Abstract
Rap1 is an essential DNA-binding factor from the yeast Saccharomyces cerevisiae involved in transcription and telomere maintenance. Its binding to DNA targets Rap1 at particular loci, and may optimize its ability to form functional macromolecular assemblies. It is a modular protein, rich in large potentially unfolded regions, and comprising BRCT, Myb and RCT well-structured domains. Here, we present the architectures of Rap1 and a Rap1/DNA complex, built through a step-by-step integration of small angle X-ray scattering, X-ray crystallography and nuclear magnetic resonance data. Our results reveal Rap1 structural adjustment upon DNA binding that involves a specific orientation of the C-terminal (RCT) domain with regard to the DNA binding domain (DBD). Crystal structure of DBD in complex with a long DNA identifies an essential wrapping loop, which constrains the orientation of the RCT and affects Rap1 affinity to DNA. Based on our structural information, we propose a model for Rap1 assembly at telomere.
INTRODUCTION
Yeast Rap1 (Repressor Activator Protein 1) is an abundant essential DNA-binding protein that plays multiple functional roles in vivo, and is found at promoters, silencers and telomeres (1,2). It is a key transcriptional activator of many coregulated genes (3), a modulator of chromatin structure at numerous yeast promoters (4), a repressor that silences transcription at HML, HMR and telomeres (5,6), and an essential telomeres component (7,8). Its association to DNA transcription sites affects nucleosome occupancy, and chromatin structure and dynamics (4,9). Rap1 activates transcription at binding sites that can be distant by >300 bp from the activated genes, in collaboration with other DNA-associated proteins (3,4,10). Rap1 tightly and independently binds the double-stranded telomeric DNA with an average frequency of one protein every 18 bp (11–13). It recruits functional partners essential and specific for either negative regulation of telomere elongation, transcriptional repression or inhibition of non-homologous end joining (NHEJ) (14, 15).
The initiation of Rap1 functions relies on its interaction with DNA. DNA consensus sequences have been published which consist of 12–14 bp (10,16,17). The prototypical consensus sequence ACACCCRYACAYM includes two tandem half-sites ACACC or ACAYC and ACATY or ACAYM at bases 1–6 and 9–13 (12,17), although thermodynamic mapping of Rap1 binding includes three additional bases located immediately at the 5′ of this sequence (18). Rap1 not only binds this sequence with an extreme stability (19,20), but also tolerates large variation in the sequence, particularly in the second half-site (2,10,20). In addition to specific DNA binding, Rap1 is able to promote modifications of the conformation of its target DNA site, including bending, untwisting and quadruplex formation (G4 DNA) (11,21,22). By its ability to promote single-strand invasion (23), Rap1 appears functionally related to the mammalian TRF2, which is known to promote t-loop in vitro (24).
Mapping studies on Rap1 have identified a N-terminal region that includes a BRCT domain, a double-Myb DNA binding domain (DBD), and a regulatory C-terminal domain (RCT), which directly interacts with several functional partners (1) (Figure 1A). In addition to these three domains, 40% of Rap1 peptidic chain corresponds to predicted unstructured regions. The first 279 amino acids of the protein, which include the BRCT domain, can be deleted without affecting any known function (3), although yeast two-hybrid experiments have shown that this region is required for the interaction with the transcription factor Gcr1 (25). The N-terminal and C-terminal parts are dispensable for chromatin opening (26), or interaction with nucleosomal binding sites (9). The unstructured linker between DBD and RCT includes a region required for transactivation (residues 630–695) (27) and a toxicity region (residues 598–616) (28). The RCT is required for the negative feedback loop that represses telomere elongation by telomerase (29,30), and for the establishment of a silent chromatin near telomeres (6,31). It interacts with the proteins Rif1, Rif2, Sir3 and Sir4. Rif1 and Rif2 are required for the inhibition of telomere elongation through different mechanisms (15,32–35). Sir3 and Sir4 establish transcriptional silencing, which also requires Sir2, a conserved histone deacetylase (36). The binding of the Rif and Sir proteins to RCT appears to be mutually exclusive (37,38). Rif2 and Sir4 are required for NHEJ inhibition, although protection against NHEJ is also observed with a DBD–RCT construct of Rap1 in the absence of Rif2 and Sir4 (15).
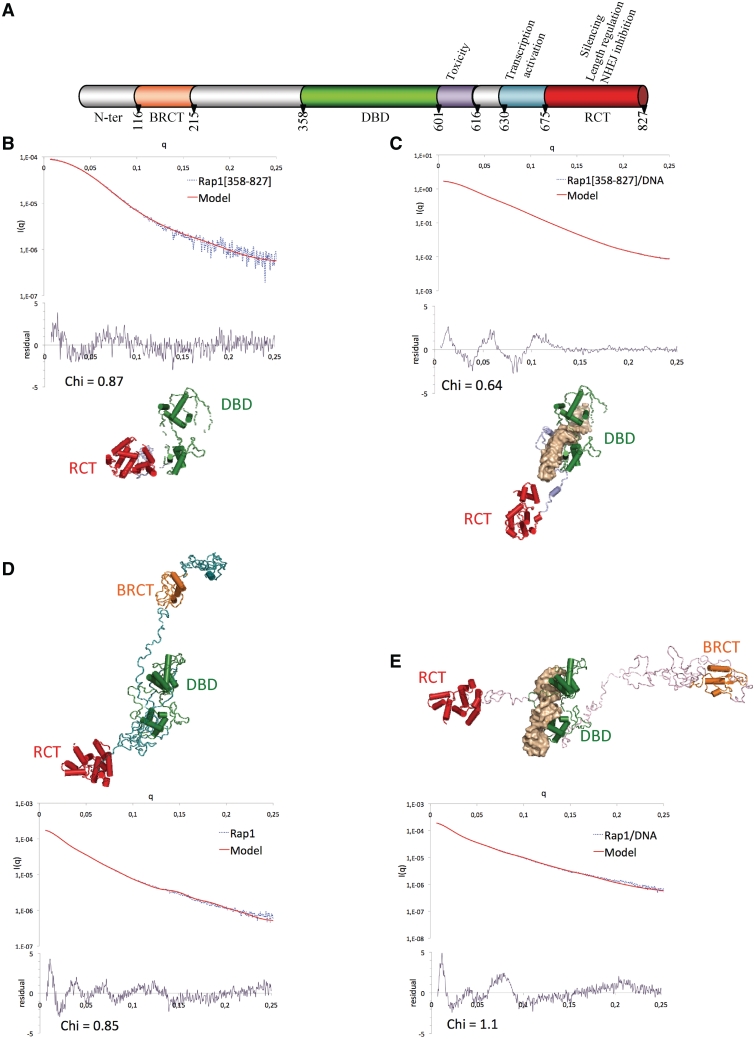
Figure 1.
SAXS analysis of Rap1 and Rap1–DNA complex. (A) Schematic boxing of Rap1 functional domains. (B–E) Cross-validated models from SAXS data, calculated with BUNCH (48), with fit and Chi residual of Rap1[358–827] (B), Rap1[358–827]/DNA (C), Rap1 (D) and Rap1/DNA (E). The color code of domains is the same than in (A). Chi = 1/(N − 1) × √(∑((Iexp − cxIth)/sigma(Iexp))2).
