Abstract
Most chloroplast mRNAs are processed from larger precursors. Several mechanisms have been proposed to mediate these processing events, including site-specific cleavage and the stalling of exonucleases by RNA structures. A protein barrier mechanism was proposed based on analysis of the pentatricopeptide repeat (PPR) protein PPR10: PPR10 binds two intercistronic regions and impedes 5′- and 3′-exonucleases, resulting in processed RNAs with PPR10 bound at the 5′- or 3′-end. In this study, we provide evidence that protein barriers are the predominant means for defining processed mRNA termini in chloroplasts. First, we map additional RNA termini whose arrangement suggests biogenesis via a PPR10-like mechanism. Second, we show that the PPR protein HCF152 binds to the immediate 5′- or 3′-termini of transcripts that require HCF152 for their accumulation, providing evidence that HCF152 defines RNA termini by blocking exonucleases. Finally, we build on the observation that the PPR10 and HCF152 binding sites accumulate as small chloroplast RNAs to infer binding sites of other PPR proteins. We show that most processed mRNA termini are represented by small RNAs whose sequences are highly conserved. We suggest that each such small RNA is the footprint of a PPR-like protein that protects the adjacent RNA from degradation.
INTRODUCTION
Gene expression in chloroplasts involves core transcription, translation and RNA turnover machineries that were acquired from the chloroplast's cyanobacterial ancestor (1). These ancient mechanisms function in concert with more recently evolved RNA processing steps that include RNA editing, the processing of mRNA termini and the protein-facilitated splicing of group II introns. In land plant chloroplasts, the majority of protein-coding genes are found in polycistronic transcription units that give rise to complex transcript populations via processing between coding regions (intercistronic processing) and upstream of the 5′ open reading frame (5′-processing). Where orthologous transcription units have been examined, the populations of processed transcripts are highly conserved between monocot, dicot and even non-vascular plants (2–4). However, the mechanisms and functional consequences of these widespread and conserved RNA processing events remain subjects of debate.
Genetic analyses have highlighted members of the pentatricopeptide repeat (PPR) family as effectors of intercistronic and 5′ RNA processing in chloroplasts. PPR proteins are defined by tandem arrays of a degenerate 35 amino acid repeating unit, which are predicted to form an elongated solenoid consisting of stacked helical repeats (5). The PPR proteins CRP1, PPR10 and HCF152 are each required for the accumulation of chloroplast RNAs with processed 5′- or 3′-ends mapping in specific intergenic regions (6–9). The underlying mechanism has been described for PPR10, which binds RNA segments in each of two intergenic regions and impedes exoribonucleases intruding from either direction (7,10). Genetic data implicate other PPR proteins as well as ‘PPR-like’ proteins with distinct helical repeat architectures in stabilizing chloroplast RNAs with specific 5′ termini (11–16). Together, these observations suggest that intercistronic RNA processing, 5′ RNA processing and 5′ RNA stabilization in chloroplasts involve similar mechanisms: in each case a helical repeat protein binds a specific RNA segment and protects the adjacent RNA by serving as a barrier to exoribonucleases.
Although there is considerable evidence that this mechanism accounts for the processing of several chloroplast mRNAs, its global impact on the chloroplast transcriptome is unknown. In fact, stable RNA structures provide an alternative mechanism for impeding the vectorial degradation of chloroplast mRNAs from both the 5′ and 3′ directions (17), and the involvement of site-specific endonucleases in intercistronic processing has typically been invoked. In this study, we provide evidence that protection by PPR or PPR-like proteins is the predominant mechanism for defining the positions of processed 5′ and intercistronic mRNA termini in land plant chloroplasts. In addition, we use the attributes of known PPR binding sites to infer likely binding sites for PPR (or PPR-like) proteins on chloroplast mRNAs for which stabilizing proteins have not been identified.
MATERIALS AND METHODS
Genome-wide mapping of 5′-termini in barley chloroplasts
Chloroplasts purified from the first leaf of 11-day-old barley seedlings were used for RNA extraction. RNA (7 µg) was treated with 7 units of Terminator™ exonuclease (TEX; Epicentre #TER51020) or in buffer alone for 60 min at 30°C. After phenol–chloroform extraction and ethanol precipitation, the RNA was further treated with 1 unit tobacco acid pyrophosphatase (Epicentre #T19100) for 1 h at 37°C to generate 5′-monophosphates for linker ligation, and again purified by organic extraction and ethanol precipitation. cDNA library preparation and 454 pyrosequencing were performed as previously described (18) but without size fractionation. Sequencing was performed on Roche 454 FLX instruments at the MPI for Molecular Genetics (Berlin, Germany). 5′-Linker and polyA-tail-clipped reads longer than 17-nt were aligned to the H. vulgare chloroplast genome (NC_008590) using WU Blast 2.0 with the following parameters: −B = 1, −V = 1, −m = 1, −n = −3, −Q = 3, −R = 3, −gspmax = 1, −hspmax = 1, −mformat = 2, −e = 0.0001. Graphs representing the number of mapped reads per nt were calculated and visualized with the Integrated Genome Browser version 6.1 (http://genoviz.sourceforge.net/).
Mapping RNA termini in maize
The termini of several chloroplast mRNAs in maize were mapped by circular RT–PCR (cRT–PCR) or primer extension according to the methods described in (7). The primers and inferred map positions are listed in Supplementary Table S1. RNA gel blots were prepared with 5 µg seedling leaf RNA per lane, and hybridized using the conditions described previously for oligonucleotide probes (7).
Mapping 3′-RNA termini in barley
3′-RACE analysis used endogenous chloroplast 16S rRNA to provide the primer binding site for reverse transcription. Primers are listed in Supplementary Table S2. RNA (1 µg) from barley chloroplasts was treated with 40 U T4 RNA ligase (Epicentre) in the presence of 1 mM ATP and 40 U of RNase Inhibitor (Fermentas) for 60 min at 37°C. RNA was purified by phenol–chloroform extraction and ethanol precipitation. Reverse-transcription reactions used a primer complementary to sequences near the 5′-end of 16S rRNA and SuperScript®III (Invitrogen) according to the manufacturer's protocol. Products were purified by organic extraction and ethanol precipitation, and used in nested PCR reactions with gene-specific primers (forward) and rrn16 (reverse) primers at an annealing temperature of 55°C in the first PCR and 58°C in the subsequent, nested PCR. PCR products were resolved on 1.5% agarose gels, excised, cloned into pGEM(®-T (Promega) and transformed into Escherichia coli TOP10 cells. Approximately 10 insert-containing clones were sequenced for each terminus mapped.
RNA gel blot analysis of sRNAs
Leaf RNA (15 µg) from 8-day-old maize seedlings was electrophoresed through small-format 15% polyacrylamide gels containing 8 M urea and 1× TBE (90 mM Tris base, 90 mM boric acid, 2 mM EDTA, pH 8). Synthetic RNA oligonucleotides (200 pg) mimicking the putative PPR10 and CRP1 footprints were analyzed in parallel, to serve as hybridization controls and as size markers. RNAs were denatured by heating in an equal volume of denaturation buffer (90% formamide, 20 mM EDTA pH 8, 20 mM Tris–HCl pH 7.5, 0.04% bromophenol blue and xylene cyanol) and electrophoresed until the bromophenol blue was ∼2 cm from the bottom of the gel. The RNA was transferred to MagnaCharge nylon (Fisher Scientific) in a mini-transblot apparatus (Bio-Rad) for 60 min at 80 V in 0.5× TBE at 4°C. Blots were prehybridized and hybridized in 7% SDS, 0.5 M Na2HPO4 at 37°C, using the following synthetic DNA oligonucleotides as probes: petB–petD sRNA probe (5′-AGCAATGAAATACCACAACCTACCCGATATG), atpH 5′-UTR sRNA probe (5′-AAAAGAAATGGTTAAGGATACAAT). Blots were rinsed two times with 0.2% SDS, 5× SSC, washed twice for 5 min in the same buffer at 37°C, and then imaged with a Storm phosphorimager (Molecular Dynamics).
Expression of recombinant HCF152
The coding sequences of mature HCF152 from Arabidopsis (At), maize (Zm) and rice (Os) were amplified from leaf DNA using Phusion DNA polymerase (New England Biolabs). AtHCF152 was amplified with primers: 5′ CACAggatccGCTAATAGCTCCGCCGAAGACCTCTCG and 5′ CACAgtcgacCTAGTCTTCTCTTGGACCTAAC. ZmHCF152 was amplified with primers 5′ CACAggatccGCTACTTCCCGCTCCAGCACACC and 5′ CACAaagcttCTAACTTAGGTCATCGCCATCC. OsHCF152 was amplified in two steps in order to delete an internal BamHI site. First, two overlapping fragments were amplified with primer pairs: (i) OsHCF152BamHI (5′ CACAggatccGCTGCTGCATCCTCCACGC) and OsHCF152 IR (5′CAGAGGAGAgaatccGGTGGAGCACG) and (ii) OsHCF152IF (5′ CGTGCTCCACCggattcTCTCCTCTG) and OsHCF152HindIII (5′ CACAaagcttctaGTTGAGGCCGTCGTCTTGG). Second, the two fragments were joined by amplification with primers Os HCF152 BamHI and Os HCF152 HindIII. The PCR products were digested with BamHI and SalI (AtHCF152) or BamHI and HindIII (ZmHCF152 and OsHCF152). These were cloned into pMAL-TEV to generate in-frame fusions with maltose-binding protein (MBP), transformed into Rosetta-2 cells and used for protein induction as described for APO1 (19). The MBP-HCF152 fusion proteins were purified by successive amylose affinity and size-exclusion chromatography as for MBP-ZmAPO1, except that the lysis buffer included 0.02% CHAPS and Buffer A included 250 mM NaCl.
