Abstract
Huntington's disease is caused by expression of a mutant form of Huntingtin protein containing an expanded polyglutamine repeat. One possible treatment for Huntington's disease may be to reduce expression of mutant Huntingtin in the brain via RNA interference. Unless the therapeutic molecule is designed to be allele-specific, both wild-type and mutant protein will be suppressed by an RNA interference treatment. A key question is whether suppression of wild-type as well as mutant Huntingtin in targeted brain regions can be tolerated and result in a net benefit to patients with Huntington's disease. Whether Huntingtin performs essential functions in the adult brain is unclear. Here, we tested the hypothesis that the adult primate brain can tolerate moderately reduced levels of wild-type Huntingtin protein for an extended period of time. A serotype 2 adeno-associated viral vector encoding for a short hairpin RNA targeting rhesus huntingtin messenger RNA (active vector) was bilaterally injected into the striatum of four adult rhesus monkeys. Four additional animals received a comparable vector encoding a scrambled control short hairpin RNA (control vector). General health and motor behaviour were monitored for 6 months. Upon termination, brain tissues were sampled and assessed blindly for (i) huntingtin messenger RNA knockdown; (ii) Huntingtin protein expression; and (iii) neuropathological changes. Reduction in wild-type huntingtin messenger RNA levels averaging ∼30% was measured in the striatum of active vector recipients 6 months post-injection. A widespread reduction in Huntingtin protein levels was also observed by immunohistochemistry in these animals, with an average protein reduction of ∼45% relative to controls measured by western blot analysis in the putamen of active vector recipients. As with control vector recipients, no adverse effects were observed behaviourally, and no neurodegeneration was found on histological examination of active vector recipients. Our results suggest that long-term partial suppression of wild-type Huntingtin may be safe, and thus if a comparable level of suppression of mutant Huntingtin is beneficial, then partial suppression of both wild-type and mutant Huntingtin may result in a net benefit in patients with heterozygous Huntington's disease.
Keywords: RNA interference, Huntington's disease, striatum, adeno-associated virus, non-human primate
Introduction
Huntington's disease is a fatal neurodegenerative disease affecting the cerebral cortex (Tabrizi et al., 2009) and subcortical brain regions (Gabery et al., 2010), with the most pronounced neuronal degeneration seen in the striatum (Aylward et al., 2004). It is caused by expression of a mutant, expanded form of Huntingtin protein. Consequently, Huntingtin itself is a target for therapeutic intervention. Huntingtin can be reduced in vivo via RNA interference using small interfering RNA (Wang et al., 2005) or equivalent short hairpin RNA (Harper et al., 2005). Also, Huntingtin can be reduced in vivo by antisense therapeutics (Sass and Aronin, 2011).
Each of these approaches is under development for clinical testing, and each has the potential to intervene at the earliest possible point in the pathogenic pathway of the disease: the expression of the mutant Huntingtin protein itself. Most of these therapeutic molecules do not selectively suppress the expression of the mutant protein, but also suppress expression of the wild-type protein. An alternative treatment strategy could consist of suppressing just the mutant allele in heterozygous patients using, for example, small interfering RNA targeted to polymorphisms in the Huntington's disease gene (van Bilsen et al., 2008; Lombardi et al., 2009). However, a non-allele-specific therapy in which both wild-type and mutant protein are suppressed is greatly preferred to avoid the challenges associated with regulatory approval of multiple therapeutic molecules (Sah and Aronin, 2011). Yet, whether suppression of both mutant and wild-type Huntingtin in targeted brain regions can be tolerated and result in a net benefit to patients with Huntington's disease cannot be predicted from what is currently known.
Although studies support a role for Huntingtin in early development (Duyao et al., 1995), its normal function in the adult brain remains unclear. Huntington's disease pathogenesis may result in part from a loss of wild-type Huntingtin function as well as from a toxic gain of function of mutant Huntingtin (Zuccato et al., 2010). Development of tissue-specific knock-out mice (Dragatsis et al., 2000) and studies in knock-in mice with reduced Hdh expression (the mouse homologue of Huntingtin) indicate that reduction or lack of Huntingtin affects brain function (Auerbach et al., 2001). However, in these animals, Hdh was absent in the early postnatal period or was reduced throughout development rather than starting in adulthood. Other studies support a role for Huntingtin in axonal transport (Colin et al., 2008), which may include adequate production and delivery of brain-derived neurotrophic factor from the cortex to the striatum (Zuccato and Cattaneo, 2007). Additionally, Huntingtin has been shown to be required for mitotic spindle orientation and neurogenesis (Godin et al., 2010).
Whether therapies that reduce both wild-type and mutant Huntingtin will be beneficial or harmful to patients depends in part on whether partial reduction of wild-type Huntingtin can be tolerated long-term in the fully developed adult brain. Conditional knock-down of Hdh in mice starting at 4.5 weeks of age results in reduced striatal volume in the animals when assessed at 24 weeks of age (Menalled et al., 2009). Conversely, non-allele specific suppression of Huntingtin for up to 9 months in rodents has not been found to cause pathology, although altered expression of various genes was observed (Boudreau et al., 2009; Drouet et al., 2009). Here, we expand these observations to adult non-human primates, reporting that bilateral delivery of an adeno-associated virus serotype 2 (AAV2) vector encoding an anti-Huntingtin short hairpin RNA into the striatum of rhesus monkeys resulted in ∼30% sustained reduction of huntingtin messenger RNA and widespread reduction of striatal wild-type Huntingtin protein levels, estimated at 45% reduction in the putamen, without detectable ill effects or marked pathology 6 months post administration.
Materials and methods
Animals and treatment groups
Eight adult female rhesus monkeys (Macaca mulatta) weighing 5–7.5 kg were obtained from Covance Inc. and housed individually in the same room on a 12-h light/12-h dark cycle. The animals were divided in two groups of four age-matched animals: 19.00 ± 1.35 years old (active vector recipients) and 19.75 ± 0.37 years old (control vector recipients), roughly equivalent to 58 years old in humans (Tigges et al., 1988; Gore and Terasawa, 1991; Andersen et al., 1999). All behavioural, molecular and histopathological data measurements were performed blindly with respect to the treatment group. All procedures were approved by the University of Kentucky Institutional Animal Care and Use Committee and were conducted in facilities accredited by the Association for Assessment and Accreditation of Laboratory Animal Care International. Animal care was supervised by veterinarians skilled in the care and maintenance of non-human primates.
Development of short hairpin RNA vectors
Twelve small interfering RNA sequences targeting both rhesus and human Huntingtin were designed and screened for their ability to reduce huntingtin messenger RNA in human (HEK293T, American Type Culture Collection, catalogue number CRL-11268) and rhesus (LLC-MK2, catalogue number CCL-7) cell lines. An effective candidate (HD5: 5′–GGAGUAUUGUGGAACUUAU–3′) was selected for development of a viral vector, and the corresponding short hairpin RNA sequence was cloned into a plasmid providing AAV2 inverted terminal repeats and the human U6 promoter. The middle 11 nt of the sequence were scrambled to create a control vector (CTRL5: 5′–GGAGUAGUCGUAAUGUUAU–3′). The remainder of the AAV transgene was filled with an inert DNA sequence to allow for efficient viral packaging. Vectors were produced from the pAAV-HD5 and pAAV-CTRL5 plasmids by Virapur Limited Liability Corporation yielding a titre of 2 × 1012 vector genomes/ml and undetectable endotoxin levels (<1 EU/ml). Vector identity was confirmed by sequencing the short hairpin RNA-expressing region.
