Abstract
The HIV-1 envelope (Env) spike (gp1203/gp413) undergoes considerable structural rearrangements to mediate virus entry into cells and to evade the host immune response. Engagement of CD4, the primary human receptor, fixes a particular conformation and primes Env for entry. The CD4-bound state, however, is prone to spontaneous inactivation and susceptible to antibody neutralization. How does unliganded HIV-1 maintain CD4-binding capacity and regulate transitions to the CD4-bound state? To define this mechanistically, we determined crystal structures of unliganded core gp120 from HIV-1 clades B, C, and E. Notably, all of these unliganded HIV-1 structures resembled the CD4-bound state. Conformational fixation with ligand selection and thermodynamic analysis of full-length and core gp120 interactions revealed that the tendency of HIV-1 gp120 to adopt the CD4-bound conformation was restrained by the V1/V2- and V3-variable loops. In parallel, we determined the structure of core gp120 in complex with the small molecule, NBD-556, which specifically recognizes the CD4-bound conformation of gp120. Neutralization by NBD-556 indicated that Env spikes on primary isolates rarely assume the CD4-bound conformation spontaneously, although they could do so when quaternary restraints were loosened. Together, the results suggest that the CD4-bound conformation represents a “ground state” for the gp120 core, with variable loop and quaternary interactions restraining unliganded gp120 from “snapping” into this conformation. A mechanism of control involving deformations in unliganded structure from a functionally critical state (e.g., the CD4-bound state) provides advantages in terms of HIV-1 Env structural diversity and resistance to antibodies and inhibitors, while maintaining elements essential for entry.
Keywords: conformational equilibrium, viral evasion, X-ray crystallography
Enveloped viruses enter host cells through a variety of mechanisms, of which the type I membrane fusion machinery, used by HIV-1, influenza virus, and respiratory syncytial virus, is among the better characterized (1, 2). Type I entry machines are synthesized as a single polypeptide, with a single membrane-spanning region. These polypeptides form homotrimers, and each chain is cleaved to produce two components: an N-terminal component generally involved in recognizing the host receptor, and a C-terminal membrane-anchored component with a hydrophobic fusion peptide sequence at the “new” N terminus created by the cleavage. The mechanism of type I viral entry can be understood in terms of three states of the postcleavage machine. In the prefusion state, the fusion peptide is occluded within the viral spike oligomer, and this state generally resembles the uncleaved state. Activation by pH changes or by receptor binding induces an intermediate state in which the fusion peptide is embedded in the membrane of the host cell, with the viral spike in an extended conformation that bridges the viral and host membranes. This intermediate state is resolved by transition to the postfusion state, in which structural rearrangements of the viral spike bring host and viral membranes into close apposition, and fusion of these membranes ensues.
With HIV-1, the trimeric Env spike is assembled from gp160 glycoprotein precursors, which are cleaved to form the gp120 exterior glycoprotein (the N-terminal receptor-binding component) and the gp41 transmembrane glycoprotein (which contains both fusion peptide and transmembrane regions) (3). In a twist on the standard type I fusion mechanism, HIV-1 entry requires two receptors (4, 5). The unliganded viral spike is first recognized by the CD4 receptor, which induces structural rearrangements that form the coreceptor-binding site and the prehairpin intermediate; coreceptor binding triggers the remaining steps in the membrane fusion process. A precise delineation, however, remains to be made between the conformational changes of the two receptor-interacting gp120 and the standard type 1 mechanism. How is the conformational equilibrium between unbound and receptor-bound states altered by the double-receptor mechanism? To what extent is the requirement for immune evasion satisfied by stepwise receptor binding? How is the extensive conformational mobility of the gp120 accommodated?