The presence in Rap1 of several domains involved in specific interactions and linked by flexible regions suggests a high structural plasticity of this molecule as part of its functional competence. Recent electron microscopy studies have provided the first information about Rap1 architecture, which adopts a pseudo-ring conformation in the absence of DNA (13,39). The X-ray structure of the DBD in interaction with a DNA fragment is available (12), as well as that of the C-terminal domain (38,40), and the NMR structure of the BRCT domain (41). Despite these studies, the structural determinants underlying the wide range of functions that Rap1 is able to fulfill are still poorly understood.
A crucial step to understand the role of Rap1 plasticity is to access to its whole architecture in complex with DNA. The size of Rap1 together with its high content of unstructured regions requires complementary approaches to characterize its 3D structure (42). Our approach integrates structural information obtained from SAXS, X-ray crystallography and NMR. We combined SAXS with analytical ultracentrifugation (AUC) using three different constructs of Rap1: DBD–RCT, BRCT–DBD–RCT and full-length protein. This enabled to step-by-step build the architecture of the whole molecule and of its complex with DNA, and revealed conformational adjustment upon DNA binding. The crystal structure of the DBD in complex with a 30 bp DNA provided the structural determinants of Rap1 conformational adjustment upon DNA binding. Finally, NMR titration of the RCT with full-length Rif2 and a Sir3 peptide highlighted partially overlapping surfaces, which remain accessible in the Rap1/DNA complex.
MATERIALS AND METHODS
Sample preparation
The 6His-tagged Rap1[1–224], Rap1[117–224], Rap1[117–352], Rap1[1–827] WT or mutant Y592A–K597A and Δ591–597, Rap1[117–827], Rap1[358–827] and Rap1[675–827] were cloned into pETM-13 expression vector and expressed in Escherichia coli BL21(DE3) Star strain grown overnight at 20°C after IPTG induction at [IPTG] = 0.5 mM. The proteins were purified using affinity chromatography with HisTrap column (GE Healthcare), cation exchange chromatography with ResourceQ columns (GE Healthcare), affinity chromatography HiTrap Heparine column (GE Healthcare) and size exclusion chromatography, using Superdex200 and Superdex75 column (GE Healthcare). Double-strand DNA were prepared using single strand oligonucleotides ordered at Eurogentec, and further purified using anion exchange chromatography with MonoQ 10/100 GL column (Amersham). Purified single-strand oligonucleotides were hybridized by mixing equimolar ratio of complementary DNA strands, heated to 96°C for 5 min, and slowly cooled overnight. Complexes between DNA and Rap1[1–827], Rap1[117–827] or Rap1[358–827] were formed by mixing equimolar ratio of protein/DNA, and purified by size exclusion chromatography using Superdex200 column. The different oligonucleotides and protein constructs used in the study are summarized in Supplementary Table S1.
SAXS data collection, analysis and models construction
Rap1[1–827], Rap1[117–827], Rap1[358–827] alone or in complex with DNA were prepared in the same storage buffer and same concentration than for AUC experiment (Supplementary Methods). All data were collected on SWING Beamline using either the sample changer or the online HPLC system (43) in the case of Rap1[1–827] or Rap1[1–827]/DNA complex (Supplementary Figure S1A and B). The data were analyzed using FOXTROT (from SWING beamline) and PRIMUS (Primary Analysis and Manipulations with Small Angle Scattering DATA) from ATSAS 2.1 (44), from which Guinier and normalized Kratky plots (45) were generated (Supplementary Figure S1A–D). From the corrected scattering curves, the pair-distribution functions were computed using GNOM (46), leading to the radius of gyration Rg, and the maximal distance Dmax (Supplementary Figure S1D, Table 1). The molecular weights of the particles were derived from the extrapolated intensity at the origin I0 (Table 1). In the cases of the proteins alones, the program DAMMIN (47) was used to generate the low-resolution ab initio shape envelops, which were superimposed and averaged using DAMAVER (44) (Figure S1E). To calculate the models, we used PDB coordinates from the RCT X-ray structure (entry 3CZ6, 38) and from our crystal structure of DBD/DNA for the three constructs, and from the BRCT NMR structure (entry 2L42, 41) for Rap1 full length and Rap1[117–827]. The models were calculated in an iterative manner with the program BUNCH (48) and cross-validated with AUC sedimentation coefficients (see Supplementary Methods for detailed procedure and Table 1).
Table 1.
Analytical ultracentrifugation, SAXS analysis and cross-validation of SAXS models
| Construct | Rap1[358–827] | Rap1[117–827] | Rap1 | Rap1[358–827]/DNA | Rap1[117–827]/DNA | Rap1/DNA | |
|---|---|---|---|---|---|---|---|
| Th. MW (kDa) | 55 | 82 | 94 | 67 | 94 | 105 | |
| Partial specific volume (ml/g) | 0.717 | 0.712 | 0.710 | 0.689 | 0.693 | 0.693 | |
| AUC | [Sample] (mg/ml) | 0.55, 1.1, 2.2 | 0.82, 1.64, 3.11 | 0.56, 0.94, 1.4, 1.88, 2.81, 3.74 | 0.67, 1.35, 2.02 | 0.56, 0.94, 1.88, 2.81 | 0.63, 1.06, 2.12, 3.17, 4.23 |
| MW (kDa) | 55 | 82 | 94 | 67 | 94 | 106 | |
| Sed coef (S0,W,20) | 3.6 | 3.8 | 3.9 | 4.2 | 4.3 | 4.2 | |
| f/f0 | 1.5 | 1.9 | 2.0 | 1.6 | 2.0 | 2.0 | |
| SAXS | [Sample] (mg/ml) | 1, 3.2 | 1.8, 2.5 | 1.1 a | 9 | 0.7, 1.4, 2.8, 4.1 | 1.2 a |
| Rg (Å) | 32.7 | 58.1 | 67.4 | 40.6 | 71.6 | 72.4 | |
| Dmax (Å) | 120 | 224 | 260 | 162 | 275 | 260 | |
| Calc. sed. Coef. | 3.6 | 3.8 | 4.0 | 4.2 | 4.3 | 4.2 |
Crystallization and structure determination
Initial crystallization conditions were obtained at PF6 (Institut Pasteur, Paris, France) and further optimized in our laboratory. The most suitable crystals were obtained using seating drop vapor diffusion at room temperature, with a concentration of Rap1[358–827]/DNA complex between 3.3 and 5.5 mg/ml, in a solution containing 100 mM Tris–HCl pH 8.0, 20% (v/v) PEGmme-550, 100 mM CaCl2 and 5% (v/v) glycerol. Native diffraction data were collected on PX1 beamline at SOLEIL synchrotron, and reduced with XDS (49). The structure was solved by molecular replacement with PHASER (50), using 1IGN pdb entry as model probe (12), and refined with BUSTER5 (51). Electron density and SDS page gel analysis revealed that Rap1[358–827] construct was partly degraded and that in addition to the 31-bp oligonucleotide, only the DBD moiety and part of DBD–RCT linker was present in the crystals. Statistics of the data collection, refinement and model validation are shown in Table 2.
