Abstract
Iron, an essential nutrient with limited bioavailability, requires specialized cellular mechanisms for uptake. Although iron uptake into the cytoplasm in the form of heme has been well characterized in many bacteria, the subsequent trafficking is poorly understood. The cytoplasmic heme-binding proteins belong to a structurally related family thought to have evolved as “induced fit” ligand-binding macromolecules. One member, Pseudomonas aeruginosa cytoplasmic protein (PhuS), has previously been shown to be important for delivering heme to the iron regulated heme oxygenase (HemO). Spectroscopic investigations of the holo-PhuS complex revealed a dynamic heme environment with overlapping but distinct heme-binding sites with alternative coordinating heme ligands, His-209 or His-212. In the present work we establish a mechanism for how heme is transferred from PhuS to its partner, HemO. Using surface plasmon resonance and isothermal titration calorimetry, we have discovered that holo-PhuS, but not apo-PhuS, forms a 1:1 complex with HemO. Sedimentation velocity and limited proteolysis experiments suggest that heme binding to PhuS induces a conformational rearrangement that drives the protein interaction with HemO. Hydrodynamic analysis reveals that the holo-PhuS displays a more expanded hydrodynamic envelope compared with apo-PhuS, and we propose that this conformational change drives the interaction with HemO. We further demonstrate that replacement of His-212 by Ala disrupts the interaction of holo-PhuS with HemO; in contrast, the His-209-Ala variant can still complex with HemO, albeit more weakly. Together, the present studies reveal a mechanism that couples a heme-dependent conformational switch in PhuS to protein–protein interaction, the subsequent free energy of which drives heme release to HemO.
Keywords: cytoplasmic heme-binding protein, heme trafficking, heme degradation, iron-acquisition
The requirement by almost all living organism for iron, a micronutrient that has limited bioavailability, requires specialized mechanisms for uptake and storage. Furthermore, the evolutionary struggle for iron between the host and invading bacterial pathogen sets up a competition that has shaped both the host's defense strategies and the iron-acquisition mechanisms deployed by the invading pathogen (1–3). This has led many pathogens to develop additional systems that use the host's heme-containing proteins as a source of iron (4–6). The opportunistic pathogen Pseudomonas aeruginosa encodes two heme uptake systems: the phu (Pseudomonas heme uptake) and has (heme assimilation system) (7). Heme uptake into the cytoplasm has been well characterized in Gram-negative organisms, and crystal structures of several heme-binding proteins have been solved (8–10). However, the fate of the heme once internalized is less well understood, and it remains unclear whether all pathogens use similar heme degradative pathways. Heme oxygenase (HemO) enzymes have been identified and characterized in Gram-positive and Gram-negative pathogens, including Corynebacterium diphtheria (11), Neisseriae spp., and P. aeruginosa (12), where the structural fold of the proteins is strikingly similar to the eukaryotic heme oxygenases (HO) (13, 14). In contrast, there are several recent reports describing nonclassical HemOs from Staphylococcus aureus (15), Bacillus anthracis (16), and Mycobacterium tuberculosis (17). Furthermore, homologs of the cytoplasmic heme-binding protein PhuS, including HemS from Yersinia entercolitica and ChuS in Escherichia coli, have previously been annotated as heme-degrading factors, and in the case of ChuS the purified protein has been biochemically characterized as a HemO (18, 19).
Recently our laboratory has demonstrated that the PhuS protein is not a HemO but rather a heme-trafficking protein that delivers heme to HemO (20–22). Previous spectroscopic characterization of the holo-PhuS protein indicated a dynamic heme that can access a range of spin-states and coordination number as a function of pH (8). A combination of site-directed mutagenesis and spectroscopic studies further confirmed the conserved His-209 to be the proximal ligand to the heme. However, spectroscopic studies of the holo-PhuS H209A mutant also identified an overlapping but mutually exclusive heme binding site provided by coordination to His-212 (8). Moreover, His-210 does not directly coordinate to the heme, but in the absence of His-209 was shown to stabilize ligand coordination through His-212 (21). In the present studies we have further investigated the role of proximal helix flexibility and dynamic heme properties in driving conformational change and protein–protein interaction. The present studies confirm that both His-212 and His-210, in addition to the proximal His-209, are required for the heme-dependent conformational change and subsequent protein interaction with HemO. Furthermore, we describe a mechanism of heme transfer for the cytoplasmic heme-binding proteins, whereby a ligand-induced conformational change facilitates protein–protein interaction, the subsequent free energy of which drives heme transfer.
Results
Heme Binding to PhuS Is Facilitated Through Noncovalent Interactions of the Porphyrin with the Protein Scaffold.
