Abstract
Caenorhabditis elegans has been used for over a decade to characterize signaling cascades controlling innate immune responses. However, what initiates these responses in the worm has remained elusive. To gain a better understanding of the initiating events we delineated genome-wide immune responses to the bacterial pathogen Pseudomonas aeruginosa in worms heavily-colonized by the pathogen versus worms visibly not colonized. We found that infection responses in both groups were identical, suggesting that immune responses were not correlated with colonization and its associated damage. Quantitative RT-PCR measurements further showed that pathogen secreted factors were not able to induce an immune response, but exposure to a non-pathogenic Pseudomonas species was. These findings raise the possibility that the C.elegans immune response is initiated by recognition of microbe-associated molecular patterns. In the absence of orthologs of known pattern recognition receptors, C. elegans may rely on novel mechanisms, thus holding the potential to advance our understanding of evolutionarily conserved strategies for pathogen recognition.
Introduction
The soil nematode Caenorhabditis elegans has been used for over a decade to study host-pathogen interactions. Such studies provided detailed information on pathogen-specific innate immune responses, and the signal transduction pathways and transcription factors that activated them (reviewed in [1]). However, what initiates innate immune responses in the worm remains unknown.
In vertebrates, innate immune responses are initiated mainly by recognition of pathogen-associated molecular patterns (PAMPs), such as lipopolysaccharide, peptidoglycan, or microbially-modified nucleic acids; PAMPs are occasionally referred to as MAMPs–microbe-associated molecular patterns, to acknowledge their existence in non-pathogenic microbes. In addition, recognition of damage-associated molecular patterns (DAMPs), such as ATP and monsodium urate crystals, can enhance activation by PAMPs, but also may be sufficient for initiation of innate immune responses [2]. The pattern recognition receptors (PRRs) responsible for recognizing PAMPs or DAMPs include members of several protein families including the Toll/Toll-like receptors (TLRs), NOD-like receptors (NLRs) and RIG-I-like nucleotide recognition receptors (RLRs) [3]. Innate immune recognition is conserved through evolution, both in terms of its general strategy as well as the proteins involved: The first TLR, Toll, originally identified as essential for Drosophila's development, was subsequently shown to also have a crucial role in innate immunity [4], [5], NLRs take part in antimicrobial defenses both in animals and in plants [6] and a RIG-I homolog was recently shown to function in C. elegans anti-viral defenses [7]. Although C. elegans mounts specific responses to infections with different pathogenic bacteria, manifested in a robust gene induction [8], it is yet unknown how it discriminates between different bacteria. C. elegans has one Toll homolog gene, tol-1, which appears to be is largely dispensable for immune protection ([9], [10], but see [11])).; Additionally, FSHR-1, a heterotrimeric G protein with leucine-rich repeats (a motif shared among all vertebrate TLRs and NLRs), is necessary for immune protection, but is equally protective against Gram-negative and–positive pathogens, suggesting that it might not function in the proximal events of pathogen recognition. [12]. The inability to identify C. elegans PRRs based on orthology raises the possibility that C. elegans may use novel modes of pathogen recognition. Alternatively, C. elegans may respond to pathogen-specific damage caused in the course of infection. To discriminate between these two possibilities we used an infection model of C. elegans intestinal colonization by Pseudomonas aeruginosa [13].
Methods
Strains
Worms were of the N2 wild-type strain. Bacterial strains included E. coli OP50-1, the clinical isolate Pseudomonas aeruginosa strain PA14, or a PA14 derivative expressing GFP off a stable plasmid [13]. Pseudomonas mendocina, a non-pathogenic environmental Pseudomonad was isolated from worms grown on soil (Montalvo-Katz, unpublished).
Worm infection and sorting
Synchronized populations of wild-type worms, grown under standard conditions, were transferred at day two of adulthood either to E. coli, or to PA14-GFP. After eighteen hours, worms presented a wide-range of colonization reflected by accumulation of GFP-expressing bacteria in their intestine. Colonized (intensely green) and non-colonized (dark) worms were separated, either using the COPASTM BIOSORT worm sorter (Union Biometrica; two experiments) or by picking >100 worms of each group under a fluorescent stereoscope (one experiment, serving as a control for the automatic sorting).
