Abstract
The threonine endopeptidase Taspase1 has a critical role in cancer cell proliferation and apoptosis. In this study, we developed and evaluated small molecule inhibitors of Taspase1 as a new candidate class of therapeutic modalities. Genetic deletion of Taspase1 in the mouse produced no overt deficiencies, suggesting the possibility of a wide therapeutic index for use of Taspase1 inhibitors in cancers. We defined the peptidyl motifs recognized by Taspase1 and conducted a cell-based dual-fluorescent proteolytic screen of the NCI diversity library to identify Taspase1 inhibitors (TASPINs). Based on secondary and tertiary screens the 4-[(4-arsonophenyl)methyl]phenyl]arsonic acid NSC48300 was determined to be the most specific active compound. Structure-activity relationship studies indicated a crucial role for the arsenic acid moiety in mediating Taspase1 inhibition. Additional FRET-based kinetic analysis characterized NSC48300 as a reversible, non-competitive inhibitor of Taspase1 (KI = 4.22 μM). In the MMTV-neu mouse model of breast cancer and the U251 xenograft model of brain cancer NSC48300 produced effective tumor growth inhibition. Our results offer an initial preclinical proof of concept to develop TASPINs for cancer therapy.
Introduction
Site-specific proteolysis offers spatiotemporal controls over fundamental aspects of organismal and cellular physiology (1–9). Accordingly, the identification and characterization of regulatory proteases in the context of human diseases has fueled the discovery of therapeutic interventions targeted at respective proteases (10). The best examples are the use of angiotensin-converting enzyme (ACE) inhibitors, HIV protease inhibitors, and 26S proteasome inhibitors to treat hypertension, AIDS, and multiple myeloma, respectively (2, 11–12).
Taspase1 (threonine aspartase) encodes a highly conserved 50 kDa α-β proenzyme that undergoes autoproteolysis, generating a mature α28/β22 heterodimeric protease that displays an overall α/β/β/α structure (13–14). Taspase1 was initially purified as the protease that cleaves MLL to regulate the expression of HOX genes (13, 15). Subsequent studies identified additional Taspase1 substrates, including MLL2 (also known as MLL4 in the GenBank database) (8), TFIIAα-β and ALF (TFIIA like factor) (16). The cloning of Taspase1 founded a novel class of endopeptidases that employs conserved amino-terminal threonine of the mature β subunit to cleave peptide bonds after P1 aspartate (13).
Taspase1 is the only protease within the family of enzymes that possesses an asparaginase_2 (PF01112) homology domain, whereas other members, including L-asparaginase and glycosylasparaginase, participate in the metabolism of asparagines and the ordered breakdown of N-linked glycoproteins, respectively (13, 17). Taspase1-mediated cleavage follows a distinct aspartate residue of a conserved QXD/GXDD motif (15), suggesting that Taspase1 evolved from hydrolyzing asparagines and glycosylasparagines to cleaving polypeptides after aspartates (13). In addition to MLL, MLL2, TFIIA and ALF, Taspase1 also proteolyzes Drosophila HCF (dHCF) whereas mammalian HCF is cleaved by O-GlcNAc transferase due to the loss of GXD/GXDD motif during the evolution (18–19).
Initial characterization of Taspase1−/− mice discovered a critical role of Taspase1 in cell cycle control (8). In the absence of Taspase1, cell cycle is disrupted with decreased expression of Cyclins and increased expression of CDK inhibitors (CDKIs) (8). Consequently, Taspase1−/− mouse embryonic fibroblasts (MEFs) are resistant to oncogenic transformation (8). Furthermore, Taspase1 is over-expressed in primary human cancers and required for tumor maintenance in many cancer cell lines (20). Knockdown of Taspase1 disrupts proliferation in the majority of cancer cells within which a subset of cell lines also displays enhanced apoptosis (20). Of note, Taspase1 is expressed at high levels in many cancer cells (8, 21–22) and in general increased expression positively correlates with the cellular dependence on Taspase1 (8, 20). These data suggest that Taspase1 is co-opted to promote and sustain tumorigenesis. Therefore, inhibition of Taspase1 may offer a new anticancer strategy. Here, we present our endeavors in (1) establishing the safety of Taspase1 inactivation in adult mammals using a genetically well-defined mouse model, (2) characterizing the consensus cleavage motif of Taspase1, (3) developing an in vivo, dual fluorescent, Taspase1 proteolytic screen, (4) screening, confirming and characterizing a small molecule TASPIN NSC48300, and (5) examining the efficacy of NSC48300 in treating cancers using two different preclinical mouse tumor models.
Materials and Methods
Animal studies
Animal studies were approved by the Animal Studies Committee at Washington University School of Medicine. Mice carrying straight and conditional knockout alleles of Taspase1(8), MMTV-neu (23), and MMTV-wnt (24) transgenes have been described. Tumor mass followed by bioluminescence imaging using an IVIS 100 system has been previously described (25).
Constructs, recombinant proteases, cell lines, cell culture, knockdown, and Western blot analyses
The DFPR was constructed by sequentially inserting cDNA encoding eGFP, 2XNES of MAPKK, aa 2,400–2,900 of hMLL, 3XNLS of SV40 large T antigen, and dsRED2 into the pMSCVpuro (Clontech) vector. Amphotropic retrovirus was produced as described (26) and utilized to infect 293T cells. The generation of recombinant Taspase1 and Caspase8 has been described (13, 27). NCI60 cell lines were obtained from NCI DTP. BT-474 was obtained from American Type Culture Collection. Taspase1−/− MEFs have been described (20) and were authenticated by both PCR-genotyping and Western blot analysis. All of the cell lines were expanded, frozen, and used for no more than 2 months after the resuscitation of frozen aliquots. Cell culture, Taspase1 knockdown, and Western blot analyses were performed as previously described (28).