The electronic absorption spectra of the wild-type, H209A, H210A, and H212A mutant PhuS proteins have previously been reported (21). The spectrum of holo-PhuS H209/210/212A is provided in Fig. S1. Initial circular dichroism (CD) analysis revealed no overall change in secondary structure in the apo- or holo-PhuS His-mutants, confirming the structural integrity of the proteins (Fig. S2). Binding affinities (KDs) were determined by fluorescence quenching, and as previously reported for the wild type, the His-mutants bind one heme per monomer (Table 1 and Fig. S3). Interestingly, according to the fluorescence quenching experiments the His-mutants had apparent KDs similar to the wild-type protein. Perhaps most surprisingly, the H209/210/212A mutant seems to bind one heme molecule with an affinity similar to the wild-type protein, despite the loss of both His-ligands. Although we cannot be certain that heme is binding at the same site, previous resonance Raman studies suggest that the heme environment in the holo-PhuS H209/212A complex, which also lacks both potential coordinating His-residues, is similar to that of the wild-type holo-PhuS complex (21).
Table 1.
Spectroscopic and binding parameters for the holo-PhuS wild-type and His-mutant proteins.
| Protein (PhuS) | Soret band (nm) | Q-bands (nm) | Spin-state (pH 7.8)* | Proximal His-ligand* | Kd (μM) heme† | KD (μM) heme-PhuS:HemO† |
| Wild type | 411 | 545, 570 | 6CHS/LS | His-209 | 0.41 ± 0.05 | 1.23 ± 0.05 |
| H209A | 410 | 532, 566 | 6CLS | His-212 | 1.6 ± 0.70 | 7.0 ± 0.6 |
| H210A | 412 | 540, 578 | 6CHS/LS | His-209 | 0.48 ± 0.1 | ND |
| H212A | 407 | 544, 580 | 6CHS/5CHS | His-209 | 1.0 ± 0.3 | ND |
| H209/210/212A† | 398, 368 (sh) | 398, 600 | 5CHS | None | 1.2 ± 0.3 | 8.3 ± 0.8 |
ND, no binding observed.
*Previously determined (21).
†Data obtained in the present study as described in Experimental Procedures.
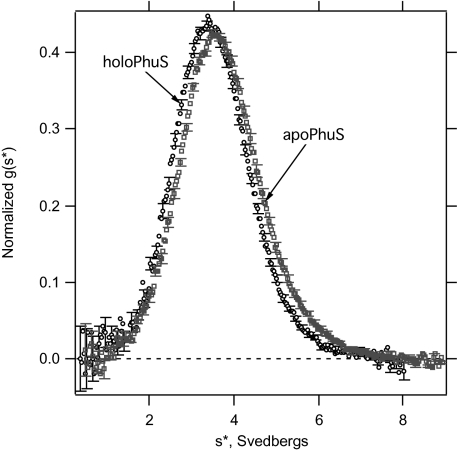
Sedimentation velocity experiments were performed to further evaluate the oligomeric state and overall shape of the wild-type apo- and holo-PhuS proteins. The sedimentation coefficient distributions (Fig. 1) show that apo- and holo-PhuS each sedimented as a single boundary with slightly different sedimentation coefficient distributions. Although the differences are small, the concentration dependences (Fig. S4) show that the sedimentation properties of apo- and holo-PhuS are reproducibly distinct from one another. To eliminate the possibility that this could be due to sample heterogeneity, we first confirmed that both samples were monodisperse by fitting the sedimentation coefficient distributions to single Gaussian functions in DCDT+. For homogeneous samples, this procedure should return the molecular weight, which we obtained for all concentrations of both samples. The average deviation of the fitted from the true molecular weight was 4%. In addition, the concentration dependences of the weight average sedimentation coefficients show a small but negative slope, characteristic of single ideal species in solution (Fig. S4). Because both proteins behave in a monodisperse manner, the difference in the sedimentation coefficients must reflect distinct hydrodynamic shapes for apo- and holo-Phus, with the holo form of the protein displaying a more expanded hydrodynamic envelope compared with apo-PhuS. Because the effect of the heme on both the molecular weight and the partial specific volume are minimal, we propose that the conformational changes associated with heme binding drive a conformational change required for interaction with HemO.
Fig. 1.
Normalized g(s*) distributions of holo- and apo-PhuS. Representative data are shown for 3.88 μM holo-PhuS (○) and 3.88 μM apo-PhuS (□). For clarity, every fifth error bar is plotted for the sedimentation coefficient distribution functions. The observed weight average sedimentation coefficients for these data equal 3.57 S and 3.73 S for holo- and apo-Phus, respectively.