Testing the effects of the P. aeruginosa secretome on C. elegans immune responses
Three approaches were employed for testing potential contribution of P. aeruginosa secreted factors independently of the secreting bacteria; the results presented were obtained using the first and third of those: in the first approach, P. aeruginosa was grown on a 0.2 µM mixed cellulose esters filter (Millipore) placed on modified NGM plates at 37°C for 24 hours, at the end of which the underlying agar was blue due to secreted pyocyanin; the filter (containing bacteria) was then removed, E. coli added as food, and worms laid on plates; since filters may absorb some of the secreted molecules, particularly proteins, the second approach involved exposing worms to supernatants obtained from saturated P. aeurigonsa cultures; supernatants were cleared of bacteria by repeated centrifugation/transfer (microcentrifuge, 14 K RPM, 10 minutes each, 8 times), and following testing for absence of bacteria, 100 µl supernatant was added to lawns of dead E. coli; to account for molecules possibly secreted only on solid medium, in the third approach we submerged P. aeruginosa lawns in 1.2 ml M 9 solution, let it sit for an hour at room temperature to allow secreted factors to diffuse out of the agar and lawn, then collected supernatant, removed bacteria by repeated centrifugation as above, and added to lawns of dead E. coli. All three methods resulted in the same results.
RNA extraction, microarrays and qRT-PCR
RNA was extracted from 100–700 worms per group/time-point using Trizol (Invitrogen). For microarray experiments, RNA was amplified using the MessageAmp™ II aRNA Amplification Kit (Ambion), labeled with the ULS™ aRNA Labeling Kit (Kreatech) and co-hybridized to Epoxy (Corning) microarrays spotted with 60-mer oligonucleotides (Washington University Genome Sequencing Center) with a similarly amplified and labeled reference RNA sample [14].
For (q)RT-PCR measurements, gene-specific threshold cycle (Ct) values were normalized to the respective actin values, and presented as fold change over the time = 0 point.
PCR Primers
pan-actin forward TCGGTATGGGACAGAAGGAC
pan-actin reverse CATCCCAGTTGGTGACGATA
F55G11.2 forward TGGTTCTCCAGACGTGTTCA
F55G11.2 reverse CAGCCTTGCCTTTACTGACA
lys-2 forward CCAATATCAAGCTGGCAAGG
lys-2 reverse GTTGGATTGTTTGGCCAGTT
Statistical analysis
Gene expression profiles obtained with microarrays were analyzed by a multi-class t-test using Significance Analysis of Microarrays (SAM; [15]), implemented as part of the TMEV software package. Based on T statistics the test retrieves genes with a T value above a cutoff score estimated to give the desired false discovery rate (selected to be 10%). This analysis was used to identify C. elegans genes differentially expressed in either one of the three analyzed groups: worms exposed to E. coli, worms exposed and colonized by P. aeruginosa PA14-GFP, or worms exposed to PA14-GFP, but not colonized. Since no difference was observed between expression profiles in worms collected manually or with the worm sorter, data from the three repeats for each of the three experimental groups were pooled. The resulting list of genes responding to PA14 contained 359 genes.
Results
The PA14 strain of Pseudomonas aeruginosa, made to express GFP (PA14-GFP from here on), can be followed as it colonizes the worm intestine, leading to death within three days. This colonization requires live bacteria and depends on bacterial regulators of virulence [16]. Furthermore, once reaching a significantly visible level of colonization (e.g. following 18 hours of exposure), most worms remain colonized, even when transferred to E. coli (65% of N = 96). This suggests active interactions between the pathogen and its host, which enable the pathogen to persist in most cases. When a genetically-homogenous and age-synchronized population of worms is exposed to PA14-GFP, significant heterogeneity is seen in colonization by the pathogen (Fig. 1A). We reasoned that if colonization-associated damage elicited immune responses in C. elegans, then immune responses would be correlated to the degree of colonization. To test this, we chose the extreme case of comparing significantly colonized worms with worms that were not visibly colonized. We exposed wild-type worms to PA14-GFP under standard conditions (or to E. coli control) and following 18 hours, separated significantly colonized worms (green) from visibly non-colonized (dark). Subsequently, gene expression was examined using microarrays in both groups as well as in those exposed to the E. coli control. Genes responding to P. aeruginosa were identified using a multi-class t-test with a false discovery rate of 10%. This analysis identified gene classes previously reported as being induced by P. aeruginosa, including lysozymes, lectins [14], [17], several neuropeptide-like genes, and detoxification genes [17] (Fig. 1B and Table S1). The analyzed dataset is attached as Table S2. Importantly, infection responses were largely independent of the degree of colonization. Thus, although worm death from PA14 infection is correlated with colonization, the responses against it were not.