Results
Acute deletion of Taspase1 in adult mice does not incur overt toxic phenotypes
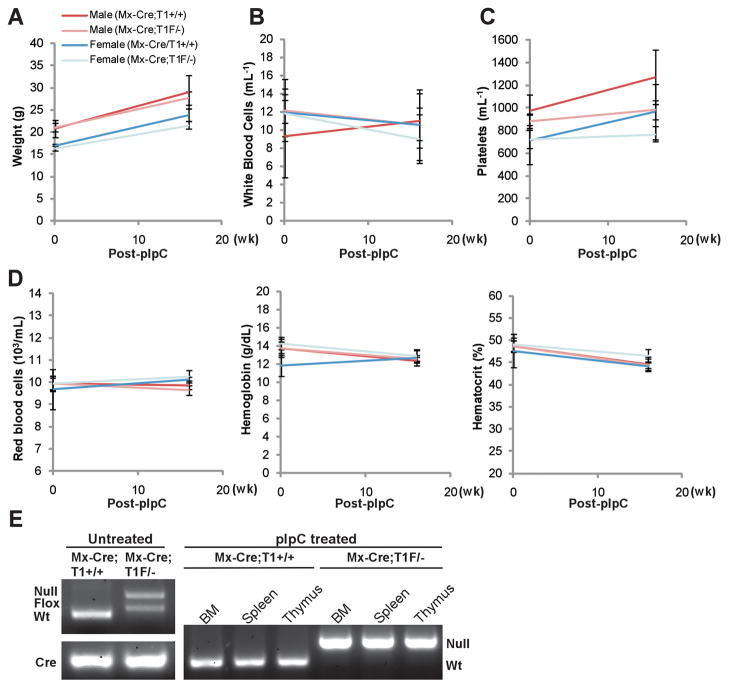
Our in vitro studies using Taspase1 knockout and knockdown cells demonstrated an important role of Taspase1 in tumor initiation and maintenance, suggesting that Taspase1 inhibitors (TASPINs) may be developed and utilized in cancer therapy (20). Since Taspase1 plays critical roles in mouse embryonic development, the application of TASPINs in children and pregnant women would be inadvisable (8). On the other hand, the few Taspase1−/− mice alive at weaning age went on to live a normal lifespan (Westergard et al., unpublished data), implicating the safe use of TASPINs in adults. To evaluate whether Taspase1 can be safely inactivated in developed mammals, we induced a global deletion of the Taspase1 gene in 7 week-old Mx-Cre;Taspase1F/− mice by administering polyinosine-polycytidine (pIpC) (29). No discernible toxicities were observed in these Taspase1−/− mice (Fig. 1A, B, C and D). The efficiency of Taspase1 deletion in bone marrow, spleen, and thymus was determined (Fig. 1E). These results indicate that inactivation of Taspase1 through pharmacological means is likely to be tolerated by adult mammals.
Figure 1.
Induced genetic deletion of Taspase1 is well-tolerated in adult mice. A, Mice bearing an interferon-inducible cre recombinase (Mx-Cre) transgene and the indicated alleles of Taspase1 (+, wild-type; −, knockout; F, conditional allele) at 7 weeks of age were subjected to 5 doses of polyinosine-polycytidine (pIpC) injection. Baseline measurements were obtained at 6 weeks of age. Mice were sacrificed 16 weeks after the last dose of pIpC to obtain post-treatment measurements and harvest bone marrow (BM), spleen, and thymus for genotypic analysis. B, C, and D, Genetic deletion of Taspase1 dose not affect the parameters of white blood cell (B), platelet (C), and red blood cell counts (D). E, Genotyping of the indicated tissues demonstrates a complete deletion of Taspase1 after the pIpC treatment.
Critical amino acid residues within the Taspase1 cleavage consensus motif
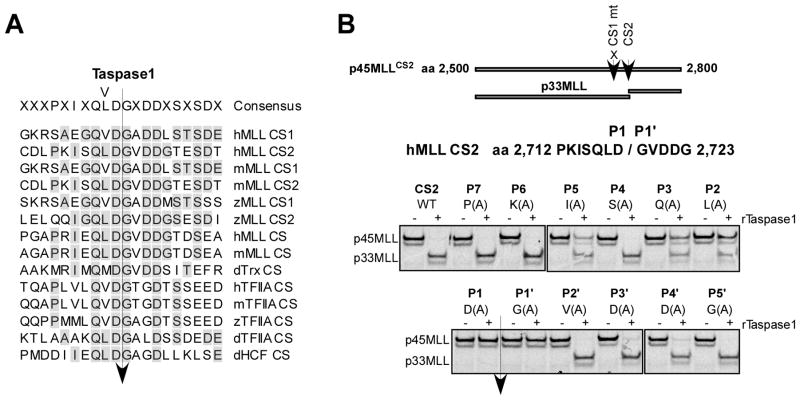
To provide mechanistic insights concerning how Taspase1 recognizes its substrates, we characterized the Taspase1 cleavage motif. MLL contains two Taspase1 cleavage sites (CS1 and CS2) that are positioned 53 amino acids (aa) apart, among which CS2 is more conserved and more efficiently processed (Fig. 2A) (13, 30). In vitro transcribed/translated (IVTT), 35S-methionine labeled, human MLL aa 2,500–2,800 fragment containing a mutated CS1 site was employed as a CS2-specific cleavage reporter (p45MLLCS2). Alanine scanning mutagenesis across the CS2 site (PKISQLD/GVDDG) of p45MLLCS2 was performed and the cleavage of individual reporters by recombinant Taspase1 (rTaspase1) was examined. P1 aspartate and P1′ glycine are essential, P2 leucine, P3 glutamine, and P5 isoleucine are important, and P3′ and P4′ aspartates are dispensable (Fig. 2B). These data are consistent with the fact that P1 aspartate and P1′ glycine are absolutely conserved, P2, P3 and P5 hydrophobic residues are highly conserved, and P4 is variable, among known Taspase1 cleavage motifs (Fig. 2A). Surprisingly, highly conserved P3′ and P4′ aspartate residues are dispensable, indicating that these residues may play an indirect role in presenting substrates for Taspase1 recognition. In summary, the IXQL(V)D/G sequence represents the best Taspase1 cleavage recognition motif.
Figure 2.
Characterization of the Taspase1 cleavage site consensus motif. A, Alignment of known Taspase1 cleavage sites (CS). h, m, z, and d denote human, mouse, zebra fish, and Drosophila, respectively. B, Alanine scanning mutagenesis of the CS2 region of human MLL. The indicated p45MLLCS2 (aa 2,500 to 2,800) cleavage reporters were generated by single alanine substitution of individual amino acid across P7 to P5′ of CS2, where the CS1 was invariably mutated to enable assessment of a single cleavage site. The individual IVTT, 35S-methionine labeled, p45MLLCS2 reporters were incubated with 15 ng of rTaspase1 (recombinant Taspase1) for 30 minutes at 30°C, resolved by SDS-PAGE, and monitored by autoradiography.
Accordingly, primitive peptidomimetic Taspase1 inhibitors, derivatives of ISQLD carrying common protease-inactivating functional groups such as aldehyde and chloromethyl ketone (cmk), were synthesized and tested for their ability to inhibit Taspase1-mediated cleavage of p75MLL (aa 2,400–2,900). Although aldehyde and cmk are effective warheads against general classes of proteases (31), ISQLD-aldehyde and -cmk were minimally active in inhibiting Taspase1 (IC50 > 100 μM) (Supplementary Fig. S1A). Of all proteases in mammals, the β subunit of the 20S proteasome (32) is the only known N-terminal threonine endopeptidase other than Taspase1 (13). Bortezomib—a boronic acid containing chemotherapeutic drug that targets the active site threonine of the 26S proteasome (11)—has no activity against Taspase1 (Supplementary Fig. S1B), which is consistent with the lack of activity of ISQLD-boronate in inhibiting Taspase1 (33). Altogether, these data highlight unique mechanistic processes concerning Taspase1-mediated proteolysis.
