Abstract
Vsx-1 is a paired-like:CVC homeobox gene whose expression is linked to bipolar cell differentiation during zebrafish retinogenesis. We used a yeast two-hybrid screen to identify proteins interacting with Vsx-1 and isolated Ubc9, an enzyme that conjugates the small ubiquitin-like modifier SUMO-1. Despite its interaction with Ubc9, we show that Vsx-1 is not a substrate for SUMO-1 in COS-7 cells or in vitro. When a yeast two-hybrid assay is used, deletion analysis of the interacting domain on Vsx-1 shows that Ubc9 binds to a nuclear localization signal (NLS) at the NH2 terminus of the homeodomain. In SW13 cells, Vsx-1 localizes to the nucleus and is excluded from nucleoli. Deletion of the NLS disrupts this nuclear localization, resulting in a diffuse cytoplasmic distribution of Vsx-1. In SW13 AK1 cells that express low levels of endogenous Ubc9, Vsx-1 accumulates in a perinuclear ring and colocalizes with an endoplasmic reticulum marker. However, NLS-tagged STAT1 protein exhibits normal nuclear localization in both SW13 and SW13 AK1 cells, suggesting that nuclear import is not globally disrupted. Cotransfection of Vsx-1 with Ubc9 restores Vsx-1 nuclear localization in SW3 AK1 cells and demonstrates that Ubc9 is required for the nuclear localization of Vsx-1. Ubc9 continues to restore nuclear localization even after a C93S active site mutation has eliminated its SUMO-1-conjugating ability. These results suggest that Ubc9 mediates the nuclear localization of Vsx-1, and possibly other proteins, through a nonenzymatic mechanism that is independent of SUMO-1 conjugation.
As regulators of retinal progenitor cell proliferation and cell specification, homeobox proteins are one set of transcription factors that are essential to vertebrate eye development (1–5). Of these, the paired-like homeobox proteins (3, 6, 7) are particularly important during retinogenesis. Vsx-1 and Vsx-2 are two paired-like homeobox proteins that were originally discovered in the regenerating visual pathway of goldfish (8–10) and further studied in the developing zebrafish (11, 12). During retinal development, the complementary expression of Vsx-1 and Vsx-2 mRNA suggests that Vsx-1 is involved in bipolar cell differentiation whereas Vsx-2 is involved in cell proliferation (12).
Vsx-1 and Vsx-2 are denoted as paired-like:CVC homeobox proteins because their primary structures contains a conserved 54-aa CVC domain that is downstream and adjacent to the homeodomain. Orthologs of Vsx-1 and Vsx-2 have been isolated and characterized from other organisms, including Caenorhabditis elegans (13) and chicken (14), as well as mammals (15–17). More specifically, Chx10 is the mouse and human ortholog of Vsx-2 (18), whereas the bovine Vsx-1, named RINX/Vsx-1, has recently been identified (16). The functional importance of these genes is clear because a human CHX10 gene mutation results in microphthalmia (19). Furthermore, a homozygous null allele for Chx10 gives rise to an ocular retardation phenotype in mice that can be partially rescued (20).
Such a functional role in retinogenesis suggests that these transcription factors are likely to be temporally and spatially regulated within the cell. Several mechanisms probably contribute to such tight regulatory control. For example, in developing zebrafish, Vsx-1 and Vsx-2 mRNAs are expressed in a precisely coordinated sequential pattern (12). In addition to its transcriptional regulation, Vsx-1 protein is ubiquitinated and subsequently degraded by the 26S proteasome in vitro and in vivo. Thus, we have proposed that Vsx-1 is developmentally regulated by ubiquitin-mediated proteolysis (21).
Here, we show that Vsx-1 is also regulated by an interacting protein, ubiquitin-conjugating enzyme 9 (Ubc9). Ubc9 is similar to other ubiquitin-conjugating enzymes (E2) but conjugates an ubiquitin-like protein that is designated as Smt3c in yeast (22) and SUMO-1 in vertebrate cell culture systems (23, 24). Unlike polyubiquitination, SUMO-1 conjugation of target proteins (sumoylation) does not signal proteolysis. Instead, recent results indicate that sumoylation has roles in subcellular protein translocation [RanGAP1 (25)] and in nuclear body formation [PML (26) and TEL (27)]. Sumoylation also has been shown to protect proteins from ubiquitin-mediated proteolysis [IκBα (28) and Mdm2 (29)] and to regulate transcriptional activity [Dorsal (30), IE2-p86 (31), HIPK2 (32), p53 (33), androgen receptor (34), and c-Jun (35)]. Other reports indicate that Ubc9 mediates transcriptional activity independent of SUMO conjugation [e.g., ETS1 (36), TEL (37), and androgen receptor (38)]. We now show that, despite its interaction with Ubc9, Vsx-1 is not a substrate for SUMO-1 conjugation. Rather, Ubc9 binds to a NLS on Vsx-1 and facilitates nuclear localization of Vsx-1. Therefore, in addition to its SUMO-conjugating function, Ubc9 may have a role in protein translocation and may have a capacity to operate nonenzymatically.
Materials and Methods
Reagents.
Yeast strains L40 and AMR70 (39) as well as associated two-hybrid constructs were generously provided by R. Sternglanz [State University of New York (SUNY), Stony Brook, NY]. The anti-LexA antibody was purchased from CLONTECH. We also used the following generously provided reagents: anti-Myc ascites (9e10), D. Bar-Sagi (SUNY, Stony Brook, NY); guinea pig anti-human Ubc9 antibody, E. Golub (Yale University, New Haven, CT); Lens lectin and anti-calnexin (8211–1) antibody, J. Trimmer (SUNY, Stony Brook, NY); and recombinant glutathione S-transferase (GST)-SUMO-1 and Ubc9 (Xenopus) protein, H. Saitoh (Picower Institute for Medical Research, Manhasset, NY). Easytag Express Protein Labeling Mix (35S-labeled methionine and cysteine) was purchased from NEN.
Plasmids and Cloning.