Figure 1. C. elegans immune responses to Pseudomonas aeruginosa are independent of colonization.
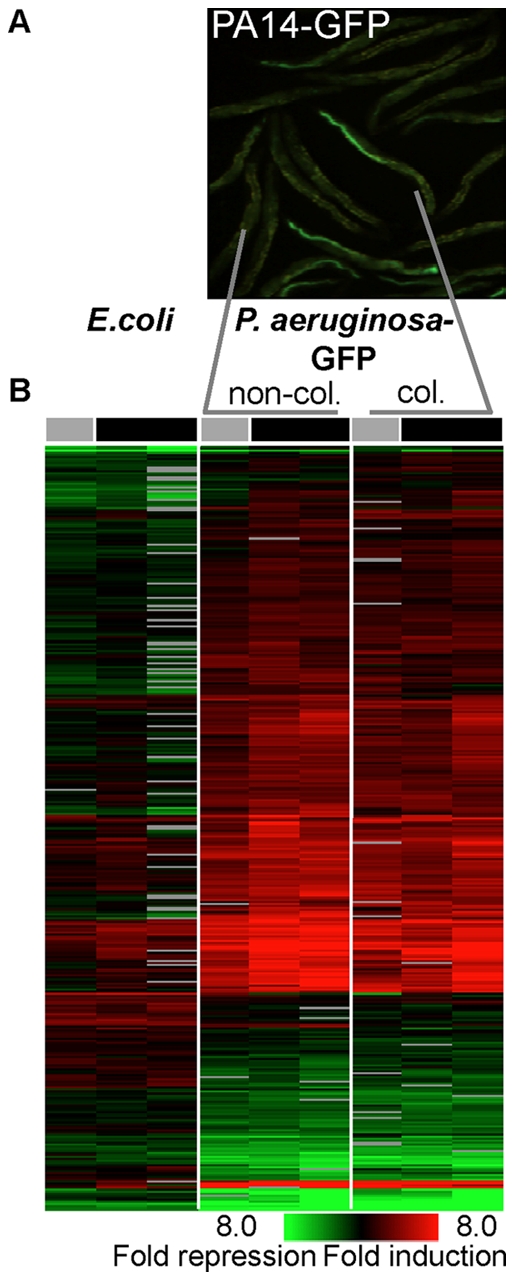
A) 2-day old adult C.elegans exposed to GFP-expressing P. aeruginosa for 18 hrs show variability in colonization, allowing isolation of colonized and non-colonized worms. B) Gene expression profiles of C. elegans fed with E. coli or with GFP-expressing P. aeruginosa (colonized and non-colonized) for 18 hours; separation was achieved either by picking under a fluorescent stereoscope (1 experiment; grey bar), or using the COPASTM Worm Sorter (two independent experiments ; black bar). Shown are genes responding to the pathogen, as identified with a multi-class t-test analysis (10% false discovery rate).
Several possible explanations may account for immune responses without visible colonization. The first that we wished to be able to rule out was that worms that are not visibly colonized were previously colonized, but managed to clear the infection. To test this possibility under similar conditions as those of the experiment we picked visibly colonized worms (N = 80) following a 12-hour exposure to PA14-GFP, and continuing the exposure to PA14-GFP on new plates, we examined whether any of these worms appeared dark by the time of collection (6 hours later); this turned out not to be the case, as all worms remained green. This does not mean that ‘dark’ worms are completely non-colonized, as they may be colonized with an undetectably small number of bacteria, but the similarity between their immune responses and those of fully-colonized worms implies that the immune response is not correlated with the degree of colonization.