A cell-based, dual-fluorescent, proteolytic screen identifies small molecule TASPINs in the NCI Diversity Set library
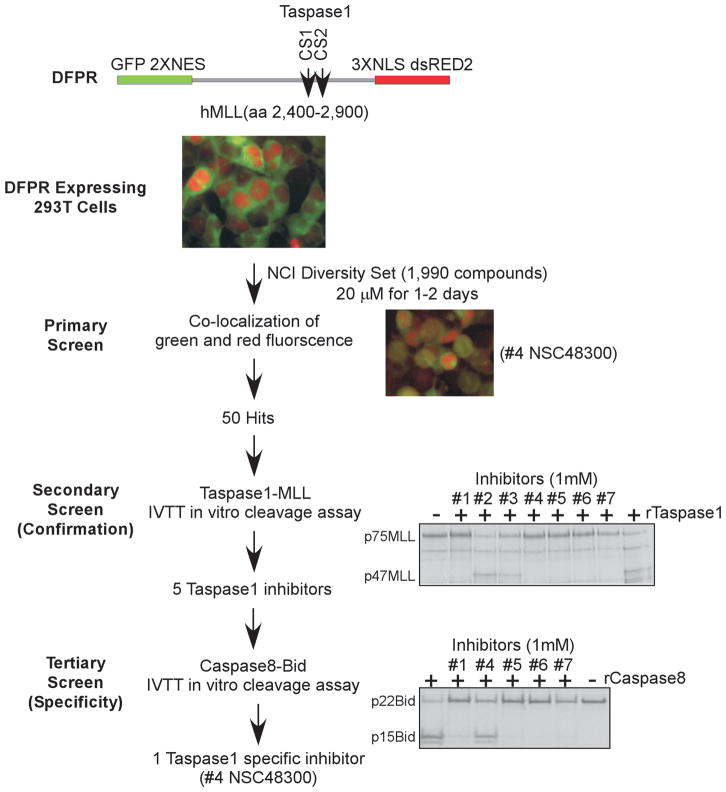
To identify bioactive, small molecule inhibitors of Taspase1, we developed an in vivo screen in which 293T HEK (human embryonic kidney) cells were engineered to stably express a dual fluorescent Taspase1 proteolytic reporter (DFPR, Fig. 3). The DFPR (eGFP-2XNES-p75MLL-3XNLS-dsRED2) consists of the CS1 & CS2 spanning human MLL polypeptide (aa 2,400–2,900, p75MLL) that is flanked by amino-terminal eGFP-NES (nuclear export signal) and carboxy-terminal NLS (nuclear localization signal)-dsRED2 fusions. Taspase1-mediated cleavage of DFPR results in the distribution of green signal (eGFP/NES-p47MLL) in the cytosol and red signal (p28MLL-NLS/dsRED2) in the nucleus. Inhibition of Taspase1 would prevent cleavage of newly synthesized DFPR and result in the accumulation of yellow fluorescence. DFPR-expressing cells were screened against the NCI Diversity Set library, which identified 50 candidate inhibitors that induce varying degrees of co-localization of red-green fluorescence (Fig. 3). A secondary confirmation screen using an established Taspase1-based in vitro cleavage assay was performed (13). Among candidate inhibitors, 5 demonstrated appreciable inhibition of Taspase1-mediated cleavage of p75MLL in vitro (Fig. 3; Supplementary Table S1). To further characterize these small molecule TASPINs, a tertiary specificity screen was employed, in which Caspase8-mediated cleavage of p22Bid was utilized to identify dual Taspase1-Caspase8 inhibitors (27). Caspase8 was chosen because Caspases, like Taspase1, proteolyze their substrates after P1 aspartate. The IVTT-based, Caspase8-p22Bid in vitro cleavage assay was optimized (Supplementary Fig. S2). Interestingly, four out of the five TASPINs were dual Taspase1-Caspase8 inhibitors and compound #4 (NSC48300) specifically targeted Taspase1 (Fig. 3). Taken together, our cell-based screen, followed by in vitro confirmation and specificity assays, identified NSC48300, [4-[(4-arsonophenyl)methyl]phenyl] arsonic acid, as a specific TASPIN.
Figure 3.
A Novel, cell-based, dual fluorescent Taspase1 proteolytic reporter (DFPR) identified small molecule TASPINs. Schematics depict the design of our three-tiered screen for TASPINs. In the primary screen, DFPR expressing 293T HEK cells were treated with 20 μM of the NCI DTP Diversity Set compounds for 2 days, and the fluorescence was determined at day 1 and 2. Pictures of DFPR expressing 293T HEK cells, either mock treated (DMSO) as a negative control (top middle color picture) or compound #4 NSC48300 treated as a positive illustration (lower right color picture), are presented. In the secondary (confirmation) screen, 15 ng of rTaspase1 was incubated with 1 mM of the indicated compounds for 30 minutes before adding the radiolabeled p75MLL (aa 2,400–2,900) cleavage reporter for additional 30 minutes at 30°C. In the tertiary (specificity) screen, 250 ng of rCaspase8 was incubated with 1 mM of the indicated compounds for 30 minutes before adding the radiolabeled p22Bid cleavage reporter for 2 hours at 30°C. Cleavage was assessed by SDS-PAGE and autoradiography.
Characteristics and structure-activity relationships of TASPIN NSC48300
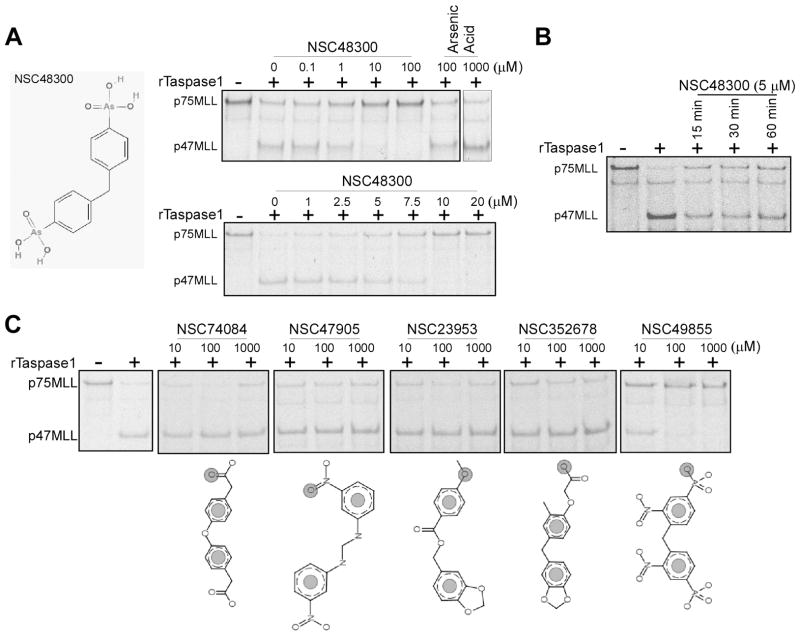
The activity of TASPIN NSC48300 was evaluated using the IVTT, 35S methionine- labeled p75MLL reporter, which demonstrated an IC50 around 7.5 μM (Fig. 4A). The mechanisms by which NSC48300 inhibits Taspase1 were further investigated. We first determined whether it effects as a reversible or irreversible inhibitor. Pre-incubation of Taspase1 with NSC48300 for extended periods of time did not enhance inhibition, favoring a reversible mechanism (Fig. 4B). As NSC48300 is an arsonic acid, we assessed whether free arsenic acid can inactivate Taspase1. Up to 1 mM of arsenic acid was utilized and no detectable inhibition of Taspase1 was observed (Fig. 4A). Nevertheless, the arsenic acid moiety of NSC48300 appears to be essential in that its analogues NSC74084, NSC47905, NSC23953 and NSC352678, have no demonstrable activity (Fig. 4C). Additionally, modifications of the benzene ring, as illustrated in arylarsonic acid NSC49855, resulted in a 10 fold decrease of the inhibitory activity (Fig. 4C). In summary, NSC48300 functions as a reversible TASPIN and arsenic acid may serve as an active functional group against Taspase1 when correctly conjugated onto appropriate chemical backbones.