All gene names refer to cDNAs. Plasmid pYVsx-1a was generated by inserting a blunt-ended Sau3A fragment of goldfish Vsx-1 that encoded the NLS through the putative PEST site (amino acids 135–326) into a blunt-ended BamHI site of pBTM116 ADE2. This plasmid was used as the target for the initial yeast two-hybrid library screen. Subsequent two-hybrid experiments used pSTT91, a derivative of pBTM116, as a vector. The following inserts were amplified by PCR and cloned into EcoRI and BamHI restriction enzyme sites: zVsx-1(ORF); zVsx-1ΔC-term (deletes amino acids 211–344); zVsx-1ΔCVC (deletes amino acids 211–268); zVsx-1ΔN-term1 (deletes amino acids 1–150); zVsx-1ΔNterm2 (deletes amino acids 1–146); zVsx-1ΔNLS/homeo domain (HD) (deletes amino acids 147–210) ; and zVsx-1ΔNLS (deletes amino acids 147–153). For immunocytochemistry, the Vsx-1 variants listed above were cloned into the EcoRI and XbaI sites of pCS2MT vector (generous gift from D. L. Turner and R. A. W. Rupp, Fred Hutchinson Cancer Research Center, Seattle): zVsx-1, zVsx-1ΔC- term, zVsx-1ΔCVC, zVsx-1ΔN-term1, zVsx-1ΔNterm2, zVsx-1ΔNLS/HD, and zVsx-1ΔNLS. Plasmid pcDNA 3.1+A.S.hUbc9 was constructed by amplifying hUbc9 from pTY316-hUbc9wt (generous gift from J. Benson, Harvard Medical School, Boston) and cloning in reverse orientation into the EcoRI and XbaI sites of pcDNA 3.1 (Invitrogen). For rescue experiments, plasmid pcDNA3.1+zUbc9 was generated by subcloning zUbc9 as a EcoRI/NotI fragment from pBS+zUbc9, and pCMV-Flag-Ubc9 and pBK-CMV-UBC9 (C93S) were generous gifts from G. Nucifora (Loyola University Medical Center, Maywood, IL). pEGFP-N1+STAT1+NLS+GFP was a generous gift from K. McBride and N. Reich (SUNY, Stony Brook, NY). Sumoylation experiments used pCI-RanGAP1, pET-28a(+)-RanGAP1, pEGFP-N1-SUMO-1, and pEGFP-N1-SUMO-2, which were generous gifts from H. Saitoh. All plasmids were verified by DNA sequencing.
Cell Culture.
COS-7 cells were propagated in DMEM supplemented with 10% FBS, 1 mM sodium pyruvate, 1% glutamine, 100 units/ml penicillin G, and 100 μg/ml streptomycin sulfate. SW-13 c1.1 Vim+ (SW-13) cells, provided by R. Evans (University of Colorado Health Sciences Center, Denver), were maintained as described (40). SW-13 AK1 cells were generated by transfecting SW-13 cells with pcDNA 3.1+A.S.hUbc9 (neoR). Stable transfectants were maintained in a 1:1 mixture of Dulbecco's modified Eagle's medium and Ham's F12 (GIBCO/BRL), supplemented with 5% FBS, 100 units/ml penicillin G sodium, 100 μg/ml streptomycin sulfate, and 300 μg/ml G418.
Immunocytochemistry.
Plasmid DNA, as noted in the figure legends, was transfected into SW-13 cells with FuGene 6 (Roche Molecular Biochemicals) according to the manufacturer's instructions. Proteins were detected 24 h after transfection by using the indirect immunofluorescence method (40). Myc tag (MT)-zVsx-1 fusion proteins were detected by using 1:250 9e10 and 1:500 anti-mouse IgG1-FITC (Southern Biotechnology Associates). Calnexin was detected with 1:50 anti-calnexin and 1:1,000 anti-rabbit IgG2b-tetramethylrhodamine B isothiocyanate (Southern Biotechnology Associates), with the nucleus labeled by using 2 μg/ml Hoechst 33258 for 10 min. NLS-STAT1-GFP was detected by washing cells twice in PBS, fixing in MeOH at −20°C for 10 min, and washing in PBS four times for 5 min each.
Yeast Two-Hybrid Screen.
The two-hybrid screening was done essentially as previously described (39). However, using AMR70 for mating, specificity testing used six nonspecific proteins: histone H4 (amino acids 1–30), lamin, Saccharomyces cerevisiae topoisomerase 1, Xenopus topoisomerase 1, alcohol dehydrogenase 1, and the yeast-silencing gene SIR1. The cDNA library was constructed from random-primed and oligo(dT)-primed mRNA from zebrafish. Library cDNAs were cloned into the EcoRI site of pGAD10 (CLONTECH). To screen for interacting proteins, we cotransformed 5 μg of pYVsx-1a or pSTT91+zVsx-1, 5 μg of the zebrafish cDNA two-hybrid library, and 25 μg of denatured sheared salmon sperm DNA in each 20-ml culture of logarithmic phase L40 yeast. In subsequent two-hybrid tests between the zUbc9 clone and zVsx-1 deletions, quantification of β-galactosidase activity used the CPRG (chlorophenol red-β-d-galactopyranoside) liquid culture assay (64).
Immunoblotting and Fluorography.
Proteins were fractionated on Tris–glycine SDS/PAGE minigels and electrophoretically transferred onto poly(vinylidene difluoride) membranes (PVDF; Amersham Pharmacia) in 3-(cyclohexylamino)-1-propanesulfonic acid (CAPS; Sigma) buffer (10 mM CAPS/10% methanol) at 400 mA for 35 min. Gradient gels were purchased from Bio-Rad. Membranes were incubated for 1 h at room temperature in TBS-T wash buffer (Tris-buffered saline, pH 7.4/0.2% Tween 20) containing 5% nonfat dry milk. Membranes were probed with 1:2500 9e10 ascites to detect MT-Vsx-1 fusion proteins and also with 1:7000 guinea pig anti-HsUbc9 antibody, 1:2000 rabbit anti-RanGAP1, and 1:1000 mouse anti-SUMO-1 antibody (Zymed). Secondary antibodies were conjugated with horseradish peroxidase. Proteins were visualized by enhanced chemiluminescence (ECL; Amersham Pharmacia). Radioactively labeled proteins were fractionated by SDS/PAGE. Gels were fixed in 10% acetic acid and 30% methanol overnight and incubated in EN3HANCE (DuPont) fluorography solution according to the manufacturer's protocol. To reduce the risk of gels cracking during drying, gels were incubated in a solution of 7% acetic acid, 7% methanol, and 1% glycerol for 5 min before drying.
Results
Vsx-1 Interacts with Ubc9.
