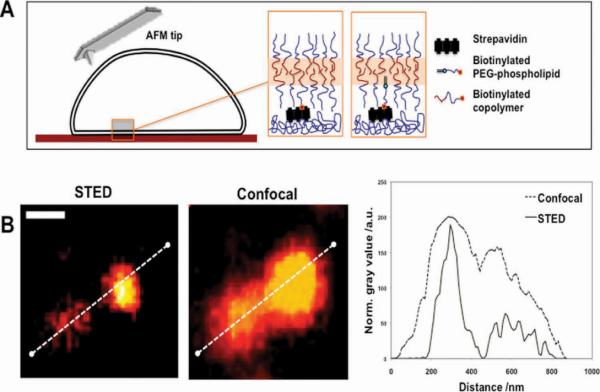
Figure 1.
A) Schematics of the biotinylated polymersome binding to a streptavidin surface. B) Confocal and STED imaging of 200 nm fluorescently (ATTO label)-labelled polymersomes immobilized by biotinylated PEG–phospholipid confirm the enhanced STED resolution by examining the intensity profile (scale bar = 500 nm).
Biotin/Rhodamine-labelled polymersomes were incubated with streptavidin-functionalized, plasma-polymerized acrylic acid coated silicon wafers. After only approx. 10 min the surfaces showed substantial fouling due to adsorbed polymersomes, as demonstrated in the Supporting Information (SI), Figure S1, by confocal laser scanning micrographs. This time was independent from the type of functionalization. Traditional confocal microscopy, however, can only give an estimation of the immobilization of the polymersomes and possibly gather functional information using fluorescence readouts. In order to gather more structural information with higher spatial resolution, we performed STED microscopy using ATTO640-functionalized polymersomes. ATTO dyes possess optimal fluorescence properties that enable high spatial resolution to be achieved in STED.[13] Figure 1B shows a comparison between STED images and the corresponding conventional confocal laser scanning microscopy (CLSM) images of polymersomes decorated with biotinylated phospholipid dispersed in PBS and immobilized on a streptavidin-decorated surface. The enhanced STED resolution reveals the presence of two clustered polymersomes that are otherwise not distinguishable, as shown by the fluorescent intensity profiles. According to the STED images (Figure 1B), the mean polymersome diameter is about half of the diameter suggested by CLSM allowing a more accurate measurement and very similar to size measured by dynamic light scattering (DLS) and AFM (Figure 2D).
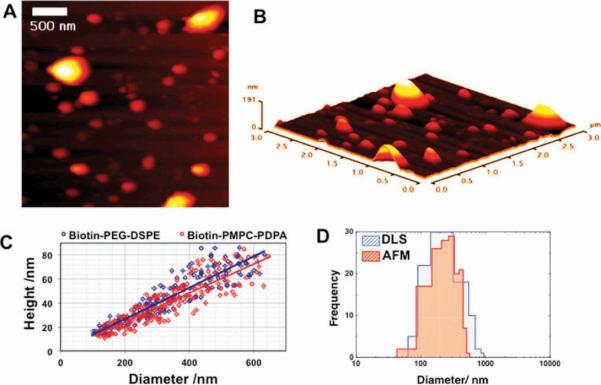
Figure 2.
A) Tapping-mode liquid AFM height image of biotinylated polymersomes immobilized by Biotinylated PEG-phospholipid onto Si substrate coated with streptavidin. B) The corresponding topographic 3D rendered image showing the distribution of polymersome heights. C) AFM-measured polymersome height versus diameter for Biotin–PMPC–PDPA and Biotin–PEG–DSPE immobilized PMPC–PDPA polymersomes. D) AFM- and DLS-measured particle size histograms.