Figure 4.
Characterization of NSC48300 and determination of its structure-activity relationship. A, Chemical structure of NSC48300. 5 ng of rTaspase1 was incubated with the indicated concentrations of either NSC48300 or arsenic acid for 30 minutes at room temperature before adding the radiolabeled p75MLL reporter for 30 minutes at 30°C. B, 5 ng of rTaspase1 was incubated with 5 μM of NSC48300 for the indicated periods of time at room temperature before adding the radiolabeled p75MLL reporter for 30 minutes at 30°C. C, 5 ng of rTaspase1 was incubated with the indicated concentrations of individual NSC48300 analogues for 30 minutes at room temperature before adding the radiolabeled p75MLL reporter for 30 minutes at 30°C. Cleavage was assessed by SDS-PAGE and autoradiography.
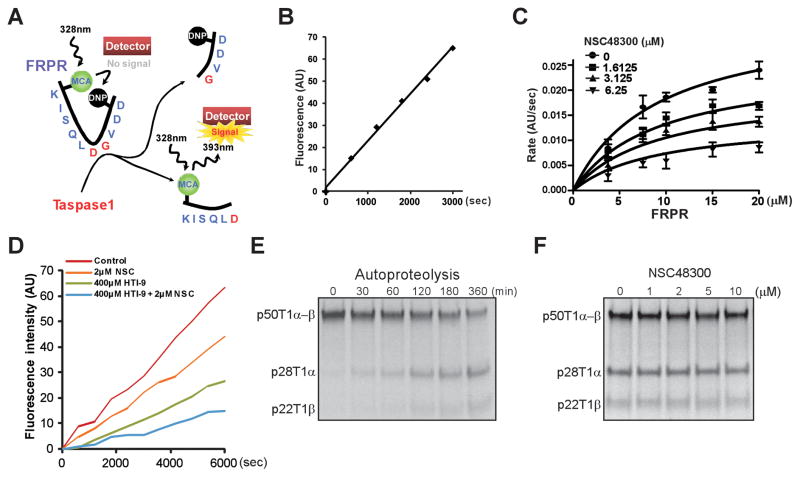
Kinetic analysis of NSC48300 using a FRET-based cleavage reporter
To enable detailed kinetic analysis of TASPINs, we modified an in vitro FRET (Fluorescence Resonance Energy Transfer)-based Taspase1 cleavage assay (14). The FRET-based Taspase1 proteolytic reporter (FRPR, MCA-KISQLDGVDD-DNP) consists of the 10 aa CS2 consensus sequence conjugated with a fluorogenic coumarin (MCA) group and a quenching 2,4-dinitrophenyl (DNP) group at the amino- and carboxy-terminus, respectively (Fig. 5A). Upon Taspase1-mediated cleavage, MCA is no longer quenched by DNP, resulting in a linear increase of fluorescence emission that is excited at 328 nm and detected at 393 nm (Fig. 5B). The apparent KM of FRPR is 9.06 ± 2.80 μM (Fig. 5C). By incubating varying concentrations of FRPR and NSC48300, we demonstrated NSC48300 as a non-competitive TASPIN (KI = 4.22 ± 0.46 μM) (Fig. 5C). To confirm the non-competitive nature of NSC48300 in inhibiting Taspase1, we employed HTI-9 (ISQLAGVDD), a weak, CS2-based, competitive peptide inhibitor of Taspase1 (Supplementary Fig. S3A and B). The fact that NSC48300 and HTI-9 cooperated to inhibit Taspase1 indicates that these two inhibitors function at distinct sites. It confirms NSC48300 as a non-substrate competitive inhibitor and reveals the presence of a yet to be characterized allosteric site on Taspase1 (Fig. 5D).
Figure 5.
Enzyme kinetics analyses of NSC48300. A, Diagram depicts the FRPR. B, 15 μM of FRPR was incubated with 100 nM of rTaspase1 and the fluorescence was monitored. AU denotes absolute fluorescence units. C, Non-linear regression of the reaction curves revealed that NSC48300 is a non-competitive inhibitor of Taspase1. Increasing concentrations of FRPR were incubated with 100 nM of rTaspase1 that was pretreated with the indicated concentrations of NSC48300. Data presented are mean ± SD of three independent experiments. D, 100nM of Taspase1 was pre-incubated for 30 minutes with the indicated concentrations of NSC48300 (NSC) and/or HTI-9 before the addition of FRPR. Reaction progress curves are shown. E, Precursor Taspase1 (p50T1α-β) undergoes autoproteolysis to generate 28kD α (p28T1α) and 22kD β (p22T1β) subunits. 0.5 μl of radiolabeled, 50kD precursor Taspase1 (p50T1α-β) was incubated at 30°C for the indicated periods of time. F, 0.5 μl of radiolabeled p50T1α-β was incubated at 30°C for 6 hours in the presence of indicated concentrations of NSC48300.
NSC48300 does not inhibit the intramolecular autoproteolysis of Taspase1
Our data thus far support a working model in which NSC48300 targets an allosteric site and thereby non-competitively inhibits Taspase1. However, it remains plausible that NSC48300 can further function by interfering the maturation step of Taspase1, i.e., autoproteolysis. Taspase1 is translated as a non-processed α-β precursor enzyme which undergoes autoproteolysis to generate mature α/β protease (13). Taspase1 precursors can be activated by intramolecular autoproteolysis. However, whether these precursors can also be activated by mature Taspase1 through intermolecular autoproteolysis remains undetermined. To probe into the mechanism by which Taspase1 matures, radiolabeled Taspase1 precursor (p50T1α-β) was incubated in cleavage buffer alone. A slow rate of autoproteolysis with ~50% maturation after 6 hours of incubation was observed (Fig. 5E). If p50T1α-β can be activated by intermolecular autoproteolysis, the addition of purified rTaspase1 would greatly expedite the maturation of labeled p50T1α-β. No enhanced cleavage of p50T1α-β was observed upon the addition of up to 100 ng of rTaspase1, indicating the lack of intermolecular autoproteolysis (Supplementary Fig. S4). Altogether, Taspase1 precursors undergo intramolecular, but not intermolecular, autoproteolysis to generate mature Taspase1 enzyme. With this information in hand, increasing amounts of NSC48300 were added to the autoproteolysis reaction and no impact on the Taspase1 autoproteolysis was observed (Fig. 5F), excluding the possibility that NSC48300 inhibits Taspase1 through disrupting autoproteolysis.