Table 2.
Statistics of the diffraction data collection, refinement and model validation
| Data collection | |
|---|---|
| Wavelength (Å) | 0.98 |
| Space-group | P212121 |
| Diffraction limits (last shell) | 2.95 Å (3.13 Å–2.95 Å) |
| Unit cells (axbxcxαxβxγ) | 40.6 × 102.9 × 116.8 |
| 90 × 90 × 90 | |
| Rmerge | 0.138 (0.688) |
| Number of unique reflections | 9831 |
| I/σ | 14.32 (3.39) |
| Completeness | 0.901 (0.688) |
| Molecular replacement | LLG = 1305.76 |
| Refinement | Buster5 |
| Resolution | 2.95 Å |
| Rwork | 0.1923 |
| Rfree | 0.2650 |
| Figure of merit | 0.869 |
| Number of residues | 223 |
| Number of bases | 62 |
| Number of water molecules | 6 |
| RMSD bond | 0.0010 |
| RMSD angles | 1.48 |
| Average B-factor (protein, Å2) | 62 |
| Average B-factor (DNA, Å2) | 100 |
| Average B-factor (residues 675–601, Å2) | 69 |
| PDB entry | 3UKG |
Directed-mutagenesis for the in vivo experiments
RAP1 sequence (from 676 bp before the ATG to 427 bp after the STOP codon) was cloned into pRS414 plasmid. Introduced mutations were Y592A (GCT instead of TAT) and K597A (GCT instead of AAG) for the double mutant, a 21-bp deletion that lead to the absence of residues 591–597 for the deletion mutant and R580A (GCT instead of AGG) for the single mutant. These plasmids, together with a plasmid encoding wild-type Rap1, were introduced in yeast strain Lev559 (W303-1a background MATα ade2-1 trp1-1 ura3-1 leu2-3,112 his-3-11,15 can1-100 RAD5 rap1-(Δ)::KANr pACE1-UBR1 pACE1-ROX1) (52).
Western blot
Exponentially growing cells in synthetic medium lacking tryptophan and supplemented or not with 400 µM CuSO4 during 6 h were subjected to protein extraction. Proteins were extracted at 95°C in urea 8 M, SDS 2% and Tris–HCl pH 7.5 100 mM followed by vortexing at 2500 r.p.m. with glass beads. About 3 µg of total extract were loaded on 4–12% NuPage gel (Invitrogen™), and then blotted on a Hybond-P membrane (Amersham). Rap1 was detected with RAP1-Y-300 rabbit polyclonal IgG from Santa Cruz Biotechnology.
RESULTS
Orientation of Rap1 RCT is modulated upon DNA binding
The overall structural organization of free Rap1 was compared with that of Rap1 bound to DNA by using analytical ultracentrifugation and small angle X-ray scattering techniques. Three different constructs were tested: Rap1[358–827], Rap1[117–827] and Rap1 full-length (Rap1). Rap1[358–827] includes the DBD, the toxicity region, the transcription activation region and the RCT domain (Figure 1A), and corresponds to the required and sufficient region of Rap1 for telomere associated functions (53). The long N-terminal moiety Rap1[1–357] is predicted as flexible, and was built in two steps using the construct Rap1[117–827] (including BRCT, DBD and RCT domains), and Rap1 full-length (Rap1). The molecular weights derived from Guinier analyzes of the SAXS data and the sedimentation coefficient distribution profiles from analytical ultracentrifugation experiments reveal no sample aggregation, and confirmed that these three constructs of Rap1 are monomeric in solution and that Rap1/DNA complexes have a one to one stoechiometry (Supplementary Figures S1C, S2 and Table 1).
The use of Kratky plots to detect macromolecular flexibility from SAXS data is well established and extensively used (54–56). The bell shapes of Kratky plots calculated for Rap1[358–827] free and in complex with DNA reflect their compact organization (Supplementary Figure S1D). The pair distribution profile corresponds to a globular molecule in the case of free Rap1[358–827], and to an elongated molecule in the case of its complex with DNA (Supplementary Figure S1E). Average structures were calculated with BUNCH (48), and structures consistent with SAXS and AUC data were further selected (Supplementary Methods, Table 1). These structures of Rap1[358–827] and Rap1[358–827]/DNA are characterized by residual chi values of 0.87 and of 0.64, respectively (Figure 1B and C). Interestingly, the RCT orientation with regard to the DBD is different when the molecule is alone or in complex with DNA; this implies a conformational adjustment of Rap1[358–827] upon DNA binding.
Unlike in the case of Rap1[358–827], the Kratky plots of both Rap1[117–827] and Rap1, free or in complex with DNA, correspond to partially unstructured molecules (Supplementary Figure S1D), and pair distribution profiles are characteristic of elongated molecules (Supplementary Figure S1E). Calculated ab initio envelopes reveal a compact shape for Rap1[358–827], and a progressive elongation for Rap1[117–827] and Rap1 (Supplementary Figure S1F). Superposition of the three envelops highlights structural conservation of a compact region that could be attributed to region [358–827] (Supplementary Figure S1F). The frictional ratios measured by AUC arise from 1.5 to 1.9 and 2.0 in Rap1[358–827], Rap1[117–827] and Rap1, respectively, showing the transition from a compact structure for Rap1[358–827] to elongated structures for Rap1[117–827] and Rap1 (Table 1, Supplementary Figure S2). The average structures of Rap1[117–827] and of Rap1, free or in complex with DNA, remain compact in region 358–827, the N-terminal half is extended in all models (Supplementary Figures S3A and B, S1D–E), and the overall shapes of the molecules free and in complex with DNA are similar (Figure S3C–D). We calculated theoretical SAXS curves using region [358–827] from Rap1[117–827] and Rap1 models free or in complex with DNA, and compared them respectively with experimental Rap1[358–827] or Rap1[358–827]/DNA SAXS curve. We observed a better fit between theoretical and experimental curves in the case of DNA complexes, although superposition of region [358–827] from free Rap1[358-827], Rap1[117-827] and Rap1 reveals similar overall shapes (Supplementary Figure S4A and B). Superposition of region 358–827 on the DBD domain from the three Rap1 constructs shows that the relative orientation of RCT with regard to DBD fluctuates when the protein is free, whereas it is confined to a limited area in the DNA complex (Supplementary Figure S4C–D). Moreover, the orientation of the RCT domain does not depend on the presence of the N-terminal moiety of the molecule.
In summary, comparison of the average structures from our three constructs highlights two important points: (i) the compact conformation of region 358–827 does not depend on the presence of the N-terminal moiety; (ii) the interaction with DNA is associated with a reorientation of the RCT and a decreased flexibility.