Stereotaxic surgical procedures
All surgical procedures were conducted under isoflurane anaesthesia (1–3%) and sterile field conditions. Using MRI-guided techniques, Hamilton syringes (100 µl, model 1710) fitted to 26G side-port needles and loaded with either AAV2-CTRL5 or AAV2-HD5 were inserted bilaterally through small burr holes into the caudate nucleus and left in place for 10 min. Then, 30 µl of AAV was injected into each target at 2 µl/min, using a stereotaxic nanopump (model 310 Plus, Stoelting Co.). Upon completion, the needles were left in place for 20 min then slowly retracted. The syringes were then fitted to new 26G side-port needles and loaded with 60 µl of the same AAV2 solution for bilateral injection into the putamen, 3–4 mm caudal from the caudate nucleus injection site. Two injections of 30 µl each were made dorsoventrally in the putamen, 3-mm apart along a single needle tract. Finally, two additional injections of 30 µl each were made dorsoventrally along a single needle tract into the putamen, 3–4 mm caudal from the first putamenal injection site. Injection site coordinates and volumes are summarized in Table 1. An MRI was taken immediately postoperatively to verify injection placement. Buprenorphine (0.01 mg/kg) was administered subcutaneously pre- and postoperatively, every 12 h for 48 h.
Table 1.
The 10 injection site coordinates and volumes
| Structure targeted | Coordinates |
Left hemisphere |
Right hemisphere |
|||
|---|---|---|---|---|---|---|
| Anterior–posterior (mm) | Dorsal–ventral (mm) | Medial–lateral (mm) | Volume injected (µl) | Medial–lateral (mm) | Volume injected (µl) | |
| Caudate nucleus | 25.1 ± 0.5 | 17.1 ± 0.2 | +5.3 ± 0.1 | 30 | −5.3 ± 0.1 | 30 |
| Rostral putamen (ventral) | 22.3 ± 0.3 | 21.5 ± 0.3 | +10.9 ± 0.2 | 30 | −10.9 ± 0.2 | 30 |
| Rostral putamen (dorsal) | 22.3 ± 0.3 | 18.5 ± 0.3 | +10.9 ± 0.2 | 30 | −10.9 ± 0.2 | 30 |
| Caudal putamen (ventral) | 19.1 ± 0.3 | 21.2 ± 0.4 | +13.2 ± 0.3 | 30 | −13.2 ± 0.3 | 30 |
| Caudal putamen (dorsal) | 19.1 ± 0.3 | 17.8 ± 0.4 | +13.2 ± 0.3 | 30 | −13.2 ± 0.3 | 30 |
General health assessment
Baseline body weights were obtained by averaging the last two weight measurements taken within 45 days of AAV delivery. Postoperatively, body weights were measured every other week for 6 months. Food consumption was measured daily for 2 weeks preoperatively and 6 months postoperatively by counting the number of primate biscuits consumed (Harlan 2050 Teklad Global 20% protein diet).
Motor function assessment
Home-cage activity levels were measured pre- and post-AAV delivery using an Actical accelerometer (MiniMitter) mounted on a collar worn by the animal, and scored as day time (6 am to 6 pm) and night time (6 pm to 6 am) activity. Upper limb motor function was evaluated pre- and post-AAV delivery using a task consisting of retrieving miniature marshmallows from an automated receptacle chamber attached to the home-cage (Walton et al., 2008). Motor performance was electronically recorded over 12 trials as the time (to within 10 ms) for the animal to retrieve the food from a platform in the receptacle chamber. As previously reported (Grondin et al., 2003), the automated food retrieval task can detect changes in motor function as a result of changes in basal ganglia function in rhesus macaques, particularly in the caudate nucleus and putamen. To assess for motor memory preservation, testing was performed monthly post-AAV administration, without intervening practice between test sessions (Walton et al., 2008).
Necropsy and brain tissue processing
Six months post-surgery, animals were deeply anaesthetized and euthanized by transcardial saline perfusion. The brain was removed and both hemispheres cut into consecutive 2-mm thick slabs spanning the striatum. Using a biopsy needle (2-mm outside diameter), tissue punches were taken bilaterally in the caudate and putamen for quantification of short hairpin RNA and huntingtin messenger RNA expression. The adjacent slabs were processed so that 40 µm coronal sections could be cut by frozen sliding microtome, and stored in cryoprotectant solution at −20°C until processed for histopathological evaluations.
Relative quantification of short hairpin RNA and huntingtin messenger RNA expression
Total RNA was isolated from AAV2-treated brain tissue punches using the mirVana™ mitochondrial RNA isolation kit (Applied Biosystems). Total RNA was also isolated from four caudate and four putamen punches from each of four age-matched, non-AAV-treated rhesus monkeys. These samples were pooled separately and used as a control in the short hairpin RNA and huntingtin messenger RNA expression analyses of the AAV2-recipient animals. Complementary DNA was generated using short hairpin RNA-specific primers and the TaqMan® MicroRNA reverse transcription kit. Custom TaqMan® small RNA assays detected the processed HD5 and CTRL5 short hairpin RNAs (Applied Biosystems). Endogenous U18 small nuclear RNA was used to normalize the expression of the short hairpin RNAs (i.e. quantify relative to the U18 small nuclear RNA) across samples. A custom TaqMan® assay spanning the junction between exons 14 and 15 of rhesus Huntingtin was used to measure Huntingtin expression. Glyceraldehyde 3-phosphate dehydrogenase (GAPDH) expression was used to normalize the level of Huntingtin gene expression across samples.
Histopathological examination
One in every twelve 40 -µm thick coronal sections throughout the entire striatum was processed for haematoxylin and eosin and for Nissl staining. Adjacent sections were processed by immunohistochemistry, using procedures described previously (Ai et al., 2003), for the following proteins: (i) glial fibrillary acidic protein (GFAP 1:5000, Chemicon, catalogue number MAB360); (ii) the human leukocyte antigen HLA-DR (1:200, Becton Dickinson Biosciences, catalogue number 347360); (iii) Huntingtin (1:2000, Chemicon, catalogue number MAB2174); and (iv) dopamine- and cAMP-regulated neuronal phosphoprotein (DARPP-32 1:1000, Santa Cruz, catalogue number Sc-11365), a cytosolic neuronal phosphoprotein expressed in GABAergic, medium-sized spiny neurons in the striatum.
Analysis of all injection sites in the caudate (one needle tract per hemisphere) and putamen (two needle tracts per hemisphere) in each animal was performed blindly with respect to AAV vectors by a board-certified neuropathologist (n = 3–4 sections per brain per type of stain). The severity of the histopathological abnormalities seen on the Nissl and haematoxylin/eosin stains was evaluated using a semi-quantitative scale ranging from 0 to 5 as described in Supplementary Table 1. The severity of astrocytosis or microglial activation seen on GFAP or HLA-DR stained sections, respectively, was rated using a separate semi-quantitative scale (Supplementary Table 1). The lateral-to-medial width and dorsal-to-ventral height of the abnormalities as seen under the microscope were measured by the neuropathologist to assess the size of disruption in tissue around the injection tract (visualized with Nissl), the area of abnormal astrocytosis (visualized with GFAP), the area of abnormal microglial response (visualized with HLA-DR) and any area of reduced immunoreactivity of Huntingtin or DARPP-32.
Western blot analysis
Protein was isolated and quantified from a tissue punch taken from the right and left putamen from each monkey. Thirty micrograms of protein were loaded in each lane of a Tris–acetate gel (Criterion XT, Biorad, catalogue number 345-0129) and electrophoresed at 200 V for 45 min. The protein was transferred to membranes using the Invitrogen iBlot® transfer system (catalogue number IB101). For detection of Huntingtin, membranes were exposed to primary antibody (1:1000, Millipore, catalogue number MAB2166), washed, then exposed to secondary antibody (1:50 000, Millipore peroxidase-conjugated goat-anti-Mouse IgG, catalogue number AP124P), with each antibody incubation performed for 1 h at room temperature with gentle agitation. Protein bands were detected using peroxidase-conjugated goat anti-mouse IgG (1:5000, Millipore catalogue number AP124P) and the ECL Prime Western Blotting Detection kit (Amersham catalogue number PRN2108). Chemiluminescence was captured by direct exposure of the membranes to a charge-coupled device camera for 30 min. Membranes were also re-probed for alpha tubulin (Abcam catalogue number ab7291) to confirm comparable amounts of protein loading across lanes.