Gel mobility shift assays
Gel mobility shift assays were performed as previously described (10). Briefly, synthetic RNAs (IDT) were 5′-end labeled with [γ-32P]-ATP and T4 polynucleotide kinase. RNAs were purified by denaturing gel electrophoresis, followed by phenol–chloroform extraction and ethanol precipitation. Binding reactions contained 180 mM NaCl, 25 mM Tris–HCl pH 7.5, 4 mM DTT, 0.1 mg/ml BSA, 0.5 mg/ml heparin, 10% glycerol, 0.4 units RNAsin (Promega), 15 pM radiolabeled RNA and protein concentrations as indicated. Reactions were incubated for 20 min at 25°C and resolved on 5% native polyacrylamide gels. Results were visualized on a STORM phosphorimager.
Multiple sequence alignments and RNA structure predictions
Multiple sequence alignments were made with Clustal W and included adjacent coding regions for anchors. The aligned sequences come from chloroplast genomes of barley (Hv; NC_008590), maize (Zm; NC_001666), rice (Os; NC_001320), poplar (Pa; NC_008235), Arabidopsis (At; NC_000932), tobacco (Nt; NC_001879) and moss (Pp; NC_005087). Secondary structures were predicted with the Mfold server at http://mfold.rna.albany.edu/?q=mfold/ using default parameters.
RESULTS
The positions of RNA termini in the clpP–rps12 intergenic region provide evidence for RNA processing via a blockade to exonucleolytic decay
We previously mapped the RNA termini in four intergenic regions in maize chloroplasts (atpI–atpH, psaJ–rpl33, psbH–petB, petB–petD); in each case, the processed 5′-end from the downstream gene maps ∼25-nt upstream of the processed 3′-end from the upstream gene (6,7). This organization is not consistent with intercistronic processing via a single site-specific cleavage. We showed further that RNAs with termini in the atpI–atpH and psaJ–rpl33 intergenic regions are generated by nucleases that degrade precursors back from the 5′- or 3′-directions until halted by a PPR10 molecule bound in each intergenic region (7,10).
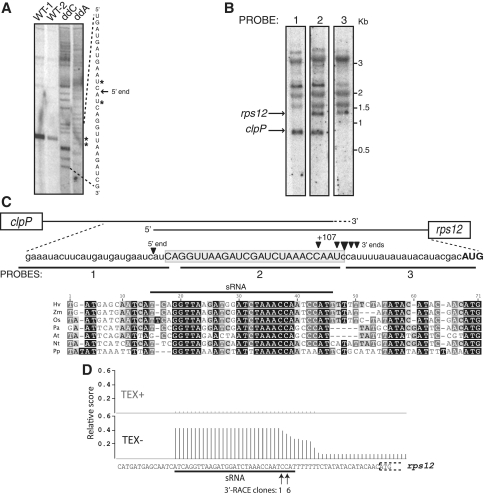
A PPR protein in moss, PPR38, binds in the clpP–rps12 intergenic region and stabilizes a processed clpP 3′-end (20,21). However, neither the PPR38-dependent 3′-end nor the PPR38 binding site had been mapped precisely, so it was not possible to evaluate whether PPR38 acts as does PPR10, by directly impeding an exonuclease. To gain insight into RNA processing in the clpP–rps12 intergenic region, we mapped the processed RNA termini in this region in maize by cRT–PCR and primer extension (Figure 1). The processed clpP 3′-ends are heterogeneous and span 7-nt (Figure 1C), similar to the PPR10-dependent atpH and psaJ 3′-termini (7). The processed rps12 5′-end maps ∼29-nt upstream of the clpP 3′-termini (Figure 1A and C). This ‘overlapping’ arrangement of processed rps12 and clpP transcripts was confirmed by RNA gel blot hybridizations using three closely spaced oligonucleotide probes (Figure 1B): the outer probes (1 and 3) each detected one unique transcript (marked with arrows), whereas Probe 2 in between detected the combined population of transcripts. The orthologous termini map to similar positions in barley (Figure 1D). The processed rps12 5′-ends in moss and Arabidopsis map at analogous positions (20,22) and the sequence corresponding to the overlap between the processed clpP and rps12 mRNAs shows striking conservation between angiosperms and moss (Figure 1C). These results support the view that a conserved protein, possibly the PPR38 ortholog, binds to the clpP–rps12 intergenic region and blocks RNA degradation from both directions.
Figure 1.
Mapping RNA termini in the maize and barley rps12–clpP intergenic region. (A) Primer extension analysis of the rps12 5′-end in maize. The ddC and ddA sequencing ladders identify the positions of G and U residues in the RNA template. Two RNA samples were analyzed (WT-1 and WT-2). (B) RNA gel blot hybridizations, using maize seedling leaf RNA and the three oligonucleotide probes diagramed in (C). The panels came from adjacent lanes of the same gel. Kb-RNA size markers. (C) Maize RNA sequence annotated with the 5′- and 3′-termini determined by primer extension and cRT-PCR, respectively. A multiple sequence alignment of the same region is annotated with the position of a small chloroplast RNA. (D) Histogram of barley transcriptome sequence reads mapping to the rps12 5′-region. TEX+ data are derived from a library that had been treated with Terminator Exonuclease, which will degrade processed 5′-termini. TEX- data were derived from an untreated library, and represent both processed and unprocessed 5′-ends. The plateau illustrates an sRNA that matches the sequence at the overlapping 5′- and 3′-termini in the clpP–rps12 intergenic region, and that matches a region of high conservation shown in panel (C). 3′-ends identified in barley by 3′-RACE are marked with arrows and annotated with the number of clones corresponding to each position.
The position of the HCF152 binding site supports a protein barrier mechanism for intercistronic processing in the psbH–petB intergenic region
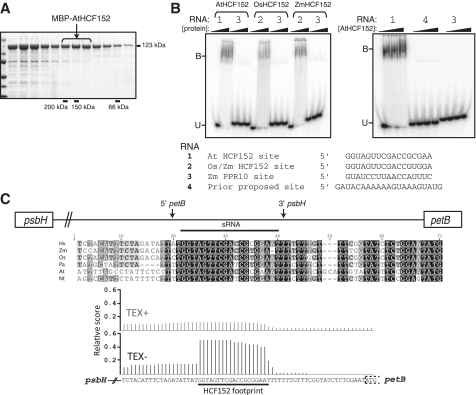
HCF152 is a PPR protein that is required for the accumulation of RNAs with processed 5′- or 3′-ends mapping between psbH and petB in Arabidopsis chloroplasts (8). The HCF152-dependent 5′- and 3′-RNA termini have an overlapping arrangement analogous to those in the intergenic regions discussed earlier: the 3′-end of processed psbH RNA maps ∼25-nt downstream of the 5′-end of processed petB RNA (7). These observations suggest that HCF152 acts analogously to PPR10, by binding in the psbH–petB intergenic region and stabilizing the upstream and downstream RNA segments by impeding 5′- and 3′-exonucleases. This model predicts that the HCF152 binding site should map to the overlap between the upstream and downstream HCF152-dependent transcript forms. However, in apparent contradiction to this model, this region was not identified among the binding sites reported for recombinant HCF152 (8,23). To resolve this issue, we tested the sequence-specificity of recombinant HCF152 from maize (ZmHCF152), rice (OsHCF152) and Arabidopsis (AtHCF152). Each protein was expressed as a fusion to MBP and purified by successive amylose affinity and gel filtration chromatography (Figure 2A). RNA binding activities were tested with gel mobility shift assays (Figure 2B). In accord with our hypothesis, each protein bound with high affinity to an RNA oligonucleotide corresponding to the sequence that is shared by processed psbH and petB RNAs (Figure 2B). Furthermore, they did not bind significantly to two other RNAs of similar length: the PPR10 binding site and the site between psbH and petB that was previously reported to bind HCF152 (8,23). The different conclusion in the prior study may have arisen from the use of a UV-cross-linking assay to monitor sequence specificity, a technique that is not well suited for that purpose (24). AtHCF152 bound its cognate binding site with very high affinity (equilibrium Kd of ∼1 nM; Supplementary Figure S1). These results provide strong evidence that HCF152 binds to the 5′-or 3′-terminus of the processed RNAs that fail to accumulate in its absence. Therefore, it is very likely that HCF152 functions analogously to PPR10, and defines the positions of the processed RNA termini in the psbH–petB intergenic region by blocking exonucleases.
Figure 2.