These questions could be answered by atomic-level structures of the functional viral Env spike in its various states. Despite extensive effort, the atomic-level structure of the functional HIV-1 spike has resisted analysis. A number of ∼15- to 25-Å electron-microscopy structures of the HIV-1 spike have nevertheless been determined (6–9) as well as atomic-level structures of the postfusion state of the gp41 ectodomain (10, 11), of an HIV-1 minimal gp120 core (coremin) in complex with CD4 and/or other ligands (including the N terminus of the CCR5 coreceptor) (12–16), and of an unliganded gp120 coremin from simian immunodeficiency virus (SIV) (17). Missing from this pantheon of gp120 structures has been the unliganded structure of HIV-1 gp120. Here we determine crystal structures of more-extended gp120 cores (coree) from diverse strains of HIV-1, primarily in the unliganded state, and use a variety of techniques to explore the implications of these structures for understanding the conformational equilibrium of gp120, its control, and contribution to immune evasion.
Results
Structures of Unliganded gp120 coree.
We were unable to obtain crystals of the unliganded HIV-1 gp120 coremin, which contained deletions of the first three variable loops (V1–V3) and of the gp41-interactive region at the N and C termini (18), but found that alterations in variable loop truncations and inclusion of the N terminus allowed crystallization (SI Appendix, Figs. S1–S3 and Table S1). We obtained structures of this extended core (coree) for primary HIV-1 isolates from clade B (strain YU2), clade C (strains C1086 and ZM109), and clade E (strain 93TH057) at 3.4-, 2.8-, 4.0-, and 1.9-Å resolution, respectively (Fig. 1 and SI Appendix, Figs. S4 and S5 and Table S2).
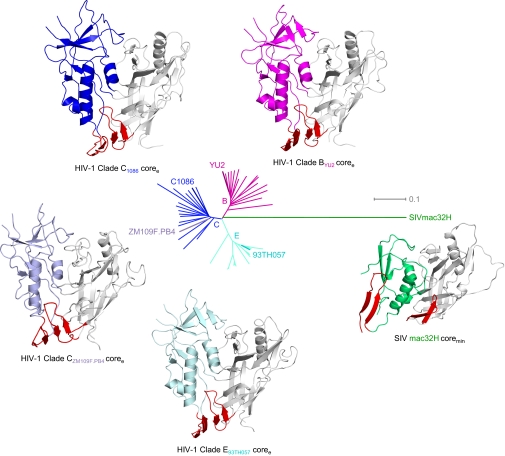
Fig. 1.
Unliganded structures of HIV-1 gp120 core. Crystal structures are displayed as Cα-ribbon diagrams with outer domains in gray and inner domains in magenta, blue or light blue, cyan, and green for HIV-1 clades B, C, and E, and SIV, respectively, and the region that in the CD4-bound conformation makes up the bridging sheet in red. The evolutionary relationships of these Env glycoproteins are represented by a dendrogram where the length of connections is proportional to evolutionary distance. [The four HIV-1 structures were determined here; the SIV structure was determined previously by Chen et al. (17).]
The four unliganded HIV-1 gp120 coree structures showed pairwise rmsds in Cα-atom positions of 1.2–1.9 Å, indicating highly similar structures, despite substantially different packing arrangements (Fig. 1 and SI Appendix, Figs. S6–S8). In contrast, similar pairwise comparisons with the unliganded SIV coremin structure (17) showed rmsds of 9.5–9.8 Å, indicating substantial differences (Fig. 1). Although many of the secondary structural elements were retained in both the HIV-1 and SIV unliganded gp120 core structures, differences in orientation, positioning, and packing of secondary structural features were apparent. Such differences could reflect evolutionary changes between HIV-1 and SIV (the HIV-1 structures are ∼threefold less evolutionarily divergent from each other than from SIV; Fig. 1) or differences between coree and coremin (SI Appendix, Figs. S1 and S3).
Comparison of Unliganded HIV-1 gp120 Core to Previously Determined gp120 Structures.