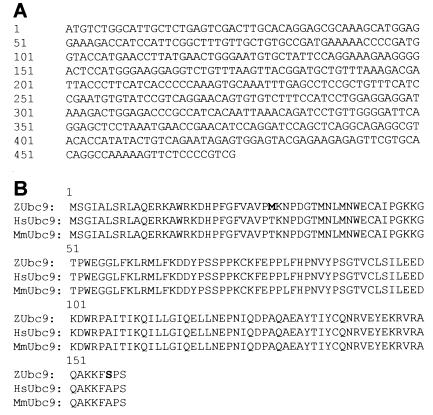
To identify proteins interacting with Vsx-1, we used the yeast two-hybrid system (41). The two-hybrid cDNA library was created from a 1-mo-old zebrafish and was designed to produce Gal-4 activation domain fusion proteins. pYVsx-1a encoded a fusion protein, which joined the LexA-binding domain to a region of goldfish Vsx-1 that encompassed the NLS, HD, CVC domain, and putative PEST site (21). By using this portion of Vsx-1 as the target, an initial screening of the library identified one clone that specifically interacted with the target. After sequencing, this clone (GenBank accession no. AF128240; Fig. 1A) showed highest homology [blast p(n) = 5.9e−57, high score = 435] to Ubc9, a SUMO-1-conjugating enzyme from mouse (42) and from human (43). The predicted amino acid sequence for zebrafish Ubc9 (zUbc9) differed from its mouse and human orthologs by only two amino acids (shaded; Fig. 1B). Additional two-hybrid library screenings replicated our initial finding and identified a single clone that was identical in size and sequence to the original zUbc9 clone. Screening the library with full-length Vsx-1 identified the same zUbc9 clone as well as an allelic variant of zUbc9 (zUbc9–2, GenBank accession no. AF332623). Experiments to test nonspecific interactions with other proteins (see Materials and Methods) demonstrated a specific association between Vsx-1 and Ubc9 in this system (data not shown).
Figure 1.
A region of goldfish Vsx-1 encoding the NLS-HD-CVC-PEST and full-length zVsx-1 was used to screen a zebrafish cDNA yeast two-hybrid library and detected an interaction with zUbc9. (A) zUbc9 cDNA sequence. (B) Amino acid comparison of zUbc9 to human and to mouse Ubc9 show differences at M29 and S156 (bolded).
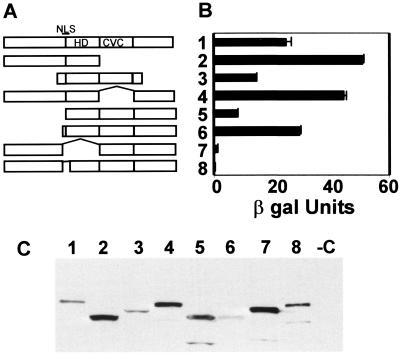
To identify sequences on Vsx-1 that interact with zUbc9, we determined whether Vsx-1 deletion constructs would bind to zUbc9 in the two-hybrid system. Deletion analysis showed that COOH-terminal amino acids of Vsx-1 are not required for interaction with zUbc9. Because the function of the CVC domain is unknown, we determined whether this domain contributed to the interaction. Results show that the CVC domain does not participate in Ubc9 binding (Fig. 2A) or in other interactions with proteins represented in the present cDNA library. Furthermore, in the yeast two-hybrid system, zUbc9 did not interact with the HD alone (data not shown). zUbc9 interacted strongly with the NH2 terminus and the HD. This interaction was eliminated solely by deleting the seven amino acids (QKRKKRR) that comprised the putative NLS region at the NH2 terminus of the homeodomain (Fig. 2A). Because the vector, pSTT91, contains an NLS within the LexA-binding domain (Y. Rhee, SUNY, Stony Brook, NY, unpublished data), this difference in the interaction of Vsx-1ΔNLS with zUbc9 should not be because of a failure of Vsx-1ΔNLS to enter the nucleus. Protein expression in the yeast was confirmed by Western blot analysis using the anti-LexA antibody (Fig. 2B).
Figure 2.
zUbc9 interacts with a NLS at the NH2 terminus of the homeodomain. (A) Vsx-1 deletion mutants: 1, zVsx-1(ORF); 2, zVsx-1ΔC-term; 3, goldfish Vsx-1 target; 4, zVsx-1ΔCVC; 5, zVsx-1ΔN-term1; 6, zVsx-1ΔN-term2; 7, zVsx-1ΔNLS/HD; and 8, zVsx-1ΔNLS. (B) Liquid culture assay using CPRG as substrate (CLONTECH) was used to calculate β-galactosidase activity. One unit of β-galactosidase equals the amount that hydrolyzes 1 μmol of CPRG to chlorophenol red and d-galactose per minute per cell. In the graph, the means (±SEM) are the result of samples processed in triplicate. (C) Anti-LexA antibody Western blot of yeast lysates shows expression of the LexA- binding domain fusion proteins noted in A.
Vsx-1 Is Not a Substrate for SUMO-1 Conjugation.
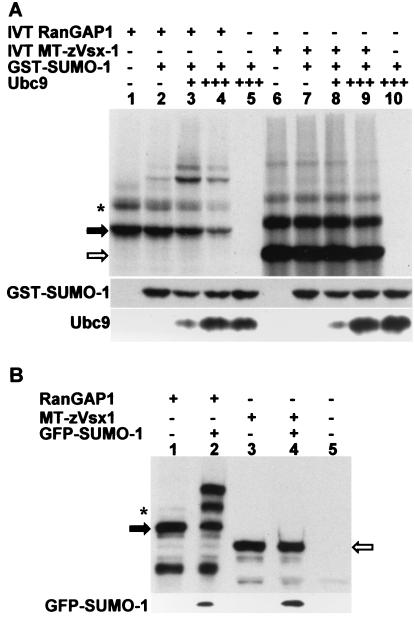
Interaction between Vsx-1 and zUbc9 suggests that Vsx-1 may be a target for SUMO-1 conjugation. Therefore, we determined whether SUMO-1 could be conjugated to Vsx-1 in vitro by using rabbit reticulocyte lysate, a system that efficiently sumoylates RanGAP1 (44). Fig. 3A, lane 1 shows that RanGAP1 is efficiently translated and appears as two bands, a 70-kDa unconjugated band and a 90-kDa sumoylated form (45–47). When GST-SUMO-1 is added to the reaction, a new higher molecular weight band appears that corresponds to the additional 25-kDa GST tag (lane 2). As expected, addition of recombinant Ubc9 to the reaction increased the intensity of the GST-SUMO-1-conjugated form of RanGAP1 (lane 3). Although expression of RanGAP1 is decreased in lane 4, additional Ubc9 increased the proportion of the 90-kDa form relative to the 70-kDa form. Unlike lanes containing RanGAP1, MT-Vsx-1 lanes display multiple high molecular weight bands [shown to be ubiquitin conjugates (21)]. Comparison of lanes 7 and 2 (Fig. 3A), shows that adding GST-SUMO-1 does not induce a size-shifted band. Addition of recombinant Ubc9 to the transcription/translation reaction (lanes 8 and 9) failed to yield GST-SUMO-1-conjugated MT-Vsx-1. Immunoblot analysis confirms the presence of GST-SUMO-1 and Ubc9 proteins in the reactions (Fig. 3A). These experiments show that Vsx-1 was not conjugated with SUMO-1 in vitro and that Ubc9 did not induce SUMO-1 modification of Vsx-1.