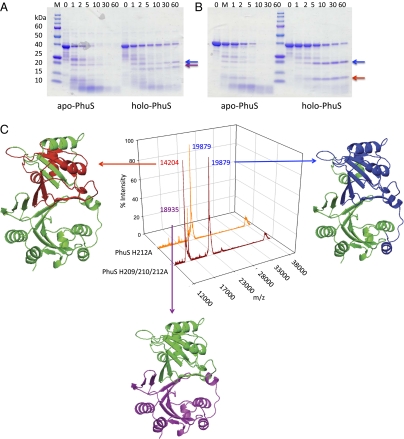
The role of heme coordination in driving a conformational change in holo-PhuS was further investigated by limited proteolysis and MALDI-MS analysis. Limited proteolysis over time revealed that holo-PhuS was protected from extensive degradation, whereas apo-PhuS was rapidly degraded within the first 10 min (Fig. 2A). After a 30-min exposure to trypsin no detectable protein bands were observed in the apo-PhuS sample, whereas for the holo-PhuS a significant percentage of full-length protein (40% by density measurement) remained. Interestingly, the proteolysis profile of apo- and holo-PhuS show distinct bands, with holo-PhuS yielding two major bands at ≈20 and 19 kDa. In contrast, the apo-PhuS shows an early ≈24-kDa core band that is rapidly degraded with the appearance of the ≈19-kDa band and a new band at ≈14 kDa (Fig. 2A). Analysis of the apo- and holo-PhuS H210A and H212A mutants, which retain His-209 as the proximal ligand, gave profiles similar to that of the wild-type protein (Fig. S5 A and C). In contrast, the holo-PhuS H209A and H209/210/212A mutants yielded proteolysis profiles more typical of the apo-PhuS proteins (Fig. S5B and Fig. 2B, respectively). Interestingly, although heme binding to the H209A and H209/210/212A PhuS proteins seems to afford some protection from rapid proteolysis, no evidence of the ≈19-kDa band is observed in the holo-PhuS proteins lacking the proximal His-209.
Fig. 2.
Limited proteolysis and MALDI-MS of the wild-type and mutant apo- and holo-PhuS proteins. SDS/PAGE of (A) wild-type apo- and holo-PhuS and (B) apo- and holo-PhuS H209/210/212A. Samples (10 μL) at various time intervals as marked were separated on 8–20% Tris•HCl gradient gels. (C) MALDI-MS spectra of holo-PhuS H212A and holo-PhuS H209/210/212A after 60-min limited proteolysis. The major peptide fragments are highlighted and color coded according to the homology model analysis. The peak at ≈39 kDa corresponds to intact PhuS. The homology model of PhuS was generated from the apo-ChuS crystal structure (Protein Data Bank file 1U9T) using Swiss-Model as previously described (8).
The 20- and 19-kDa fragments observed on SDS/PAGE after cleavage of the holo-PhuS wild type, H210A and H212A proteins were assigned by MALDI-MS to the N-terminal core domain (Ala-23 to Arg-206; m/z 19879) and C-terminal core domain (Ala-183 to Leu-354; m/z 18935), respectively (Fig. 2C). The peaks surrounding the main peak at m/z 19879, a result of cleavage at Arg-22, arise from additional cleavage sites at Arg-25 and Arg-31. In contrast, MALDI-MS analysis of the holo-PhuS H209/210/212A protein yielded the N-terminal core domain (m/z 19879), a smaller N-terminal fragment at m/z 14204 (Met-1 to Arg-130), but no evidence of the C-terminal helical domain (m/z 18935) (Fig. 2B). Furthermore, MALDI-MS analysis of the holo-PhuS H209A and H209/210/212A mutants at earlier time points did not show any evidence of the C-terminal core domain (Fig. S5). MALDI-MS analysis of the apo-PhuS proteins after exposure to trypsin for 2 min yielded only the smaller N-terminal fragment (m/z 14204) previously observed in the holo-PhuS proteins lacking the proximal His-209 (Fig. S5). The present data indicate that heme coordination through His-209 stabilizes the helical C-terminal domain, allowing for conformational rearrangement and optimal interactions of the heme with the N-terminal distal face. Interestingly, the N-terminal core domain (m/z 19879) seems to be stabilized on heme coordination even in the absence of the proximal ligand, consistent with significant nonbonding interactions of the heme with the protein scaffold.
Heme Binding to PhuS Induces a Conformational Rearrangement Required for Protein–Protein Interaction with HemO.
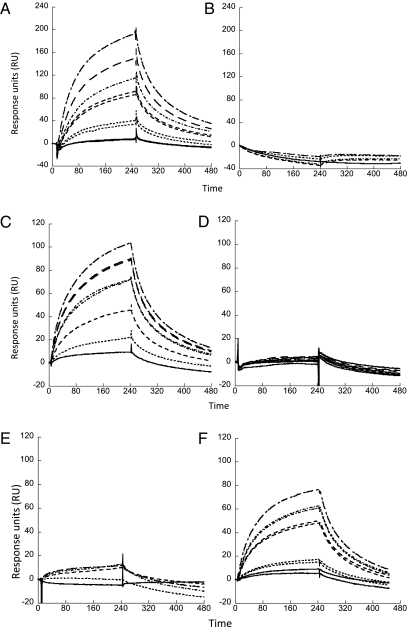
Our previous studies have shown that holo-PhuS interacts with and transfers heme to apo-HemO (22). The role of heme coordination through H209A and H212A in determining interactions with apo-HemO was further investigated by surface plasmon resonance (SPR). Increasing concentrations of wild-type holo-PhuS passed over immobilized apo-HemO gave a concentration-dependent increase in plasmon resonance response and a calculated binding affinity (Kd) of 1.23 μM (Fig. 3A and Table 1). As expected, the apo-PhuS complex gave no evidence of a response at concentrations 10-fold higher than those used in the holo-PhuS titration (Fig. 3B). Therefore, the data suggest that the “heme-induced fit” and conformational changes observed in the sedimentation and proteolysis experiments drive protein–protein interaction with HemO (23).