Mapping the binding site of recombinant HCF152. (A) Elution of affinity-purified MBP-AtHCF152 from a gel filtration column. Aliquots of consecutive fractions were analyzed by SDS–PAGE and staining with Coomassie Blue. The bracketed fractions were pooled and used for RNA binding assays. The elution positions of globular size standards are shown below. The elution profiles of MBP-OsHCF152 and MBP-ZmHCF152 were similar to that for MBP-AtHCF152 (data not shown). (B) Gel mobility shift assays demonstrating sequence-specificity of MBP-HCF152. The proteins indicated were used in RNA binding assays with the radiolabeled RNA oligonucleotides shown below. RNA 4 corresponds to the sequence proposed previously to bind HCF152 (23). Protein concentrations were 0, 35 and 75 nM (left panel), or 0, 12, 25 and 75 nM (right panel). Bound (B) and unbound (U) RNAs were separated by native gel electrophoresis. (C) Genomic context and evolutionary conservation of the HCF152 binding site. The positions of the HCF152-dependent 5′- and 3′-termini (7,8) are marked. The HCF152 binding site is represented by a small RNA in barley chloroplasts, as shown by the histogram of sequence reads below.
The binding sites of several characterized PPR proteins are marked by small chloroplast RNAs
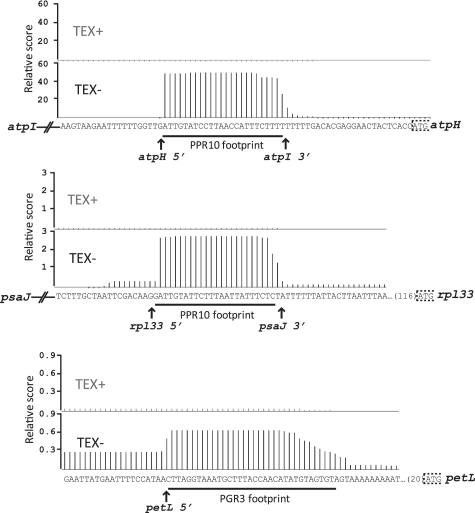
We noted previously (7) that PPR10's binding sites were detected as small RNAs (sRNAs) in large-scale sRNA data sets from rice and maize (25,26). Likewise, the HCF152 binding site defined here is among the sRNAs reported in tobacco (27), rice and maize (see http://sundarlab.ucdavis.edu/smrnas/). We proposed that these sRNAs are the in vivo footprints of PPR10 and HCF152—i.e. metastable degradation intermediates that are protected by the protein from ribonuclease attack. Indeed, the boundaries of the sRNAs harboring the PPR10 binding site correspond with the positions at which recombinant PPR10 blocks 5′- and 3′-exonucleases in vitro (10). sRNAs exist at orthologous positions in Arabidopsis (22, 28), and the binding site of PGR3, a PPR protein that stabilizes the petL 5′-terminus in Arabidopsis (14,29), is represented by an sRNA in these same datasets.
sRNAs at orthologous positions in barley can be inferred from data collected during our recent genome-wide analysis of 5′-transcript termini in barley chloroplasts (P. Zhelyazkova and T. Börner manuscript submitted): the PPR10, HCF152 and PGR3 binding sites appear as plateaus of sequence reads spanning ∼25 nt (Figures 2C and 3); these plateaus are eliminated by treatment of the RNA with Terminator Exonuclease (TEX), a 5′ → 3′ exonuclease that is inhibited by a 5′-terminal triphosphate. Therefore, the 5′-ends represented by these plateaus are produced by processing. It is reasonable to conclude that each such plateau represents a set of sRNAs that accumulate due to protection by the cognate PPR protein.
Figure 3.
Barley chloroplast transcriptome data documenting sRNAs matching known binding sites of PPR proteins. The binding sites of PPR10 and PGR3, and the RNA termini that are stabilized by these proteins are marked (7,10,14,29). The positions on the atpH RNA at which PPR10 blocks exonucleases in vitro (10) correspond with the borders of the sRNA, providing evidence that the sRNA is PPR10's in vivo footprint.
sRNAs are also apparent at the sites of action of several PPR proteins that have been genetically characterized, but for which direct binding sites have not been reported (Table 1). For example, CRP1 is required for the accumulation of processed 5′- and 3′-termini in the maize petB–petD intergenic region (6,9). Abundant sRNAs in barley (Supplementary Figure S2), rice and maize (http://sundarlab.ucdavis.edu/smrnas/) correspond to the sequence that is shared by the 5′- and 3′-ends of the CRP1-dependent transcripts; these are likely to be the in vivo footprint of CRP1 or a CRP1-dependent complex. An orthologous sRNA is not apparent in Arabidopsis (22), correlating with the absence of monocistronic petD mRNA in that species (8). Another sRNA corresponds to the site of action of the Arabidopsis PPR protein MRL1 (11). MRL1 is required for the accumulation of a processed rbcL 5′-end; sRNAs matching this end—the putative MRL1 footprint—can be inferred from our barley transcriptome data (Supplementary Figure S2) and appear in Arabidopsis and rice sRNA datasets as well (22). Furthermore, an abundant sRNA from the clpP–rps12 intergenic region has boundaries that match the processed 5′- and 3′-termini mapped here (Figure 1D). We suggest that this sRNA marks the binding site for a PPR protein, possibly PPR38, which defines and stabilizes the processed 5′- and 3′-termini in the clpP–rps12 intergenic region.
Table 1.
Chloroplast sRNAs matching known or predicted binding sites of characterized PPR proteins
| Genomic region | Speciesa | Proteinb | Sequence in Barleyc (U residues are indicated by T) | Corresponding RNA terminid |
|---|---|---|---|---|
| atpI–atpH intergenic | Hv, Zm, At, Os | PPR10 | ATTGTATCCTTAACCATTTCTTTT | atpI 3′ |
| atpH 5′ (−49) | ||||
| psaJ–rpl33 intergenic | Hv, Os | PPR10 | ATTGTATTCTTTAATTATTTCTCT | psaJ 3′ |
| rpl33 5′ (−161) | ||||
| petL 5′ | Hv,Os,At | PGR3 | CTTAGGTAAATGCTTTACCAACATATGTAGT | petL 5′ (−66) |
| psbH–petB intergenic | Hv, At, Zm,Os, Nt | HCF152 | GGTAGTTCGACCGCGGAATT | psbH 3′ |
| petB 5′ (−44) | ||||
| clpP–rps12 intergenic | Hv,Os,At, Zm | PPR38 (putative) | ATCAGGTTAAGATGGATCTAAACCAATCCATTTTT | clpP 3′ |
| rps12 5′ (−52) | ||||
| psbT–psbH intergenic | Hv,Os, Zm, At | HCF107 (putative) | AGTATACAAAGTCAACACCAATGATT | psbH 5′ (−37) |
| petB–petD intergenic | Hv, Zm, Os | CRP1 (putative) | CATATCGGGTAGGTTGTGGTATTTCATTGCT | petB 3′ |
| petD 5′ (−149) | ||||
| rps7–ndhB intergenic | Hv,Zm,Os, Nt, At | CRR2 (putative) | ATGCAGTTACTAATTCATGATCTGGCATGT | rps7 3′ |
| ndhB 5′ (−16, −70)e | ||||
| rbcL 5′ | Hv,Os, At | MRL1 (putative) | CATCGAGTAGACCCTGTTATTGTGAGAATT | rbcL 5′ (−59) |
aSpecies in which the sRNA has been reported. Hv—Hordeum vulgare (data from this study); Zm—Zea mays (data from http://sundarlab.ucdavis.edu/smrnas/); Os—Oryza sativa and At—Arabidopsis thaliana [data from (22)]; Nt—Nicotiana tabacum [data from (27)].
bProtein that has been shown to bind to this sequence, or that is hypothesized to do so based on genetic data (putative). Citations are provided in the ‘Results’ section.
cEach sRNA represents a population of molecules with ends mapping within several nucleotides of the sequence shown.
dThe indicated 5′- and 3′-RNA termini match the 5′- and 3′-termini of the corresponding sRNA. The position of the sRNA 5′-end with respect to the downstream start codon in barley is shown in parentheses.
ePositions relative to the annotated (−16) and proposed (−70) start codons.
PPR10, HCF152, PGR3, CRP1, PPR38 and MRL1 belong to the ‘P’ subfamily of PPR proteins, and harbor long tandem arrays of canonical PPR motifs (30). Proteins of this nature may be particularly effective at blocking exonucleases (and thus yielding sRNA footprints) due to their extensive, high-affinity interface with RNA (10). However, there is genetic evidence that other types of helical repeat proteins can mediate similar effects. Examples in land plants include HCF107 and CRR2. HCF107 consists largely of ‘HAT’ repeats, a variant of the tetratricopeptide repeat (16,31,32). HCF107 is required for the accumulation of RNAs with a processed 5′-end upstream of psbH. The HCF107-dependent 5′-end is marked by an sRNA in barley (Supplementary Figure S2), maize (http://sundarlab.ucdavis.edu/smrnas/), Arabidopsis and rice (22). We postulate that this sRNA contains the binding site for HCF107 and that HCF107 defines the position of the processed psbH 5′-terminus by blocking 5′ → 3′ degradation.
