Abstract
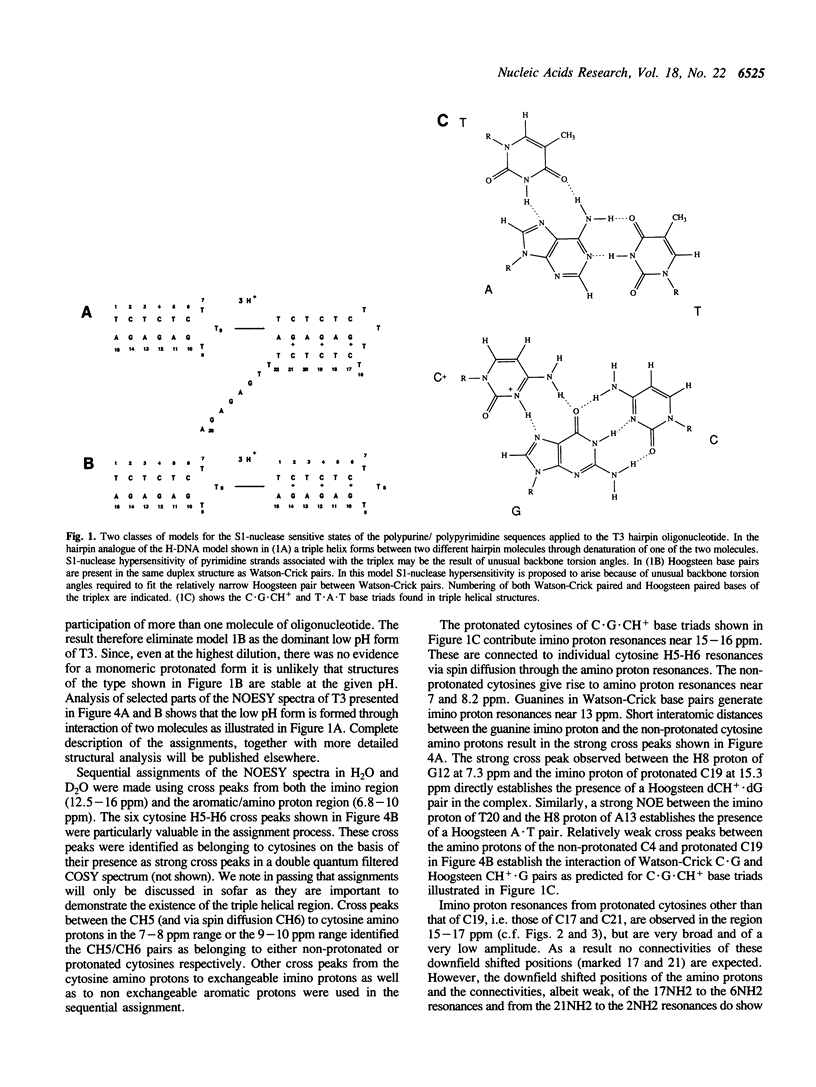
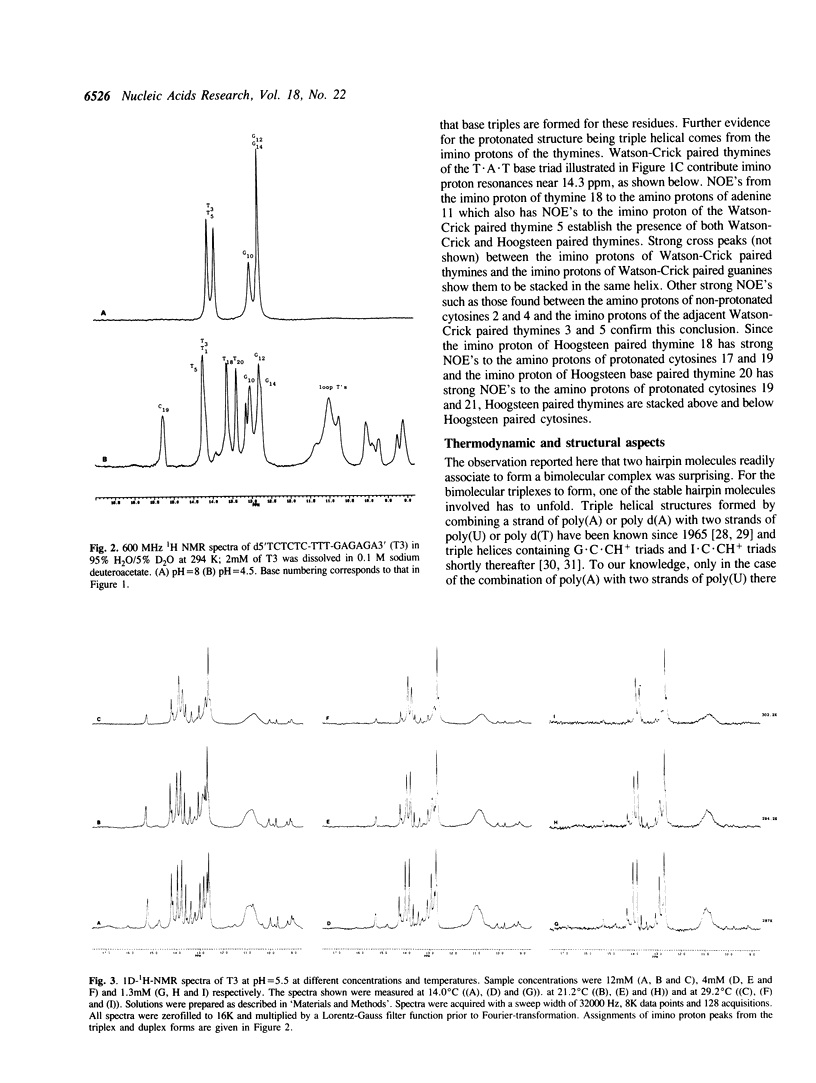
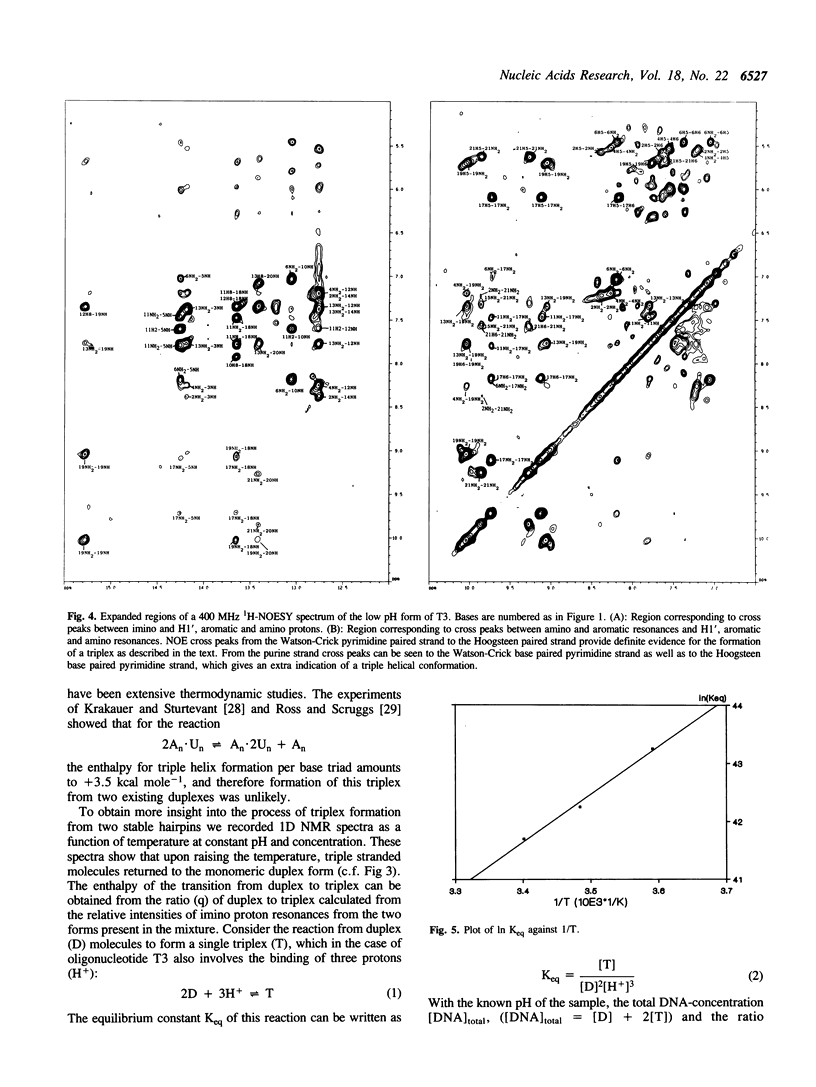
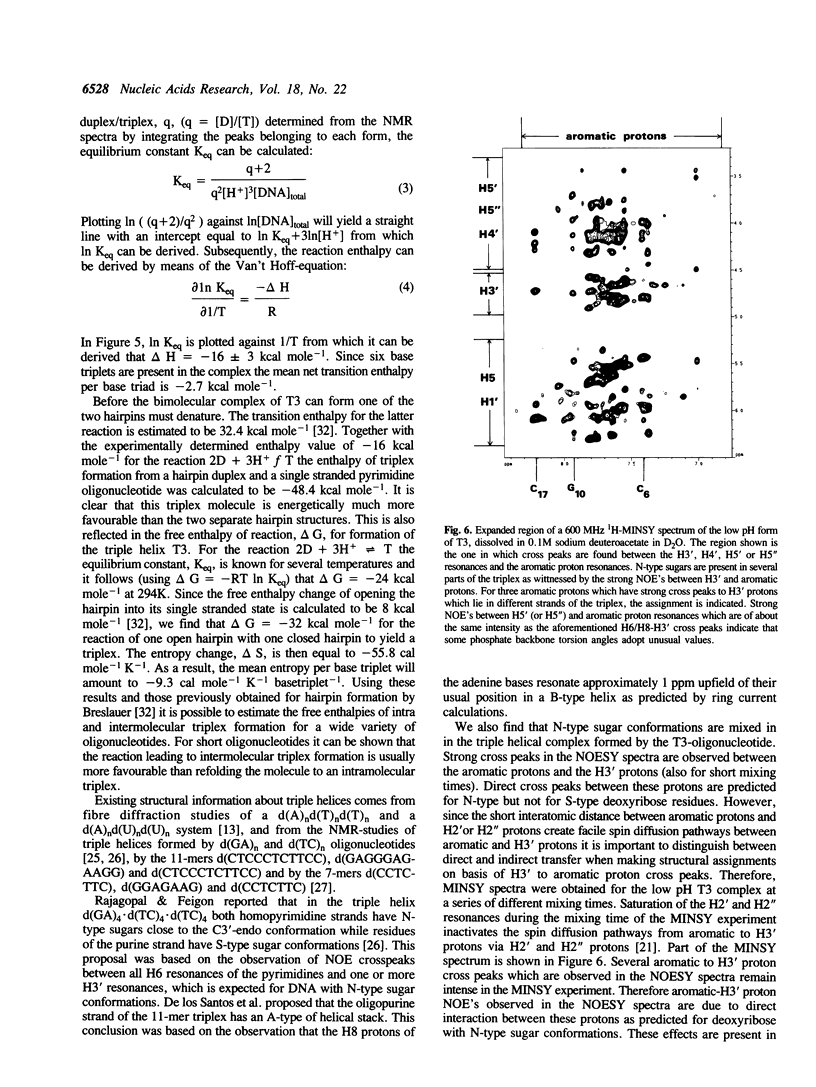
1D and 2D NMR investigations of the 15 residue deoxynucleotide sequence d(TCTCTC-TTT-GAGAGA) show that above pH = 6.5 the molecule adopts a B-form hairpin conformation. As the pH is lowered below 6.5 molecules progressively associate in pairs to form a partially triple helical, partially single stranded structure in which the bases of the oligopyrimidine d(TC)3 tract from one molecule form Hoogsteen pairs with the d(G-A)3 tract of the other. Imino protons of protonated cytosines can be observed at very low field (approximately 15 ppm). The enthalpy of triplex formation was estimated by NMR techniques to be -16 kcal mol-1. Intense H6 to H3' cross peaks from residues in all three strands suggest the presence of N-type sugars at some but not at all possible sites. Surprisingly strong cross peaks between H5' or H5" and non-exchangeable base protons are also observed. These suggest that certain of the O5'-C5'-C4'-C3' phosphate backbone torsion angles (gamma) are unusual.
Full text
PDF


Selected References
These references are in PubMed. This may not be the complete list of references from this article.