Fig. 3.
Kinetic analysis by SPR of the PhuS–HemO interaction. PhuS samples injected over Ni-NTA immobilized apo-HemO at 0.5 μM (______), 1.0 μM (…………), 2.5 μM (- - - - -), 5.0 μM (- · - · - · -), 7.5 μM (__ __ __), and 10 μM (____ _ ___). (A) Holo-PhuS, (B) Apo-PhuS, (C) Holo-PhuS H209A, (D) Holo-PhuS H210A, (E) Holo-PhuS H212A, and (F) Holo-PhuS H209/210/212A.
Proximal Helix His-210 and His-212 Residues Are Required for Protein–Protein Interaction with HemO.
Compared with wild-type holo-PhuS the heme-loaded PhuS His mutants were significantly compromised in their ability to interact with apo-HemO. Previous resonance Raman spectroscopic studies have clearly shown that mutation of the proximal His-209 to an Ala results in an alternate heme-binding site provided through His-212 (21). Interestingly, the holo-PhuS H209A mutant gave a detectable SPR response, and a binding affinity approximately sixfold lower than that of wild-type holo-PhuS, suggesting that heme coordination through His-212 induces a protein conformation competent for interaction with apo-HemO, albeit with a lower affinity (Table 1 and Fig. 3C). As expected, the apo-PhuS H209A protein gave no plasmon resonance response. In contrast, the holo-PhuS H212A and H210A mutants gave no detectable change in plasmon resonance up to concentrations 10-fold higher than the maximum response observed for holo-PhuS (Fig. 3 D and E and Table 1). The lack of a detectable protein–protein interaction between the holo-PhuS H210A or H212A mutants and apo-HemO, despite retention of the proximal His-209, suggests that both His-210 and 212 are required for protein–protein interaction.
Interestingly, the holo-PhuS H209/210/212A protein in which both coordinating ligands are absent has a binding affinity for HemO similar to that of the alternate His-212 coordinated holo-PhuS H209A protein (Fig. 3F and Table 1). It is feasible that the absence of either His-210 or His-212 disrupts the ability of the His-209 coordinated heme to be seated within the binding cleft in a conformation critical for interaction of the heme with the N-terminal distal face. In contrast, the lack of a proximal ligand in the PhuS H209/210/212A mutant may allow for the nonbonding side chain interactions between the heme and the N-terminal domain, inducing a conformational change with a lower affinity for apo-HemO. Heme binding within the binding site in the absence of His-209 and His-212 is supported by previous resonance Raman studies, which suggest that the heme is bound in an environment similar to that in the wild-type holo-PhuS (21). Heme binding within the ligand site is further supported by the stabilization of the N-terminal domain in limited proteolysis experiments of holo-PhuS H209/210/212A compared with its apo-PhuS counterpart (Fig. 2B). Furthermore, apo-PhuS H209/210/212A on passage over apo-HemO gives no detectable plasmon resonance response, confirming that heme binding to holo-PhuS H209/210/212A induces a conformational change allowing interaction with HemO.
Thermodynamics of Holo-PhuS:HemO Association.
To further characterize the interaction of the wild-type and H209A holo-PhuS proteins with HemO we performed isothermal titration calorimetry (ITC) studies. Titration of wild-type holo-PhuS with apo-HemO gave an enthalpic signal with a 1:1 stoichiometry (Fig. S6A). The apparent Ka for the interaction of holo-PhuS with apo-HemO of 2.3 μM is consistent with that obtained by SPR (Table 1). Similar analysis by ITC of apo-PhuS with apo-HemO gave no detectable signal or heat of exchange (Fig. S6B). The apparent Ka for holo-PhuS H209A with apo-HemO (9.6 μM) is approximately fourfold lower than that obtained for the wild-type protein and similar to that obtained by SPR (Table 1 and Fig. S6C). The relatively weak association of the PhuS and HemO proteins is consistent with the transient nature of the protein complex and previously reported heme transfer rate of 0.1 s−1 (20).
The observed heat (ΔHobs) generated upon mixing holo-PhuS with apo-HemO is a sum of the heat due to the protein–protein interaction (ΔHprotein:protein) and heme transfer from PhuS to HemO (ΔHhemetransfer), where ΔHobs = ΔHprotein:protein + ΔHhemetransfer. The interaction of holo-PhuS wild type (ΔHobs = −4.4 kcal mol−1) or H209A (ΔHobs = −7.5 kcal mol−1) with HemO is enthalpy driven. In the case of the H209A mutant the moderate increase in enthalpy is reflected in the reduced entropy (TΔS = 2.0 kcal mol−1) compared with the wild-type PhuS (TΔS = 4.0 kcal mol−1). However, the overall favorable free energy for the interaction and transfer of heme from holo-PhuS wild type (−8.3 kcal mol−1) or holo-PhuS H209A (−9.5 kcal mol−1) to HemO indicates that coordination through His-209 does not play a significant role in protein association and heme transfer. We were unable to obtain thermodynamic parameters for the interaction of holo-PhuS H209/210/212A with apo-HemO because of protein precipitation during the titration.