CRR2 is a member of the PLS-DYW subfamily of PPR proteins, which consist of alternating canonical (P), ‘long’ (L) and ‘short’ (S) PPR motifs followed by a DYW motif. Most PLS-DYW proteins are site-specificity factors for RNA editing (33). CRR2, however, is required for the accumulation of RNAs with a processed end upstream of ndhB (34). The CRR2-dependent 5′-end is represented by an abundant sRNA in barley (Supplementary Figure S2), tobacco (27), Arabidopsis and rice (22). It is likely, therefore, that CRR2 or a CRR2-dependent protein binds to this sequence and stabilizes ndhB RNA by blocking 5′ → 3′ degradation. Furthermore, 3′-RACE of barley and Arabidopsis chloroplast RNA detected rps7 3′-termini matching the 3′-boundaries of this sRNA (Supplementary Figure S2; 22), providing evidence for bidirectional RNA stabilization by a protein bound to this site. This putative protein footprint is unusual in that it overlaps the annotated ndhB start codon (Supplementary Figure S2). Because, the stable binding of a protein to this region would preclude ribosome binding, we suggest that the ndhB start codon is misannotated, and that the true start codon or an alternative start codon lies downstream. This view is supported by a phylogenetic argument in the accompanying paper (22).
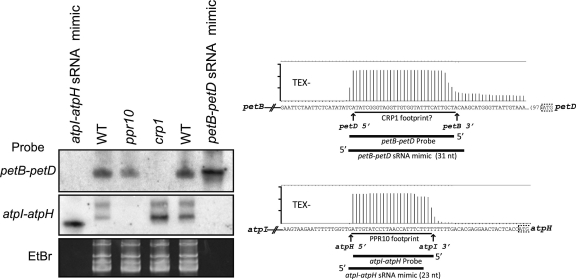
PPR10- and CRP1-dependent accumulation of sRNAs: confirmation of sRNA biogenesis via PPR protection
If the sRNAs discussed earlier are, in fact, in vivo footprints of PPR proteins that stabilize mRNA termini, then these sRNAs should fail to accumulate in the absence of the cognate PPR protein. To test this prediction, we monitored the abundance of the proposed sRNA footprints of PPR10 and CRP1 in maize ppr10 and crp1 mutants (Figure 4). Duplicate RNA gel blots were probed with oligodeoxynucleotides complementary to the sRNA in the atpH 5′-untranslated region (UTR) (the putative PPR10 footprint) or in the petB–petD intergenic region (the putative CRP1 footprint). Synthetic RNAs that mimic the sRNA from each region were included in adjacent lanes, to serve as hybridization controls and as size markers.
Figure 4.
RNA gel blots demonstrating PPR-dependent accumulation of two sRNAs. Total leaf RNA (15 µg) of the indicated genotypes was fractionated in denaturing polyacrylamide gels and electrophoretically transferred to charged nylon membrane. Wild-type (WT) samples came from phenotypically normal siblings grown in parallel. The ppr10 and crp1 mutants were described previously and were shown to be null alleles (6,7,10). Duplicate blots were hybridized with oligodeoxynucleotide probes that are diagrammed in the context of the barley transcriptome data to the right. Synthetic RNA oligonucleotides that mimic each sRNA were included in adjacent lanes, and are also diagrammed. The maize and barley sequences are identical in the regions encoding these sRNAs. A portion of the ethidium bromide (EtBr) stain of one of the gels is shown below to illustrate equal sample loading.
A probe complementary to the PPR10 binding site in the atpH 5′-UTR detected two sRNAs in wild-type and crp1 mutant plants; both were missing in ppr10 mutant plants, as predicted if they accumulate due to stabilization by bound PPR10 (Figure 4). The smaller of the two bands matches the size expected for the sRNA in the atpH region based on the barley sequencing data. The upper band may have arisen from cross-hybridization to the related sRNA in the psaJ 3′-UTR, which also harbors a PPR10 binding site. Analogous results were obtained for the sRNA in the petB–petD intergenic region, which we propose to be the CRP1 footprint. In this case, a single sRNA of the expected size was detected in wild-type and in ppr10 mutant plants but not in crp1 mutants. Together, these results provide strong support for the view that sRNAs matching the genetically defined targets of characterized PPR proteins are metastable degradation intermediates that are protected by the cognate protein (i.e. they are in vivo footprints of PPR proteins). By extrapolation, sRNAs with similar features mapping elsewhere in the genome are also likely to be footprints of PPR proteins or of ‘PPR-like’ proteins with other repeat architectures.
sRNAs with hallmarks of PPR footprints map to most processed mRNA 5′-termini in barley chloroplasts
Stable RNA structures can block both 5′ → 3′ and 3′ → 5′ RNA degradation in chloroplasts, and are known to define the 3′-ends of several chloroplast mRNAs (17). The relative contribution of intrinsic RNA structure versus bound proteins as a means to define and stabilize chloroplast mRNA termini is not known. Our genome-wide mapping of the 5′-termini of barley chloroplast RNAs (P. Zhelyazkova and T. Börner manuscript submitted) provides an opportunity to address this issue by correlating the positions of mRNA termini with the positions of sRNAs whose features resemble those that mark the PPR–RNA interactions summarized earlier. During the course of this study, we also mapped several mRNA termini in maize chloroplasts (Supplementary Table S1); these confirmed several 5′-termini inferred from the barley data and mapped several 3′-termini.
To identify potential binding sites for uncharacterized PPR proteins (or PPR-like proteins such as HCF107), we mined our barley chloroplast transcriptome data for sRNAs with hallmarks of PPR footprints. In the ensuing discussion, the term ‘PPR-like’ will be used to refer both to canonical PPR proteins and to proteins with other repeating units that may bind RNA in a similar manner.
Candidate footprints of PPR-like proteins were selected based on the following criteria. (i) We consider only sRNAs that can be inferred from the barley chloroplast transcriptome data. Because these data were derived from purified chloroplast RNA, this rules out the possibility that the sRNAs arise from chloroplast DNA remnants found in the nuclear genome. (ii) sRNAs that we propose to be PPR footprints lack strong secondary structure and have little propensity to form stable structures with neighboring sequences. This criterion distinguishes sRNAs that accumulate due to protection by a bound protein from those that are ribonuclease resistant due to their structure. Furthermore, current data support the view that PPR tracts bind RNA in single-stranded form along a surface formed by stacked repeating units (10,35), so highly structured RNA segments seem unlikely to serve as binding sites for PPR-like proteins. Finally, we consider only sRNAs mapping near protein coding genes, as sRNAs corresponding to tRNAs or rRNAs may arise in a different manner. Most of the barley sRNAs meeting these criteria map to 5′- or 3′-UTRs. The accompanying manuscript presents a global view of sRNAs in Arabidopsis and rice chloroplasts, and supports the view that sRNAs are, in fact, strongly biased toward UTRs (22). Blocks of sequence conservation demarcate most of the sRNAs we selected (see below), suggesting that these not only mark protein binding sites, but that the interface with the cognate protein is unusually long, as is anticipated for binding sites of PPR-like proteins. It has been noted previously that PPR binding sites are evolutionarily constrained in comparison to their flanking sequences (36).
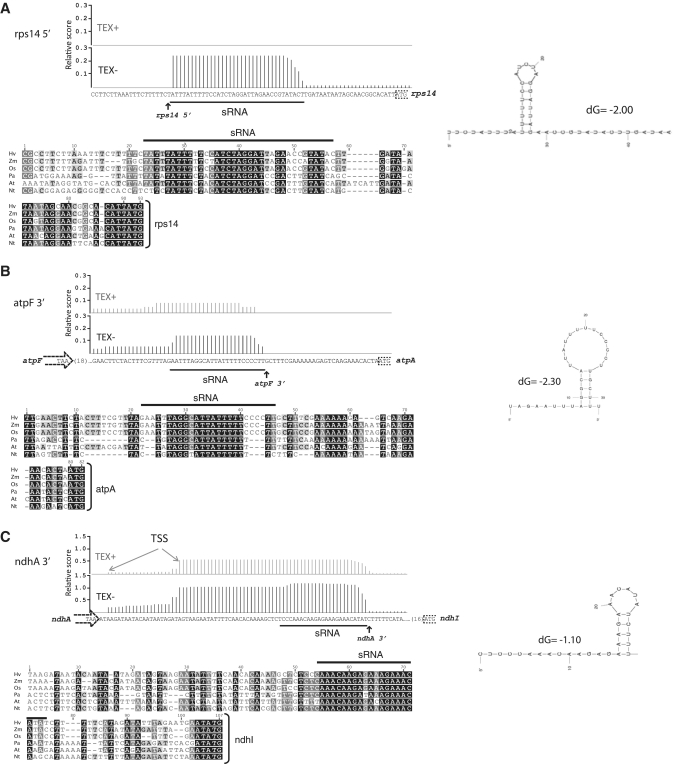
This analysis revealed that the first ∼25 nt of the vast majority of processed mRNAs in barley chloroplasts correspond with sRNAs suggestive of PPR footprints. The data are summarized in Tables 1 and 2, with supporting barley transcriptome data, sequence alignments and secondary structure predictions in Supplementary Figure S3. Of the 20 processed mRNA 5′-termini that were mapped unambiguously in barley, 19 have corresponding sRNAs. Eighteen of these 19 are predicted to lack strong secondary structure and so are excellent candidates for the footprints of PPR-like proteins that stabilize the downstream RNA. These include the sRNAs discussed earlier, for which candidate protective proteins have already been identified (Table 1: sRNAs mapping upstream of atpH, rpl33, petL, petB, rps12, psbH, petD, ndhB, rbcL), and sRNAs that we suggest to be footprints of PPR-like proteins, although a corresponding protein has not been identified (Table 2: sRNAs mapping upstream of rps16, ycf3, psbB, psaC, rps14, rps2, psbC, ndhK). Abundant sRNAs at orthologous sites have been mapped in at least one other species for each of these except for rps14 and ndhK. The sequence encoding the rps14 sRNA is highly conserved among monocot and dicot plants (Figure 5A), suggesting that this sRNA (and its putative protein partner) exist also in Arabidopsis (and other angiosperms). The ndhK sRNA sequence is likewise conserved, but this is uninformative because it maps within the upstream ndhC coding region and is subject to other constraints.