The amount of Huntingtin protein per lane was quantified by densitometry using the ‘Gels’ macro provided by the National Institutes of Health ImageJ software (version 1.42q) following the method described in the user manual (Ferreira and Rasband, 2011). Briefly, the background was digitally subtracted from the image using the macro provided, and then the lanes were manually designated with identically sized rectangles adequate to fully enclose the Huntingtin band in each lane. The 1D lane profile plots were generated by the software (representing the density of the image summed over horizontal raster lines of each rectangle), then the area under each lane's plot at the peak corresponding to the Huntingtin band was computed. These values were normalized by dividing by the value for the density of the tubulin band in the same lane obtained by the same method.
Statistical analyses
Tests of the null hypotheses of no differences in short hairpin RNA or huntingtin messenger RNA expression, changes in body weight, food consumption, upper limb motor function and home-cage activity levels over time were performed by repeated measures analysis of variance (ANOVA). Tests of the null hypotheses of no difference in Huntingtin protein quantification by western blot were performed by two-way (hemisphere × treatment) ANOVA. Semi-quantitative histopathological findings (0–5 scale) were analysed using a non-parametric Mann–Whitney U test. Quantitative histopathological measures (width and height) were analysed using a one-tailed, unpaired t-test. A P < 0.05 was considered significant in all analyses.
Results
Short hairpin RNA development
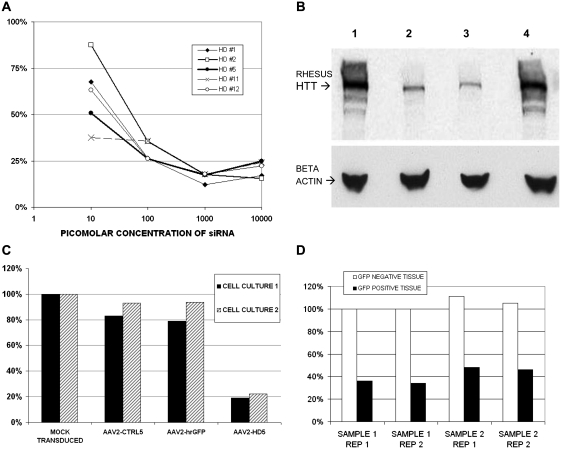
Twelve candidate small interfering RNA sequences targeting Huntingtin at regions of 100% rhesus and human sequence homology were screened in vitro in HEK293T (human) and LLC-MK2 (rhesus) cell lines. The candidate selected for production of AAV-based short hairpin RNA (HD5) suppressed huntingtin messenger RNA by ∼50% in vitro in LLC-MK2 cells at a concentration of 10 pM and >75% at higher concentrations (Fig. 1A). By western blot, HD5 small interfering RNA suppressed rhesus Huntingtin protein expression in LLC-MK2 cells by 76.7% compared with cells transfected with a scrambled, control small interfering RNA (Fig. 1B).
Figure 1.
(A) Level of rhesus huntingtin messenger RNA expression (normalized to GAPDH) in LLC-MK2 cells transfected with various concentrations of candidate anti-huntingtin small interfering RNA (siRNA), relative to levels in mock transfected cells. (B) Western blot of rhesus Huntingtin (HTT) and beta-actin proteins in lysates from LLC-MK2 cells transfected with anti-huntingtin small interfering RNA candidate HD1 or HD5. Lanes: (1) untreated cells, (2) cells transfected with HD1, (3) cells transfected with HD5, (4) cells transfected with a scrambled control small interfering RNA. (C) Level of huntingtin messenger RNA expression in HEK293T cells transduced with AAV2-CTRL5, AAV2-GFP or AAV2-HD5, relative to levels in mock transduced cells. Data from two separate cell experiments are shown. (D) Level of rhesus huntingtin messenger RNA expression in striatal brain tissue from two samples of GFP-positive versus two samples of GFP-negative cells obtained by laser capture microdissection from the pilot monkey co-infused with AAV2-HD5 and AAV2-GFP. Data from two replicate assays are shown.
Verification of vector function in vitro and in vivo
The short hairpin RNA sequence corresponding to HD5 was cloned into a plasmid providing AAV2 inverted terminal repeats and the human U6 promoter. The middle 11 nt of the sequence were scrambled to create control vector AAV2-CTRL5. Neither AAV2-CTRL5 nor an AAV2 vector encoding for green fluorescent protein (AAV2-GFP) suppressed Huntingtin messenger RNA when transduced into HEK293T cells, while AAV2-HD5 virus suppressed Huntingtin in these cells by ∼80% (Fig. 1C).
Using the surgical methods described above, we conducted a 28-day pilot study in one rhesus monkey in which AAV2-HD5 was co-infused into the striatum at a 1:1 ratio with AAV2-GFP for a total of 1.5 × 1011 vector genomes of each vector infused per hemisphere. Extensive GFP transgene expression was observed by fluorescence microscopy in serial brain tissue sections spanning the caudate and putamen (Supplementary Fig. 1) over an anterior–posterior posterior distance of 12 mm. The proportion of this monkey's putamen that was positive for GFP was quantified by volumetric reconstruction from every 12th 40 -µm tissue section using BioQuant software and found to be ∼64% of the putamenal volume. Huntingtin messenger RNA expression was reduced by an average of 60.6% in tissue samples captured by laser microdissection from GFP-positive regions, compared with adjacent GFP-negative regions (Fig. 1D).
Surgical targeting accuracy
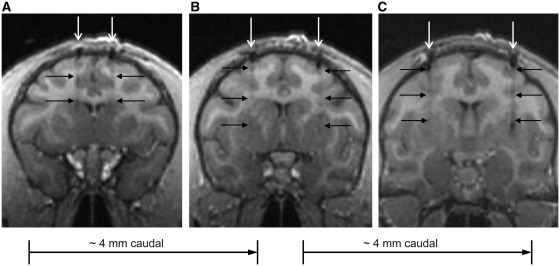
A representative example of needle placement in each site is shown in Fig. 2. Needle placements were confirmed by same-day post-surgical MRI to be within a radius of 2 mm from the planned targets in all animals. Bilateral AAV delivery into multiple striatal sites was well tolerated in all eight animals, as supported by the absence of observable adverse effects such as anorexia, infection, seizures or vomiting postoperatively.
Figure 2.
Representative T1-weighted MRI showing bilateral needle placement in (A) the caudate nucleus, (B) rostral putamen and (C) caudal putamen. White arrows indicate needle trajectories. Black arrows point out needle tracks in brain tissue.
Short hairpin RNA expression
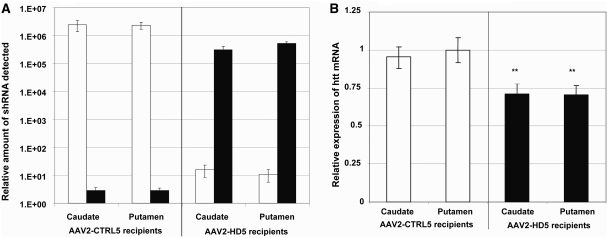
RNA was isolated from a tissue punch from the caudate and two punches from the putamen of each hemisphere of each monkey. Quantitative reverse transcription–polymerase chain reaction assays were performed using primers that directly detect the complementary DNA form of the processed hairpin transcript of CTRL5 or HD5, respectively. HD5 short hairpin RNA was detected above background only in monkeys receiving the AAV2-HD5 vector (P < 0.0001), and CTRL5 short hairpin RNA was detected above background only in monkeys receiving the AAV2-CTRL5 vector (P < 0.0001, Fig. 3A).