Structural determinants of Rap1 organization upon DNA binding
The reorientation of the RCT domain with regard to the DBD upon DNA binding suggests that the overall conformation of Rap1 is driven by important structural determinants located in the DBD–DNA moiety. The available high-resolution crystal structure of DBD in complex with a 19 bp double-stranded DNA shows that the two Myb-domains are tandemly arranged on the DNA so that each is aligned to and makes similar contacts with the GGTGT sequences of Rap1 binding site (12,57,58). However, due to crystal packing interactions, the available structure leaves uncertainty concerning the conformation of the C-terminal loop of DBD (12), which limits our knowledge about the RCT orientation with respect to the DBD. To clarify this information, we solved the X-ray structure of Rap1DBD in complex with a 31 bp double stranded DNA oligonucleotide (Table 2). The oligonucleotide length enlarges the binding interface, and reduces the crystal packing constraints that tighten the DNA conformation. The superimposition of our structure with the available Rap1DBD/DNA complex coordinates (PDB entry 1IGN, 12) leads to a rms deviation of 1.77 Å. As expected, Rap1DBD exclusively binds the two hemi-sites spaced by three nucleotides; the additional DNA region remains free of protein (Figure 2A and B).
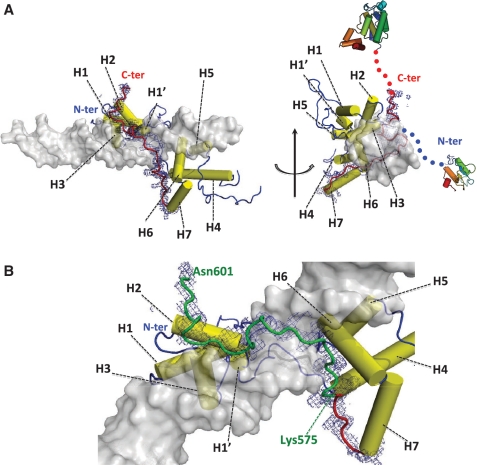
Figure 2.
Structural determinants of Rap1–DNA interaction. (A) Overall structure of Rap1–DBD in complex with DNA, with 1sigma level electron density map in blue around the region corresponding to the wrapping loop (residues 565–601). H1–H7 labels highlights alpha-helices in sequential order. The blue and red arrows indicate the respective directions of N- and C-terminal ends. (B) Zoom on Rap1 wrapping loop from Lys575 to Asn601 (in green).
The main novelty of our structure lies in the quality of the definition of loop [565–601]. The 1IGN structure lacks three protein regions: loops [482–512], [565–571] and [579–586], and ends at position 594. In our structure, the loop corresponding to [482–512] is still disordered between residues 480–498. The [565–601] region (seven residues longer than in 1IGN) is well defined, and wraps around the DNA molecule. It runs along DNA major groove with no secondary structure element, and brings the N and C-terminus extremities nearby, although they are orientated in opposite directions (Figure 2A). In addition, this wrapping loop is deeply engaged in protein/protein and protein/DNA interactions, with a total buried surface of 1508 Å2, corresponding to 37% of its total surface. The interaction with DNA buries 2374 Å2 of the DBD, among which 642 Å2 comes from the wrapping loop. The wrapping loop starts at region 565–571, lengthens helix 7 by one helical turn, and forms a tight turn at residues 565–569 that directs the loop back toward DNA major groove (Supplementary Figure S5A). The interaction between the wrapping loop and DNA involves segments 575–583 and 589–597. Residues 575–583 interact with Ade19, Cyt20, Ade21 from C-rich strand and Gua8 from G-rich strand, and Arg580 is deeply plugged in the major groove (Supplementary Figure S5B). Region 589–597 interacts with Cyt16 and Cyt17 from C-rich strand, and residues Tyr592 and Lys 597 interact with helix 2 from Myb1. These Myb1/wrapping-loop interactions close the clamp formed by the wrapping loop (Figure 3A). Finally, the N-terminal and C-terminal extremities of DBD are oriented in opposite direction, in agreement with the observation provided by our SAXS analysis.
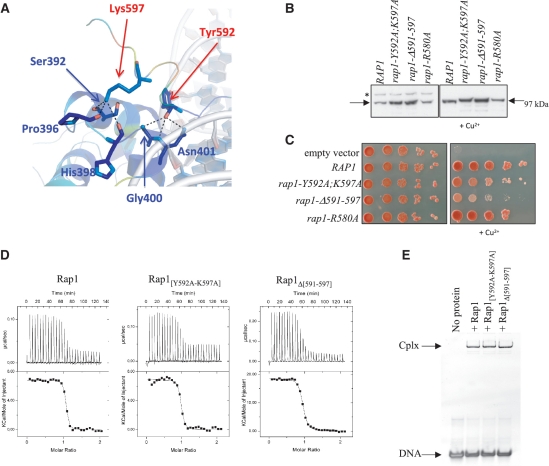
Figure 3.
Functional importance of Rap1 wrapping loop. (A) Cartoon representation of C-terminal clamp region colored from conservation analysis scores. Residues involved in C-terminal clamp are represented in sticks. Red and blue arrows highlight residues of the wrapping loop (red) that interact with residues of Myb1 (blue). (B) Western blot comparing Rap1 amount in the different strains. Rap1 level in rap1-(Δ) cells complemented with RAP1 (WT), rap1-Y592A;K597A, rap1-Δ591-597 and rap1-R580A alleles. The asterisk indicates the position of the tagged Rap1 degron, which disappears upon Cu2+ addition. (C) Ability of the rap1 alleles to complement Rap1 loss. The 10-fold serial dilution of exponentially growing cells was spotted on synthetic medium plates with or without 400 mM CuSO4. The plates were photographed after 3 days of growth at 30°C. (D) ITC titration of Rap1, Rap1[Y592A–K597A] or Rap1Δ[591–597] by DNA. (E) EMSA experiments using 8 µM DNA in the presence or not of 4 µM of Rap1, Rap1[Y592A–K597A] or Rap1Δ[591–597]. Cplx indicates the position of DNA/protein complex; DNA indicates free DNA.
Rap1 wrapping loop is important for its functional integrity and its binding to DNA
Our SAXS analysis together with the crystal structure implies that the wrapping of Rap1 around DNA has important consequences regarding its overall conformation, and its adjustment upon DNA binding. The physiological relevance of the wrapping loop was addressed with three Rap1 alleles mutated within the loop: Arg580 from the first segment was modified into alanine (rap1-R580A), residues Tyr592 and Lys597 that lock the clamp were replaced by alanines (rap1-Y592A; K597A), and the whole region 591–597 was deleted (rap1-Δ591–597). These constructs were introduced in Lev559, a strain encoding a Rap1 degron protein that will be degraded upon copper addition (14), allowing in vivo analysis of mutated Rap1 proteins. As shown in Figure 3B, rap1-R580A protein level is identical to wild-type level but rap1-Y592A–K597A and rap1-Δ591–597 levels are slightly increased. All strains display normal growth when rap1 degron is not induced. Upon Cu2+ addition, uncomplemented cells die whereas cells complemented with wild-type RAP1 or with the rap1-R580A allele grow normally (Figure 3C). Interestingly, the rap1-Y592A–K597A and rap1-Δ591–597 alleles fail to fully compensate endogenous Rap1 loss, the rap1-Δ591–597 mutant displaying a more severe growth defect. In the same way, ITC titration of Rap1 wild type or mutants Rap1[Y592A–K597A] and Rap1Δ[591–597] by DNA reveals a lower affinity by a factor of 2 or 5, respectively for each mutant as compared to the wild type (Figure 3D, Supplementary Table S3). Native gel migration profiles are similar for both wild type and mutated Rap1/DNA complexes, although the size and flexibility of the molecule suggest that changes in domain orientation remains possible (Figure 3E). These analyses indicate that the region [591–597] as well as residues 592 and/or 597 are important if not essential for Rap1 function, and highlight the importance of DBD wrapping loop and its clamp through interactions with Myb1.