The expression level of Taspase1 in breast and brain cancer cell lines correlates with sensitivity to NSC48300
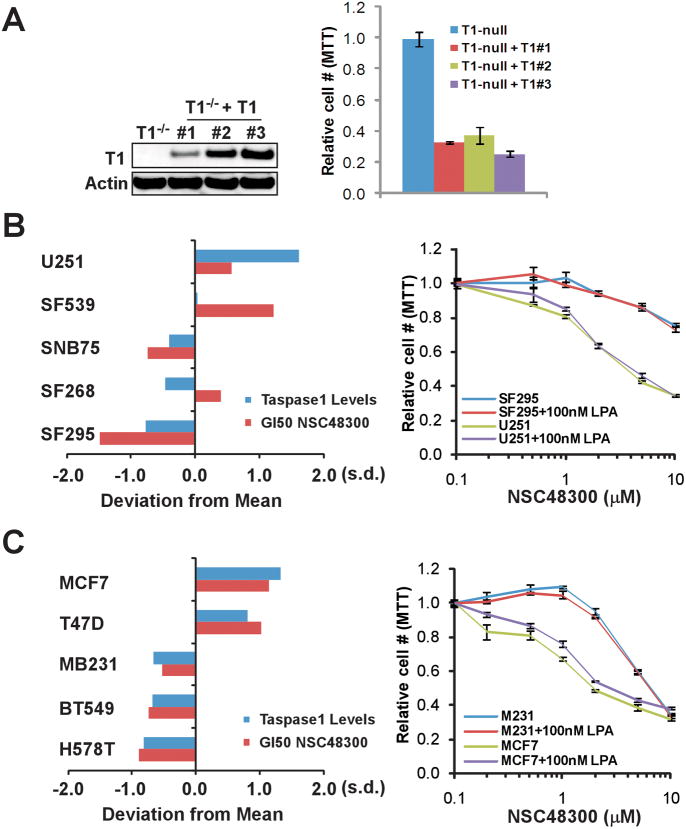
With a specific TASPIN in hand, we explored its potential application in treating cancer cells. Knockdown studies using cancer cell lines demonstrated that cells with higher expression of Taspase1 are more dependent on Taspase1 (20). Hence, we envisioned that certain cancer types might exhibit a strong correlation between their Taspase1 expression and sensitivity to NSC48300. To examine this hypothesis, we first employed SV40-transformed Taspase1−/− MEFs that are stably reconstituted with Taspase1 (Taspase1−/−;Taspase1). When compared to the congenic parental Taspase1−/− cell line, Taspase1−/−;Taspase1 cells displayed increased sensitivity to NSC48300, indicating that the presence of Taspase1 renders treatment sensitivity (Fig. 6A). We next integrated the Taspase1 protein expression profile and the NSC48300 growth inhibition database of the NCI60 cancer cell lines (8, 34). According to the NCI Developmental Therapeutic Program (DTP) database (35), NSC48300 produces a distinct pattern of growth inhibition in the NCI60 in vitro anticancer drug screen (Table S2). The sensitivity to NSC48300-mediated growth inhibition is in general agreement with the protein level of Taspase1 in many human cancer cell lines (Table S2), among which an especially tight correlation is detected in breast and brain cancer cells (Fig. 6B and C, left panels). The differential sensitivity to NSC48300 within these two cancer types was confirmed (Fig. 6B and C, right panels), supporting Taspase1 as a target of NSC48300 in treating cancer cells. However, when high concentrations of NSC48300 were utilized, non-specific toxicity became evident, which could relate to its intrinsic arsenic acid moiety or effects on other cellular enzymes. NSC48300 was recently shown to inhibit autotaxin, an extracellular enzyme that converts lysophosphatidylcholine to lipophosphatidic acid (LPA) (36). The lysophospholipase D activity of autotoxin can be inhibited by NSC48300, resulting in the disruption of LPA production and a subsequent decrease of the in vitro invasiveness of cancer cells, which can be reversed by adding extracellular LPA (36). To examine whether our observed cancer cell sensitivity to NSC48300 can be partially attributed to the inhibition of autotaxin, we treated cancer cells with NSC48300 in the presence or absence of LPA. Since the relative cell number curves basically overlaid irrespective of LPA in both NSC48300 sensitive and insensitive lines, we excluded autotaxin as a target for NSC48300-mediated growth inhibition, which is consistent with the known extracellular expression and action of autotaxin (Fig. 6B and C, right panels).
Figure 6.
The cellular sensitivity to NSC48300 correlates with the protein expression of Taspase1. A, The indicated MEFs were treated with 1uM of NSC48300 for 2 days and the relative cell number was measured by MTT assays. Values obtained from the parental Taspase1−/− MEFs after NSC48300 treatment were designated as 1. Data presented are mean ± SD of three independent experiments. B, The NSC48300 GI50 (growth inhibition 50%) data were acquired through a publically available database (35). To compare relative sensitivity among different brain tumor cell lines, log(GI50) values of each individual lines were obtained. The standard deviations of individual GI50 from the mean were plotted (positive values indicate higher sensitivity). The protein levels of Taspase1 in the indicated cell lines have been reported and are represented as a bar graph. The indicated brain cancer cell lines were treated with increasing concentration of NSC48300 for 2 days plus or minus 100nM LPA and then subjected to MTT assays. Data presented are mean ± SD of two triplicate independent experiments. C, A positive correlation of Taspase1 protein levels and cellular sensitivity to NSC48300 in the NCI60 breast tumor cell lines. Data collection, experimentations, and analyses were performed as described in B on the indicated breast cancer cell lines.
NSC48300 inhibits the growth of MMTV-neu mouse breast cancers and U251 brain tumor xenografts
Following up on the cell line data, we wished to evaluate the in vivo efficacy of NSC48300 in treating cancers. Since NSC48300 is an arsonic acid and no data concerning its in vivo safety have been reported, we determined its toxicity profile. Short term, instead of long term, tail vein injections were performed due to observed fibrotic damages of regional vessels at injection sites upon repetitive treatment of NSC48300. Histological examination of major organs did not reveal any obvious abnormalities whereas blood chemistry revealed a significant decrease in the LDH level and certain alterations in electrolytes and kidney functions (Supplementary Fig. S5 and Table S3). Complete blood counts were also performed, which demonstrated a decrease in white blood cell counts and hemoglobin levels (Supplementary Fig. S6A, B and C).