Figure 3.
(A) Expression levels of HD5 short hairpin RNA (shRNA: black bars) and CTRL5 short hairpin RNA (white bars) in tissue punches from the caudate (two per monkey, one per hemisphere, total n = 8 per group) and putamen (four per monkey, two per hemisphere, total n = 16 per group) of AAV2-HD5 or AAV2-CTRL5 recipients, relative to the minimum detectable amount by reverse transcription–polymerase chain reaction assay. Samples below the detection limit were assigned a value of zero as a raw score, and one as a log-transformed score. (B) Expression of huntingtin (Htt) messenger RNA levels in the same tissue punches and treatment groups (black bars = AAV2-HD5 recipients, white bars = AAV2-CTRL5 recipients) as a percentage of the average amount detected in monkeys receiving AAV2-CTRL5. **P < 0.015.
Huntingtin suppression
The level of huntingtin messenger RNA quantified from tissue punches (caudate n = 2 and putamen n = 4 punches per animal) was significantly reduced in monkeys receiving AAV2-HD5 compared with those receiving AAV2-CTRL5 (P < 0.05), with no significant differences related to hemisphere or punch location. Thus, data for the caudate nucleus were combined for the left and right hemispheres and data for the putamen were combined across hemispheres and location (rostral and caudal) providing a single value per brain region per animal. Compared with the monkeys receiving CTRL5, the expression of huntingtin messenger RNA was suppressed by an average of 28% [95% confidence interval (CI) = 15–41%] in the caudate and 29% (95% CI = 17–41%) in the putamen of monkeys receiving AAV2-HD5 (Fig. 3B).
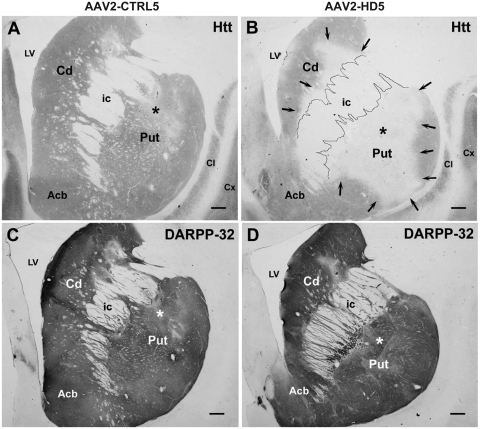
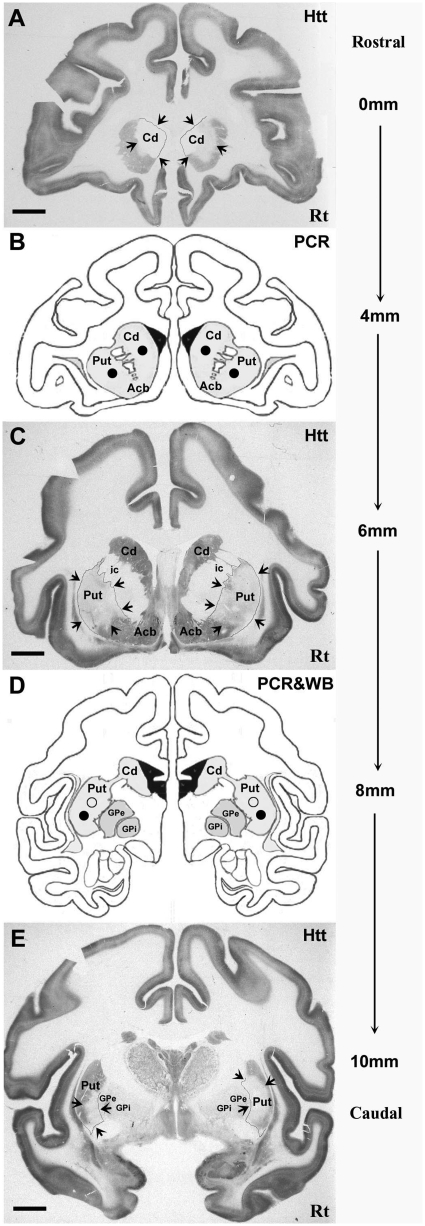
Immunostained tissue sections containing the caudate and putamen showed that Huntingtin protein was substantially reduced in monkeys receiving AAV2-HD5, including in regions not limited to the immediate site of the AAV infusion (Fig. 4A and B). Areas of decreased immunoreactivity seen at each of six injection sites averaged 5.88 ± 0.53 mm (medial–lateral axis) by 6.25 ± 1.01 mm (dorsal–ventral axis) in AAV2-HD5 recipients (P < 0.05 versus AAV2-CTRL5, Table 2). In contrast, immunostaining for DARPP-32, a marker for medium spiny neurons in the striatum, revealed no significant effect of AAV2-HD5 or CTRL5 in the same brain regions (Fig. 4C and D, Table 2). Based on the qualitative reduction in Huntingtin protein seen in immunostained sections from the slabs of caudate nucleus, rostral putamen and caudal putamen, and the quantitative reduction in huntingtin messenger RNA measured by reverse transcription–polymerase chain reaction assays of punches from adjacent 2-mm thick slabs, the Huntingtin suppression in the striatum extended at least 10 mm along the rostro-caudal axis. The anatomical extent of the Huntingtin suppression obtained in an AAV2-HD5 recipient is illustrated in Fig. 5, showing the rostral–caudal series of brain tissue sections containing the infusion sites, and intervening tissue slabs that were sources of tissue for molecular measurements.
Figure 4.
Immunohistochemistry for Huntingtin protein and DARPP-32. (A and B) Coronal tissue sections immunostained for huntingtin (Htt) (Chemicon, MAB2174) in the rostral putamen from rhesus monkeys receiving AAV2-CTRL5 (left, #RQ317) or AAV2-HD5 (right, #RQ476). Arrows highlight region of putamen in the AAV2-HD5 recipient in which staining for Huntingtin protein is reduced. Asterisks indicate approximate site of needle tip delivering the AAV2-HD5 or AAV2-CTRL5 vectors. (C and D) Coronal tissue sections immunostained for DARPP-32 in the rostral putamen from the same rhesus monkeys as in A and B, respectively. Acb = nucleus accumbens; Cd = caudate; Cl = claustrum; Cx = cortex; ic = internal capsule; LV = lateral ventricle; Put = putamen. Scale bars = 1 mm (×0.5).
Table 2.
Histopathological findings
| Markers | AAV2-CTRL5 | AAV2-HD5 | P-value |
|---|---|---|---|
| Mean ± SEM | Mean ± SEM | ||
| Scale (0–5) | |||
| Nissl/haematoxylin/eosin | 0.34 ± 0.12 | 0.88 ± 0.44 | NS |
| GFAP | 0.63 ± 0.23 | 1.23 ± 0.12 | NS |
| HLA-DR | 0.33 ± 0.10 | 0.56 ± 0.12 | NS |
| Width (mm) | |||
| Nissl/haematoxylin/eosin | 0.90 ± 0.50 | 1.52 ± 0.53 | ns |
| GFAP | 2.96 ± 1.06 | 4.92 ± 0.54 | ns |
| HLA-DR | 1.23 ± 0.55 | 2.04 ± 0.99 | ns |
| DARPP32 | 1.67 ± 0.97 | 1.77 ± 0.76 | ns |
| Huntingtin | 1.69 ± 0.73 | 5.88 ± 0.53 | <0.002 |
| Height (mm) | |||
| Nissl/haematoxylin/eosin | 0.92 ± 0.46 | 1.52 ± 0.44 | ns |
| GFAP | 3.17 ± 1.23 | 5.75 ± 0.51 | ns |
| HLA-DR | 1.06 ± 0.47 | 2.42 ± 1.14 | ns |
| DARPP32 | 2.48 ± 1.46 | 2.15 ± 1.00 | ns |
| Huntingtin | 2.69 ± 1.46 | 6.25 ± 1.01 | <0.046 |
Scale = blind semi-quantitative ratings on a 0–5 ordinal severity scale, where 0 = no or minimal pathology, and 5 = severe pathology (Supplementary Table 1).