Table 2.
Chloroplast sRNAs matching processed 5′-mRNA termini for which stabilizing proteins have not been identified
| Genomic region | Speciesa | Sequence in Barleyb (U residues are indicated by T) | Corresponding RNA terminusc | Position of 5′ end relative to start codon (Hv) |
|---|---|---|---|---|
| rps16 5′ | Hv, Os, At, Nt | AAACCAATGACTATTCATGATTCCATCCAT | rps16 5′ (Hv) | −80 |
| ycf3 5′ | Os,Hv, At | TTTGTTTTTATGTTATTTTGTGAAG | ycf3 5′ (Hv) | −62 |
| psbB 5′ | Hv,Os, At | TTTTTCAATGCGATAAAATAAAGCGACATCGTGT | psb 5′ (Zm,Hv,At) | −63 |
| psaC 5′ | Hv, At | CAAAATTCAAGTCTCTTGGCTCTTTTCACGC | psaC 5′ (Hv) | −188 |
| rps14 5′ | Hvd | ATTTATTTTTCCATCTAGGATTAGAACCGTATACT | rps14 5′ (Hv) | −59 |
| rps2 5′ | Hv, Os | ATTTATTTCAAGCTATTTCGGATCTT | rps2 5′ (Hv) | −97 |
| psbC 5′ | Hv,Os, At,Zm | ATCAGCCTCATGAAAATCTTATATA | psbC 5′ (Hv,At,Zm) | −45 (GTG) |
| ndhK 5′ | Hv | TTTCGTGCTTATCTTAGTTGTCGGTTTAGT | ndhK 5′ (Hv) | −57 |
| rps12–locus 2,5′ | Hv | CAACATAGGTCATCGAAAAGATCTCGGACAACTCA CCAAAGCA | 5′ end of second intron fragment (Hv) | Not applicable: intron sequence |
| ndhA 5′ | Hv, At | AAATTGGCTGATATCATGACGATATTAGGTAG | ndhA 5′ (Hv, Zm, At) | −67 |
The barley sequencing data, secondary structures and multiple sequence alignments are provided in Supplementary Figure S3. Evidence for orthologous sRNAs in rice and Arabidopsis is presented in the accompanying paper (22).
aSpecies in which the sRNA has been reported, as described in Table 1.
bEach sRNA represents a population of molecules with ends mapping within several nucleotides of the sequence shown.
cSpecies in which the end has been mapped are indicated in parentheses. The 5′-termini of the sRNA and corresponding mRNA match in each case.
dThe sequence corresponding to this sRNA is highly conserved among angiosperms (Figure 5), suggesting that the apparent absence of this sRNA in Arabidopsis is a false negative.
Figure 5.
Examples of sRNAs that we suggest to be footprints of uncharacterized PPR-like proteins. Histograms of barley sequence reads are annotated with the positions of mapped mRNA termini. The most stable structure predicted for the sRNA by MFold (37) is shown to the right. The predicted structures are very unstable in comparison with those that are known to stabilize 3′-termini in chloroplasts, which average approximately −25 kcal/mol (22). Each sequence alignment ends with the start codon of the downstream gene. (A) Example of an sRNA corresponding with a processed 5′-end. This sRNA was not detected in Arabidopsis, but its sequence is highly conserved. (B) Example of an sRNA corresponding with a processed 3′-end. The atpF 3′-end has been mapped in maize (7) but not in barley. (C) Example of an sRNA mapping near a transcription start site (TSS). The TSS is inferred from the position of the TEX-resistant 5′-end. The 3′-half of the plateau of sequence reads is TEX-sensitive, corresponds with a conserved sequence element, and is not predicted to form a stable structure. cRT-PCR data place an ndhA 3′-end at the 3′-end of this sRNA (Supplementary Table S1).
Several of the processed 5′-termini require special mention. (i) A barley sRNA and 5′-end map ∼680-nt upstream of rps12 exon 2 (Table 2 and Supplementary Figure S3C). These are derived from the second of two loci that yield rps12 mRNA via trans-splicing of a split group II intron. This 5′-end and sRNA mark the beginning of the 3′-section of the intron. The maize PPR protein PPR4 is required for the trans-splicing of this intron (38) and is a candidate for binding to this site. (ii) An sRNA and processed 5′-end upstream of ndhA (Table 2 and Supplementary Figure S3B) are found also in Arabidopsis (22) and match a proposed binding site for the PPR protein PGR3 (29). This sRNA differs from those above in that it is predicted to form a fairly stable RNA hairpin (dG = −10.7 kcal/mol). This structure is considerably less stable than those that stabilize the 3′-ends of abundant chloroplast mRNAs (approximately −25 kcal/mol), but it still seems an unlikely target for binding by a PPR protein. Thus, both a protein-based and RNA-structure based mechanism for stabilizing this 5′-end seem equally plausible at this time. (iii) The processed 5′-terminus mapping 135 bp upstream of the psbD gene stands out as the sole example for which stabilization by something other than a PPR-like protein seems most likely. This end is not accompanied by an sRNA and is predicted to adopt a stable structure (data not shown).
The observations above show that 18 of the 20 most prominent processed 5′-termini of mRNAs in barley chloroplasts lack strong intrinsic structure and correspond with the 5′-end of an sRNA resembling the footprint of a PPR-like protein. In most cases, orthologous sRNAs and/or RNA termini have been detected in Arabidopsis or rice (22) and, with just one exception, the sequences are highly conserved between monocot and dicot plants (the sequence of the rps2 5′-end and sRNA is conserved only among the monocots) (Supplementary Figure S3A). These results, in conjunction with the biochemical and genetic data for PPR10, PGR3 and HCF152 summarized earlier, support the view that most processed 5′-ends of mRNAs in angiosperm chloroplasts result from the stalling of 5′ → 3′ RNA degradation at the upstream edge of a bound PPR-like protein.
Evidence for protein-mediated stabilization of 3′-mRNA termini in chloroplasts
Stable stem-loop structures define the 3′-ends of some chloroplast mRNAs by blocking 3′ → 5′ exoribonucleases (17). Fourteen sRNAs with very stable stem–loops (dG < −25 kcal/mol) were detected in the barley transcriptome data; these map to 3′-UTRs and are likely the remnants of 3′-stabilizing elements that accumulate due to their intrinsic resistance to nucleases (P. Zhelyazkova and T. Börner manuscript submitted). An alternative mechanism for 3′-end stabilization–stalling of 3′ → 5′ exonucleases by a bound protein–has been shown for two PPR10-dependent 3′-ends (7,10) and is implied by analogous, but less complete data for 3′-termini that fail to accumulate in hcf152, ppr38 and crp1 mutants. In each case, an mRNA 3′-terminus corresponds with the 3′-end of an sRNA and with genetic data placing a PPR protein near that 3′-end (summarized in Table 1). The position of the HCF152 binding site at the 3′-end of an HCF152-dependent RNA (Figure 2) solidifies this interpretation. Orthologous rps7 3′-ends mapped here (barley) and in the accompanying paper (Arabidopsis) (22) have an analogous spatial relationship with an sRNA and with the CRR2-dependent ndhB 5′-end (Supplementary Figure S2), suggesting biogenesis in an analogous fashion.
To assess the prevalence of protein-mediated protection of mRNA 3′-termini in chloroplasts, we examined our barley chloroplast transcriptome data for unstructured sRNAs mapping in 3′-UTRs (Table 3). This analysis revealed seven sRNAs that are good candidates for footprints of uncharacterized PPR-like proteins that protect 3′-ends; these map downstream of the atpF, ndhA, ycf3, ndhJ, ndhE, rps4 and rps16 genes and have 3′-ends that match mapped 3′-termini. The sequences corresponding to the atpF, ndhA, ycf3, ndhJ and ndhE 3′-sRNAs are highly conserved among monocots and dicots (Figure 5B and C; Supplementary Figures S3A and S4A), whereas those for rps16 and rps4 are conserved in monocots only (Supplementary Figures S3 and S4). The putative PPR binding sites downstream of atpF and ndhA correlate with abundant 3′-ends but not with processed 5′-ends; however, these would place PPR proteins in proximity to downstream open reading frames on polycistronic RNAs (Figure 5B and C) and could potentially influence translation (see below). The sRNAs downstream of ndhJ and ycf3 are conserved in sequence but not in position in Arabidopsis (Supplementary Figure S4A). 3′-termini from these genes have not been mapped in Arabidopsis; it will be interesting to learn whether the placement of these 3′ termini mirrors that of these conserved sequence elements.
Table 3.