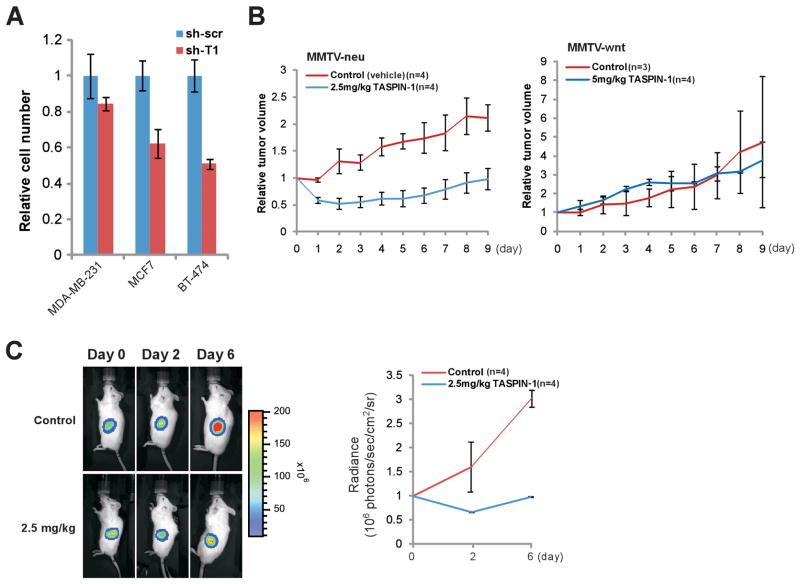
To enable a scientific selection of the most relevant breast cancer model for in vivo experiments, we expanded our breast cancer cell line repertoire to incorporate an estrogen receptor (ER) negative, Her2/neu over-expressing cancer cell line, BT474 (37), and performed genetic knockdown experiments (Supplementary Fig. S7). Among these three breast cancer cell lines, MDA-MB-231 cells, an estrogen receptor (ER) negative, Her2/neu negative cell line (38) that expresses a very low level of Taspase1, were least affected by the deficiency of Taspase1 (Fig. 7A and Supplementary Fig. S7) (20). By contrast, cells with higher levels of Taspase1, including MCF7, an ER positive, Her2/neu negative cell line (39), and BT-474 cells were more dependent on Taspase1 (Fig. 7A and Supplementary Fig. S7). Based on these in vitro data, Her2/neu-driven tumors may be more dependent on Taspase1 and would be more sensitive to NSC48300 treatment, whereas ER negative, Her2/neu negative breast cancers would be resistant. To examine this hypothesis in vivo, we treated MMTV-neu (23) or MMTV-wnt (ER-, Her2-) (24) breast cancer bearing mice with NSC48300. Indeed, NSC48300 consistently disrupted the growth of MMTV-neu breast cancers, whereas it exhibited no effects on the growth of MMTV-wnt breast cancers (Fig. 7B).
Figure 7.
NSC48300 disrupts breast and brain cancer growth in mouse cancer models. A, The indicated human breast cancer cell lines with control (sh-scr) or Taspase1 (sh-T1) knockdown were plated, and cell numbers were counted 4 days after the initial plating. The average cell number of control knockdown cells was assigned as 1. Data presented are mean ± SD of three independent experiments. B, Tumor bearing MMTV-neu or MMTV-wnt mice were treated at day 0 when de novo tumors reached 1cm at the longest dimension. NSC48300 was given at the indicated doses every other day for a total of 5 intravenous injections. Graphs represent the relative tumor volume throughout the course of treatment. The tumor size measured at day 0 immediately before treatment was assigned as 1. Data presented are mean ± SD of the indicated number of tumors of each arm. C, U251 cells transduced with a luciferase reporter were implanted into the flanks of male NOD-scid IL2Rγ−/− mice at 6–8 weeks of age (day −1). Mice were then treated on days 0, 2, and 4 with the indicated dose of NSC48300. Tumor mass was determined by bioluminescent imaging using an IVIS 100 and representative bioluminescent images are presented. The graph represents mean ± SD of 4 tumors from each arm. The absolute bioluminescence was normalized to a value of 1 at day 0.
To test the efficacy of NSC48300 in treating brain tumors, we employed a brain cancer xenograft model using U251 cells based on two reasons. First, knockdown of Taspase1 in U251 cells impaired cell proliferation (20). Second, among the NCI60 brain tumor cell lines, U251 cells express high levels of Taspase1 and are most sensitive to NSC48300 treatment in vitro. To monitor treatment response in live animals, U251 cells were first engineered to stably express firefly luciferase (fLuc) before subcutaneous implantation into mice. Bioluminescent imaging demonstrated a stasis of tumor growth in NSC48300 treated compared to vehicle treated tumors (Fig. 7C). In summary, our in vivo preclinical efficacy trials demonstrated an anti-tumor effect of NSC48300 in both MMTV-neu breast cancer and U251 GBM xenograft models.
Discussion
In the evolution of cancer, individual cells must overcome a multitude of challenges and eventually exhibit hallmarks of cancer, a process demanding multiple oncogenes and non-oncogenes that function cooperatively to achieve and maintain the aberrant, oncogenic state. The evolved reliance of oncogenes on particular sets of subordinate non-oncogenes during tumorigenesis, offering a novel anti-cancer treatment strategy aiming at tumor dependent, non-oncogenes (40). Thus far, well-characterized tumor dependent, non-oncogenic actors are scarce, including the 26S proteasome, HSF1 (41), and IRF4 (42), which handles the rapid protein turnover, mediates the stress response, and maintains expression of the MYC oncogene, respectively. The fact that the proteasome inhibitor Bortezomib is effective against multiple myeloma in human patients substantiates this new anticancer therapeutic concept (11). Successful application of this strategy in cancer therapy relies on the identification and characterization of tumor dependent, non-oncogenic factors that cancer but not normal cells heavily rely on.
Taspase1 by itself or in conjunction with MYC, RAS, or E1A fails to transform NIH/3T3 cells or primary MEFs, respectively, yet is required for efficient cancer initiation and maintenance (20). Hence, Taspase1 functions as a tumor dependent, non-oncogenic protease whose inhibition may offer an anticancer strategy. However, Taspase1 is highly conserved and known to regulate embryonic development (8). The severe perinatal lethality resulting from the embryonic loss of Taspase1 suggests that inactivation of Taspase1 by genetic or pharmacological means is inadvisable in pregnant females and children to avoid potential developmental sequelae. In contrast, inactivation of Taspase1 in fully developed adult mammals appears to be well-tolerated. Cancer commonly hijacks key developmental pathways during tumorigenesis and thus frequently exhibits unique properties, which may underlie the preferential therapeutic benefit conferred by targeting Taspase1 to treat cancers.