Figure 3.
Zebrafish Vsx-1 is not a substrate for SUMO-1 conjugation in vitro or in COS-7 cells. Open and closed arrows indicate unconjugated MT-zVsx-1 and RanGAP1, respectively. Asterisk shows RanGAP1 conjugated to endogenous SUMO-1. (A) Xenopus RanGAP1 (lanes 1–4) or MT-zVsx-1 (lanes 6–9) were in vitro translated and labeled with [35S]Met/[35S]Cys. Recombinant GST-SUMO-1 and Ubc9 were added as indicated. Ubc9 (3×) was added to lanes 4, 5, 9, and 10. Western blots using anti-SUMO-1 or anti-hsUbc9 antibodies show relative levels of GST-SUMO-1 and Ubc9. (B) COS-7 cells were transfected with expression plasmids encoding RanGAP1, MT-zVsx-1, and GFP-SUMO-1. Cell lysates were immunoblotted with anti-RanGAP1 antibody (lanes 1 and 2 in upper gel), 9e10 antibody (lanes 3–5 in upper gel), or anti-GFP antibody (lower gel). Identical results were observed when GFP-SUMO-2 was substituted for GFP-SUMO-1.
To determine whether Vsx-1 was conjugated to SUMO-1 in live cells, we cotransfected COS-7 cells with pEGFPN1+ SUMO-1 and either pCS2MT+zVsx-1 or pCI+RanGAP1. As observed in vitro, immunoblotting of cell lysates did not show GFP-SUMO-1 modification of Vsx-1 (Fig. 3B). To determine whether Vsx-1 was conjugated to an alternate SUMO protein, we repeated the COS-7 transfection experiment by using GFP-SUMO-2 instead of GFP-SUMO-1. In contrast to RanGAP1, results show that Vsx-1 was not conjugated to SUMO-2 (data not shown). Therefore, Vsx-1 is not a substrate for sumoylation in COS-7 cells.
Vsx-1 Localizes to the Nucleus.
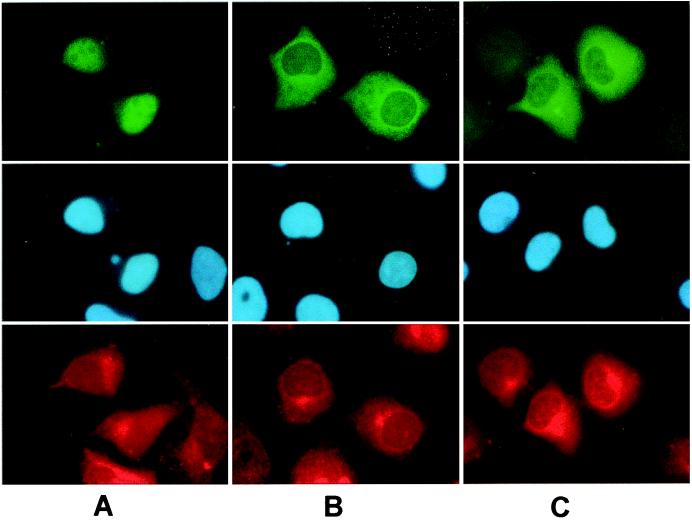
Human SW-13 cells were propagated on glass cover slips and transfected with pCS2MT+zVsx-1. Immunocytochemistry with 9e10 antibody and a fluorescent secondary antibody showed that MT-Vsx-1 localized diffusely in the nucleus, but was excluded from presumptive nucleoli (Fig. 4).
Figure 4.
MT-zVsx-1 localizes to the nucleus. SW-13 cells were transfected with plasmids expressing MT-zVsx-1 (A), MT-zVsx-1ΔNLS/HD (B), or MT-zVsx-1ΔNLS (C). (Top) Indirect immunofluorescence using the 9e10 antibody. (Middle) Hoechst 33258 staining of nuclei. (Bottom) Indirect immunofluorescence using anti-calnexin antibody 8211–1. MT-zVsx-1ΔNLS/HD and MT-zVsx-1ΔNLS show diffuse cytoplasmic staining with a slight increase around nuclei and endoplasmic reticulum.
The Putative NLS Is Required for Vsx-1 Nuclear Import.
Although the yeast two-hybrid data (Fig. 2A) suggested that Ubc9 binds to a putative NLS (QKRKKRR) amino acid sequence of Vsx-1, we hypothesized that deletions of the NLS/HD or the NLS would prevent Vsx-1 from entering the nucleus of SW-13 cells. SW-13 cells were transfected with plasmids pCS2MT+zVsx-1ΔNLS/HD or zVsx-1ΔNLS. They were probed 24 h later with anti-calnexin and with the 9e10 antibody to the MT, followed by nuclear staining with Hoechst 33258. Both of these deletions failed to enter the nucleus and were distributed evenly throughout the cytoplasm with slight increases near the nuclear membrane and the endoplasmic reticulum (Fig. 4). This result shows that the NLS is functional and is required for the nuclear import of Vsx-1.
Ubc9 Is Required for Nuclear Localization of Vsx-1.
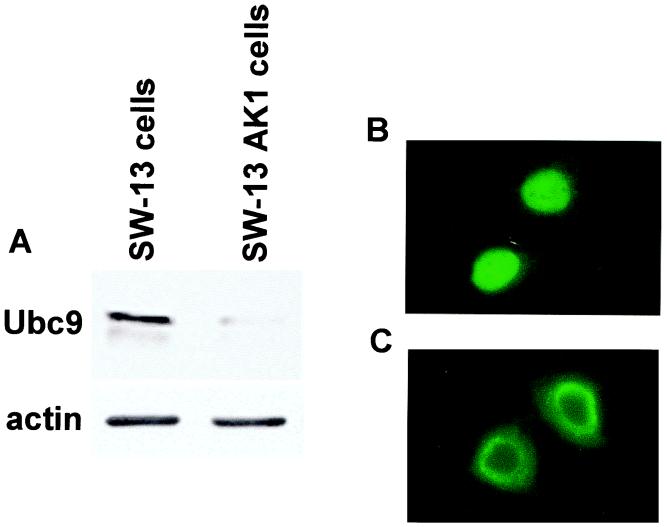
To determine whether Ubc9 influenced Vsx-1 localization, we modified SW-13 cells with a stable transfection of human antisense Ubc9 (A.S.hUbc9). Compared to SW-13 cells, these SW-13 AK1 cells expressed decreased levels of endogenous Ubc9 (Fig. 5A). Morphologically, the SW-13 AK1 cells appeared normal but grew more slowly than the SW-13 cells. As shown above, MT-zVsx-1 localized to the nucleus in SW-13 cells (Fig. 5B). However when expressed in SW-13 AK1 cells, MT-zVsx-1 accumulated in a perinuclear distribution, suggesting that Vsx-1 was unable to enter the nucleus (Fig. 5C). This result suggested a role for Ubc9 in the nuclear import of Vsx-1.