Chloroplast sRNAs mapping to 3′-UTRs
| Genomic region | Speciesa | Sequence in Barleyb (U residues are indicated by T) | Corresponding RNA terminus | Position of sRNA relative to stop codon (Hv) |
|---|---|---|---|---|
| atpF–atpA intergenic | Os, Hv | AATTTAGGCATTATTTTTCCCCTT | atpF 3′ (Zm) | +39 (−52 with respect to atpA) |
| ndhA–ndhI intergenic | Hv, Os | CCCAAACAAGAGAAAGAAACATAT | ndhA 3′ (Zm) | +52 (−49 with respect to ndhI) |
| rps16 3′ | Hv, Os, Zm | TATCGTGCCAATCCAACATAAGCCCCT | rps16 3′ (Hv) | +110 |
| ycf3 3′ | Hv, Os, Atd | AGAATTTCATTATATCCATTTCTTAT | ycf3 3′ (Hv) | +84 |
| ndhJ 3′ | Hv,Zm, Os, Atd | AACTTTGTATCGCGCACATGACT | ndhJ 3′ (Hv) | +250 |
| ndhE–psaC intergenic | Hv, At | CAAAATTCAAGTCTCTTGGCTCTTTT CACGC | ndhE 3′ (Hv) | +293 (−188 with respect to psaC) |
| rps4–ycf3 intergenic | Os,Hv, At | TTTGTTTTTATGTTATTTTGTGAAG | rps4 3′ (Hv) | +937 (−62 with respect to ycf3) |
| rps7–ndhB intergenic | Hv | GAAATCATGATCAACTAAGCCCTCTCGA GGGCTTG | rps7 3′ (At)c | +127 |
| petD 3′ | Hv, Os | ATTATTTTATTATGATCCATTTCGCG | One of two 3′-ends (Hv) | +96 |
The barley sequencing data, secondary structures and multiple sequence alignments are provided in Figure 5B and C and in Supplementary Figures S3A and S4.This table excludes sRNAs predicted to form very stable stem-loops, which are anticipated to directly block 3′ → 5′ RNA decay.
aSpecies in which the sRNA has been reported, as described in Table 1.
bEach sRNA represents a population of molecules with ends mapping within several nucleotides of the sequence shown.
cThe Arabidopsis rps7 3′-end reported in (34) maps near this position, but was not mapped to high resolution.
dThe sRNA and sequence are conserved in Arabidopsis, but the position in the 3′-UTR is not (Supplementary Figure S4A).
sRNAs mapping downstream of rps7 and petD have unusual features (Supplementary Figure S4B). The rps7 sRNA maps near a 3′-end reported in Arabidopsis (34) and may reflect a binding site for a stabilizing PPR-like protein. However, the 3′-end of this sRNA can fold into a fairly stable structure (dG = −11.8 kcal/mol), and this structure may be sufficient to position this 3′-end. An sRNA detected in barley and rice maps immediately adjacent to and downstream of a stable RNA stem–loop whose ortholog marks the mature petD 3′-terminus in spinach (39). This sRNA is predicted to be unstructured and its 3′-end matches a petD 3′-end in barley, suggesting that some petD transcripts are stabilized at their 3′ ends by a bound protein. The function of such a protein binding site is unclear, as the upstream stem–loop should be sufficient to protect the petD mRNA 3′-end.
Primary 5′ RNA termini rarely have features resembling PPR footprints
Our barley transcriptome study mapped a large number of TEX-resistant 5′-termini, which mark sites of transcription initiation (P. Zhelyazkova and T. Börner manuscript submitted). In contrast with the strong correlation between sRNAs and processed 5′-ends, only four primary 5′-termini correlate with sRNAs (Table 4). The sRNA mapping to the rps15 transcription start site is much longer than a putative PPR footprint and has the potential to form a hairpin of moderate stability at its 3′-end (Supplementary Figure S5). This sRNA seems unlikely to bind a PPR-like protein and may accumulate due to the stabilizing effects of the 5′-triphosphate and its intrinsic structure. The sRNA associated with the atpI transcription start site is also very long and is predicted to adopt considerable structure. However, an sRNA in Arabidopsis matches a highly conserved interval at the 3′-end of this region (Supplementary Figure S5). Thus, the immediate 5′-end of the primary atpI transcript may be intrinsically resistant to nucleases but a PPR-like protein may bind downstream. An analogous explanation is consistent with the data for the ndhI and rpoB transcription start sites. Both of these are associated with long ‘bimodal’ plateaus of sequence reads whose 3′-components are TEX sensitive and highly conserved (Figure 5C and Supplementary Figure S5). The TEX-sensitive components might correspond to sRNAs that are protected by PPR-like proteins. In fact, the TEX-sensitive component of the plateau near the ndhI transcription start site corresponds with the 3′-end of an RNA from the upstream gene, ndhA (Figure 5C); this suggests that a PPR-like protein binds to that site and stabilizes that 3′-end (see above). Taken together, these observations suggest that protein-mediated protection is less prevalent at primary than at processed 5′-mRNA termini, but that several transcription start sites may be followed shortly thereafter by the binding site for a PPR-like protein.
Table 4.
Chloroplast sRNAs that map to primary 5′ mRNA termini
| Transcription start site | Speciesa | Sequence in Barleyb (U residues are indicated by T) |
|---|---|---|
| rps15 | Hv | AATAAATAAATCAGCAAAATTCTTTCTACTATATTTAGATAGAAGAAACATTTC |
| atpI | Hv, Os, At | GATGTGCTTTCTTGGTATCCTAAATATCAAATTAATAGTTCAAGTTGCTGAGTTGAGAAAGAGAT GGTTGAATCAAAAGAATTC |
| ndhIc | Hv,Os | ATAGTAAGAATATTTTCAACACAAAAGCTCTCCCAAACAAGAGAAAGAAACATAT |
| rpoB | Hv,Os | GAAATACGTATGTGGAGTTCCCTAGAATTTCATGTGATTCAGTAAACAGAATA |
aSpecies in which the sRNA has been reported, as described in Table 1.
bEach sRNA represents a population of molecules with ends mapping within several nucleotides of the sequence shown. Underlined sequences have features suggestive of PPR footprints, as discussed in the text.
cThe ndhI promoter maps in the ndhA–ndhI intergenic region. The 3′ portion of this sRNA is discussed also in the context of 3′ stabilization of processed ndhA RNA (Table 3).
DISCUSSION
Results presented here provide evidence that the vast majority of processed 5′-termini and many processed 3′-termini of mRNAs in angiosperm chloroplasts result from the site-specific binding of a PPR-like protein to an RNA precursor, followed by exonucleolytic RNA degradation back to the protein barrier. The arguments underlying this conclusion are summarized below.
Abundant chloroplast sRNAs mark the binding sites of those few P-type PPR proteins for which binding sites are well defined (PPR10, HCF152 and PGR3). The termini of these sRNAs match those of the transcripts that fail to accumulate in the corresponding mutant background. These observations imply that these sRNAs accumulate as degradation intermediates due to protection by the bound PPR protein—i.e. they are ‘PPR footprints’. This view is supported by the absence of the putative PPR10 and CRP1 footprints in ppr10 and crp1 mutants, respectively.
sRNAs match several mRNA termini that are known to require PPR-like proteins for their accumulation, but for which the protein binding sites are not well-defined (CRP1, MRL1, HCF107, CRR2 and cognate RNA termini). Analogous sRNAs mark the vast majority of the processed 5′-ends of mRNAs detected in our barley transcriptome study, implying analogous biogenesis mechanisms.
These sRNAs are marked by plateaus of sequence reads in the barley transcriptome data, indicating that they are found at levels similar to, or even higher than the mRNAs whose ends they match. This implies that they are resistant to ribonucleases, despite the fact that they lack both a 5′-triphosphate and stable structure, the two intrinsic features that are predicted to be protective (1,17,40). It seems, therefore, that sRNAs lacking such protective features must be protected by bound proteins.
Chloroplasts house an abundance of RNA binding proteins with various types of RNA binding domains (1,41). We suggest, however, that it is the PPR class (and PPR-like proteins with long tracts of other repeating units) that is primarily responsible for stabilizing sRNAs and corresponding processed mRNA termini. As discussed previously (10), a long PPR tract can present an unusually long and stable interface for RNA binding, which is anticipated to be a particularly effective barrier to exonucleases. The striking sequence conservation of many sRNAs provides further evidence that they are protected by PPR-like proteins rather than by proteins with globular RNA binding domains; this feature is characteristic of known or suspected PPR binding sites [alignments herein and (10,11,36)] and implies an extensive interface for sequence-specific interaction with a protein surface.
The accompanying manuscript provides a genome-wide perspective on the occurrence of abundant chloroplast sRNAs in large-scale sRNA data sets from rice and Arabidopsis, and correlates many such sRNAs with mRNA termini (22). Our two studies are complementary and mutually supportive. Together, our findings highlight the striking positional correspondence between chloroplast sRNAs and processed mRNA termini, and show that many sRNAs (and corresponding processed termini) are conserved among multiple species. Our observations imply that a protein is bound stably to the 5′-terminal ∼25 nt of most processed mRNAs and to the 3′-terminal ∼25 nt of many processed mRNAs in angiosperm chloroplasts. Protein footprints of this length are unlikely to result from interactions with globular RNA binding proteins, so we favor the view that they result from RNA binding along the surface of PPR or ‘PPR-like’ proteins that form long surfaces for RNA interaction. These observations imply that protection of 3′-mRNA termini by a protein—most likely a PPR-like protein—is a frequent alternative to protection by stable 3′-RNA stem–loops.