Discussion
Through a series of in vitro biophysical studies we have characterized a heme-induced conformational switch in the cytoplasmic heme-binding protein PhuS that is required for interaction and heme transfer to HemO. Previous spectroscopic characterization of the holo-PhuS complex revealed an extremely dynamic heme ligand that can access a range of spin-states and coordination number as a function of pH (21, 22). This distribution of heme across multiple spin-states and coordination number is indicative of a flexible heme environment and consistent with a role as a heme-trafficking protein. The structure of the apo- and holo-HemS from Y. entercolitica reveals two homologous domains that provide a heme-binding cleft with the C-terminal domain donating the proximal ligand and the distal N-terminal domain a pair of twisted β-sheets (23). On heme coordination, residues within the pocket provide significant contributions through noncoordinating interactions, reshaping the heme-binding cleft with little or no change in overall secondary structure. The reshaping of the heme-binding cleft and subsequent conformational change observed in the holo-HemS structure is consistent with the current sedimentation analysis of the apo- and holo-PhuS proteins. Thus, the induced conformational change on heme coordination through His-209 provides the thermodynamically stable “storage complex” of holo-PhuS. The fact that coordination through His-212 in the holo-PhuS H209A mutant allows interaction with apo-HemO indicates some flexibility within the pocket can be accommodated. This is further supported by the comparable free energy (ΔG) of protein interaction and heme transfer from holo-PhuS H209A compared with the wild-type holo-PhuS complex, where heme coordination through His-209 does not play the major role in driving the protein–protein interaction.
In contrast, mutation of the proximal helix His-210 or His-212 to Ala resulted in holo-PhuS complexes that do not interact with HemO despite a proteolytic profile similar to that of the wild-type holo-PhuS. Therefore, coordination through His-209 stabilizes the C-terminal domain, but mutation of His-210 and His-212 introduce subtle effects within the proximal helix that disrupt the protein–protein interaction. Although it is not clear at present why replacement of His-210 or His-212 with Ala disrupts the interaction with apo-HemO, it is feasible that the heme seating within the binding site is altered such that the noncoordinating π-stacking interactions of the porphyrin with the N-terminal domain are compromised. Interestingly, mutation of all three His-residues (H209/210/212A) results in a “compensatory effect” whereby the lack of a proximal ligand “tether” allows rearrangement of the N- and C-terminal domains, facilitating noncoordinating π-stacking interactions of the heme with the protein. The significant interactions of the porphyrin ring with aromatic residues, and the fact that loss of the proximal ligand does not greatly affect heme affinity, would support a model whereby significant contributions are provided by noncoordinating π-stacking interactions. This mode of heme binding to PhuS is reminiscent of the mammalian heme transport protein hemopexin, in which two homologous domains form a heme-binding cleft, with aromatic residues providing a significant contribution to the binding affinity (24).
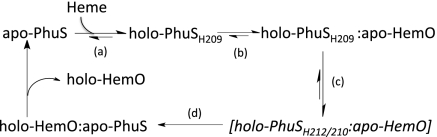
Taken together the present data suggest a model whereby heme binding to apo-PhuS through His-209 triggers a conformational switch that promotes interaction with HemO (Scheme 1 A and B). The favorable free energy derived from the interaction with HemO triggers a histidine ligand switch to His-212 (Scheme 1C). Previous resonance Raman studies indicated that His-210, although not directly involved in coordinating to the heme, was required to stabilize heme binding to the alternate His-212 site and is consistent with both His-210 and His-212 being required for protein–protein interaction and heme transfer (21). In the final step the proposed holo-PhuS His-212 intermediate specifically delivers heme to HemO (Scheme 1D). This model is consistent with the His-212 coordinated holo-PhuS H209A complex having the ability to interact with HemO, and with previous kinetic studies that indicate a spin state transition or ligand switch occurs within holo-PhuS before heme transfer to HemO (20). In summary, this is a bacterial heme-trafficking protein for which a protein partner has been identified and characterized. Because both PhuS and HemO are relatively small soluble proteins, this will facilitate further spectroscopic and structural characterization of heme transfer in this unique family of proteins, while providing insight into the general mechanism of heme transport and release across a wide range of systems with common structural motifs.
Scheme 1.
Proposed mechanism of heme transfer from holo-PhuS to apo-HemO.
Experimental Procedures
Strains and Plasmids.
Site-directed mutagenesis of the phuS gene in pET21α has previously been reported (21). Oligonucleotides were generated to have melting temperatures (Tm) in the range 65–70 °C encoding the required base changes for the respective histidine to alanine substitutions. E. coli DH5α [F’ araD(lac-proAB) rpsL ϕ80dlacZDM15 hsd R17] was routinely used for DNA manipulation and E. coli strain BL21 (DE3) plysS [F− ompT hsdSB (rB− mB−) gal dcm (DE3)] for protein expression. Bacteria were cultured in LB broth containing 100 μg/mL ampicillin.
Protein Purification.