Through proteolytic processing of nuclear transcription regulators, Taspase1 controls cellular proliferation by suppressing and activating the expression of CDKIs and Cyclins, respectively (8, 20). Although knockdown of Taspase1 affects proliferation in many cancer cell lines, candidate cancer types that may respond to Taspase1 inhibition in vivo remain unclear. Unlike activating mutations of EGFR and BCR-ABL that can serve as powerful biomarkers for selecting responsive cancers (43), Taspase1 dose not function like a classical oncogene and is not mutated in cancers (COSMIC, Sanger Institute). Data thus far indicate a positive correlation between Taspase1 protein levels and the underlying dependence, which provides a useful primary tool in identifying potentially susceptible cancers. Our prior knockdown experiments demonstrated an in vitro and in vivo reliance of U251 brain cancer cells on Taspase1 for a full cancer phenotype (20). Here, we demonstrate treatment response of U251 cells to TASPIN NSC48300 in both cell culture and xenograft models. Hence, genetic knockdown experiments in cancer cells could reflect the cellular dependence on Taspase1 and thus provide assessment of in vivo responsiveness of individual cancers to TASPINs.
In addition to brain tumors, we expanded our in vivo studies to incorporate breast cancer models. Deficiency in Taspase1 by genetic knockdown disrupts the proliferation of Her2/neu+ human BT-474 breast cancer cells, suggesting that Her2-neu+ breast cancer may be sensitive to the inactivation of Taspase1. Indeed, de novo breast cancers developed in MMTV-neu mice responded to the treatment with NSC48300, whereas MMTV-wnt breast cancers were insensitive. Consistent with the treatment effects, mammary tissue specific knockout of Taspase1 disrupts MMTV-neu-driven but not MMTV-wnt-driven breast carcinogenesis (Van Tine et al., unpublished data). Taspase1 appears to play an important role in the Her2/neu growth factor signaling pathway whereas WNT signals apparently bypass Taspase1-mediated oncogenic events. These data highlight the heterogeneity underlying individual tumorigenesis and the importance of selecting the responsive cancers that may benefit from treatment with TASPINs. Further studies concerning the involvement of Taspase1 in various oncogenic pathways and the pathogenesis of subtypes of cancers would offer a better target cancer selection for the potential use of TASPINs in the future.
Supplementary Material
Acknowledgments
We thank Ms. Hsiu-Fang Chen for her technical assistance and Dr. Shoemaker for editing this manuscript. This study is supported by NIH CA R01-119008 and American Cancer Society.
Footnotes
Disclosure of Potential Conflicts of Interest
No potential conflicts of interests were disclosed.
References
- 1.Furie B, Furie BC. Molecular and cellular biology of blood coagulation. N Engl J Med. 1992;326:800–6. doi: 10.1056/NEJM199203193261205. [DOI] [PubMed] [Google Scholar]
- 2.Zaman MA, Oparil S, Calhoun DA. Drugs targeting the renin-angiotensin-aldosterone system. Nat Rev Drug Discov. 2002;1:621–36. doi: 10.1038/nrd873. [DOI] [PubMed] [Google Scholar]
- 3.Brown MS, Ye J, Rawson RB, Goldstein JL. Regulated intramembrane proteolysis: a control mechanism conserved from bacteria to humans. Cell. 2000;100:391–8. doi: 10.1016/s0092-8674(00)80675-3. [DOI] [PubMed] [Google Scholar]
- 4.Ye Y, Fortini ME. Proteolysis and developmental signal transduction. Semin Cell Dev Biol. 2000;11:211–21. doi: 10.1006/scdb.2000.0167. [DOI] [PubMed] [Google Scholar]
- 5.Thornberry NA, Lazebnik Y. Caspases: enemies within. Science. 1998;281:1312–6. doi: 10.1126/science.281.5381.1312. [DOI] [PubMed] [Google Scholar]
- 6.Boatright KM, Salvesen GS. Mechanisms of caspase activation. Curr Opin Cell Biol. 2003;15:725–31. doi: 10.1016/j.ceb.2003.10.009. [DOI] [PubMed] [Google Scholar]
- 7.Julien E, Herr W. Proteolytic processing is necessary to separate and ensure proper cell growth and cytokinesis functions of HCF-1. Embo J. 2003;22:2360–9. doi: 10.1093/emboj/cdg242. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Takeda S, Chen DY, Westergard TD, Fisher JK, Rubens JA, Sasagawa S, et al. Proteolysis of MLL family proteins is essential for taspase1-orchestrated cell cycle progression. Genes Dev. 2006;20:2397–409. doi: 10.1101/gad.1449406. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Saklatvala J, Nagase H, Salvesen G. Proteases and the regulation of biological processes. Biochemical Society Symposium. 2002:70. [Google Scholar]
- 10.Docherty AJ, Crabbe T, O’Connell JP, Groom CR. Proteases as drug targets. Biochem Soc Symp. 2003:147–61. doi: 10.1042/bss0700147. [DOI] [PubMed] [Google Scholar]
- 11.Adams J. The proteasome: a suitable antineoplastic target. Nat Rev Cancer. 2004;4:349–60. doi: 10.1038/nrc1361. [DOI] [PubMed] [Google Scholar]
- 12.Markowitz M, Saag M, Powderly WG, Hurley AM, Hsu A, Valdes JM, et al. A preliminary study of ritonavir, an inhibitor of HIV-1 protease, to treat HIV-1 infection. The New England journal of medicine. 1995;333:1534–9. doi: 10.1056/NEJM199512073332204. [DOI] [PubMed] [Google Scholar]
- 13.Hsieh JJ, Cheng EH, Korsmeyer SJ. Taspase1: a threonine aspartase required for cleavage of MLL and proper HOX gene expression. Cell. 2003;115:293–303. doi: 10.1016/s0092-8674(03)00816-x. [DOI] [PubMed] [Google Scholar]
- 14.Khan JA, Dunn BM, Tong L. Crystal structure of human Taspase1, a crucial protease regulating the function of MLL. Structure. 2005;13:1443–52. doi: 10.1016/j.str.2005.07.006. [DOI] [PubMed] [Google Scholar]
- 15.Hsieh JJ, Ernst P, Erdjument-Bromage H, Tempst P, Korsmeyer SJ. Proteolytic cleavage of MLL generates a complex of N- and C-terminal fragments that confers protein stability and subnuclear localization. Mol Cell Biol. 2003;23:186–94. doi: 10.1128/MCB.23.1.186-194.2003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Zhou H, Spicuglia S, Hsieh JJ, Mitsiou DJ, Hoiby T, Veenstra GJ, et al. Uncleaved TFIIA is a substrate for taspase 1 and active in transcription. Mol Cell Biol. 2006;26:2728–35. doi: 10.1128/MCB.26.7.2728-2735.2006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Xu Q, Buckley D, Guan C, Guo HC. Structural insights into the mechanism of intramolecular proteolysis. Cell. 1999;98:651–61. doi: 10.1016/s0092-8674(00)80052-5. [DOI] [PubMed] [Google Scholar]