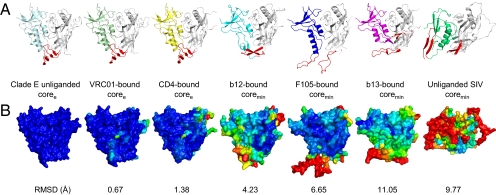
To understand the relationship between the unliganded HIV-1 gp120 structures reported here and previously described gp120 structures (12–17, 19–21), we performed detailed structural comparisons (Fig. 2 and SI Appendix, Fig. S9). The gp120 glycoprotein consists of two domains, inner and outer, with loop excursions. In the CD4-bound state, two β-hairpin excursions from the inner domain (strands β2–β3) and outer domain (β20–β21) come together to form a four-stranded “bridging sheet” minidomain (12). Mapping of ligand-induced changes revealed extensive conformational movement for these β-hairpin excursions, and much less movement for the outer domain (Fig. 2B). Quantification of these changes showed that the outer domain moves on average only ∼2 Å, the inner domain ∼4 Å, and the residues that make up the bridging sheet move ∼10 Å (SI Appendix, Fig. S9 and Tables S3–S6). Thus, despite the extraordinary diversity observed for the inner domain and bridging sheet regions of gp120, the unliganded structure of the HIV-1 gp120 coree closely resembled the CD4-bound state (average Cα-rmsds of 1.3 Å and 1.4 Å to CD4-bound coree and coremin, respectively).
Fig. 2.
Comparison of unliganded HIV-1 gp120 core to previously determined gp120 structures. Despite substantial conformational diversity of the gp120 envelope glycoprotein, unliganded HIV-1 gp120 snaps into a conformation that closely resembles the CD4-bound state. (A) Ribbon diagrams, displayed as in Fig. 1. (B) Molecular surface representation colored by structural deviations from the HIV-1 clade E unliganded gp120 structure. The color scale ranges from dark blue to red for rmsds of 0 to >10 Å, respectively. Notably, conformational changes >100 Å are observed in the bridging sheet region between the b13-bound and unliganded forms of HIV-1 gp120.
Before determining these unliganded core structures, data from thermodynamic studies of gp120 binding to CD4 (22) and to a number of CD4 binding-site antibodies (19–21), and from the SIV unliganded coremin structure (17), suggested that the unliganded gp120 structure would be substantially different from the CD4-bound conformation. We were concerned that the purification and crystallization processes might have selected for gp120s in the CD4-bound conformation. We observed, however, that the 17b-purified full-length and coree gp120 proteins bound not only CD4 and 17b, but also b12, b13, F105, and other antibodies that recognize non–CD4-bound conformations of gp120 (SI Appendix, Fig. S10). Moreover, we screened gp120s for recognition by the quaternary structure-preferring PG9 antibody (23), because gp120s identified in this manner might preferentially be stabilized in the non–CD4-bound conformation or at least be selected by a different criterion. The gp120 derived from the ZM109 strain showed optimal PG9 recognition (SI Appendix, Fig. S11); its unliganded coree structure, nevertheless, was in the CD4-bound conformation (Fig. 1).
Taken together, our four unliganded gp120 structures demonstrate that the CD4-bound conformation is a default state for the HIV-1 gp120 coree and alter dogma that achieving the CD4-bound conformation requires ligand induction. The coree proteins studied here and the coremin proteins analyzed previously likely have different conformational equilibria as a result of potentially disruptive truncations of the coremin V3 base. In the coree construct, 10 amino acids at the V3 base (SI Appendix, Fig. S1 and S2), which are missing in coremin, form hydrogen bonds and allow coree gp120 to bind with high affinity to the 17b antibody, which recognizes a CD4-induced conformation (24). The SIV coremin (25), which was designed in the same manner as the HIV-1 coremin (12), may likewise have lost the propensity to assume the CD4-bound conformation as a default state. Note that many CD4 binding-site antibodies do induce gp120 conformations that are considerably different from the CD4-bound state (Fig. 2 and SI Appendix, Fig. S9); transitions from the unliganded coree structure of HIV-1 gp120 to those observed in different liganded states are illustrated in Movies S1, S2, S3, S4, S5, and S6.
Conformational Diversity of HIV-1 gp120 in Solution.