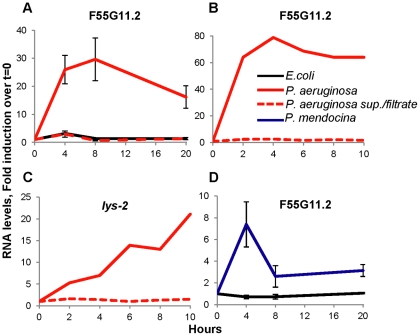
A second possibility is that molecules secreted by P. aeruginosa cause damage to the worm, which independent of colonization can induce the immune response. This is particularly plausible since P. aeruginosa is known to secrete a wide array of small-molecule and proteinaceous exotoxins [18]. To address this possibility, we used quantitative (q)RT-PCR to follow the expression of F55G11.2, a gene with a yet uncharacterized function, which was previously identified as part of the earliest responses to P. aeruginosa [14]. A closer examination showed that F55G11.2 was strongly induced within two hours of exposure to the pathogen (Fig. 2B) and as early as one hour following exposure (not shown). However, F55G11.2 was not induced when worms were exposed to a conditioned solution from a P. aeruginosa 24-hour culture (the ‘secretome’), laced onto dead E. coli (Fig. 2A). Induction of F55G11.2 was similarly missing when worms were grown on plates conditioned with P. aeruginosa grown on a filter, which was removed prior to transfer of worms and replaced with dead E. coli serving as food (Fig. 2B). Similar lack of induction was observed for lys-2 and pgp-5, two additional infection response genes that respond to P. aeruginosa, but with a slower time course than F55G11.2 (Fig. 2C and results not shown). Thus, secreted factors are not sufficient to induce immune responses against P. aeruginosa.
Figure 2. Pseudomonas secreted factors are not sufficient to induce immune responses, while conserved cell-associated factors are.
Gene expression measured by qRT-PCR, following exposure of young-adult C. elegans (at T0) to E. coli, alone or with P. aeruginosa supernatant (A) or P. aeruginosa filtrate (B,C), or to the non-pathogenic P. mendocina (D).
A third possibility that we are unable to rule out is that volatile toxins released by P. aeruginosa cause damage to the worm, which induces immune responses. P. aeruginosa has a distinctive smell produced by a combination of volatile compounds. Of these, one is hydrogen cyanide, a potent toxin. However, cyanide production was not found to take part in PA14 pathogenicity in C. elegans (unlike the PA01 strain [19]) and without any additional known toxic volatile compounds released from P. aeruginosa, this is unlikely.
The fourth possible explanation for induction of immune responses prior to detectable colonization is that C. elegans can recognize molecules associated with the pathogen (i.e. PAMPs/MAMPs), with a sensitivity that allows it to respond to a small number of bacteria. The failure of the P. aeruginosa ‘secretome’ to induce immune responses, supported this possibility. To decouple structural features of Pseudomonas from its pathogenicity, we examined immune responses to Pseudomonas mendocina, a recently identified C. elegans commensal that shows no pathogenicity, both in terms of survival/lifespan as well as with regards to early symptoms of infection (i.e. muscle function and movement) (Fig. S1 and Montalvo-Katz, unpublished results). Exposure to intact P. mendocina lead to F55G11.2 gene induction, smaller than the response to P. aeruginosa, but reproducible (Fig. 2D). This is consistent with the hypothesis that C. elegans can recognize cell-associated moieties that are shared between P. aeruginosa and P. mendocina, and that such recognition plays a role in the initiation of early immune responses. Altogether, our data suggest that C. elegans immune responses against P. aeruginosa are initiated by PAMP recognition, or, since the recognized pattern is shared with the non-pathogenic P. mendocina, MAMP recognition.