‘Width’ and ‘Height’ = measurements at the injection site related to cell loss (Nissl), astrocytosis (GFAP) or microglial response (HLA-DR). For DARPP-32 and huntingtin, ‘Width’ and ‘Height’, denote the area in terms of decreased immunoreactivity. For each marker, data are expressed as the averaged results for all six needle insertion sites per animal providing a single, overall value of the pathology in the brain for all sites combined. NS = not significant (P > 0.05), one-tailed, Mann–Whitney U-test; ns = not significant (P > 0.05), one-tailed, unpaired t-test.
Figure 5.
Immunohistochemistry for Huntingtin (Htt) protein and tissue punch location for polymerase chain reaction (PCR) and western blot (WB) analyses. (A, C and E) Full coronal tissue sections immunostained for Huntingtin in the caudate nucleus (A), rostral putamen (C) and caudal putamen (E) are shown from a rhesus monkey receiving AAV2-HD5. Arrows highlight regions in which staining for Huntingtin protein is bilaterally reduced for each brain target over a distance of at least 10 mm along the rostro-caudal axis. (B and D) Diagrams of coronal tissue sections adjacent to those immunostained for Huntingtin depicting the location of tissue punches in the caudate nucleus and putamen used to assess huntingtin messenger RNA levels by polymerase chain reaction (filled circles, B and D) and Huntingtin protein expression by western blots (open circles, D). Acb = nucleus accumbens; Cd = caudate nucleus; ic = internal capsule; GPe = external globus pallidus; GPi = internal; globus pallidus; Put = putamen; Rt = Right. Scale bars = 5 mm.
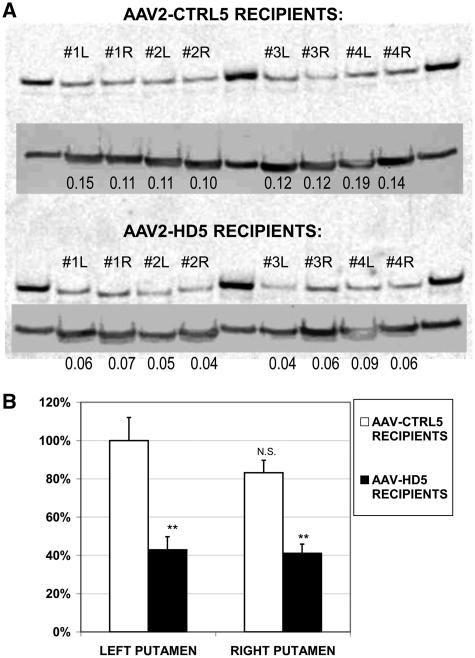
To further characterize and confirm the protein suppression observed by immunohistochemistry, protein lysates from tissue punches from the left and right putamen of each monkey (at the locations shown in Fig. 5D) were obtained and analysed by western blot. A single band of the expected molecular weight for Huntingtin protein (∼350 kDa) was detected in each sample (Fig. 6). Analysis of variance of the normalized densitometric values revealed no significant differences between left and right hemispheres (P = 0.119) nor interaction between AAV2-HD5 or CTRL5 treatment and hemisphere (P = 0.195), as expected, while the effect of AAV2-HD5 versus CTRL5 treatment was significant (P = 0.0027). Compared with the monkeys receiving CTRL5, the expression of Huntingtin protein in punches from the putamen of monkeys receiving AAV2-HD2 was suppressed by an average of 45.8% (observed range: 31.6–67.5%).
Figure 6.
(A) Western blot of Huntingtin protein in tissue punches from left (L) and right (R) putamen of the four monkeys receiving AAV-CTRL5 (top) or the four receiving AAV-HD5 (bottom) Lanes 2–5 and 7–10 (Lanes 1, 6 and 11 are protein standards). Insets show same blot re-probed and imaged (at a shorter exposure time) for tubulin protein. Numbers in each lane provide the ratio of Huntingtin to tubulin densitometry values. (B) Amount of Huntingtin protein (normalized to tubulin) in left or right putamen of each group of monkeys, relative to average amount in left putamen of monkeys receiving AAV-CTRL5, **P < 0.01 AAV-HD5 versus AAV-CTRL5 by hemisphere. NS = left versus right putamen difference in AAV-CTRL5 recipients is not statistically significant.
Body weight and food consumption
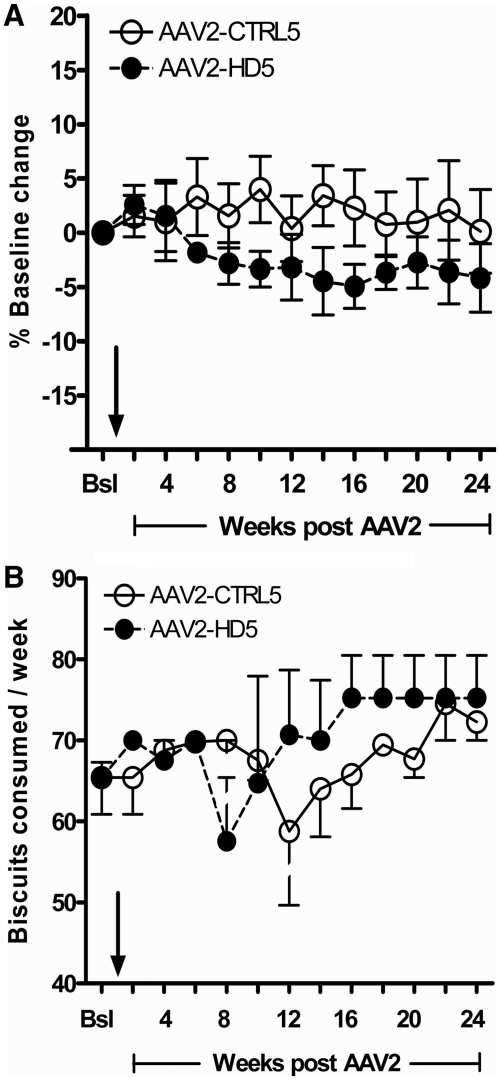
When each animal's weight at any post-surgical time point was compared with its own baseline weight, there was no difference between AAV groups (P = 0.286), nor any difference between groups as a function of time (vector by time interaction, P = 0.159) (Fig. 7A). Food consumption was comparable between AAV2-HD5 and AAV2-CTRL5 recipients preoperatively and generally remained within or above baseline level postoperatively for both treatment groups (Fig. 7B). Transient reduction in appetite was seen in one CTRL-5 (#93B652) and one HD-5 (#RQ476) animal mid-study. Animal #93B652 received veterinary care for a laceration to the mouth (left commissure) and #RQ476 was treated for gastrointestinal motility problems. Both animals resumed their normal eating habits within ∼2 weeks. The other six animals did not require any veterinary care during the study.
Figure 7.
Change in body weight (A) and food consumption (B) over time. The effect of AAV2-HD5 (anti-huntingtin short hairpin RNA) administration on body weights and food consumption was comparable to that recorded in control animals (AAV2-CTRL5). The arrows indicate AAV delivery. Bsl = baseline.