- 18.Capotosti F, Hsieh JJ, Herr W. Species selectivity of Mixed Lineage Leukemia/Trithorax and HCF proteolytic maturation pathways. Mol Cell Biol. 2007;27:7063–72. doi: 10.1128/MCB.00769-07. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Capotosti F, Guernier S, Lammers F, Waridel P, Cai Y, Jin J, et al. O-GlcNAc transferase catalyzes site-specific proteolysis of HCF-1. Cell. 2011;144:376–88. doi: 10.1016/j.cell.2010.12.030. [DOI] [PubMed] [Google Scholar]
- 20.Chen DY, Liu H, Takeda S, Tu HC, Sasagawa S, Van Tine BA, et al. Taspase1 functions as a non-oncogene addiction protease that coordinates cancer cell proliferation and apoptosis. Cancer Res. 2010;70:5358–67. doi: 10.1158/0008-5472.CAN-10-0027. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Niehof M, Borlak J. EPS15R, TASP1, and PRPF3 are novel disease candidate genes targeted by HNF4alpha splice variants in hepatocellular carcinomas. Gastroenterology. 2008;134:1191–202. doi: 10.1053/j.gastro.2008.01.027. [DOI] [PubMed] [Google Scholar]
- 22.Scrideli CA, Carlotti CG, Jr, Okamoto OK, Andrade VS, Cortez MA, Motta FJ, et al. Gene expression profile analysis of primary glioblastomas and non-neoplastic brain tissue: identification of potential target genes by oligonucleotide microarray and real-time quantitative PCR. J Neurooncol. 2008;88:281–91. doi: 10.1007/s11060-008-9579-4. [DOI] [PubMed] [Google Scholar]
- 23.Muller WJ, Sinn E, Pattengale PK, Wallace R, Leder P. Single-step induction of mammary adenocarcinoma in transgenic mice bearing the activated c-neu oncogene. Cell. 1988;54:105–15. doi: 10.1016/0092-8674(88)90184-5. [DOI] [PubMed] [Google Scholar]
- 24.Tsukamoto AS, Grosschedl R, Guzman RC, Parslow T, Varmus HE. Expression of the int-1 gene in transgenic mice is associated with mammary gland hyperplasia and adenocarcinomas in male and female mice. Cell. 1988;55:619–25. doi: 10.1016/0092-8674(88)90220-6. [DOI] [PubMed] [Google Scholar]
- 25.Gross S, Piwnica-Worms D. Real-time imaging of ligand-induced IKK activation in intact cells and in living mice. Nature methods. 2005;2:607–14. doi: 10.1038/nmeth779. [DOI] [PubMed] [Google Scholar]
- 26.Liu H, Takeda S, Kumar R, Westergard TD, Brown EJ, Pandita TK, et al. Phosphorylation of MLL by ATR is required for execution of mammalian S-phase checkpoint. Nature. 2010;467:343–6. doi: 10.1038/nature09350. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Zha J, Weiler S, Oh KJ, Wei MC, Korsmeyer SJ. Posttranslational N-myristoylation of BID as a molecular switch for targeting mitochondria and apoptosis. Science. 2000;290:1761–5. doi: 10.1126/science.290.5497.1761. [DOI] [PubMed] [Google Scholar]
- 28.Ren D, Tu HC, Kim H, Wang GX, Bean GR, Takeuchi O, et al. BID, BIM, and PUMA are essential for activation of the BAX- and BAK-dependent cell death program. Science. 2010;330:1390–3. doi: 10.1126/science.1190217. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Kuhn R, Schwenk F, Aguet M, Rajewsky K. Inducible gene targeting in mice. Science. 1995;269:1427–9. doi: 10.1126/science.7660125. [DOI] [PubMed] [Google Scholar]
- 30.Yokoyama A, Kitabayashi I, Ayton PM, Cleary ML, Ohki M. Leukemia proto-oncoprotein MLL is proteolytically processed into 2 fragments with opposite transcriptional properties. Blood. 2002;100:3710–8. doi: 10.1182/blood-2002-04-1015. [DOI] [PubMed] [Google Scholar]
- 31.Walsh CT. Enzymatic Reaction Mechanisms. W H Freeman & Company; 1978. [Google Scholar]
- 32.Coux O, Tanaka K, Goldberg AL. Structure and functions of the 20S and 26S proteasomes. Annu Rev Biochem. 1996;65:801–47. doi: 10.1146/annurev.bi.65.070196.004101. [DOI] [PubMed] [Google Scholar]
- 33.Lee JT, Chen DY, Yang Z, Ramos AD, Hsieh JJ, Bogyo M. Design, syntheses, and evaluation of Taspase1 inhibitors. Bioorganic & medicinal chemistry letters. 2009;19:5086–90. doi: 10.1016/j.bmcl.2009.07.045. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Shoemaker RH. The NCI60 human tumour cell line anticancer drug screen. Nat Rev Cancer. 2006;6:813–23. doi: 10.1038/nrc1951. [DOI] [PubMed] [Google Scholar]
- 35.Developmental Therapeutics Program [Internet] Bethesda (MD): The National Cancer Institute; c1990. [cited 2011 Nov 9]. Available from: http://dtp.nci.nih.gov/dtp_search.html. [Google Scholar]
- 36.Saunders LP, Ouellette A, Bandle R, Chang WC, Zhou H, Misra RN, et al. Identification of small-molecule inhibitors of autotaxin that inhibit melanoma cell migration and invasion. Mol Cancer Ther. 2008;7:3352–62. doi: 10.1158/1535-7163.MCT-08-0463. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Kumar R, Shepard HM, Mendelsohn J. Regulation of phosphorylation of the c-erbB-2/HER2 gene product by a monoclonal antibody and serum growth factor(s) in human mammary carcinoma cells. Mol Cell Biol. 1991;11:979–86. doi: 10.1128/mcb.11.2.979. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Kang Y, Siegel PM, Shu W, Drobnjak M, Kakonen SM, Cordon-Cardo C, et al. A multigenic program mediating breast cancer metastasis to bone. Cancer Cell. 2003;3:537–49. doi: 10.1016/s1535-6108(03)00132-6. [DOI] [PubMed] [Google Scholar]
- 39.Coezy E, Borgna JL, Rochefort H. Tamoxifen and metabolites in MCF7 cells: correlation between binding to estrogen receptor and inhibition of cell growth. Cancer Res. 1982;42:317–23. [PubMed] [Google Scholar]
- 40.Luo J, Solimini NL, Elledge SJ. Principles of cancer therapy: oncogene and non-oncogene addiction. Cell. 2009;136:823–37. doi: 10.1016/j.cell.2009.02.024. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Dai C, Whitesell L, Rogers AB, Lindquist S. Heat shock factor 1 is a powerful multifaceted modifier of carcinogenesis. Cell. 2007;130:1005–18. doi: 10.1016/j.cell.2007.07.020. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Shaffer AL, Emre NC, Lamy L, Ngo VN, Wright G, Xiao W, et al. IRF4 addiction in multiple myeloma. Nature. 2008;454:226–31. doi: 10.1038/nature07064. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Sawyers CL. The cancer biomarker problem. Nature. 2008;452:548–52. doi: 10.1038/nature06913. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.