Our surface immobilization strategy allows the use of AFM and its ability to gather other functional information. As shown in SI, Figure S2, we have performed AFM studies in both contact and tapping modes. Contact-mode imaging causes flattening of the relatively soft polymersome due to the high normal pressure exerted by the tip (see SI, Figure S2A). In contrast, with tapping mode imaging the probe is oscillated above the sample and only briefly interacts with the highly deformable polymersome surface, which allows better imaging (SI, Figure S2B). AFM confirms the presence of polymersomes immobilized on the precoated silicon wafer across large areas (SI, Figure S2C). Figure 2A shows a tapping-mode height AFM image obtained for polymersomes decorated with biotinylated PEGylated phospholipid immersed in 100 nm PBS and immobilized at the streptavidin-coated surface. Previous imaging studies in the presence of electrolyte have often been hindered by the deposition of dry salt, forcing the introduction of dilution steps to reduce the electrolyte concentration in order to minimize such artefacts. In contrast, our method works well in the presence of electrolyte and in principle allows the study of polymersomes under complex solution conditions with nanometer resolution. In addition, AFM enables the measurement of polymersome heights (Figure 2C). The binding of the anchored biotin to the surface-immobilized streptavidin leads to detectable deformation of the polymersome. It is interesting that the mean polymersome diameter scales, within experimental error, linearly with its height, with an average diameter-to-height ratio of 7.75 ± 1.31. This suggests that these surface-confined polymersomes adopt a spherical `cap'. This is in agreement with the Seifert and Lipowsky model for the deformation of strongly adherent vesicles bound to a planar surface.[14] In Figure 2C we also show the difference between the two different immobilization approaches using chemical modified Biotin–PMPC–PDPA and Biotin–PEG–DSPE functionalized PMPC–PDPA polymerosmes. The two formulations were prepared so to achieve the same number of biotin molecules on the surface (about 10% mol/mol) and interestingly the average diameter to height ratio seems very similar for both formulations indicating that the polymersomes deformation is not affected by the hydrophobic interaction between the phospholipid and the polymer membrane. The ability of measuring both polymersomes height and diameter allows the estimation of the polymersomes particle size distribution. However as polymersomes deform once bound to the substrate we calculated the unperturbed polymersome diameter assuming bound polymersomes to have a perfect spherical cap volume and deriving the equivalent spherical diameter. In Figure 2D the AFM-measured particle distribution is plotted together with DLS-measured particle distribution. The two histograms have a quite good agreement, confirming the validity of the AFM measurements.
Both STED and AFM studies confirm that the polymersomes can be immobilized at a surface and imaged in situ under water. This suggests the possibility of acquiring useful structural information as well as evaluating surface and mechanical properties via AFM. The AFM tip can be positioned in all three dimensions with sub-nanometer accuracy and can be combined with an ultrasensitive force sensor. This exquisite spatial control allows measurements in which either interaction forces are applied to the sample to investigate the response of the sample (indentation) or forces between the tip and the sample are measured (adhesion mapping).[7,11] As already shown by the contact mode imaging PMPC–PDPA polymersomes are too soft to allow tip indentation studies (data not shown). On the other hand, combining AFM with FSM enables highly sensitive measurements to be conducted. In order to interrogate the polymersome surface, we functionalized AFM tips with streptavidin and measured the binding between the tip and the polymersome surface in order to assess the mechanical properties of the polymersomes (see SI for more details). We conducted experiments involving streptavidin–biotin binding alone and also streptavidin–biotin adsorbed polymersomes. The characteristic distance observed for decomplexation is significantly greater for the latter system, which implies that the difference is due to polymersome displacement. This allows us to estimate the mechanical properties of the polymersomes calculating the polymersomes membrane line modulus of 3.9 ± 2.3 pN nm−1. We would like to point out that this value is based on a very simplistic model that does not account for the viscous components and the streptavidin/biotin binding kinetics. Current work is underway to generate more sophisticated models and measurement. However such result shows very well how the effective polymersomes immobilisation can enable a large paramount of measurements using scanning probe techniques. This is particularly demonstrated using FSM. This allows precise mapping of the polymersome surface. We demonstrate this concept by imaging polymersomes formed by a mixture of biotinylated PMPC25 –PDPA70 and poly(ethylene oxide)- co -poly(butylene oxide) (PEO16 –PBO22) at a 25:75 molar ratio.