Motor function
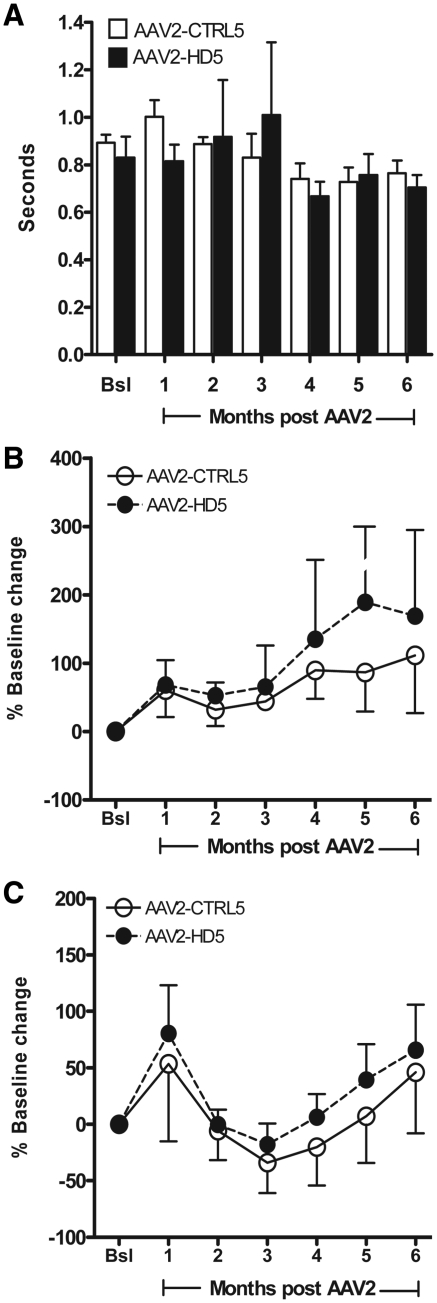
Upper limb motor performance times were not adversely affected by either AAV2-CTRL5 or AAV2-HD5 delivery (Fig. 8A). In fact, motor performance improved over time for both treatment groups. Repeated-measures ANOVA yielded a significant main effect of time (P < 0.05), but no significant effect of vector (P = 0.77) nor interaction between vector and time (P = 0.43). Motor memory preservation was not adversely affected by either AAV2-CTRL5 or HD5 delivery as all animals recalled and performed the retrieval task without intervening practice between the monthly test sessions.
Figure 8.
(A) Change in motor performance on the hand/arm-retrieval task over time. All animals recalled and were able to perform the task after AAV2 delivery. Overall, upper limb motor function was not adversely affected by AAV2-CTRL5 (open bars) or AAV2-HD5 delivery (filled bars). (B and C) No significant differences were seen in day time (B) or night time (C) home-cage activity levels between AAV2-CTRL5 (open circles) and AAV2-HD5 recipients (filled circles). Bsl = baseline.
Home-cage activity levels were quantified using accelerometers. The repeated-measures ANOVA for the average daytime activity level by month revealed no statistically significant overall or month-by-month differences between monkeys receiving AAV2-HD5 versus AAV2-CTRL5 (main effect of vector, P = 0.61, vector by month interaction, P = 0.96, main effect of month, P = 0.33, Fig. 8B). Similarly, the repeated-measures ANOVA for the average night time activity level by month revealed no statistically significant differences between treatment groups (main effect of vector, P = 0.58, vector by month interaction, P = 0.99, Fig. 8C). There were significant overall differences in average night time activity across months, regardless of the vector received by the monkey (main effect of month, P = 0.03), in part due to a transient increase in night time activity in the first month post-surgery (Fig. 8C), perhaps reflective of post-surgical restlessness that later resolved.
Neuropathology
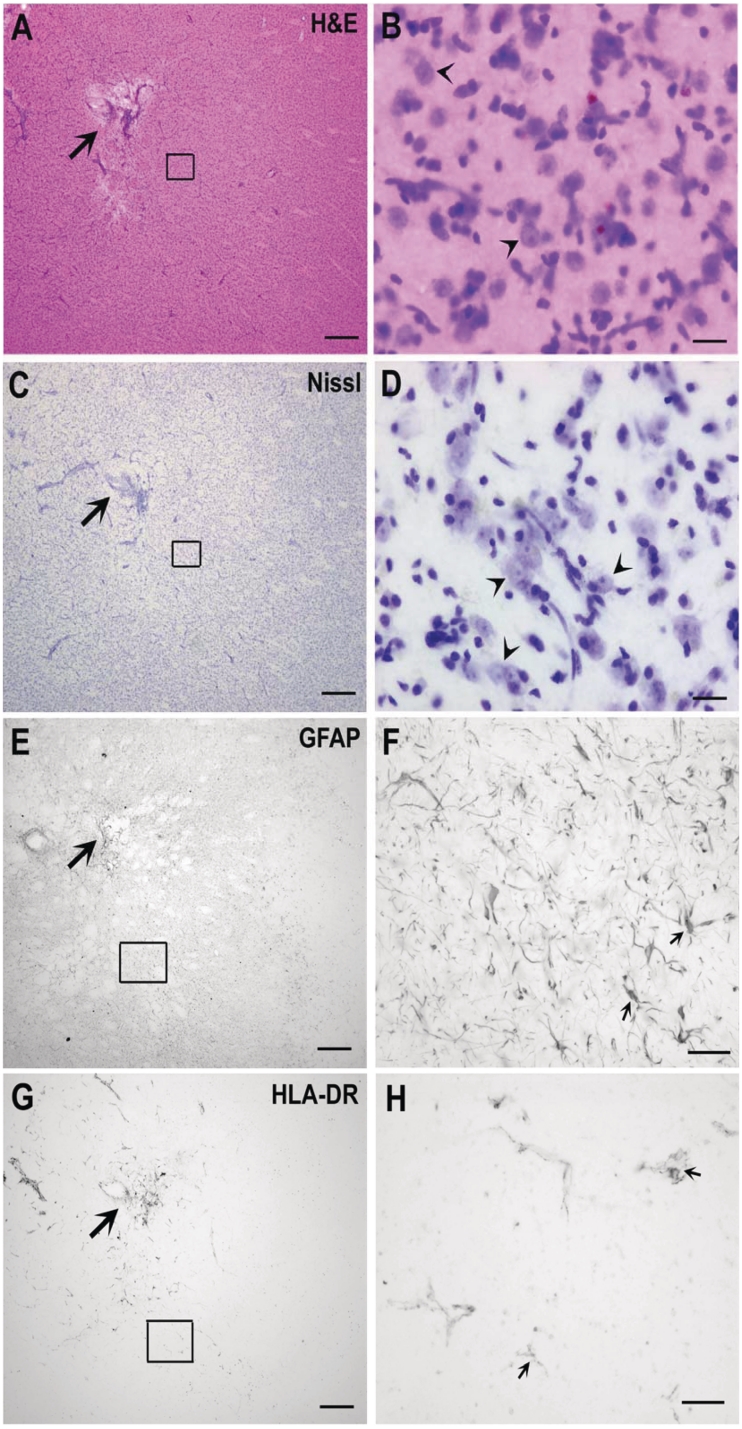
Using three to four sections per marker for each brain, histopathological examinations were conducted bilaterally on full coronal sections cut along the anterior–posterior axis at the level where each needle insertion was made in the striatum. For Nissl, GFAP and HLA-DR markers, the severity of pathological changes related to the injection sites or connected structures was low for AAV2-HD5 recipients and comparable to that seen in AAV2-CTRL5 recipients (Table 2). As illustrated in Fig. 9 for AAV2-HD5 recipient #RQ476, there was no discernible neuronal loss, or abnormal astrocytosis, nor evidence of chronic infection, or discernible haemorrhage (acute or chronic) noted on microscopic evaluation in any of the animals.
Figure 9.