- Arnott S., Selsing E. Structures for the polynucleotide complexes poly(dA) with poly (dT) and poly(dT) with poly(dA) with poly (dT). J Mol Biol. 1974 Sep 15;88(2):509–521. doi: 10.1016/0022-2836(74)90498-7. [DOI] [PubMed] [Google Scholar]
- Breslauer K. J., Frank R., Blöcker H., Marky L. A. Predicting DNA duplex stability from the base sequence. Proc Natl Acad Sci U S A. 1986 Jun;83(11):3746–3750. doi: 10.1073/pnas.83.11.3746. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Evans T., Efstratiadis A. Sequence-dependent S1 nuclease hypersensitivity of a heteronomous DNA duplex. J Biol Chem. 1986 Nov 5;261(31):14771–14780. [PubMed] [Google Scholar]
- Haasnoot C. A., Hilbers C. W. Effective water resonance suppression in 1D- and 2D-FT-1H-NMR spectroscopy of biopolymers in aqueous solution. Biopolymers. 1983 May;22(5):1259–1266. doi: 10.1002/bip.360220502. [DOI] [PubMed] [Google Scholar]
- Haasnoot C. A., de Bruin S. H., Berendsen R. G., Janssen H. G., Binnendijk T. J., Hilbers C. W., van der Marel G. A., van Boom J. H. Structure, kinetics and thermodynamics of DNA hairpin fragments in solution. J Biomol Struct Dyn. 1983 Oct;1(1):115–129. doi: 10.1080/07391102.1983.10507429. [DOI] [PubMed] [Google Scholar]
- Hanvey J. C., Klysik J., Wells R. D. Influence of DNA sequence on the formation of non-B right-handed helices in oligopurine.oligopyrimidine inserts in plasmids. J Biol Chem. 1988 May 25;263(15):7386–7396. [PubMed] [Google Scholar]
- Hanvey J. C., Shimizu M., Wells R. D. Intramolecular DNA triplexes in supercoiled plasmids. Proc Natl Acad Sci U S A. 1988 Sep;85(17):6292–6296. doi: 10.1073/pnas.85.17.6292. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hilbers C. W., Haasnoot C. A., de Bruin S. H., Joordens J. J., van der Marel G. A., van Boom J. H. Hairpin formation in synthetic oligonucleotides. Biochimie. 1985 Jul-Aug;67(7-8):685–695. doi: 10.1016/s0300-9084(85)80156-5. [DOI] [PubMed] [Google Scholar]
- Htun H., Dahlberg J. E. Single strands, triple strands, and kinks in H-DNA. Science. 1988 Sep 30;241(4874):1791–1796. doi: 10.1126/science.3175620. [DOI] [PubMed] [Google Scholar]
- Htun H., Dahlberg J. E. Topology and formation of triple-stranded H-DNA. Science. 1989 Mar 24;243(4898):1571–1576. doi: 10.1126/science.2648571. [DOI] [PubMed] [Google Scholar]
- Johnston B. H. The S1-sensitive form of d(C-T)n.d(A-G)n: chemical evidence for a three-stranded structure in plasmids. Science. 1988 Sep 30;241(4874):1800–1804. doi: 10.1126/science.2845572. [DOI] [PubMed] [Google Scholar]
- Kohwi Y., Kohwi-Shigematsu T. Magnesium ion-dependent triple-helix structure formed by homopurine-homopyrimidine sequences in supercoiled plasmid DNA. Proc Natl Acad Sci U S A. 1988 Jun;85(11):3781–3785. doi: 10.1073/pnas.85.11.3781. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Krakauer H., Sturtevant J. M. Heats of the helix-coil transitions of the poly A-poly U complexes. Biopolymers. 1968 Apr;6(4):491–512. doi: 10.1002/bip.1968.360060406. [DOI] [PubMed] [Google Scholar]
- Larsen A., Weintraub H. An altered DNA conformation detected by S1 nuclease occurs at specific regions in active chick globin chromatin. Cell. 1982 Jun;29(2):609–622. doi: 10.1016/0092-8674(82)90177-5. [DOI] [PubMed] [Google Scholar]