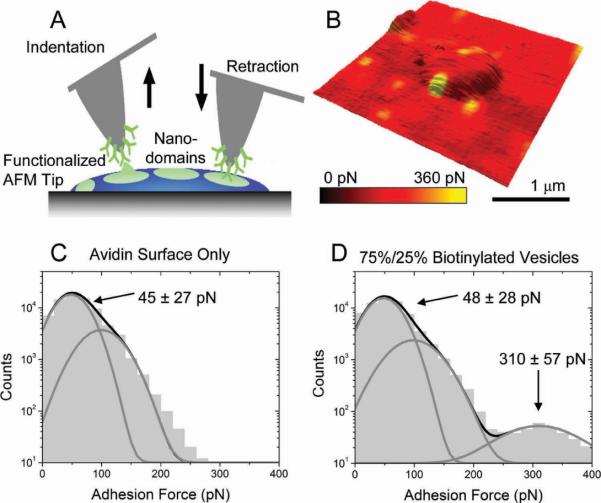
We have previously shown that mixing such polymersome-forming copolymers[5] leads to the formation of nanoscopic domains on the polymersome surface. Here we utilize FSM conducted under liquid so as to detect the biotinylated poly mer domains within the polymersomes. FSM uses a functionalized AFM tip that is brought into contact with an adhesive surface and then retracted (Figure 3A; top), resulting in force spectrograms where one can observe adhesion events as the bond ruptures (bottom).[11] By performing indentations at high lateral resolution down to 20 nm (28 force curves μ m−2 for images shown here), small adhesive changes in the polymersome surface can be detected. When these `patchy' polymersomes were deposited onto a streptavidin-coated silicon wafer, a streptavidin-coated tip was used to resolve the phase-separated domains formed by bio tinylated PMPC25 –PDPA70, as shown in Figure 3B. Interaction between the tip and the vesicle indicate that roughly 25% of the polymersome surface was adhesive. Previously measured rupture forces for biotin–avidin[17] are in good agreement with those measured here (160 ± 55 pN; Figure 3D). Note that The regions without polymersomes were not blocked by BSA or another protein to minimize nonspecific adhesion as is common practice when adhesion forces are lower than those seen here.[11] However, the number of `false positive' measurements is less than 1% as shown in Figure 3C and D.
Figure 3.
A) Force spectroscopy mapping-mode schematic. B) FSM map overlaid onto topographic images to illustrate the spatial location of an adhesive event on the polymersome surface immobilized by biotin–PMPC–PDPA copolymers. Histograms of streptavidin surface alone (C) and polymersomes (D) illustrate the differences between nonspecific adhesion of the tip (single peak in C and first peak in D) and the specific adhesion event observed in D.
In conclusion, biotinylation of PMPC25 –PDPA70 polymersomes was successfully performed either by mixing this diblock copolymer with a biotinylated phospholipid or by functionalizing the block copolymer with biotin prior to polymersome formation. The decoration of polymersomes with biotin allowed i) their immobilization on streptavidin-coated surfaces and ii) their subsequent high-resolution imaging in liquid using advanced techniques such as STED, AFM, and FSM. In particular, it was possible to characterize neutral polymersomes in situ at the nanoscale, thus avoiding any artifacts due to either drying or freezing. Finally, FSM studies conducted on immobilized polymersomes prepared by mixing two polymersome-forming diblock copolymers revealed the presence of the expected phase-separated nanodomains, thus opening up new perspectives for the study of nanostructured soft matter.
Experimental Section
Materials
1,2-distearoyl-sn-glycero-3-phosphoethanolamine-N -[biotinyl(polyethylene glycol)-2000] (DSPE–PEG–Biotin) was purchased from Avanti Polar Lipids (USA). Methanol, chloroform, and phosphate-buffered saline (PBS) tablets were purchased from Sigma Aldrich (UK).
2-(Methacryloyloxy)ethyl phosphorylcholine monomer (MPC, 99.9% purity) was kindly donated by Biocompatibles Ltd. 2-(Diisopropylamino)ethyl methacrylate (DPA) was obtained from Scientific Polymer Products and the polymerization inhibitor was removed prior to use by passing the monomer through a column as recommended by the manufacturer. 2-Bromoisobutyryl bromide (98%), anhydrous methanol (MeOH 99.8%), copper(I) bromide [Cu(I)Br, 99.999%], 2,2′ -bipyridine (bpy, 99%) and N -biotinoyl-N′ -(6-maleimidohexanoyl)hydrazide (Biotin–maleimide, ≥ 95%) were purchased from Sigma Aldrich (Dorset, UK) and were used as received. The silica gel 60 (0.063–0.200 μ m) used to remove the spent ATRP catalyst was purchased from E. Merck (Darmstadt, Germany) and was also used as received. Bis[2-(2-bromoisobutyryloxy)ethyl] disulfide ((BiBOE)2 S2) was prepared according to a previously published procedure.[15]
Tri- n -butyl phosphine (Bu3 P, 95%) was obtained from Acros Organics (Geel, Belgium). Triethylamine (Et3 N) (laboratory reagent grade), dichloromethane, chloroform and methanol (all HPLC-grade) were obtained from Fisher Scientific (Loughborough, UK) and used as received. Regenerated cellulose dialysis membranes (molecular weight cut off (MWCO) 1000 Da or 3500 Da) was purchased from Spectra/Por. The HABA/Avidin Reagent was purchased from Sigma Aldrich (Dorset, UK) and used according to the manufacturer's directions.