Orientation of C-terminal domain within Rap1/DNA complex is compatible with its interaction with functional partners Rif2 and Sir3
Two significant features come from our analysis: (i) the reorientation of the RCT domain with regard to the DBD upon DNA binding, (ii) the location of Rap1 N- and C-terminal parts on opposite sides of the DNA (Figure 1E). Numerous studies have shown that RCT domain plays a crucial role in Rap1 partners recruitment (6,15,29–35). We therefore checked if the conformation of Rap1/DNA complex and the orientation of RCT was compatible with its interaction with functional partners in solution. We performed NMR titrations of 15N-labeled Rap1[675–827] with a Sir3[450–487] interacting peptide designed on the basis of previous double-hybrid experiments (59) and Rif2 full length (Supplementary Methods, Supplementary Figure S6). After assigning the NMR spectra of Rap1[675–827] (Supplementary Methods, Figure S6A), we identified residues whose peaks were affected upon titration with Sir3[450–487] and Rif2. The resulting surfaces are consistent with previous studies (38,40), and are partly overlapping (Supplementary Figure S7A). In addition, these Sir3 and Rif2 interacting surfaces on Rap1 RCT are still accessible when positioned onto the average structure of Rap1 full length in complex with DNA (Supplementary Figure S7B).
DISCUSSION
Interaction of Rap1 with DNA is associated to structural adjustments
The integrated structural biology study presented here allows for the first time to build a complete molecular image of the average architecture of Rap1 in solution free and in complex with DNA, despite its modular organization and flexible character. In the absence of DNA the molecule is elongated and partially disordered. The N-terminal part is particularly extended and poorly organized, whereas the DBD–RCT is more compact. The combined analysis of the three different constructs confirms the modular character of the molecule and also shows that the relative orientation of the globular domains fluctuates one with respect to the others. Comparison of our results with available electron microscopy images of Rap1 is of particular interest since our approach describes the average extended conformation, whereas high-resolution microscopy highlights the more frequent pseudo-ring (13,39) or V-shape (22) conformations. NMR titrations, ITC and gel filtration experiments confirmed that there is no preferential interaction between the N-terminal part of Rap1 and the RCT domain or DBD–RCT region that could stabilize a pseudo-ring structure of Rap1 (details in Supplementary Methods and Supplementary Figure S8). The extended and modular arrangement of free Rap1, as well as the accessibility of the Myb domains, is favorable to its interaction with DNA. However, modeling a DNA/Myb interaction using the domain organization of free Rap1 leads to sterical hindrance between DNA and RCT domain (Figure 4A). This shows that the average domains organization of Rap1 needs to adjust upon DNA binding, in agreement with the conformation observed for Rap1 in complex with DNA (Figures 1E and Supplementary Figure S3B). The conformational adjustment of Rap1 upon DNA binding leads to an orientation of the N- and C-terminal parts of the molecule in opposite directions almost perpendicular to the DNA axis (Figure 1E). Our crystal structure of DBD/DNA reveals that such conformational adjustment is probably driven by a loop located at position 575–601 of DBD that wraps DNA molecule (Figure 2A). We observed a reorientation of the RCT relatively to the DBD due to DNA binding, and the orientation of the RCT is more constrained with regard to the DBD. Furthermore, NMR titration of 15N-labeled Rap1[675-827] with the full length protein Rif2, and a Sir3[450-483] peptide leads to interacting surfaces, which remain solvent-exposed in our average structure of Rap1/DNA complex and thus accessible to Rif2 and Sir3 (Figures S6 and S7). Therefore, we propose that the orientation of the RCT in Rap1/DNA complex may facilitate its interaction with its functional partners, and the corresponding telomeric functions.
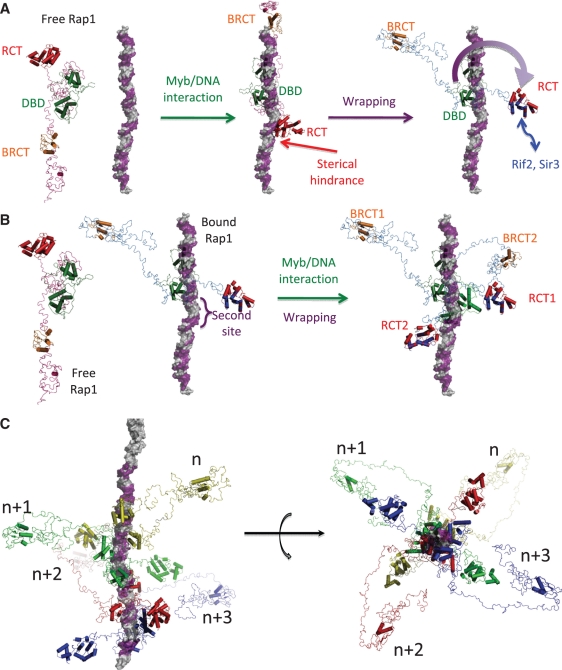
Figure 4.
Architecture of telomere first shell macro-assembly. (A) Complete architecture of free Rap1, model of free Rap1 in interaction with DNA and Rap1/DNA complex in interaction with a telomeric DNA, with the RCT surface involved in functional partners interactions, based on the combination of SAXS analysis, crystal structure and NMR titration. DNA is represented in surface mode colored in grey and telomere hemi-site in purple, BRCT in orange, DBD in green and RCT in red (with overall surface involved in Rif2 and Sir3 interaction in blue). The path of the wrapping loop is indicated with a purple arrow. (B) Modeling of the interaction of a second Rap1 molecule along telomere. (C) Reconstitution of a four Rap1 sites telomere along or in front of DNA fiber. Bound Rap1 molecules are represented in cartoon mode with different colors. Rap1 sites in violet are spaced every 18 bp.
Remodeling of Rap1 involves its wrapping around DNA
The wrapping loop located at position 575–601 of DBD is involved in extensive interactions with DNA, and increases the surface involved in DNA binding by 25%. In addition, its interaction with the first Myb of the DBD clamps the molecule around DNA through two residues (Tyr592 and Lys597) located at the tip of the loop (Figures 2A and 3A). In vivo experiments using yeast strains expressing mutants of Tyr592 and Lys597 or deletion of 591–597 segment highlight the functional significance of these residues (Figure 3B and C). Importance of region 583–596 in Rap1 DNA binding was previously observed using band shift assays (60). In this publication, Kingsman's group observed DNA binding with Rap1 region 302–596, but not with region 302–583, and concluded that the DNA binding domain ends within segment 583–596. In our study, we observe that mutation of Rap1 at Tyr592 and Lys597, or deletion of 591–597 segment affect its affinity for DNA by a factor of two and five respectively, although DNA interaction is not fully abolished (Figure 3D and E). Altogether, our results suggest that (i) the wrapping, (ii) the clamp of the molecule at a specific position on DNA molecule and/or (iii) the orientation of N- and C-terminal part in opposite direction are essential for Rap1 functional integrity. In fact, in the case of telomere protection, chromatin opening, or interaction with nucleosomal binding sites, the tight wrapping of Rap1DBD is probably sufficient since these functions can occur in the absence of N- and C-terminal domains of the molecule (9,26,29,36).