- Lee J. S., Johnson D. A., Morgan A. R. Complexes formed by (pyrimidine)n . (purine)n DNAs on lowering the pH are three-stranded. Nucleic Acids Res. 1979 Jul 11;6(9):3073–3091. doi: 10.1093/nar/6.9.3073. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lyamichev V. I., Mirkin S. M., Frank-Kamenetskii M. D. A pH-dependent structural transition in the homopurine-homopyrimidine tract in superhelical DNA. J Biomol Struct Dyn. 1985 Oct;3(2):327–338. doi: 10.1080/07391102.1985.10508420. [DOI] [PubMed] [Google Scholar]
- Lyamichev V. I., Mirkin S. M., Frank-Kamenetskii M. D. Structures of homopurine-homopyrimidine tract in superhelical DNA. J Biomol Struct Dyn. 1986 Feb;3(4):667–669. doi: 10.1080/07391102.1986.10508454. [DOI] [PubMed] [Google Scholar]
- Mirkin S. M., Lyamichev V. I., Drushlyak K. N., Dobrynin V. N., Filippov S. A., Frank-Kamenetskii M. D. DNA H form requires a homopurine-homopyrimidine mirror repeat. Nature. 1987 Dec 3;330(6147):495–497. doi: 10.1038/330495a0. [DOI] [PubMed] [Google Scholar]
- Morgan A. R., Wells R. D. Specificity of the three-stranded complex formation between double-stranded DNA and single-stranded RNA containing repeating nucleotide sequences. J Mol Biol. 1968 Oct 14;37(1):63–80. doi: 10.1016/0022-2836(68)90073-9. [DOI] [PubMed] [Google Scholar]
- Pulleyblank D. E., Haniford D. B., Morgan A. R. A structural basis for S1 nuclease sensitivity of double-stranded DNA. Cell. 1985 Aug;42(1):271–280. doi: 10.1016/s0092-8674(85)80122-7. [DOI] [PubMed] [Google Scholar]
- Rajagopal P., Feigon J. NMR studies of triple-strand formation from the homopurine-homopyrimidine deoxyribonucleotides d(GA)4 and d(TC)4. Biochemistry. 1989 Sep 19;28(19):7859–7870. doi: 10.1021/bi00445a048. [DOI] [PubMed] [Google Scholar]
- Rajagopal P., Feigon J. Triple-strand formation in the homopurine:homopyrimidine DNA oligonucleotides d(G-A)4 and d(T-C)4. Nature. 1989 Jun 22;339(6226):637–640. doi: 10.1038/339637a0. [DOI] [PubMed] [Google Scholar]
- Ross P. D., Scruggs R. L. Heat of the reaction forming the three-stranded poly (A + 2U) complex. Biopolymers. 1965;3(4):491–496. doi: 10.1002/bip.1965.360030410. [DOI] [PubMed] [Google Scholar]
- Sklenár V., Brooks B. R., Zon G., Bax A. Absorption mode two-dimensional NOE spectroscopy of exchangeable protons in oligonucleotides. FEBS Lett. 1987 Jun 1;216(2):249–252. doi: 10.1016/0014-5793(87)80699-3. [DOI] [PubMed] [Google Scholar]
- Thiele D., Guschlbauer W. Polynucléotides protonés. VII. Transitions thermiques entre differents complexes de l'acide polyinosinique et de l'acide polycytidylique en milieu acide. Biopolymers. 1969;8(3):361–378. doi: 10.1002/bip.1969.360080307. [DOI] [PubMed] [Google Scholar]
- Thiele D., Guschlbauer W. Protonated polynucleotide structures. IX. Disproportionation of poly (G)-poly (C) in acid medium. Biopolymers. 1971;10(1):143–157. doi: 10.1002/bip.360100111. [DOI] [PubMed] [Google Scholar]
- Voloshin O. N., Mirkin S. M., Lyamichev V. I., Belotserkovskii B. P., Frank-Kamenetskii M. D. Chemical probing of homopurine-homopyrimidine mirror repeats in supercoiled DNA. Nature. 1988 Jun 2;333(6172):475–476. doi: 10.1038/333475a0. [DOI] [PubMed] [Google Scholar]
- Wells R. D., Collier D. A., Hanvey J. C., Shimizu M., Wohlrab F. The chemistry and biology of unusual DNA structures adopted by oligopurine.oligopyrimidine sequences. FASEB J. 1988 Nov;2(14):2939–2949. [PubMed] [Google Scholar]
- de los Santos C., Rosen M., Patel D. NMR studies of DNA (R+)n.(Y-)n.(Y+)n triple helices in solution: imino and amino proton markers of T.A.T and C.G.C+ base-triple formation. Biochemistry. 1989 Sep 5;28(18):7282–7289. doi: 10.1021/bi00444a021. [DOI] [PubMed] [Google Scholar]