Brain tissue pathology. Representative coronal tissue sections stained for haematoxylin and eosin (H&E; A and B), Nissl (C and D), glial fibrillary acidic protein (GFAP; E and F) and HLA-DR (G and H) in the region of the rostral putamen from a rhesus monkey receiving AAV2-HD5 (#RQ476). Histopathological changes in AAV2-HD5 recipients were comparable to those seen in the AAV2-CTRL5 recipients (Table 2). There was no discernible neuronal loss or abnormal astrocytosis noted on microscopic evaluations. Arrows indicate approximate site of needle tip delivering the AAV2-HD5 vectors. Scale bars: A, C, E and G = 400 µm (×2). Scale bars for the higher magnification inset panels from the left column = 20 µm (×40, B and D) and 50 µm (×20, F and H).
Discussion
To test the hypothesis that the adult brain can tolerate partially reduced levels of wild-type Huntingtin protein, we developed an AAV2 vector encoding for a short hairpin RNA (HD5) targeting rhesus huntingtin messenger RNA. To our knowledge, this is the first report of a study investigating the effects of suppressing wild-type Huntingtin for as long as 6 months in the adult primate brain. Six months following bilateral infusion into the caudate nucleus and putamen of adult rhesus monkeys, we detected HD5 short hairpin RNA transcripts in the targeted brain regions and a corresponding suppression of huntingtin messenger RNA averaging ∼30% compared with AAV2-CTRL5 animals. We measured a corresponding suppression of Huntingtin protein in the putamen averaging ∼45% bilaterally compared with the putamen of AAV2-CTRL5 animals. These results are not inconsistent, as a 1:1 ratio of huntingtin messenger RNA to Huntingtin protein suppression is not necessarily expected. Also, the results are not inconsistent with our in vitro data from the development of AAV2-HD5 or the in vivo data from the pilot animal, because a higher level of messenger RNA and protein suppression may be expected to be measured in a homogenous cell culture or tissue isolated for transgene expression by laser microdissection.
Although data on huntingtin messenger RNA and protein reduction were only obtained at termination, our pilot data (Kaemmerer et al., 2006) as well as other studies utilizing AAV2 vectors in rodents and rhesus monkeys (McCarty et al., 2003; Sanftner et al., 2005) indicate that expression of the AAV-delivered short hairpin RNA occurs within a few weeks post-surgery. Therefore, striatal expression of wild-type huntingtin messenger RNA and corresponding protein in the AAV2-HD5 recipients was reduced for at least 5 months without causing motor dysfunction or marked pathology, supporting the interpretation that the adult primate brain can tolerate sustained partial suppression of wild-type Huntingtin. Other studies using short hairpin RNA to suppress Huntingtin expression in the mouse striatum using AAV1 vectors have observed toxicity after 15 weeks as revealed by a reduction in immunostaining for DARPP-32 (McBride et al., 2008). These authors found that this toxicity can be avoided by utilizing mitochondrial RNA-like sequences to encode the active molecule, rather than short hairpin RNA sequences such as those used in our study. Nevertheless, we did not observe toxicity or a significant reduction in DARPP-32 immunostaining resulting from expression of either AAV2-HD5 or AAV2-CTRL5 short hairpin RNA in our study.
Prior studies investigating Huntingtin reduction in the brain have utilized normal (Drouet et al., 2009) or transgenic rodents (Harper et al., 2005; Wang et al., 2005) or have investigated Huntingtin reduction starting in early development (Dragatsis et al., 2000). There are legitimate concerns that reduction of wild-type Huntingtin expression in the brain of patients with Huntington's disease, concomitant with reduction in the disease-causing mutant protein, may have negative as well as beneficial effects (Sah and Aronin, 2011). However, the results of our study in adult rhesus monkeys suggest that the negative effects might be minimal relative to the benefit that could result from reduction of the mutant protein, if partial reduction of the mutant protein ranging from 40% to 50% is beneficial in patients.
There are a number of limitations to the interpretation of our results. First, the length of our study was 6 months, roughly equivalent to 18 months in humans considering that rhesus monkeys age at a rate approximately three times faster (Tigges et al., 1988; Gore and Terasawa, 1991; Andersen et al., 1999). In contrast, the time required for mutant Huntingtin to result in overt pathology in humans is at least a few decades. It is possible that the time required for a reduction in wild-type Huntingtin to have ill effects may also be measured in years. Also, our sample size may not have allowed detection of mildly negative effects. Since Huntingtin is not a secreted protein, the reduction in Huntingtin protein levels was limited to those cells transduced by the AAV2 vector, (predominantly neurons; Tenenbaum et al., 2004), which would be <100% of the neurons in any given region. Conversely, since we measured huntingtin messenger RNA reduction in tissue punches, not in individual cells (as even with laser microdissection, there is no standard against which to determine absolute Huntingtin reduction in a single cell), it is possible that we achieved substantially >30% reduction of huntingtin messenger RNA in individual neurons (e.g. up to 100% reduction in 30% of the neurons). Nevertheless, neuronal loss was not observed. However, it is possible that reduction of wild-type Huntingtin protein in astrocytes, as well as neurons, could have different effects than observed in our study. Finally, it is possible that suppression of wild-type Huntingtin has negative effects on other factors that we did not measure, such as the rate of neurogenesis (Godin et al., 2010). Despite these limitations, the results of our study in adult rhesus monkeys extend those obtained in rodents (Boudreau et al., 2009; Drouet et al., 2009) and suggest that non-allele-specific, partial reduction of Huntingtin expression in striatal brain regions may be a viable approach for the treatment of Huntington's disease. Because use of anti-Huntingtin RNA interference offers the possibility of a disease-modifying treatment, it may be one of the more promising therapies under development for this devastating disease for the foreseeable future.
Funding
Medtronic, Inc. [to R.G., Y.A. and Z.Z.]; University of Kentucky College of Medicine Start-up Funds [to R.G.].
Supplementary material
Supplementary material is available at Brain online.
Acknowledgements
The expert advice of Dr Don M. Gash during the conduct of this study, and the assistance of Christos Kolaris (QPS Pharmacogenomics) and Karen Hayes in performing western blotting is gratefully acknowledged. The authors also thank Dr Peter Hardy for providing MRI acquisition expertise, and Hamed Haghnazar and Daisy Ramos for their excellent technical assistance.