Toward a molecular model of telomere assembly
Combination of NMR results, crystal structure and SAXS analysis allows us to build the overall architecture of Rap1 in interaction or not with double-stranded DNA. These results highlight the modular character of Rap1, its structural plasticity that may facilitate its binding to DNA, and a complete and tight wrapping of the molecule around DNA that constraints the orientation of the C-terminal fragment and may favor its interaction with its functional partners (Figure 4A).
In a telomeric context, Rap1 binds independently to each recognition site (11, 13). In the same way, our interaction studies show neither oligomerization of Rap1 (Supplementary Figure S2, Table 1), nor interdomain interaction. In addition, the extended conformation of the molecule bound to DNA, with its axis of elongation almost perpendicular to the axis of the DNA molecule, leaves accessible the nearby sites, and is compatible with the binding and remodeling of a second molecule (Figure 4B). On the basis of these data, we reconstituted a molecular model of an 80 base pairs telomere with its first shell bound Rap1 molecules every 18 base pairs (11) (Figure 4C). We observe that the 18 bp spacing leads to a high molecular density. Because of the use of a short DNA in our study, the limit in detectable affinities, and the tight binding of Rap1–DBD at DNA, one cannot exclude that transient interactions might occur between adjacent molecules. The proximity between Rap1 molecules may also favor particular macromolecular assemblies at the second shell of telomere structure that involve Rap1 functional partners. The formation of such molecular scaffolding now needs to be documented at a structural level.
SUPPLEMENTARY DATA
Supplementary Data are available at NAR Online: Supplementary Methods, Supplementary Figures S1–S8, Supplementary Tables S1–S3 and Supplementary References [37,38,40,48,56,59,61–66]
FUNDING
Commissariat à l'Energie Atomique; Centre National de la Recherche Scientifique; French ‘Agence National pour la Recherche’ (ANR-06-BLAN-0076). Funding for open access charge: Commissariat à l'Energie Atomique.
Conflict of interest statement. None declared.
Supplementary Material
ACKNOWLEDGEMENTS
The authors thank beamline scientists Drs P. Legrand and A. Thompson from Proxima1 at SOLEIL synchrotron for their help in data collection. They thank Drs Stéphane Marcand, Marie-Josèphe Giraud-Panis, and Eric Gilson, for scientific discussions, and for their careful reading of the article. They thank Dr Raphaël Guérois for its reading of the article. Authors contribution: B.M., Y.V.L.B. and S.G. cloned, expressed and purified proteins; B.M., Y.V.L.B. and B.C. purified oligonucleotides; B.M., Y.V.L.B., J.P., G.D. and M.H.L.D. performed SAXS studies; Y.V.L.B. and B.R. conducted AUC experiments; S.M. conducted ITC experiments; Y.V.L.B. performed EMSA analysis; B.M., S.M. and S.Z.J. performed and NMR studies; B.M., P.W. and M.H.L.D. performed the crystallographic study ; R.L. performed in vivo experiments; S.G. and M.H.L.D. planned the project; M.H.L.D. integrated the data and wrote the article; all authors commented on the article.
REFERENCES
- 1.Morse RH. RAP, RAP, open up! New wrinkles for RAP1 in yeast. Trends Genet. 2000;16:51–53. doi: 10.1016/s0168-9525(99)01936-8. [DOI] [PubMed] [Google Scholar]
- 2.Piña B, Fernández-Larrea J, García-Reyero N, Idrissi FZ. The different (sur)faces of Rap1p. Mol. Genet. Genomics. 2003;268:791–798. doi: 10.1007/s00438-002-0801-3. [DOI] [PubMed] [Google Scholar]
- 3.Shore D. RAP1: a protean regulator in yeast. Trends Genet. 1994;10:408–412. doi: 10.1016/0168-9525(94)90058-2. [DOI] [PubMed] [Google Scholar]
- 4.Ganapathi M, Palumbo MJ, Ansari SA, He Q, Tsui K, Nislow C, Morse RH. Extensive role of the general regulatory factors, Abf1 and Rap1, in determining genome-wide chromatin structure in budding yeast. Nucleic Acids Res. 2011;39:2032–2044. doi: 10.1093/nar/gkq1161. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Lustig AJ, Kurtz S, Shore D. Involvement of the silencer and UAS binding protein RAP1 in regulation of telomere length. Science. 1990;250:549–553. doi: 10.1126/science.2237406. [DOI] [PubMed] [Google Scholar]
- 6.Kyrion G, Liu K, Liu C, Lustig AJ. RAP1 and telomere structure regulate telomere position effects in Saccharomyces cerevisiae. Genes Dev. 1993;7:1146–1159. doi: 10.1101/gad.7.7a.1146. [DOI] [PubMed] [Google Scholar]
- 7.Conrad MN, Wright JH, Wolf AJ, Zakian VA. RAP1 protein interacts with yeast telomeres in vivo: overproduction alters telomere structure and decreases chromosome stability. Cell. 1990;63:739–750. doi: 10.1016/0092-8674(90)90140-a. [DOI] [PubMed] [Google Scholar]
- 8.Gilson E, Geli V. How telomeres are replicated. Nat. Rev. Mol. Cell Biol. 2007;8:825–838. doi: 10.1038/nrm2259. [DOI] [PubMed] [Google Scholar]
- 9.Rossetti L, Cacchione S, De Menna A, Chapman L, Rhodes D, Savino M. Specific interactions of the telomeric protein Rap1p with nucleosomal binding sites. J. Mol. Biol. 2001;306:903–913. doi: 10.1006/jmbi.2001.4458. [DOI] [PubMed] [Google Scholar]
- 10.Yarragudi A, Parfrey LW, Morse RH. Genome-wide analysis of transcriptional dependence and probable target sites for Abf1 and Rap1 in Saccharomyces cerevisiae. Nucleic Acids Res. 2007;35:193–202. doi: 10.1093/nar/gkl1059. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Gilson E, Roberge M, Giraldo R, Rhodes D, Gasser SM. Distortion of the DNA double helix by RAP1 at silencers and multiple telomeric binding sites. J. Mol. Biol. 1993;231:293–310. doi: 10.1006/jmbi.1993.1283. [DOI] [PubMed] [Google Scholar]
- 12.König P, Giraldo R, Chapman L, Rhodes D. The crystal structure of the DNA-binding domain of yeast RAP1 in complex with telomeric DNA. Cell. 1996;85:125–136. doi: 10.1016/s0092-8674(00)81088-0. [DOI] [PubMed] [Google Scholar]