Figure 5.
Ubc9 is required for nuclear localization of zVsx-1. (A) Western blot of fractionated cell lysates from SW-13 cells and SW-13 AK1 cells after probing with anti-hsUbc9. SW-13 AK1 cells express A.S.hsUbc9 and show decreased levels of endogenous Ubc9. (B and C) Indirect immunofluorescence using the 9e10 antibody in MT-zVsx-1-transfected SW-13 cells (B) and SW-13 AK1 cells (C).
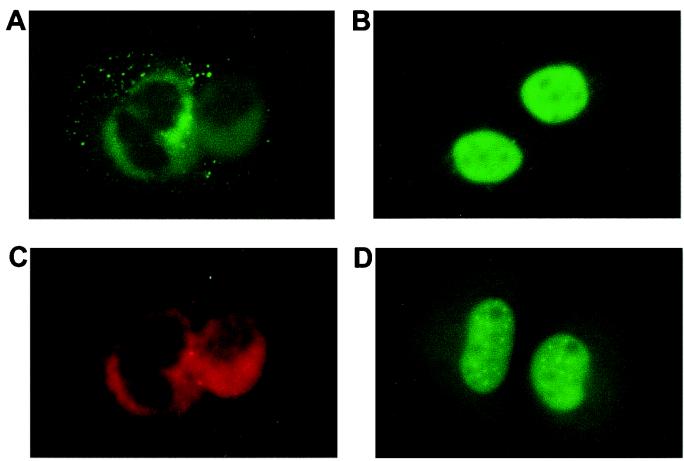
To confirm the role of Ubc9 in the nuclear import of Vsx-1, we performed the following rescue experiment. By using the SW-13 AK1 cells that express low levels of endogenous Ubc9, mammalian plasmids expressing Ubc9 (pcDNA3.1+zUbc9 or pCMV-Flag-Ubc9) were cotransfected with pCS2MT+zVsx-1. In these experiments, Vsx-1 localized to the nucleus (Fig. 6B). As a parallel control, MT-zVsx-1 localized around the nucleus when cotransfected with the vector pcDNA3.1 (Fig. 6A). Thus, we conclude that Ubc9 is required for nuclear localization of Vsx-1 in cell culture and may have a similar role during retinogenesis.
Figure 6.
Ubc9 restores nuclear localization of zVsx-1 in SW-13 AK1 cells. (A) Indirect immunofluorescence using the 9e10 antibody in SW-13 AK1 cells cotransfected with MT-zVsx-1 and vector. (B) Indirect immunofluorescence using the 9e10 antibody in SW-13 AK1 cells cotransfected with MT-zVsx-1 and Flag-Ubc9. (C) Indirect immunofluorescence with anti-calnexin antibody in SW-13 AK1 cells labels the endoplasmic reticulum. (D) STAT1-NLS-GFP localization in fixed SW-13 AK1 cells.
To determine the specific nonnuclear localization of Vsx-1 in SW13 AK1 cells, we costained these cells with anti-calnexin antibody to label the endoplasmic reticulum and with Lens lectin (data not shown) to label the Golgi apparatus. In all experiments, Vsx-1 colocalized with calnexin, suggesting that Vsx-1 is retained at the ER membrane in cells expressing low levels of Ubc9 (Fig. 6C).
We next transfected SW-13 AK1 cells with an expression plasmid encoding NLS-tagged STAT1-GFP (pEGFPN1+STAT1+NLS+GFP) to determine whether decreased concentrations of Ubc9 blocked nuclear import, in general. STAT1-NLS-GFP fusion protein normally localizes to the nucleus and concentrates in nuclear dots (48). In SW-13 AK1 cells, NLS-STAT1-GFP exhibited a normal nuclear staining (Fig. 6D). Because nuclear import is otherwise normal in these cells, we conclude that the effects of zUbc9 on nuclear localization are specific to Vsx-1.
Vsx-1 Localizes to the Nucleus Without SUMO-1 Conjugation.
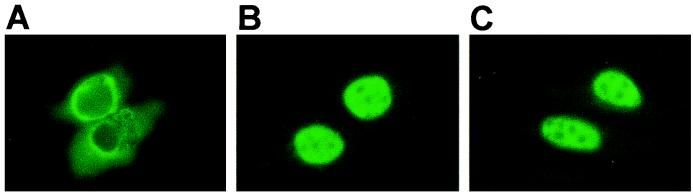
To evaluate whether SUMO-1 is involved in the nuclear import of MT-Vsx-1, we tested whether the addition of pBK-CMV-UBC9 (C93S) to SW-13 AK1 cells would normalize the localization of Vsx-1. Because Ubc9 C93S is mutated at the active site cysteine and does not form a thioester bond with the C terminus of SUMO-1 (37), it is unable to catalyze sumoylation. When hUbc9 (C93S) and MT-Vsx-1 were cotransfected into SW-13 AK1 cells, MT-Vsx-1 localized to the nucleus with high efficiency (Fig. 7). This result suggests that SUMO-1 is not involved in Vsx-1 nuclear localization. Furthermore, it suggests that Ubc9 mediates nuclear localization of Vsx-1 through a mechanism other than sumoylation.
Figure 7.
Ubc9 C93S also restores zVsx-1 nuclear localization. Indirect immunofluorescence with 9e10 antibody of SW-13 AK1 cells transfected with MT-zVsx-1 and vector control (A), MT-zVsx-1 and Flag-Ubc9 (B), and MT-zVsx-1 and Ubc9C93S (C).