Our findings also have implications with regard to mechanisms of intercistronic mRNA processing in chloroplasts. This characteristic feature of gene expression in land plant chloroplasts had long been assumed to be mediated by site-specific endonucleolytic cleavage events. The site-specific barrier mechanism documented for PPR10 offered an alternative view (7,10), but the generality of this mechanism had been unclear. The mapping of additional intercistronic mRNA termini here and in the accompanying paper (22), and our demonstration that the HCF152 binding site matches the sequences shared at the 5′- and 3′-termini of HCF152-dependent RNAs provide evidence that intercistronic processing is generally accomplished via a mechanism akin to that of PPR10: binding of a PPR-like protein to an intergenic RNA segment, in conjunction with exonucleolytic RNA degradation back to the protein barrier. The lack of evidence for the adjacent processed RNA termini predicted by the site-specific cleavage model adds further support to the view that the contribution of site-specific cleavage to intercistronic processing is minor.
Positional bias of PPR footprints in 5′-UTRs
When PPR10 binds the atpH 5′-UTR, it is placed in close proximity to the initiating ribosome and maintains the ribosome binding region in a single-stranded conformation (10). This can account for the ability of PPR10 to enhance atpH translation. This proximity may also enhance the stability of atpH mRNA by minimizing the length of RNA in the 5′-UTR that is accessible to nuclease attack. The positional distribution of the known and putative PPR footprints described here and in the accompanying study (22) suggest that these are general themes of PPR action. The majority of putative PPR footprints in 5′-UTRs are placed such that the 3′-edge of the footprint maps between ∼20- and 60-nt upstream of the start codon [Tables 1 and 2 and data in (22)]. This spacing avoids interference with ribosome binding, while also minimizing ‘excess’ UTR sequence that could provide a target for nucleases. In some cases, sequences within the putative PPR footprint are predicted to pair with (and thus inhibit) the ribosome binding region (see psbC and ndhK 5′-UTRs in Supplementary Figure S3B). The binding of a PPR protein to such sites is anticipated to enhance translation by preventing this inhibitory interaction. Even where there is not a strong potential for interaction between the ribosome binding and (putative) PPR binding region, the binding of a PPR-like protein proximal to the ribosome binding region may enhance translation by reducing local transient RNA–RNA interactions. This may be especially important for those mRNAs lacking a Shine–Dalgarno element, for which the local absence of RNA structure is a critical determinant of start codon usage (42).
Organellar sRNAs as markers of potential PPR binding sites
More than 100 P-type PPR proteins are predicted to localize to chloroplasts in angiosperms (43), but few of these have been characterized. Findings in this and the accompanying manuscript (22) provide a reservoir of candidate PPR binding sites to consider in future studies. However, abundant sRNAs clearly do not capture all binding sites for PPR-like proteins. For example, the PPR5 binding site in the trnG–UCC group II intron (35,44) is not marked by an obvious sRNA, possibly because the adjacent intron sequences are highly structured and prevent nuclease access to the PPR5 binding region. The sites of CRP1 interaction in the petA and psaC 5′-UTRs, where CRP1 activates translation but does not stabilize 5′-ends (6,45), are also not marked by abundant sRNAs. PPR proteins involved in RNA editing, which are expected to interact only transiently with RNA, seem unlikely to leave sRNA footprints; indeed, edited sites are not apparent as sRNAs in the datasets we have examined. Nonetheless, the cataloging of organellar sRNAs with features suggestive of PPR footprints can be expected to enhance efforts to link uncharacterized PPR proteins in plants with specific RNA ligands and functions.
SUPPLEMENTARY DATA
Supplementary Data are available at NAR online: Supplementary Tables 1 and 2 and Supplementary Figures 1–5.
FUNDING
National Science Foundation (grant MCB-0940979 to A.B.); Deutsche Forschungsgemeinschaft (grant SFB 429 to T.B.); Helmholtz Graduate School ‘Molecular Cell Biology’ at the Max Delbruck Center for Molecular Medicine and Humboldt University, Berlin, Germany (fellowship to P.Z.); European Molecular Biology Organization (fellowship to K.H.) and Consejo Nacional de Ciencia y Tecnología (CONACYT) (fellowship to M.V.S.). Funding for open access charge: NSF (grant MCB-0940979).
Conflict of interest statement. None declared.
Supplementary Material
ACKNOWLEDGEMENTS
We are grateful to Cynthia M. Sharma and Jörg Vogel (University of Würzburg, Germany) for support in RNA sequencing, Dylan Udy (University of Oregon) for providing the RNA gel blot data shown in Figure 1, Hannes Ruwe and Christian Schmitz-Linneweber (Humboldt University of Berlin) for valuable input and Kenny Watkins (University of Oregon) for commenting on the manuscript.
REFERENCES
- 1.Barkan A. Expression of plastid genes: organelle-specific elaborations on a prokaryotic scaffold. Plant Physiol. 2011;155:1520–1532. doi: 10.1104/pp.110.171231. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Barkan A. Proteins encoded by a complex chloroplast transcription unit are each translated from both monocistronic and polycistronic mRNAs. EMBO J. 1988;7:2637–2644. doi: 10.1002/j.1460-2075.1988.tb03116.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Westhoff P, Herrmann RG. Complex RNA maturation in chloroplasts. Eur. J. Biochem. 1988;171:551–564. doi: 10.1111/j.1432-1033.1988.tb13824.x. [DOI] [PubMed] [Google Scholar]
- 4.Kohchi T, Yoshida T, Komano T, Ohyama K. Divergent mRNA transcription in the chloroplast psbB operon. EMBO J. 1988;7:885–891. doi: 10.1002/j.1460-2075.1988.tb02892.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Small I, Peeters N. The PPR motif - a TPR-related motif prevalent in plant organellar proteins. Trends Biochem. Sci. 2000;25:46–47. doi: 10.1016/s0968-0004(99)01520-0. [DOI] [PubMed] [Google Scholar]
- 6.Barkan A, Walker M, Nolasco M, Johnson D. A nuclear mutation in maize blocks the processing and translation of several chloroplast mRNAs and provides evidence for the differential translation of alternative mRNA forms. EMBO J. 1994;13:3170–3181. doi: 10.1002/j.1460-2075.1994.tb06616.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Pfalz J, Bayraktar O, Prikryl J, Barkan A. Site-specific binding of a PPR protein defines and stabilizes 5′ and 3′ mRNA termini in chloroplasts. EMBO J. 2009;28:2042–2052. doi: 10.1038/emboj.2009.121. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Meierhoff K, Felder S, Nakamura T, Bechtold N, Schuster G. HCF152, an Arabidopsis RNA binding pentatricopeptide repeat protein involved in the processing of chloroplast psbB-psbT-psbH-petB-petD RNAs. Plant Cell. 2003;15:1480–1495. doi: 10.1105/tpc.010397. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Fisk DG, Walker MB, Barkan A. Molecular cloning of the maize gene crp1 reveals similarity between regulators of mitochondrial and chloroplast gene expression. EMBO J. 1999;18:2621–2630. doi: 10.1093/emboj/18.9.2621. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Prikryl J, Rojas M, Schuster G, Barkan A. Mechanism of RNA stabilization and translational activation by a pentatricopeptide repeat protein. Proc. Natl Acad. Sci. USA. 2011;108:415–420. doi: 10.1073/pnas.1012076108. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Johnson X, Wostrikoff K, Finazzi G, Kuras R, Schwarz C, Bujaldon S, Nickelsen J, Stern DB, Wollman FA, Vallon O. MRL1, a conserved Pentatricopeptide repeat protein, is required for stabilization of rbcL mRNA in Chlamydomonas and Arabidopsis. Plant Cell. 2010;22:234–248. doi: 10.1105/tpc.109.066266. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Loiselay C, Gumpel NJ, Girard-Bascou J, Watson AT, Purton S, Wollman FA, Choquet Y. Molecular identification and function of cis- and trans-acting determinants for petA transcript stability in Chlamydomonas reinhardtii chloroplasts. Mol. Cell. Biol. 2008;28:5529–5542. doi: 10.1128/MCB.02056-07. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Vaistij F, Boudreau E, Lemaire S, Goldschmidt-Clermont M, Rochaix J. Characterization of Mbb1, a nucleus-encoded tetratricopeptide repeat protein required for expression of the chloroplast psbB/psbT/psbH gene cluster in Chlamydomonas reinhardtii. Proc. Natl Acad. Sci. USA. 2000;97:14813–14818. doi: 10.1073/pnas.97.26.14813. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Yamazaki H, Tasaka M, Shikanai T. PPR motifs of the nucleus-encoded factor, PGR3, function in the selective and distinct steps of chloroplast gene expression in Arabidopsis. Plant J. 2004;38:152–163. doi: 10.1111/j.1365-313X.2004.02035.x. [DOI] [PubMed] [Google Scholar]