Protein expression and purification of the PhuS proteins were carried out as previously described (21, 22). Heme loading of PhuS was carried out by addition of an equimolar ratio of heme to protein, followed by gel filtration. Heme was prepared in 0.1 N NaOH and the pH adjusted with the identical buffer used to prepare the PhuS protein. All heme solutions were used within 20 min of preparation. Heme concentrations and stoichiometry of the final holo-PhuS complexes were determined by pyridine hemochrome (25). Before use the holo-PhuS, wild-type and mutant proteins were further purified by FPLC over a Sephacryl S-200 (0.5 × 30 cm) to ensure that the protein was monomeric. The native HemO protein was purified as previously described (12). The C-terminal His-tagged HemO used in SPR experiments was purified over nickel-nitriloacetic acid (NTA) agarose. The peak fractions as judged by SDS/PAGE were pooled and dialyzed against 20 mM Tris·HCl (pH 7.5) or 10 mM Hepes (pH 7.4) containing 150 mM NaCl.
Tryptic Digest Experiments.
Apo- or holo-PhuS wild-type and His-mutant proteins (30 μM) in 20 mM Tris·HCl (pH 8.0) were incubated at 37 °C with sequencing grade trypsin (25:1). Samples (10 μL) were removed at timed intervals and the reaction terminated by the addition of SDS/PAGE sample buffer containing 1 mM PMSF. The fragmentation pattern was analyzed by SDS/PAGE (8–20% Tris·HCl gradient gel).
MALDI-MS Analyses.
Quenched samples were removed at timed intervals and mixed with an equal volume of sinnapinic acid matrix in 50% acetonitrile, 0.1% TFA. Samples (2 μL) were spotted on the target plate and allowed to air-dry. MALDI-MS analyses were performed on a Bruker AmaZon X ion trap in a linear positive ion mode with laser energy 94%, pulse voltage 1,950 V, detector voltage 2,500 V, and mass suppression 4,000 Da. Calibration was performed with a ProteoMass Protein MALDI-MS Calibration Kit (Sigma). MALDI-MS data were acquired for 50 s per spot, sampling a minimum of 10 laser positions. Data were analyzed using the Bruker Daltonics flexAnalysis 2.4 software package.
CD Spectroscopy.
CD spectra of the holo-PhuS proteins were recorded on a JASCO J-810 spectropolarimeter. All samples were recorded in 1 mM potassium phosphate (pH 7.4) at 25 °C from 190 to 260 nm, 0.2-mm resolution, and 1.0 cm−1 bandwidth. The mean residue ellipticity (deg cm2 dmol−1) was calculated using CDPRO software as recommended by JASCO.
Analytical Ultracentrifugation.
Sedimentation velocity experiments were carried out using a Beckman-Coulter XL-A analytical ultracentrifuge. Samples of the indicated concentrations were centrifuged at 50,000 rpm in two-sector cells equipped with sapphire windows at 25 °C. The protein radial distributions were observed using absorbance optics at 280 nm. Sedimentation velocity data were analyzed using the time derivative method of Stafford as implemented in DCDT+ (26). The solvent density (1.00058 g mL−1) and the protein molecular weight and partial specific volume (0.739 mL g−1) of apo-PhuS were calculated using Sednterp (27) and the partial specific volume values of Cohn and Edsall (28). The molecular weight of holo-PhuS was calculated by adding 616 to the molecular weight of the protein to account for the heme. A partial specific volume of holo-PhuS of 0.7411 mL g−1 was estimated by taking the weighted sum of the partial specific volumes for apo-PhuS and oxygenated heme (29).
Fluorescence Spectrophotometry.
Titrations of wild-type and His-mutant PhuS proteins with heme were monitored by steady-state fluorescence emission. Samples were excited at 295 nm, and the fluorescence emission monitored at 337 nm. Solutions of 1.0 μM PhuS in 20 mM Tris·HCl (pH 7.8) were titrated with 1 μL aliquots (0.1–50 μM) heme at 25 °C. All stock heme solutions were freshly prepared as described above and used within 30 min. The binding constant (Kd) was fit to a one-site binding model based on the decrease in Trp fluorescence intensity at 337 nm as a function of increasing heme.
Surface Plasmon Resonance.
Kinetic evaluation of wild-type and mutant holo-PhuS binding to immobilized His-tagged HemO were carried out on a Biacore 3000 (GE Healthcare). All experiments were performed on nickel-NTA. The NTA sensor chip was activated with a 1-min injection of 500 μM NiCl2 (pH 7.4). HemO (50 nM) in 10 mM Hepes (pH 7.4) containing 150 mM NaCl was passed over the chip at 5 μL/min for 3 min, followed by 7 min stabilization. Resonance units (200–500) of HemO were immobilized. Holo-PhuS samples were prepared in degassed, filter-sterilized 10 mM Hepes (pH 7.4), 150 mM NaCl, and 0.005% Surfactant P20. Holo-PhuS proteins 0.5–10 μM were passed over the sensor chip (20 μL/min) for 4-min association, followed by a 6-min dissociation. The chip was regenerated with 10 mM Hepes (pH 8.0) 150 mM NaCl, 0.005% Surfactant P20, and 350 mM EDTA at 15 μL/min for 45 s; followed by 1 μM NiCl2 (90 s) and a 2-min stabilization. Data were fit to a 1:1 Langmuir model using the BIAevaluation software provided by BIAcore.