PMPC25 –PDPA70 copolymer was synthesized using a standard ATRP protocol. Briefly, ME–Br initiator (250.30 mg, 0.894 mmol), 2,2-bipyridine (279.40 mg, 1.789 mmol) and MPC (6.602 g, 22.36 mmol) were placed in a flask and degassed using standard Schlenk techniques. Methanol (7.0 mL) was added and the mixture was stirred while purging with nitrogen for 30 min. Cu(I)Br (128.30 mg, 0.894 mmol) was added and the solution turned brown and progressively more viscous, indicating the onset of polymerization. After 80 min, 1H NMR indicated 99% MPC mono mer conversion. At this point a previously deoxygenated solution of DPA (13.35 g, 62.6 mmol) in methanol (15 mL) was transferred into the flask via cannula. After 42 h, 1H NMR indicated > 99% DPA monomer conversion. To quench the reaction, the solution was exposed to the atmosphere and 20 mL methanol was added, followed by 10 mL chloroform. Purification was achieved by dialysis against deionized water (MWCO 1000 Da), changing the water twice daily. After one week, the copolymer was lyophilized from water overnight. GPC analysis of the resulting white powder indicated an number-averaged molecular weight Mn of 31 000 Da and weight-average molecular weight Mw of 36 890 Da corresponding to a polydispersity index of Mw / Mn = 1.19 (3:1 v/v chloroform/methanol eluent versus poly(methyl methacrylate) (PMMA) standards).
Preparation of Biotinylated Polymersomes
Biotinylated polymersomes were prepared by rehydration and mixing of PMPC25 –PDPA70 films containing 10 mol% of a biotinylated phospholipid, 1,2-dioleoyl-sn-glycero-3-phosphoethanolamine- N -(cap biotinyl) (sodium salt) (Biotinyl Cap PE). The polymer/phospholipid mixture was solubilized in a 3:1 chloroform/methanol solution and the organic solvent was evaporated by placing the solution at room temperature under dynamic vacuum for 2 h followed by static vacuum overnight. A 0.1 m PBS solution (pH 7.4) was added to the resulting film at a concentration of 5.0 g L−1 stirring for 24 h. The polymersome dispersion was then sonicated for 10 min in order to produce unilamellar polymersomes. Alternatively, biotinylated poly mersomes (in which the biotin is covalently attached to the end of the copolymer chains) were prepared by rehydration and mixing of biotin–PMPC25 −PDPA70 films, as described in the SI.
Streptavidin Coating of Supports
Supports for imaging (either silicon wafers or petri dishes) were exposed to a stream of condensed air to remove any adherent dust or silicon particles, before being immersed in isopropyl alcohol (IPA) and subjected to ultrasonics for 30 min. Supports were then removed from the IPA, dried using a stream of high-purity nitrogen gas and subsequently placed within a custom-built stainless steel plasma polymerization reactor. The samples were then coated with plasma-polymerized acrylic acid (ppAAc), using a standard methodology previously published.[1] In brief, the monomer used was acrylic acid, the plasma power used was 10 W, the deposition time was 25 min and the monomer flow rate used was 2.5 sccm. A streptavidin solution was prepared by dispersing the streptavidin in phosphate buffered saline (PBS, pH 7.4, 150 m m) at a concentration of 0.5 mg mL−1. Supports were removed from the plasma polymerization reactor and placed in 2 mL of this solution on a gently rocking platform for 45 min at room temperature to allow streptavidin immobilization on the ppAAc surface. Supports were finally rinsed very gently with deionised H2 O to remove any excess streptavidin from the surface.
Confocal Laser Scanning Microscopy
Biotinylated polymersomes were prepared using biotinylated phospholipid (Biotinyl Cap PE) as described above using a PMPC25 –PDPA70 copolymer labeled with rhodamine-6G prepared as reported elsewhere[5a] Fluorescently-labeled polymersome dispersions were diluted to a concentration of 0.5 g L−1 and placed in a streptavidin-coated 24 well-plate. Confocal laser scanning microscope (CLSM) images using an LSM 510 Zeiss instrument equipped with an Achroplan 100× immersion objective and exciting the sample at 543 nm with a HeNe laser. Images recorded over time showed that after 10 min the nanoparticles were gently attached onto the surface, retaining sufficient freedom to oscillate slightly around the binding site (see SI, Figure S1).