- 13.Williams TL, Levy DL, Maki-Yonekura S, Yonekura K, Blackburn EH. Characterization of the yeast telomere nucleoprotein core: Rap1 binds independently to each recognition site. J. Biol. Chem. 2010;285:35814–35824. doi: 10.1074/jbc.M110.170167. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Pardo B, Marcand S. Rap1 prevents telomere fusions by nonhomologous end joining. EMBO J. 2005;24:3117–3127. doi: 10.1038/sj.emboj.7600778. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Marcand S, Pardo B, Gratias A, Cahun S, Callebaut I. Multiple pathways inhibit NHEJ at telomeres. Genes Dev. 2008;22:1153–1158. doi: 10.1101/gad.455108. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Graham IR, Chambers A. Use of a selection technique to identify the diversity of binding sites for the yeast RAP1 transcription factor. Nucleic Acids Res. 1994;22:124–130. doi: 10.1093/nar/22.2.124. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Lieb JD, Liu X, Botstein D, Brown PO. Promoter-specific binding of Rap1 revealed by genome-wide maps of protein-DNA association. Nat. Genet. 2001;28:327–334. doi: 10.1038/ng569. [DOI] [PubMed] [Google Scholar]
- 18.Del Vescovo V, De Sanctis V, Bianchi A, Shore D, Di Mauro E, Negri R. Distinct DNA elements contribute to Rap1p affinity for its binding sites. J. Mol. Biol. 2004;338:877–893. doi: 10.1016/j.jmb.2004.03.047. [DOI] [PubMed] [Google Scholar]
- 19.Vignais ML, Woudt LP, Wassenaar GM, Mager WH, Sentenac A, Planta RJ. Specific binding of TUF factor to upstream activation sites of yeast ribosomal protein genes. EMBO J. 1987;6:1451–1457. doi: 10.1002/j.1460-2075.1987.tb02386.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Idrissi FZ, Piña B. Functional divergence between the half-sites of the DNA-binding sequence for the yeast transcriptional regulator Rap1p. Biochem. J. 1999;341:477–482. [PMC free article] [PubMed] [Google Scholar]
- 21.Giraldo R, Rhodes D. The yeast telomere-binding protein RAP1 binds to and promotes the formation of DNA quadruplexes in telomeric DNA. EMBO J. 1994;13:2411–2420. doi: 10.1002/j.1460-2075.1994.tb06526.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Müller T, Gilson E, Schmidt R, Giraldo R, Sogo J, Gross H, Gasser SM. Imaging the asymmetrical DNA bend induced by repressor activator protein 1 with scanning tunneling microscopy. J. Struct. Biol. 1994;113:1–12. doi: 10.1006/jsbi.1994.1027. [DOI] [PubMed] [Google Scholar]
- 23.Gilson E, Müller T, Sogo J, Laroche T, Gasser SM. RAP1 stimulates single- to double-strand association of yeast telomeric DNA: implications for telomere–telomere interactions. Nucleic Acids Res. 1994;22:5310–5320. doi: 10.1093/nar/22.24.5310. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Amiard S, Doudeau M, Pinte S, Poulet A, Lenain C, Faivre-Moskalenko C, Angelov D, Hug N, Vindigni A, Bouvet P, et al. A topological mechanism for TRF2-enhanced strand invasion. Nat. Struct. Mol. Biol. 2007;14:147–154. doi: 10.1038/nsmb1192. [DOI] [PubMed] [Google Scholar]
- 25.Mizuno T, Kishimoto T, Shinzato T, Haw R, Chambers A, Wood J, Sinclair D, Uemura H. Role of the N-terminal region of Rap1p in the transcriptional activation of glycolytic genes in Saccharomyces cerevisiae. Yeast. 2004;21:851–866. doi: 10.1002/yea.1123. [DOI] [PubMed] [Google Scholar]
- 26.Yu L, Sabet N, Chambers A, Morse RH. The N-terminal and C-terminal domains of RAP1 are dispensable for chromatin opening and GCN4-mediated HIS4 activation in budding yeast. J. Biol. Chem. 2001;276:33257–33264. doi: 10.1074/jbc.M104354200. [DOI] [PubMed] [Google Scholar]
- 27.Hardy CF, Balderes D, Shore D. Dissection of a carboxy-terminal region of the yeast regulatory protein RAP1 with effects on both transcriptional activation and silencing. Mol. Cell. Biol. 1992;12:1209–1217. doi: 10.1128/mcb.12.3.1209. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Freeman K, Gwadz M, Shore D. Molecular and genetic analysis of the toxic effect of RAP1 overexpression in yeast. Genetics. 1995;141:1253–1262. doi: 10.1093/genetics/141.4.1253. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Kyrion G, Boakye KA, Lustig AJ. C-terminal truncation of RAP1 results in the deregulation of telomere size, stability, and function in Saccharomyces cerevisiae. Mol. Cell Biol. 1992;12:5159–5173. doi: 10.1128/mcb.12.11.5159. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Marcand S, Gilson E, Shore D. A protein-counting mechanism for telomere length regulation in yeast. Science. 1997;275:986–990. doi: 10.1126/science.275.5302.986. [DOI] [PubMed] [Google Scholar]
- 31.Buck SW, Shore D. Action of a RAP1 carboxy-terminal silencing domain reveals an underlying competition between HMR and telomeres in yeast. Genes Dev. 1995;9:370–384. doi: 10.1101/gad.9.3.370. [DOI] [PubMed] [Google Scholar]
- 32.Wotton D, Shore D. A novel Rap1p-interacting factor, Rif2p, cooperates with Rif1p to regulate telomere length in Saccharomyces cerevisiae. Genes Dev. 1997;11:748–760. doi: 10.1101/gad.11.6.748. [DOI] [PubMed] [Google Scholar]
- 33.Levy DL, Blackburn EH. Counting of Rif1p and Rif2p on Saccharomyces cerevisiae telomeres regulates telomere length. Mol. Cell Biol. 2004;24:10857–10867. doi: 10.1128/MCB.24.24.10857-10867.2004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Teixeira MT, Arneric M, Sperisen P, Lingner J. Telomere length homeostasis is achieved via a switch between telomerase extendible and nonextendible states. Cell. 2004;117:323–335. doi: 10.1016/s0092-8674(04)00334-4. [DOI] [PubMed] [Google Scholar]
- 35.Anbalagan S, Bonetti D, Lucchini G, Longhese MP. Rif1 supports the function of the CST complex in yeast telomere capping. PLoS Genet. 2011;7:e1002024. doi: 10.1371/journal.pgen.1002024. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Moretti P, Freeman K, Coodly L, Shore D. Evidence that a complex of SIR proteins interacts with the silencer and telomere-binding protein RAP1. Genes Dev. 1994;8:2257–2269. doi: 10.1101/gad.8.19.2257. [DOI] [PubMed] [Google Scholar]
- 37.Palladino F, Laroche T, Gilson E, Axelrod A, Pillus L, Gasser SM. SIR3 and SIR4 proteins are required for the positioning and integrity of yeast telomeres. Cell. 1993;75:543–555. doi: 10.1016/0092-8674(93)90388-7. [DOI] [PubMed] [Google Scholar]