Discussion
We found that C. elegans immune response occurs prior to any visible colonization. Death, and presumably damage due to P. aeruginosa infection, is correlated with the extent of colonization. That this correlation does not hold for immune responses suggests sensitive detection of molecular patterns, apparently cell-associated and furthermore, shared among pathogenic and non-pathogenic Pseudomonas species. Recognition of MAMPs by C. elegans does not exclude the possibility that it can also respond to additional types of stimuli. In fact, the smaller magnitude of the response to P. mendocina compared to P. aeruginosa may be indicative of multiple signals, some cell-associated, but others associated with pathogenesis (perhaps DAMPs), leading to a full-blown immune response. The results described here provide evidence for MAMP recognition in C. elegans, but the nature of these MAMPs remains to be identified. Furthermore, since no PRR orthologs have been identified to date in C. elegans, these results further encourage a search for C. elegans PRRs, as they may represent novel mechanisms of pathogen recognition.
Supporting Information
Pseudomonas mendocina is a non-pathogenic species. (A) Lifespan analysis of worms grown on P. mendocina shows comparable lifespan to that of worms grown on the normal food bacteria E. coli (N = 90–93 worms for each group). Differences between curves were evaluated statistically using Kaplan Meier survival analysis followed by the Logrank test (p = 0.3483). (B) Muscle function decline, represented by the rate of defecation, a coordinated muscle program [20], becomes apparent following 20 hours of exposure to the pathogen P. aeruginosa, but not to E. coli or P. mendocina (at 25°C). Dots represent average interval between defecations (n = 10 cycles, or less, when intervals exceeded four minutes) in individual young-adults; green bars represent medians. *p = 0.002 (t-test). The general speed of worm movement also decreased in P. aeruginosa but not in P. mendocina (not shown).
(TIF)
Genes responding to P. aeruginosa.
(XLS)
Full data set used for microarray analysis.
(XLS)
Acknowledgments
We wish to thank Gordon Lithgow and Matthew Gill of the Buck Institute for the use and help with the worm sorter, and Terry Machen from UC Berkeley for insightful discussions.
Footnotes
Competing Interests: The authors have declared that no competing interests exist.
Funding: This work was supported by the Ellison Medical Foundation. KTB was additionally supported by an NSERC Postgraduate Scholarship. The funders had no role in study design, data collection and analysis, decision to publish, or preparation of the manuscript.
References
- 1.Irazoqui JE, Urbach JM, Ausubel FM. Evolution of host innate defence: insights from Caenorhabditis elegans and primitive invertebrates. Nat Rev Immunol. 2010;10:47–58. doi: 10.1038/nri2689. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Chen GY, Nunez G. Sterile inflammation: sensing and reacting to damage. Nat Rev Immunol. 2010;10:826–837. doi: 10.1038/nri2873. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Kumar H, Kawai T, Akira S. Pathogen recognition by the innate immune system. Int Rev Immunol. 2011;30:16–34. doi: 10.3109/08830185.2010.529976. [DOI] [PubMed] [Google Scholar]
- 4.Rosetto M, Engstrom Y, Baldari CT, Telford JL, Hultmark D. Signals from the IL-1 receptor homolog, Toll, can activate an immune response in a Drosophila hemocyte cell line. Biochem Biophys Res Commun. 1995;209:111–116. doi: 10.1006/bbrc.1995.1477. [DOI] [PubMed] [Google Scholar]
- 5.Lemaitre B, Nicolas E, Michaut L, Reichhart JM, Hoffmann JA. The dorsoventral regulatory gene cassette spatzle/Toll/cactus controls the potent antifungal response in Drosophila adults. Cell. 1996;86:973–983. doi: 10.1016/s0092-8674(00)80172-5. [DOI] [PubMed] [Google Scholar]
- 6.Ting JP, Willingham SB, Bergstralh DT. NLRs at the intersection of cell death and immunity. Nat Rev Immunol. 2008;8:372–379. doi: 10.1038/nri2296. [DOI] [PubMed] [Google Scholar]