Atomic Force Microscopy
AFM studies were carried out under liquid water (PBS, pH 7.4) using a Digital Instruments (Cambridge, UK) MultiMode AFM with NanoScope III controller operated in tapping mode. Measurements were carried out using standard silicon nitride NP–S probes with a nominal spring constant of about 0.2 N m−1. Height and phase data were collected simultaneously. Biotinylated polymersomes prepared using biotinylated phospholipid (Biotinyl Cap PE) were deposited onto streptavidin-coated Si wafers by spin-coating at 2000 rpm from their native solution. Immediately after their deposition, the samples were rehydrated and then imaged under liquid water.
Stimulated Emission Depletion Microscopy
STED images were acquired using a Leica TCS STED using a 100× objective. Polymersomes were prepared using ATTO-conjugated PMPC25 –PDPA70 in the presence of Biotinyl Cap PE and were imaged in liquid water (0.1 m PBS, pH 7.4) in a streptavidin-coated 24-well plate. Samples were excited with a 635 nm pulsed (80 MHz) laser diode (titanium sapphire) (9 mW) with a tuning range of 725–850 nm operating at 770–780 nm. A combination of dynamic spectral photodetectors (PMTs) and ultra-sensitive Avalanche Photo Diodes (APDs) enhanced the definition of the final micrograph during image acquisition. The corresponding CLSM micrographs were acquired in the traditional confocal mode by simply switching off the depletion laser.
Force Spectroscopy Mapping
AFM-based adhesion force mapping was performed as detailed elsewhere.[11] Briefly, gold-coated, pyramid-shape tips mounted on SiN cantilevers with ks p = 20 pN nm−1 (TR400PB; Olympus; Center Valley, PA) were functionalized (SI, Figure S1) with streptavidin using a previously established ethanolamine–HCL and bis[sulfosuccinimidyl] suberate method.[16] Biotinylated polymersomes (PEO16 –PBO22 and biotinylated PMPC25 –PDPA70) were immobilized on avidin-coated coverslips in PBS and placed on an MFP-3D-BIO atomic force microscope (Asylum Research; Santa Barbara, CA). Using custom software written in Igor Pro (Wavemetrics; Portland, OR), samples indented in a regular array of points with 20 nm lateral resolution or 2500 indentations μ m−2 over a scan area of 4 μ m2 and with an indentation velocity of 5 μ m s−1 (≈ 100 nN s−1 loading rate). To promote biotin-avidin binding, a dwell time of 3 s was added between tip indentation (SI, Figure S1C; red) and retraction cycles (SI, Figure 1C; blue). Knowing the resulting deflection and cantilever spring constant and assuming Hookean behavior for the cantilever, the deflection versus cantilever position data could be converted into force-indentation spectrographs.[17] Data were then analyzed to determine the maximum adhesive force, i.e., the greatest difference between the retraction curve and baseline. Using the x - and y-position for each force measurement, data were then plotted onto a map of the surface and interpolated to generate a force spectroscopy map.
Acknowledgements
The authors acknowledge funding from EPSRC (EP/E03103X/1) and Biocompatibles, a Royal Thai Government Fellowship (to SC) and a grant from NIH #1DP02OD10322765 (to AJE). CLP New Address: NBE Delivery Formulation Merck Serono SpA Via Einaudi 11 00012 Guidonia Montecelio (Rome), Italy.
Footnotes
Supporting Information Supporting Information is available from the Wiley Online Library or from the author. Available information includes materials and methods, block copolymer functionalization protocols, and CLSM images.
This Communication is part of the Special Issue dedicated to Chad Mirkin in celebration of 20 years of influential research at Northwestern University.