- 38.Feeser EA, Wolberger C. Structural and functional studies of the Rap1 C-terminus reveal novel separation-of-function mutants. J. Mol. Biol. 2008;380:520–531. doi: 10.1016/j.jmb.2008.04.078. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Papai E, Tripathi MK, Ryhlmann C, Layer JH, Weil PA, Shultz P. TFIIA and the transactivator Rap1 cooperate to commit TFIID for transcription initiation. Nature. 2010;465:956–960. doi: 10.1038/nature09080. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Chen Y, Rai R, Zhou ZR, Kanoh J, Ribeyre C, Yang Y, Zheng H, Damay P, Wang F, Tsujii H, et al. A conserved motif within RAP1 has diversified roles in telomere protection and regulation in different organisms. Nat. Struct. Mol. Biol. 2011;18:213–221. doi: 10.1038/nsmb.1974. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Zhang W, Zhang J, Zhang X, Xu C, Tu X. Solution structure of Rap1 BRCT domain from Saccharomyces cerevisiae reveals a novel fold. Biochem. Biophys. Res. Commun. 2011;404:1055–1059. doi: 10.1016/j.bbrc.2010.12.109. [DOI] [PubMed] [Google Scholar]
- 42.Perry JJ, Cotner-Gohara E, Ellenberger T, Tainer JA. Structural dynamics in DNA damage signaling and repair. Curr. Opin. Struct. Biol. 2010;20:283–294. doi: 10.1016/j.sbi.2010.03.012. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.David G, Pérez J. Combined sampler robot and HPLC: a fully automated system for biological SAXS experiments at the Synchrotron SOLEIL Swing beamline. J. Appl. Cryst. 2009;42:892–900. [Google Scholar]
- 44.Konarev PV, Petoukhov MV, Volkov VV, Svergun DI. ATSAS 2.1, a program package for small-angle scattering data analysis. J. Appl. Cryst. 2006;39:277–286. doi: 10.1107/S0021889812007662. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Durand D, Vivès C, Cannella D, Perez J, Pebay-Peyroula E, Vachette P, Fieschi F. NADPH oxidase activator p67phox behaves in solution as a multidomain protein with semi-flexible linkers. J. Struct. Biol. 2010;169:45–53. doi: 10.1016/j.jsb.2009.08.009. [DOI] [PubMed] [Google Scholar]
- 46.Svergun DI. Determination of the regularization parameter in indirect-transform methods using perceptual criteria. J. Appl. Crystallogr. 1992;25:495–503. [Google Scholar]
- 47.Svergun DI. Restoring low resolution structure of biological macromolecules from solution scattering using simulated annealing. Biophys. J. 1999;76:2879–2886. doi: 10.1016/S0006-3495(99)77443-6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Petoukhov MV, Svergun DI. Global rigid body modeling of macromolecular complexes against small-angle scattering data. Biophys. J. 2005;89:1237–1250. doi: 10.1529/biophysj.105.064154. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Kabsch W. Integration, scaling, space-group assignment and post-refinement. Acta Crystallogr. D Biol. Crystallogr. 2010;66:133–144. doi: 10.1107/S0907444909047374. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.McCoy AJ, Grosse-Kunstleve RW, Adams PD, Winn MD, Storoni LC, Read RJ. Phaser crystallographic software. J. Appl. Cryst. 2007;40:658–674. doi: 10.1107/S0021889807021206. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Bricogne G, Blanc E, Brandl M, Flensburg C, Keller P, Paciorek W, Roversi P, Smart OS, Vonrhein C, Womack TO. BUSTER, Version 2.8.0. United Kingdom: Global Phasing Ltd, Cambridge; 2009. [Google Scholar]
- 52.Pobiega S, Marcand S. Dicentric breakage at telomere fusions. Genes Dev. 2010;24:720–733. doi: 10.1101/gad.571510. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Sussel L, Shore D. Separation of transcriptional activation and silencing functions of the RAP1-encoded repressor/activator protein 1: isolation of viable mutants affecting both silencing and telomere length. Proc. Natl Acad. Sci. USA. 1991;88:7749–7753. doi: 10.1073/pnas.88.17.7749. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Glatter O, Kratky O, editors. Small Angle X-ray Scattering. London: Academic Press; 1982. [Google Scholar]
- 55.Pérez J, Vachette P, Russo D, Desmadril M, Durand D. Heat-induced unfolding of neocarzinostatin, a small all-beta protein investigated by small-angle X-ray scattering. J. Mol. Biol. 2001;308:721–743. doi: 10.1006/jmbi.2001.4611. [DOI] [PubMed] [Google Scholar]
- 56.Rambo AP, Tainer JA. Bridging the solution divide: comprehensive structural analyses of dynamic RNA, DNA, and protein assemblies by small angle X-ray scattering. Curr. Opin. Struct. Biol. 2010;20:128–137. doi: 10.1016/j.sbi.2009.12.015. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57.Taylor H, O'Reilly M, Leslie A, Rhodes D. How the multifunctional yeast Rap1p discriminates between DNA target sites: a crystallographic analysis. J. Mol. Biol. 2000;303:693–707. doi: 10.1006/jmbi.2000.4161. [DOI] [PubMed] [Google Scholar]
- 58.Rhodes D, Fairall L, Simonsson T, Court R, Chapman L. Telomere architecture. EMBO Rep. 2002;3:1139–1145. doi: 10.1093/embo-reports/kvf246. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59.Moretti P, Shore D. Multiple interactions in sir protein recruitment by Rap1p at silencers and telomeres in yeast. Mol. Cell. Biol. 2001;21:8081–8094. doi: 10.1128/MCB.21.23.8082-8094.2001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60.Henry YA, Chambers A, Tsang JS, Kingsman AJ, Kingsman SM. Characterisation of the DNA binding domain of the yeast RAP1 protein. Nucleic Acids Res. 1990;18:2617–2723. doi: 10.1093/nar/18.9.2617. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 61.Schuck P. Size distribution analysis of macromolecules by sedimentation velocityultracentrifugation and Lamm equation modeling. Biophys. J. 2000;78:1606–1619. doi: 10.1016/S0006-3495(00)76713-0. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 62.Eswar N, Eramian D, Webb B, Shen MY, Sali A. Protein structure modeling with MODELLER. Methods Mol. Biol. 2008;426:145–159. doi: 10.1007/978-1-60327-058-8_8. [DOI] [PubMed] [Google Scholar]
- 63.Svergun DI, Barberato C, Koch MHJ. CRYSOL—a program to evaluate X-ray solution scattering of biological macromolecules from atomic coordinates. J. Appl. Cryst. 1995;28:768–773. [Google Scholar]
- 64.García De La Torre J, Huertas ML, Carrasco B. Calculation of hydrodynamicproperties of globular proteins from their atomic-level structure. Biophys. J. 2000;78:719–730. doi: 10.1016/S0006-3495(00)76630-6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 65.Jung YS, Zweckstetter M. Mars—robust automatic backbone assignment of proteins. J. Biomol. NMR. 2004;30:11–23. doi: 10.1023/B:JNMR.0000042954.99056.ad. [DOI] [PubMed] [Google Scholar]
- 66.Cornilescu G, Delaglio F, Bax A. Protein backbone angle restraints from searching adatabase for chemical shift and sequence homology. J. Biomol. NMR. 1999;13:289–302. doi: 10.1023/a:1008392405740. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.