Isothermal Titration Calorimetry.
Titrations were performed at 25 °C using a MicroCal MCS titration calorimeter. All protein and heme solutions were in 20 mM sodium phosphate (pH 7.5). Samples were degassed before use and injections carried out at 5-min intervals. The heat of dilution of the ligand was measured by injecting into the buffer alone. The value obtained was subtracted from the heat of reaction to give the effective heat of binding. For all titration experiments the concentration of the protein ranged from 10 to 20 μM, and the ligand was set to 10–20 times this value. The ITC data were fitted with the Origin software package provided by MicroCal, which uses a nonlinear least-squares algorithm (minimization of χ2) and the concentrations of the titrant and the sample to fit the heat flow per injection to an equation corresponding to an equilibrium binding model, which provides best-fit values for the stoichiometry (nITC), change in enthalpy (ΔHITC), and binding constant (KITC). KD and Gibbs free energy were calculated according to Kd = 1/Ka and ΔG = −RT ln Ka. The thermodynamic relationship ΔG = ΔH − TΔS was used to find the entropic contribution to binding. The data were averaged from three independent ITC experiments.
Supplementary Material
Acknowledgments
This work was supported by National Institutes of Health Grants AI-055912 and AI-085535 (to A.W.) and GM-079440 (to K.G.F.) and National Science Foundation Grant MCB0919868.
Footnotes
The authors declare no conflict of interest.
This article is a PNAS Direct Submission.
This article contains supporting information online at www.pnas.org/lookup/suppl/doi:10.1073/pnas.1121549109/-/DCSupplemental.
References
- 1.Collins HL. Withholding iron as a cellular defence mechanism—friend or foe? Eur J Immunol. 2008;38:1803–1806. doi: 10.1002/eji.200838505. [DOI] [PubMed] [Google Scholar]
- 2.Flo TH, et al. Lipocalin 2 mediates an innate immune response to bacterial infection by sequestrating iron. Nature. 2004;432:917–921. doi: 10.1038/nature03104. [DOI] [PubMed] [Google Scholar]
- 3.Goetz DH, et al. The neutrophil lipocalin NGAL is a bacteriostatic agent that interferes with siderophore-mediated iron acquisition. Mol Cell. 2002;10:1033–1043. doi: 10.1016/s1097-2765(02)00708-6. [DOI] [PubMed] [Google Scholar]
- 4.Wandersman C, Delepelaire P. Bacterial iron sources: From siderophores to hemophores. Annu Rev Microbiol. 2004;58:611–647. doi: 10.1146/annurev.micro.58.030603.123811. [DOI] [PubMed] [Google Scholar]
- 5.Wilks A, Burkhard KA. Heme and virulence: How bacterial pathogens regulate, transport and utilize heme. Nat Prod Rep. 2007;24:511–522. doi: 10.1039/b604193k. [DOI] [PubMed] [Google Scholar]
- 6.Wilks A, Barker KD. Mechanism of heme uptake and utilization in bacterial pathogens. In: Kadish KM, Smith KM, Guilard R, editors. Handbook of Porphyrin Science, Biochemistry of Tetrapyrroles. 1st Ed. Vol 15. Singapore: World Scientific; 2011. pp. 357–398. [Google Scholar]
- 7.Ochsner UA, Johnson Z, Vasil ML. Genetics and regulation of two distinct haem-uptake systems, phu and has, in Pseudomonas aeruginosa. Microbiology. 2000;146:185–198. doi: 10.1099/00221287-146-1-185. [DOI] [PubMed] [Google Scholar]
- 8.Cobessi D, Meksem A, Brillet K. Structure of the heme/hemoglobin outer membrane receptor ShuA from Shigella dysenteriae: Heme binding by an induced fit mechanism. Proteins. 2010;78:286–294. doi: 10.1002/prot.22539. [DOI] [PubMed] [Google Scholar]
- 9.Krieg S, et al. Heme uptake across the outer membrane as revealed by crystal structures of the receptor-hemophore complex. Proc Natl Acad Sci USA. 2009;106:1045–1050. doi: 10.1073/pnas.0809406106. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Ho WW, et al. Holo- and apo-bound structures of bacterial periplasmic heme-binding proteins. J Biol Chem. 2007;282:35796–35802. doi: 10.1074/jbc.M706761200. [DOI] [PubMed] [Google Scholar]
- 11.Wilks A, Schmitt MP. Expression and characterization of a heme oxygenase (Hmu O) from Corynebacterium diphtheriae. Iron acquisition requires oxidative cleavage of the heme macrocycle. J Biol Chem. 1998;273:837–841. doi: 10.1074/jbc.273.2.837. [DOI] [PubMed] [Google Scholar]