Glossary
Abbreviations
- AAV
adeno-associated virus
- GFP
green fluorescent protein
References
- Ai Y, Markesbery W, Zhang Z, Grondin R, Elseberry D, Gerhardt GA, et al. Intraputamenal infusion of GDNF in aged rhesus monkeys: distribution and dopaminergic effects. J Comp Neurol. 2003;461:250–61. doi: 10.1002/cne.10689. [DOI] [PubMed] [Google Scholar]
- Andersen AH, Zhang Z, Zhang M, Gash DM, Avison MJ. Age-associated changes in rhesus CNS composition identified by MRI. Brain Res. 1999;829:90–8. doi: 10.1016/s0006-8993(99)01343-8. [DOI] [PubMed] [Google Scholar]
- Auerbach W, Hurlbert MS, Hilditch-Maguire P, Wadghiri YZ, Wheeler VC, Cohen SI, et al. The HD mutation causes progressive lethal neurological disease in mice expressing reduced levels of huntingtin. Hum Mol Genet. 2001;10:2515–23. doi: 10.1093/hmg/10.22.2515. [DOI] [PubMed] [Google Scholar]
- Aylward EH, Sparks BF, Field KM, Yallapragada V, Shpritz BD, Rosenblatt A, et al. Onset and rate of striatal atrophy in preclinical Huntington disease. Neurology. 2004;63:66–72. doi: 10.1212/01.wnl.0000132965.14653.d1. [DOI] [PubMed] [Google Scholar]
- Boudreau RL, McBride JL, Martins I, Shen S, Xing Y, Carter BJ, et al. Nonallele-specific silencing of mutant and wild-type huntingtin demonstrates therapeutic efficacy in Huntington's disease mice. Mol Ther. 2009;17:1053–63. doi: 10.1038/mt.2009.17. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Colin E, Zala D, Liot G, Rangone H, Borrell-Pages M, Li XJ, et al. Huntingtin phosphorylation acts as a molecular switch for anterograde/retrograde transport in neurons. EMBO J. 2008;27:2124–34. doi: 10.1038/emboj.2008.133. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dragatsis I, Levine MS, Zeitlin S. Inactivation of Hdh in the brain and testis results in progressive neurodegeneration and sterility in mice. Nat Genet. 2000;26:300–6. doi: 10.1038/81593. [DOI] [PubMed] [Google Scholar]
- Drouet V, Perrin V, Hassig R, Dufour N, Auregan G, Alves S, et al. Sustained effects of nonallele-specific Huntingtin silencing. Ann Neurol. 2009;65:276–85. doi: 10.1002/ana.21569. [DOI] [PubMed] [Google Scholar]
- Duyao MP, Auerbach AB, Ryan A, Persichetti F, Barnes GT, McNeil SM, et al. Inactivation of the mouse Huntington's disease gene homolog Hdh. Science. 1995;269:407–10. doi: 10.1126/science.7618107. [DOI] [PubMed] [Google Scholar]
- Ferreira T, Rasband W. 2011. ImageJ User Guide IJ 1.45m. http://rsb.info.nih.gov/ij/docs/guide/userguide-27.html#toc-Subsection-27.13. (6 August 2011, date last accessed) [Google Scholar]
- Gabery S, Murphy K, Schultz K, Loy CT, McCusker E, Kirik D, et al. Changes in key hypothalamic neuropeptide populations in Huntington disease revealed by neuropathological analyses. Acta Neuropathol. 2010;120:777–88. doi: 10.1007/s00401-010-0742-6. [DOI] [PubMed] [Google Scholar]
- Godin JD, Colombo K, Molina-Calavita M, Keryer G, Zala D, Charrin BC, et al. Huntingtin is required for mitotic spindle orientation and mammalian neurogenesis. Neuron. 2010;67:392–406. doi: 10.1016/j.neuron.2010.06.027. [DOI] [PubMed] [Google Scholar]
- Gore AC, Terasawa E. A role for norepinephrine in the control of puberty in the female rhesus monkey, Macaca mulatta. Endocrinology. 1991;129:3009–17. doi: 10.1210/endo-129-6-3009. [DOI] [PubMed] [Google Scholar]
- Grondin R, Cass WA, Zhang Z, Stanford JA, Gerhardt GA, Gash DM. GDNF increases stimulus-evoked dopamine release and motor speed in aged rhesus monkeys. J Neurosci. 2003;23:1974–80. doi: 10.1523/JNEUROSCI.23-05-01974.2003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Harper SQ, Staber PD, He X, Eliason SL, Martins IH, Mao Q, et al. RNA interference improves motor and neuropathological abnormalities in a Huntington's disease mouse model. Proc Natl Acad Sci USA. 2005;102:5820–5. doi: 10.1073/pnas.0501507102. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kaemmerer WF, Burright EN, Kaytor MD, Blum JL, Heisel J, Thakker DR, et al. The 36th Annual Meeting of the Society for Neuroscience; 2006. Atlanta, GA: 2006. A pilot study of the safety of suppressing wildtype huntingtin in the primate striatum. [Google Scholar]
- Lombardi MS, Jaspers L, Spronkmans C, Gellera C, Taroni F, Di Maria E, et al. A majority of Huntington's disease patients may be treatable by individualized allele-specific RNA interference. Exp Neurol. 2009;217:312–9. doi: 10.1016/j.expneurol.2009.03.004. [DOI] [PubMed] [Google Scholar]
- McBride JL, Boudreau RL, Harper SQ, Staber PD, Monteys AM, Martins I, et al. Artificial miRNAs mitigate shRNA-mediated toxicity in the brain: implications for the therapeutic development of RNAi. Proc Natl Acad Sci USA. 2008;105:5868–73. doi: 10.1073/pnas.0801775105. [DOI] [PMC free article] [PubMed] [Google Scholar]
- McCarty DM, Fu H, Monahan PE, Toulson CE, Naik P, Samulski RJ. Adeno-associated virus terminal repeat (TR) mutant generates self-complementary vectors to overcome the rate-limiting step to transduction in vivo. Gene Ther. 2003;10:2112–8. doi: 10.1038/sj.gt.3302134. [DOI] [PubMed] [Google Scholar]
- Menalled LB, Miller S, Fitzpatrick J, Kudwa A, Morin C, Patry M, et al. The 39th Annual Meeting of the Society for Neuroscience; 2009. Chicago, IL: 2009. Conditional knock-down of endogenous huntingtin in mice: Behavioral characterization. [Google Scholar]
- Sah DW, Aronin N. Oligonucleotide therapeutic approaches for Huntington disease. J Clin Invest. 2011;121:500–7. doi: 10.1172/JCI45130. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sanftner LM, Sommer JM, Suzuki BM, Smith PH, Vijay S, Vargas JA, et al. AAV2-mediated gene delivery to monkey putamen: evaluation of an infusion device and delivery parameters. Exp Neurol. 2005;194:476–83. doi: 10.1016/j.expneurol.2005.03.007. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sass M, Aronin N. Neurobiology of Huntington's Disease: Applications to Drug Discovery. 2nd edn. Boca Raton, Florida: CRC Press; 2011. RNA- and DNA- Based Therapies for Huntington's Disease. [Google Scholar]
- Tabrizi SJ, Langbehn DR, Leavitt BR, Roos RA, Durr A, Craufurd D, et al. Biological and clinical manifestations of Huntington's disease in the longitudinal TRACK-HD study: cross-sectional analysis of baseline data. Lancet Neurol. 2009;8:791–801. doi: 10.1016/S1474-4422(09)70170-X. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tenenbaum L, Chtarto A, Lehtonen E, Velu T, Brotchi J, Levivier M. Recombinant AAV-mediated gene delivery to the central nervous system. J Gene Med. 2004;6(Suppl 1):S212–22. doi: 10.1002/jgm.506. [DOI] [PubMed] [Google Scholar]
- Tigges J, Gordon TP, McClure HM, Hall EC, Peters A. Survival rate and life span of rhesus monkeys at the Yerkes regional primate research center. American Journal of Primatology. 1988;15:263–73. doi: 10.1002/ajp.1350150308. [DOI] [PubMed] [Google Scholar]
- van Bilsen PH, Jaspers L, Lombardi MS, Odekerken JC, Burright EN, Kaemmerer WF. Identification and allele-specific silencing of the mutant huntingtin allele in Huntington's disease patient-derived fibroblasts. Hum Gene Ther. 2008;19:710–9. doi: 10.1089/hum.2007.116. [DOI] [PubMed] [Google Scholar]
- Walton A, Scheib JL, McLean S, Zhang Z, Grondin R. Motor memory preservation in aged monkeys mirrors that of aged humans on a similar task. Neurobiol Aging. 2008;29:1556–62. doi: 10.1016/j.neurobiolaging.2007.03.016. [DOI] [PubMed] [Google Scholar]
- Wang YL, Liu W, Wada E, Murata M, Wada K, Kanazawa I. Clinico-pathological rescue of a model mouse of Huntington's disease by siRNA. Neurosci Res. 2005;53:241–9. doi: 10.1016/j.neures.2005.06.021. [DOI] [PubMed] [Google Scholar]
- Zuccato C, Cattaneo E. Role of brain-derived neurotrophic factor in Huntington's disease. Prog Neurobiol. 2007;81:294–330. doi: 10.1016/j.pneurobio.2007.01.003. [DOI] [PubMed] [Google Scholar]
- Zuccato C, Valenza M, Cattaneo E. Molecular mechanisms and potential therapeutical targets in Huntington's disease. Physiol Rev. 2010;90:905–81. doi: 10.1152/physrev.00041.2009. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.