References
- [1].a) Discher DE, Eisenberg A. Science. 2002;297:967–973. doi: 10.1126/science.1074972. [DOI] [PubMed] [Google Scholar]; b) Discher BM, Won YY, Ege DS, Lee JC, Bates FS, Discher DE, Hammer DA. Science. 1999;284:1143–1146. doi: 10.1126/science.284.5417.1143. [DOI] [PubMed] [Google Scholar]
- [2].LoPresti C, Lomas H, Massignani M, Smart T, Battaglia G. J. Mater. Chem. 2009;19:3576–3590. [Google Scholar]
- [3].a) Christian NA, Milone MC, Ranka SS, Li G, Frail PR, Davis KP, Bates FS, Therien MJ, Ghoroghchian PP, June CH, Hammer DA. Bioconjugate Chem. 2007;18:31–40. doi: 10.1021/bc0601267. [DOI] [PubMed] [Google Scholar]; b) Pang Z, Lu W, Gao H, Hu K, Chen J, Zhang C, Gao X, Jiang X, Zhu C. J. Control. Release. 2008;128:120–127. doi: 10.1016/j.jconrel.2008.03.007. [DOI] [PubMed] [Google Scholar]
- [4].a) Sauer M, Haefele T, Graff A, Nardin C, Meier W. Chem. Commun. 2001:2452–2453. doi: 10.1039/b107833j. [DOI] [PubMed] [Google Scholar]; b) Graff A, Sauer M, Van Gelder P, Meier W. Proc. Natl. Acad. Sci. USA. 2002;99:5064–5068. doi: 10.1073/pnas.062654499. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [5].a) Massignani M, LoPresti C, Blanazs A, Madsen J, Armes SP, Lewis AL, Battaglia G. Small. 2009;5:2424–2432. doi: 10.1002/smll.200900578. [DOI] [PubMed] [Google Scholar]; b) Lopresti C, Massignani M, Fernyhough C, Blanazs A, Ryan AJ, Madsen J, Warren NJ, Armes SP, Lewis AL, Chirasatitsin S, Engler AJ, Battaglia G. ACS Nano. 2011;5:1775–1784. doi: 10.1021/nn102455z. [DOI] [PubMed] [Google Scholar]
- [6].Hell SW. Science. 2007;316:1153–1158. doi: 10.1126/science.1137395. [DOI] [PubMed] [Google Scholar]
- [7].Bonnell D. ACS Nano. 2008;2:1753–1759. doi: 10.1021/nn8005575. [DOI] [PubMed] [Google Scholar]
- [8].Domes S, Filiz V, Nitsche J, Fromsdorf A, Forster S. Langmuir. 2010;26:6927–6931. doi: 10.1021/la904175u. [DOI] [PubMed] [Google Scholar]
- [9].Grzelakowski M, Onaca O, Rigler P, Kumar M, Meier W. Small. 2009;5:2545–2548. doi: 10.1002/smll.200900603. [DOI] [PubMed] [Google Scholar]
- [10].a) Kepczynski M, Lewandowska J, Romek M, Zapotoczny S, Ganachaud F, Nowakowska M. Langmuir. 2007;23:7314–7320. doi: 10.1021/la063442i. [DOI] [PubMed] [Google Scholar]; b) Li F, Ketelaar T, Stuart MAC, Sudholter EJR, Leermakers FAM, Marcelis ATM. Langmuir. 2008;24:76–82. doi: 10.1021/la702546b. [DOI] [PubMed] [Google Scholar]
- [11].Chirasatitsin S, Engler AJ. J. Phys.—Condens. Mat. 2010;22:P194102. doi: 10.1088/0953-8984/22/19/194102. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [12].a) Massignani M, Sun T, Blanazs A, Hearnden V, Canton I, Desphande P, Armes S, MacNeil S, Lewis A, Battaglia G. PLoS One. 2010;5:e10459. doi: 10.1371/journal.pone.0010459. [DOI] [PMC free article] [PubMed] [Google Scholar]; b) Lomas H, Canton I, MacNeil S, Du J, Armes SP, Ryan AJ, Lewis AL, Battaglia G. Adv. Mater. 2007;19:4238. [Google Scholar]
- [13].Harke B, Keller J, Ullal CK, Westphal V, Schönle A, Hell SW. Opt. Expr. 2008;16:4154–4162. doi: 10.1364/oe.16.004154. [DOI] [PubMed] [Google Scholar]
- [14].Seifert U, Lipowsky R. Phys. Rev. A. 1990;42:4768–4771. doi: 10.1103/physreva.42.4768. [DOI] [PubMed] [Google Scholar]
- [15].Madsen J, Armes SP, Bertal K, Lomas H, Macneil S, Lewis AL. Biomacromolecules. 2008;9:2265–2275. doi: 10.1021/bm8005006. [DOI] [PubMed] [Google Scholar]
- [16].Bonanni B, Kamruzzahan ASM, Bizzarri AR, Rankl C, Gruber HJ, Hinterdorfer P, Cannistraro S. Biophys. J. 2005;89:2783–2791. doi: 10.1529/biophysj.105.064097. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [17].Rotsch C, Jacobson K, Radmacher M. Proc. Natl. Acad. Sci. USA. 1999;96:921–926. doi: 10.1073/pnas.96.3.921. [DOI] [PMC free article] [PubMed] [Google Scholar]