- 12.Ratliff M, Zhu W, Deshmukh R, Wilks A, Stojiljkovic I. Homologues of neisserial heme oxygenase in gram-negative bacteria: Degradation of heme by the product of the pigA gene of Pseudomonas aeruginosa. J Bacteriol. 2001;183:6394–6403. doi: 10.1128/JB.183.21.6394-6403.2001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Friedman J, Lad L, Li H, Wilks A, Poulos TL. Structural basis for novel δ-regioselective heme oxygenation in the opportunistic pathogen Pseudomonas aeruginosa. Biochemistry. 2004;43:5239–5245. doi: 10.1021/bi049687g. [DOI] [PubMed] [Google Scholar]
- 14.Schuller DJ, Zhu W, Stojiljkovic I, Wilks A, Poulos TL. Crystal structure of heme oxygenase from the gram-negative pathogen Neisseria meningitidis and a comparison with mammalian heme oxygenase-1. Biochemistry. 2001;40:11552–11558. doi: 10.1021/bi0110239. [DOI] [PubMed] [Google Scholar]
- 15.Skaar EP, Gaspar AH, Schneewind O. IsdG and IsdI, heme-degrading enzymes in the cytoplasm of Staphylococcus aureus. J Biol Chem. 2004;279:436–443. doi: 10.1074/jbc.M307952200. [DOI] [PubMed] [Google Scholar]
- 16.Skaar EP, Gaspar AH, Schneewind O. Bacillus anthracis IsdG, a heme-degrading monooxygenase. J Bacteriol. 2006;188:1071–1080. doi: 10.1128/JB.188.3.1071-1080.2006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Chim N, Iniguez A, Nguyen TQ, Goulding CW. Unusual diheme conformation of the heme-degrading protein from Mycobacterium tuberculosis. J Mol Biol. 2010;395:595–608. doi: 10.1016/j.jmb.2009.11.025. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Suits MD, et al. Identification of an Escherichia coli O157:H7 heme oxygenase with tandem functional repeats. Proc Natl Acad Sci USA. 2005;102:16955–16960. doi: 10.1073/pnas.0504289102. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Stojiljkovic I, Hantke K. Transport of haemin across the cytoplasmic membrane through a haemin-specific periplasmic binding-protein-dependent transport system in Yersinia enterocolitica. Mol Microbiol. 1994;13:719–732. doi: 10.1111/j.1365-2958.1994.tb00465.x. [DOI] [PubMed] [Google Scholar]
- 20.Bhakta MN, Wilks A. The mechanism of heme transfer from the cytoplasmic heme binding protein PhuS to the δ-regioselective heme oxygenase of Pseudomonas aeruginosa. Biochemistry. 2006;45:11642–11649. doi: 10.1021/bi060980l. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Block DR, et al. Identification of two heme-binding sites in the cytoplasmic heme-trafficking protein PhuS from Pseudomonas aeruginosa and their relevance to function. Biochemistry. 2007;46:14391–14402. doi: 10.1021/bi701509n. [DOI] [PubMed] [Google Scholar]
- 22.Lansky IB, et al. The cytoplasmic heme-binding protein (PhuS) from the heme uptake system of Pseudomonas aeruginosa is an intracellular heme-trafficking protein to the δ-regioselective heme oxygenase. J Biol Chem. 2006;281:13652–13662. doi: 10.1074/jbc.M600824200. [DOI] [PubMed] [Google Scholar]
- 23.Schneider S, Sharp KH, Barker PD, Paoli M. An induced fit conformational change underlies the binding mechanism of the heme transport proteobacteria-protein HemS. J Biol Chem. 2006;281:32606–32610. doi: 10.1074/jbc.M607516200. [DOI] [PubMed] [Google Scholar]
- 24.Baker HM, Anderson BF, Baker EN. Dealing with iron: Common structural principles in proteins that transport iron and heme. Proc Natl Acad Sci USA. 2003;100:3579–3583. doi: 10.1073/pnas.0637295100. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Fuhrop JH, Smith KM, editors. Porphyrins and Metalloporphyrins. Amsterdam: Elsevier; 1975. pp. 804–807. [Google Scholar]
- 26.Philo JS. Improved methods for fitting sedimentation coefficient distributions derived by time-derivative techniques. Anal Biochem. 2006;354:238–246. doi: 10.1016/j.ab.2006.04.053. [DOI] [PubMed] [Google Scholar]
- 27.Laue TM. Computer-aided interpretation of analytical sedimentation data for proteins. In: Harding S, Rowe A, Hoarton J, editors. Analytical Ultracentrifugation in Biochemistry and Polymer Science. Cambridge, UK: Royal Society of Chemistry; 1992. p. 90. [Google Scholar]
- 28.Cohn E, Edsall J. Density and apparent specific volume of proteins. In: Cohn E, Edsall J, editors. Proteins, Amino Acids and Peptides. New York: Reinhold Publishing; 1943. pp. 370–381. [Google Scholar]
- 29.DeMoll E, Cox DJ, Daniel E, Riggs AF. Apparent specific volume of human hemoglobin: effect of ligand state and contribution of heme. Anal Biochem. 2007;363:196–203. doi: 10.1016/j.ab.2007.01.035. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.