Discussion
Our two-hybrid screens using the NLS-HD-CVC-PEST region of goldfish Vsx-1 uncovered only one specific interacting protein, zUbc9. Subsequent screenings using full-length zebrafish Vsx-1 showed interaction only with zUbc9 and an allelic variant, zUbc9–2. Because many mammalian chromosomal segments occur as duplicates in the zebrafish genome (49), it is not surprising that we isolated a second Ubc9 with our full-length zVsx-1 screen. Although these two clones differ significantly at the nucleotide level, zUbc9–2 and our original zUbc9 clone differ by only two amino acids at the protein level. At this time, we do not know whether zUbc9 and zUbc9–2 serve differing, complementary or overlapping functions.
The amino acid sequence of zUbc9 is also quite similar to those of mouse and of human Ubc9 (42, 43), differing by only 2 amino acids (Fig. 1B). M29 (threonine in HsUbc9) resides in the first β-strand. The second amino acid change, S156 (alanine in HsUbc9) resides at the COOH terminus. Both M29 and S156 are distal to the active site and uncharged (50). Consequently, these amino acid changes are unlikely to alter the target protein binding or catalytic activity of zUbc9 across species.
Although the two-hybrid library used in this study represented mRNA from a 1-mo-old zebrafish, we would anticipate finding the same interaction in a library from embryonic tissue. The retina of teleosts, such as zebrafish, display continuous growth throughout life (51). In mature teleosts, this embryonic-like growth persists at the retinal margin where cell proliferation and differentiation occur (52, 53). Consequently, in regions of retinal growth, we would expect the same proteins to regulate Vsx-1.
In screening the two-hybrid library, a major impetus was to determine the function of the CVC domain, which was, and remains, unknown. Although a goal was to test whether the CVC domain functions in protein–protein interaction, our results suggest that this is not the case. However, because the CVC domain is adjacent to the DNA-binding homeodomain in paired-like:CVC homeobox proteins, it most likely mediates the specificity or strength of DNA binding and essentially replaces the function of the paired box seen in paired, but not paired-like:CVC homeobox proteins (54).
In addition to showing that zUbc9 and Vsx-1 interact in a yeast two-hybrid system, we also have demonstrated a functional requirement for Ubc9 in the nuclear localization of Vsx-1 in cell culture. In the SW13 AK1 cells, which have reduced levels of endogenous Ubc9, the nuclear localization of Vsx-1 is disrupted. However, Vsx-1 will again localize to the nucleus when levels of Ubc9 are normalized by cotransfection with Ubc9-expressing plasmids.
Through its SUMO-conjugating activity, Ubc9 is known to mediate the nuclear localization of the Rel family protein, Dorsal (30) and the localization of RanGAP1 to the nuclear pore complex (25). However, it is clear that SUMO-1 conjugation does not facilitate protein targeting of all sumoylated proteins. For example, localization of the Ubc9-interacting proteins, Sp100 (55) and IE2-p86 (31), are not dependant on their sumoylation status. Furthermore, fewer than one-half of the known Ubc9-interacting proteins have been shown to be substrates for sumoylation (56).
Several lines of direct and indirect evidence suggest that Vsx-1 is not sumoylated by Ubc9. In our yeast two-hybrid screens, we did not isolate SUMO-1 from our library as interacting with zVsx-1. This result seems unlikely to be caused by difficulties in detecting SUMO-1 protein interactions because SUMO-1 has been isolated from other yeast two-hybrid screens (e.g., see refs. 57 and 58). In addition, exact SUMO-1 conjugation consensus sites (28, 59) are not found in Vsx-1. More directly, we now show that Vsx-1 is not conjugated with SUMO-1 either in vitro or in COS-7 cells. Furthermore, Ubc9 C93S rescues the nuclear localization of Vsx-1 in cells that have reduced Ubc9 levels, even though it is incapable of conjugating SUMO-1. Together these results suggest that Ubc9 mediates the nuclear localization of Vsx-1 without catalyzing the covalent modification of Vsx-1 by SUMO-1. For proteins that are sumoylated within the nucleus, such nonenzymatic mediation of nuclear import may facilitate eventual SUMO conjugation because the Ubc9-substrate is imported in an already complexed form.
Ubc9 may be capable of binding both positively and negatively charged regions on target proteins. Because the β sheet domain forms a patchy positively charged noncatalytic face (50), this distinct electrostatic potential has been postulated to account for the interaction of Ubc9 with a variety of proteins (50, 56, 60). However, Ubc9 also interacts with basic amino acid regions. Putative electrostatic interactions between Ubc9 and these basic amino acids would likely occur at negatively charged patches of Ubc9 rather than at the positively charged face. For example, Ubc9 binds the basic region of the bipartite NLS of the androgen receptor (38). With Vsx-1, our deletion analysis shows that Ubc9 interacts with a basic NLS located at the NH2 terminus of the Vsx-1 homeodomain.
For proteins containing a “classical” monopartite NLS, such as the simian virus 40 large T antigen NLS (PKKKRKV) (61), nuclear import is facilitated by the heterodimeric complex of importin α and β (62, 63). Because Ubc9 binds such an NLS sequence, we propose three potential mechanisms for the role of Ubc9 in nuclear import: (i) Ubc9 may bind the NLS, subsuming the role of the importins; (ii) Ubc9 may present Vsx-1 to the importin complex, and (iii) the importins may present proteins to Ubc9 and, thereby, permit Ubc9 to enter the nucleus with the target. Regardless of the specific mechanism, however, we propose that Ubc9 mediates nuclear import of Vsx-1 by interacting with the NLS signal of Vsx-1. In so doing, Ubc9 may play a role in the subcellular localization of this protein that is nonenzymatic and distinct from SUMO-1 conjugation. Further in vivo experiments will determine whether Ubc9 is an integral component of the system that regulates Vsx-1 during retinal development and growth.
Acknowledgments
We thank Drs. Laura Fochtmann and Aaron Nieman for critical review of this manuscript. We thank Drs. Rolf Sternglanz, Michael A. Frohman, and William Asch for helpful discussions. We thank Dr. Hisato Saitoh for reagents and advice. This research was supported by Grant EY05212 from the National Institutes of Health (to N.S.).