- 15.Boudreau E, Nickelsen J, Lemaire SD, Ossenbuhl F, Rochaix JD. The Nac2 gene of Chlamydomonas encodes a chloroplast TPR-like protein involved in psbD mRNA stability. EMBO J. 2000;19:3366–3376. doi: 10.1093/emboj/19.13.3366. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Sane AP, Stein B, Westhoff P. The nuclear gene HCF107 encodes a membrane-associated R-TPR (RNA tetratricopeptide repeat)-containing protein involved in expression of the plastidial psbH gene in Arabidopsis. Plant J. 2005;42:720–730. doi: 10.1111/j.1365-313X.2005.02409.x. [DOI] [PubMed] [Google Scholar]
- 17.Stern DB, Goldschmidt-Clermont M, Hanson MR. Chloroplast RNA metabolism. Annu. Rev. Plant Biol. 2010;61:125–155. doi: 10.1146/annurev-arplant-042809-112242. [DOI] [PubMed] [Google Scholar]
- 18.Sharma CM, Hoffmann S, Darfeuille F, Reignier J, Findeiss S, Sittka A, Chabas S, Reiche K, Hackermuller J, Reinhardt R, et al. The primary transcriptome of the major human pathogen Helicobacter pylori. Nature. 2010;464:250–255. doi: 10.1038/nature08756. [DOI] [PubMed] [Google Scholar]
- 19.Watkins K, Rojas M, Friso G, Wijk Kv, Meurer J, Barkan A. APO1 promotes the splicing of chloroplast group II introns and harbors a plant-specific zinc-dependent RNA binding domain. Plant Cell. 2011;23:1082–1092. doi: 10.1105/tpc.111.084335. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Hattori M, Miyake H, Sugita M. A pentatricopeptide repeat protein is required for RNA processing of clpP pre-mRNA in moss chloroplasts. J. Biol. Chem. 2007;282:10773–10782. doi: 10.1074/jbc.M608034200. [DOI] [PubMed] [Google Scholar]
- 21.Hattori M, Sugita M. A moss pentatricopeptide repeat protein binds to the 3′ end of plastid clpP pre-mRNA and assists with mRNA maturation. FEBS J. 2009;276:5860–5869. doi: 10.1111/j.1742-4658.2009.07267.x. [DOI] [PubMed] [Google Scholar]
- 22.Ruwe K, Schmitz-Linneweber C. Short non-coding RNA fragments accumulating in chloroplasts: footprints of RNA binding proteins? Nucleic Acid Res. 2012;40:3106–3116. doi: 10.1093/nar/gkr1138. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Nakamura T, Meierhoff K, Westhoff P, Schuster G. RNA-binding properties of HCF152, an Arabidopsis PPR protein involved in the processing of chloroplast RNA. Eur. J. Biochem. 2003;270:4070–4081. doi: 10.1046/j.1432-1033.2003.03796.x. [DOI] [PubMed] [Google Scholar]
- 24.Barkan A. Studying the structure and processing of chloroplast transcripts. In: Jarvis P, editor. Chloroplast research in Arabidopsis: Methods and Protocols, Methods in Molecular Biology. 1st edn. Vol. 774. New York: Humana Press; 2011. pp. 183–197. [DOI] [PubMed] [Google Scholar]
- 25.Morin RD, Aksay G, Dolgosheina E, Ebhardt HA, Magrini V, Mardis ER, Sahinalp SC, Unrau PJ. Comparative analysis of the small RNA transcriptomes of Pinus contorta and Oryza sativa. Genome Res. 2008;18:571–584. doi: 10.1101/gr.6897308. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Johnson C, Bowman L, Adai AT, Vance V, Sundaresan V. CSRDB: a small RNA integrated database and browser resource for cereals. Nucleic Acids Res. 2007;35:D829–D833. doi: 10.1093/nar/gkl991. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Lung B, Zemann A, Madej MJ, Schuelke M, Techritz S, Ruf S, Bock R, Huttenhofer A. Identification of small non-coding RNAs from mitochondria and chloroplasts. Nucleic Acids Res. 2006;34:3842–3852. doi: 10.1093/nar/gkl448. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Gregory BD, O'Malley RC, Lister R, Urich MA, Tonti-Filippini J, Chen H, Millar AH, Ecker JR. A link between RNA metabolism and silencing affecting Arabidopsis development. Dev. Cell. 2008;14:854–866. doi: 10.1016/j.devcel.2008.04.005. [DOI] [PubMed] [Google Scholar]
- 29.Cai W, Okuda K, Peng L, Shikanai T. PROTON GRADIENT REGULATION 3 recognizes multiple targets with limited similarity and mediates translation and RNA stabilization in plastids. Plant J. 2011;67:318–327. doi: 10.1111/j.1365-313X.2011.04593.x. [DOI] [PubMed] [Google Scholar]
- 30.Lurin C, Andres C, Aubourg S, Bellaoui M, Bitton F, Bruyere C, Caboche M, Debast C, Gualberto J, Hoffmann B, et al. Genome-wide analysis of Arabidopsis pentatricopeptide repeat proteins reveals their essential role in organelle biogenesis. Plant Cell. 2004;16:2089–2103. doi: 10.1105/tpc.104.022236. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Felder S, Meurer J, Meierhoff K, Klaff P, Bechtold N, Westhoff P. The nucleus-encoded HCF107 gene of Arabidopsis provides a link between intercistronic RNA processing and the accumulation of translation-competent psbH transcripts in chloroplasts. Plant Cell. 2001;13:2127–2141. doi: 10.1105/TPC.010090. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Champion EA, Kundrat L, Regan L, Baserga SJ. A structural model for the HAT domain of Utp6 incorporating bioinformatics and genetics. Protein Eng. Des. Sel. 2009;22:431–439. doi: 10.1093/protein/gzp022. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Fujii S, Small I. The evolution of RNA editing and pentatricopeptide repeat genes. New Phytol. 2011;191:37–47. doi: 10.1111/j.1469-8137.2011.03746.x. [DOI] [PubMed] [Google Scholar]
- 34.Hashimoto M, Endo T, Peltier G, Tasaka M, Shikanai T. A nucleus-encoded factor, CRR2, is essential for the expression of chloroplast ndhB in Arabidopsis. Plant J. 2003;36:541–549. doi: 10.1046/j.1365-313x.2003.01900.x. [DOI] [PubMed] [Google Scholar]
- 35.Williams-Carrier R, Kroeger T, Barkan A. Sequence-specific binding of a chloroplast pentatricopeptide repeat protein to its native group II intron ligand. RNA. 2008;14:1930–1941. doi: 10.1261/rna.1077708. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Hayes ML, Mulligan RM. Pentatricopeptide repeat proteins constrain genome evolution in chloroplasts. Mol. Biol. Evol. 2011;28:2029–2039. doi: 10.1093/molbev/msr023. [DOI] [PubMed] [Google Scholar]
- 37.Zuker M. Mfold web server for nucleic acid folding and hybridization prediction. Nucleic Acids Res. 2003;31:3406–3415. doi: 10.1093/nar/gkg595. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Schmitz-Linneweber C, Williams-Carrier RE, Williams-Voelker PM, Kroeger TS, Vichas A, Barkan A. A Pentatricopeptide Repeat Protein Facilitates the trans-Splicing of the Maize Chloroplast rps12 Pre-mRNA. Plant Cell. 2006;18:2650–2663. doi: 10.1105/tpc.106.046110. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Chen H, Stern DB. Specific binding of chloroplast proteins in vitro to the 3′-untranslated region of spinach chloroplast petD mRNA. Mol. Cell. Biol. 1991;11:4380–4388. doi: 10.1128/mcb.11.9.4380. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Condon C. What is the role of RNase J in mRNA turnover? RNA Biol. 2010;7:316–321. doi: 10.4161/rna.7.3.11913. [DOI] [PubMed] [Google Scholar]
- 41.Tillich M, Beick S, Schmitz-Linneweber C. Chloroplast RNA-binding proteins: repair and regulation of chloroplast transcripts. RNA Biol. 2010;7:172–178. doi: 10.4161/rna.7.2.11090. [DOI] [PubMed] [Google Scholar]
- 42.Scharff LB, Childs L, Walther D, Bock R. Local Absence of Secondary Structure Permits Translation of mRNAs that Lack Ribosome-Binding Sites. PLoS Genet. 2011;7:e1002155. doi: 10.1371/journal.pgen.1002155. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.O'Toole N, Hattori M, Andres C, Iida K, Lurin C, Schmitz-Linneweber C, Sugita M, Small I. On the expansion of the pentatricopeptide repeat gene family in plants. Mol. Biol. Evol. 2008;25:1120–1128. doi: 10.1093/molbev/msn057. [DOI] [PubMed] [Google Scholar]
- 44.Beick S, Schmitz-Linneweber C, Williams-Carrier R, Jensen B, Barkan A. The pentatricopeptide repeat protein PPR5 stabilizes a specific tRNA precursor in maize chloroplasts. Mol. Cell. Biol. 2008;28:5337–5347. doi: 10.1128/MCB.00563-08. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Schmitz-Linneweber C, Williams-Carrier R, Barkan A. RNA immunoprecipitation and microarray analysis show a chloroplast pentatricopeptide repeat protein to be associated with the 5′-region of mRNAs whose translation it activates. Plant Cell. 2005;17:2791–2804. doi: 10.1105/tpc.105.034454. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.