- 7.Lu R, Yigit E, Li WX, Ding SW. An RIG-I-Like RNA helicase mediates antiviral RNAi downstream of viral siRNA biogenesis in Caenorhabditis elegans. PLoS Pathog. 2009;5:e1000286. doi: 10.1371/journal.ppat.1000286. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Wong D, Bazopoulou D, Pujol N, Tavernarakis N, Ewbank JJ. Genome-wide investigation reveals pathogen-specific and shared signatures in the response of Caenorhabditis elegans to infection. Genome Biol. 2007;8:R194. doi: 10.1186/gb-2007-8-9-r194. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Kanzok SM, Hoa NT, Bonizzoni M, Luna C, Huang Y, et al. Origin of Toll-like receptor-mediated innate immunity. J Mol Evol. 2004;58:442–448. doi: 10.1007/s00239-003-2565-8. [DOI] [PubMed] [Google Scholar]
- 10.Pujol N, Link EM, Liu LX, Kurz CL, Alloing G, et al. A reverse genetic analysis of components of the Toll signaling pathway in Caenorhabditis elegans. Curr Biol. 2001;11:809–821. doi: 10.1016/s0960-9822(01)00241-x. [DOI] [PubMed] [Google Scholar]
- 11.Tenor JL, Aballay A. A conserved Toll-like receptor is required for Caenorhabditis elegans innate immunity. EMBO Rep. 2008;9:103–109. doi: 10.1038/sj.embor.7401104. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Powell JR, Kim DH, Ausubel FM. The G protein-coupled receptor FSHR-1 is required for the Caenorhabditis elegans innate immune response. Proc Natl Acad Sci U S A. 2009;106:2782–2787. doi: 10.1073/pnas.0813048106. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Tan MW, Mahajan-Miklos S, Ausubel FM. Killing of Caenorhabditis elegans by Pseudomonas aeruginosa used to model mammalian bacterial pathogenesis. Proc Natl Acad Sci U S A. 1999;96:715–720. doi: 10.1073/pnas.96.2.715. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Shapira M, Hamlin BJ, Rong J, Chen K, Ronen M, et al. A conserved role for a GATA transcription factor in regulating epithelial innate immune responses. Proc Natl Acad Sci U S A. 2006;103:14086–14091. doi: 10.1073/pnas.0603424103. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Tusher VG, Tibshirani R, Chu G. Significance analysis of microarrays applied to the ionizing radiation response. Proc Natl Acad Sci U S A. 2001;98:5116–5121. doi: 10.1073/pnas.091062498. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Tan MW, Rahme LG, Sternberg JA, Tompkins RG, Ausubel FM. Pseudomonas aeruginosa killing of Caenorhabditis elegans used to identify P. aeruginosa virulence factors. Proc Natl Acad Sci U S A. 1999;96:2408–2413. doi: 10.1073/pnas.96.5.2408. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Troemel ER, Chu SW, Reinke V, Lee SS, Ausubel FM, et al. p38 MAPK regulates expression of immune response genes and contributes to longevity in C. elegans. PLoS Genet. 2006;2:e183. doi: 10.1371/journal.pgen.0020183. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Lau GW, Hassett DJ, Britigan BE. Modulation of lung epithelial functions by Pseudomonas aeruginosa. Trends Microbiol. 2005;13:389–397. doi: 10.1016/j.tim.2005.05.011. [DOI] [PubMed] [Google Scholar]
- 19.Gallagher LA, Manoil C. Pseudomonas aeruginosa PAO1 kills Caenorhabditis elegans by cyanide poisoning. J Bacteriol. 2001;183:6207–6214. doi: 10.1128/JB.183.21.6207-6214.2001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Branicky R, Hekimi S. What keeps C. elegans regular: the genetics of defecation. Trends Genet. 2006;22:571–579. doi: 10.1016/j.tig.2006.08.006. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Pseudomonas mendocina is a non-pathogenic species. (A) Lifespan analysis of worms grown on P. mendocina shows comparable lifespan to that of worms grown on the normal food bacteria E. coli (N = 90–93 worms for each group). Differences between curves were evaluated statistically using Kaplan Meier survival analysis followed by the Logrank test (p = 0.3483). (B) Muscle function decline, represented by the rate of defecation, a coordinated muscle program [20], becomes apparent following 20 hours of exposure to the pathogen P. aeruginosa, but not to E. coli or P. mendocina (at 25°C). Dots represent average interval between defecations (n = 10 cycles, or less, when intervals exceeded four minutes) in individual young-adults; green bars represent medians. *p = 0.002 (t-test). The general speed of worm movement also decreased in P. aeruginosa but not in P. mendocina (not shown).
(TIF)
Genes responding to P. aeruginosa.
(XLS)
Full data set used for microarray analysis.
(XLS)