Abbreviations
- Vsx-1
visual system homeobox-1
- zUbc9
zebrafish Ubc9
- CVC
Chx10/Vsx/Ceh10
- MT
Myc tag
- SUMO
small ubiquitin-like modifier
- GST
glutathione S-transferase
- A.S.hUbc9
human antisense Ubc9
- HD
homeo domain
- PEST
proline-glutamate-serine-threonine
Footnotes
References
- 1.Beebe D C. Invest Ophthalmol Visual Sci. 1994;35:2897–2900. [PubMed] [Google Scholar]
- 2.Cvekl A, Piatigorsky J. BioEssays. 1996;18:621–630. doi: 10.1002/bies.950180805. [DOI] [PubMed] [Google Scholar]
- 3.Freund C, Horsford D, McInnes R. Hum Mol Genet. 1996;5:1471–1488. doi: 10.1093/hmg/5.supplement_1.1471. [DOI] [PubMed] [Google Scholar]
- 4.Zuber M E, Perron M, Philpott A, Bang A, Harris W A. Cell. 1999;98:341–352. doi: 10.1016/s0092-8674(00)81963-7. [DOI] [PubMed] [Google Scholar]
- 5.Barbieri A M, Lupo G, Bulfone A, Andreazzoli M, Mariani M, Fougerousse F, Consalez G G, Borsani G, Beckmann J S, Barsacchi G, et al. Proc Natl Acad Sci USA. 1999;96:10729–10734. doi: 10.1073/pnas.96.19.10729. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Furukawa T, Kozak C A, Cepko C L. Proc Natl Acad Sci USA. 1997;94:3088–3093. doi: 10.1073/pnas.94.7.3088. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Mathers P H, Grinberg A, Mahon K A, Jamrich M. Nature (London) 1997;387:603–607. doi: 10.1038/42475. [DOI] [PubMed] [Google Scholar]
- 8.Levine E M, Hitchcock P F, Glasgow E, Schechter N. J Comp Neurol. 1994;348:596–606. doi: 10.1002/cne.903480409. [DOI] [PubMed] [Google Scholar]
- 9.Levine E M, Schechter N. Proc Natl Acad Sci USA. 1993;90:2729–2733. doi: 10.1073/pnas.90.7.2729. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Levine E M, Passini M, Hitchcock P F, Glasgow E, Schechter N. J Comp Neurol. 1997;387:439–448. [PubMed] [Google Scholar]
- 11.Passini M A, Levine E M, Canger A K, Raymond P A, Schechter N. J Comp Neurol. 1997;388:495–505. doi: 10.1002/(sici)1096-9861(19971124)388:3<495::aid-cne11>3.0.co;2-l. [DOI] [PubMed] [Google Scholar]
- 12.Passini M A, Kurtzman A L, Canger A K, Asch W S, Wray G A, Raymond P A, Schechter N. Dev Genet (Amsterdam) 1998;23:128–141. doi: 10.1002/(SICI)1520-6408(1998)23:2<128::AID-DVG5>3.0.CO;2-8. [DOI] [PubMed] [Google Scholar]
- 13.Svendsen P C, McGhee J D. Development (Cambridge, UK) 1995;121:1253–1262. doi: 10.1242/dev.121.5.1253. [DOI] [PubMed] [Google Scholar]
- 14.Chen C M A, Cepko C L. Mech Dev. 2000;90:293–297. doi: 10.1016/s0925-4773(99)00251-8. [DOI] [PubMed] [Google Scholar]
- 15.Semina E V, Mintz-Hittner H A, Murray J C. Genomics. 2000;63:289–293. doi: 10.1006/geno.1999.6093. [DOI] [PubMed] [Google Scholar]
- 16.Hayashi T, Huang J, Deeb S S. Genomics. 2000;67:128–139. doi: 10.1006/geno.2000.6248. [DOI] [PubMed] [Google Scholar]
- 17.Burmeister M, Novak J, Liang M Y, Basu S, Ploder L, Hawes N L, Vidgen D, Hoover F, Goldman D, Kalnins, et al. Nat Genet. 1996;12:376–384. doi: 10.1038/ng0496-376. [DOI] [PubMed] [Google Scholar]
- 18.Passini M A, Raymond P A, Schechter N. Brain Res Dev Brain Res. 1998;109:129–135. doi: 10.1016/s0165-3806(98)00069-8. [DOI] [PubMed] [Google Scholar]
- 19.Ferda Percin E, Ploder L A, Yu J J, Arici K, Horsford D J, Rutherford A, Bapat B, Cox D W, Duncan A M, Kalnins V I, et al. Nat Genet. 2000;25:397–401. doi: 10.1038/78071. [DOI] [PubMed] [Google Scholar]
- 20.Bone-Larson C, Basu S, Radel J D, Liang M Y, Perozek T, Kapousta-Bruneau N, Green D G, Burmeister M, Hankin M H. J Neurobiol. 2000;42:232–247. doi: 10.1002/(sici)1097-4695(20000205)42:2<232::aid-neu7>3.0.co;2-4. [DOI] [PubMed] [Google Scholar]
- 21.Kurtzman A L, Gregori L, Haas A L, Schechter N. J Neurochem. 2000;75:48–55. doi: 10.1046/j.1471-4159.2000.0750048.x. [DOI] [PubMed] [Google Scholar]
- 22.Johnson E S, Blobel G. J Biol Chem. 1997;272:26799–26802. doi: 10.1074/jbc.272.43.26799. [DOI] [PubMed] [Google Scholar]
- 23.Desterro J M P, Thomson J, Hay R T. FEBS Lett. 1997;417:297–300. doi: 10.1016/s0014-5793(97)01305-7. [DOI] [PubMed] [Google Scholar]
- 24.Gong L, Kamitani T, Fujise K, Caskey L S, Yeh E T. J Biol Chem. 1997;272:28198–28201. doi: 10.1074/jbc.272.45.28198. [DOI] [PubMed] [Google Scholar]
- 25.Mahajan R, Gerace L, Melchior F. J Cell Biol. 1998;140:259–270. doi: 10.1083/jcb.140.2.259. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Müller S, Matunis M J, Dejean A. EMBO J. 1998;17:61–70. doi: 10.1093/emboj/17.1.61. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Chakrabarti S R, Sood R, Nandi S, Nucifora G. Proc Natl Acad Sci USA. 2000;97:13281–13285. doi: 10.1073/pnas.240315897. . (First Published November 14, 2000; 10.1073/pnas.240315897) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Desterro J M P, Rodriguez M S, Hay R T. Mol Cell. 1998;2:233–239. doi: 10.1016/s1097-2765(00)80133-1. [DOI] [PubMed] [Google Scholar]
- 29.Buschmann T, Fuchs S Y, Lee C G, Pan Z Q, Ronai Z. Cell. 2000;101:753–762. doi: 10.1016/s0092-8674(00)80887-9. [DOI] [PubMed] [Google Scholar]
- 30.Bhaskar V, Valentine S A, Courey A J. J Biol Chem. 2000;275:4033–4040. doi: 10.1074/jbc.275.6.4033. [DOI] [PubMed] [Google Scholar]
- 31.Hofmann H, Floess S, Stamminger T. J Virol. 2000;74:2510–2524. doi: 10.1128/jvi.74.6.2510-2524.2000. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Kim Y H, Choi C Y, Kim Y. Proc Natl Acad Sci USA. 1999;96:12350–12355. doi: 10.1073/pnas.96.22.12350. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Rodriguez M S, Desterro J M P, Lain S, Midgley C A, Lane D P, Hay R T. EMBO J. 1999;18:6455–6461. doi: 10.1093/emboj/18.22.6455. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Poukka H, Karvonen U, Janne O A, Palvimo J J. Proc Natl Acad Sci USA. 2000;97:14145–14150. doi: 10.1073/pnas.97.26.14145. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Müller S, Berger M, Lehembre F, Seeler J S, Haupt Y, Dejean A. J Biol Chem. 2000;275:13321–13329. doi: 10.1074/jbc.275.18.13321. [DOI] [PubMed] [Google Scholar]
- 36.Hahn S L, Criqui P, Wasylyk B. Oncogene. 1997;15:1489–1495. doi: 10.1038/sj.onc.1201301. [DOI] [PubMed] [Google Scholar]
- 37.Chakrabarti S R, Sood R, Ganguly S, Bohlander S, Shen Z Y, Nucifora G. Proc Natl Acad Sci USA. 1999;96:7467–7472. doi: 10.1073/pnas.96.13.7467. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Poukka H, Aarnisalo P, Karvonen U, Palvimo J J, Janne O A. J Biol Chem. 1999;274:19441–19446. doi: 10.1074/jbc.274.27.19441. [DOI] [PubMed] [Google Scholar]
- 39.Hollenberg S M, Sternglanz R, Cheng P F, Weintraub H. Mol Cell Biol. 1995;15:3813–3822. doi: 10.1128/mcb.15.7.3813. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Asch W S, Schechter N. J Neurochem. 2000;75:1475–1486. doi: 10.1046/j.1471-4159.2000.0751475.x. [DOI] [PubMed] [Google Scholar]
- 41.Chien C T, Bartel P L, Sternglanz R, Fields S. Proc Natl Acad Sci USA. 1991;88:9578–9582. doi: 10.1073/pnas.88.21.9578. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Scott L M, Mueller L, Collins S J. Blood. 1996;88:2517–2530. [PubMed] [Google Scholar]
- 43.Yasugi T, Howley P M. Nucleic Acids Res. 1996;24:2005–2010. doi: 10.1093/nar/24.11.2005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Saitoh H, Sparrow D B, Shiomi T, Pu R T, Nishimoto T, Mohun T J, Dasso M. Curr Biol. 1998;8:121–124. doi: 10.1016/s0960-9822(98)70044-2. [DOI] [PubMed] [Google Scholar]
- 45.Matunis M J, Wu J, Blobel G. J Cell Biol. 1998;140:499–509. doi: 10.1083/jcb.140.3.499. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Matunis M J, Coutavas E, Blobel G. J Cell Biol. 1996;135:1457–1470. doi: 10.1083/jcb.135.6.1457. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Mahajan R, Delphin C, Guan T L. Cell. 1997;88:97–107. doi: 10.1016/s0092-8674(00)81862-0. [DOI] [PubMed] [Google Scholar]
- 48.McBride K M, McDonald C, Reich N C. EMBO J. 2000;19:6196–6206. doi: 10.1093/emboj/19.22.6196. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Postlethwait J H, Yan Y L, Gates M A, Horne S, Amores A, Brownlie A, Donovan A, Egan E S, Force A, Gong Z, et al. Nat Genet. 1998;18:345–349. doi: 10.1038/ng0498-345. [DOI] [PubMed] [Google Scholar]
- 50.Tong H, Hateboer G, Perrakis A, Bernards R, Sixma T K. J Biol Chem. 1997;272:21381–21387. doi: 10.1074/jbc.272.34.21381. [DOI] [PubMed] [Google Scholar]
- 51.Marcus R C, Delaney C L, Easter S S., Jr Visual Neurosci. 1999;16:417–424. doi: 10.1017/s095252389916303x. [DOI] [PubMed] [Google Scholar]
- 52.Johns P R, Easter S S., Jr J Comp Neurol. 1977;176:331–341. doi: 10.1002/cne.901760303. [DOI] [PubMed] [Google Scholar]
- 53.Julian D, Ennis K, Korenbrot J I. J Comp Neurol. 1998;394:271–282. [PubMed] [Google Scholar]
- 54.Jun S, Desplan C. Development (Cambridge, UK) 1996;122:2639–2650. doi: 10.1242/dev.122.9.2639. [DOI] [PubMed] [Google Scholar]
- 55.Sternsdorf T, Jensen K, Reich B, Will H. J Biol Chem. 1999;274:12555–12566. doi: 10.1074/jbc.274.18.12555. [DOI] [PubMed] [Google Scholar]
- 56.Yeh E T H, Gong L M, Kamitani T. Gene. 2000;248:1–14. doi: 10.1016/s0378-1119(00)00139-6. [DOI] [PubMed] [Google Scholar]
- 57.Minty A, Dumont X, Kaghad M, Caput D. J Biol Chem. 2000;275:36316–36323. doi: 10.1074/jbc.M004293200. [DOI] [PubMed] [Google Scholar]
- 58.Boddy M N, Howe K, Etkin L D, Solomon E, Freemont P S. Oncogene. 1996;13:971–982. [PubMed] [Google Scholar]
- 59.Rodriguez M S, Desterro J M P, Lain S, Lane D P, Hay R T. Mol Cell Biol. 2000;20:8458–8467. doi: 10.1128/mcb.20.22.8458-8467.2000. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60.Giraud M F, Desterro J M, Naismith J H. Acta Crystallogr D. 1998;54:891–898. doi: 10.1107/s0907444998002480. [DOI] [PubMed] [Google Scholar]
- 61.Wychowski C, Benichou D, Girard M. EMBO J. 1986;5:2569–2576. doi: 10.1002/j.1460-2075.1986.tb04536.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 62.Weis K. Trends Biochem Sci. 1998;23:185–189. doi: 10.1016/s0968-0004(98)01204-3. [DOI] [PubMed] [Google Scholar]
- 63.Nakielny S, Dreyfuss G. Cell. 1999;99:677–690. doi: 10.1016/s0092-8674(00)81666-9. [DOI] [PubMed] [Google Scholar]
- 64.Clontech. Yeast Protocols Handbook. Palo Alto, CA: Clontech; 2000. p. 66. [Google Scholar]