A substantial body of evidence indicates a high degree of conformational diversity in the solution state of unliganded HIV-1 gp120 in full-length and coremin proteins (22, 26, 27). To reconcile these data with our observation that the HIV-1 gp120 coree crystallized in the CD4-bound conformation, we hypothesized that alterations in the termini or of the variable loops might affect the ensemble of conformations available to the unliganded state. To assess the solution behavior of unliganded gp120, we measured the thermodynamics of interaction between full-length or coree versions of HIV-1 gp120 and three ligands—CD4, antibody VRC01, and antibody 17b—that stabilize the CD4-bound conformation and exhibit large entropic changes upon binding to gp120 (Fig. 3A and SI Appendix, Figs. S12 and S13 and Table S7) (22, 28, 29). For gp120 from clade B and clade C HIV-1 strains, the gp120 coree proteins exhibited an average entropy change upon binding these ligands that was roughly half of that observed with full-length gp120 (Fig. 3A). We also tested a full-length version of YU2 gp120 with a change—S375W—that predisposes gp120 to sample the CD4-bound conformation (30). The entropy change upon ligand interaction of the S375W mutant was also about half of that observed for the full-length, wild-type gp120 (SI Appendix, Fig. S14). These results indicate that, relative to wild-type gp120, the unliganded gp120 coree pays a lower entropic penalty when binding to ligands that induce the CD4-bound conformation.
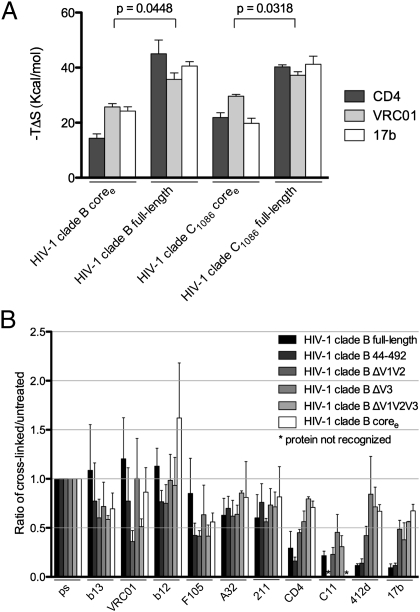
Fig. 3.
Conformational diversity of HIV-1 gp120 in solution. The conformational diversity of gp120 in solution is sensitive to the presence of the variable loops. (A) Entropy of ligand interactions with full-length and truncated gp120s (for measurements involving the HIV-1 clade B gp120, the YU2 strain was used; for measurements involving the HIV-1 clade C gp120, the C1086 strain was used). (B) Conformational fixation followed by ligand selection. The ratio of ligand binding to cross-linked vs. untreated gp120s is shown for full-length and truncated forms of gp120. ps, recognition by pooled sera from HIV-1-infected individuals, which was used for normalization.
Conformational Diversity of gp120 Assessed by Conformational Fixation Followed by Ligand Recognition.
To investigate the conformation of the unliganded state, we used conformational fixation coupled to ligand selection (Fig. 3B and SI Appendix, Fig. S15) (26). In this procedure, glutaraldehyde cross-linking is used to fix the conformation of gp120, and then the relative occupancy of gp120 conformations is assessed by quantifying the binding of ligands with different requirements for specific gp120 conformations. Five different variants of YU2 gp120 were tested, with deletions of the V1/V2 loops, of the V3 loop, and/or of the termini. The results were ligand dependent (Fig. 3B and SI Appendix, Fig. S15). Ligands that specifically recognize the CD4-bound conformation (e.g., CD4 and CD4i antibodies such as 412d and 17b) in the functional viral spike showed dramatically reduced recognition of the cross-linked full-length gp120 relative to truncated versions. In contrast, ligands that induce gp120 conformations distinct from the CD4-bound state (e.g., antibodies F105 and b13) (20) exhibited better recognition of the cross-linked full-length gp120 than of the truncated variants. Ligands that recognize multiple gp120 conformations (e.g., antibodies b12 and VRC01) or that have only modest conformational preference (e.g., antibodies A32, 211c, and C11) precipitated the cross-linked full-length and variable-loop–deleted proteins similarly; for some of these, there appeared to be an effect of the termini on the stability of the unliganded state. The results are consistent with our hypothesis that removal of the V1/V2 and V3 loops alters the conformational equilibrium of the unliganded gp120 ensemble to favor the CD4-bound state.
A more remote possibility is that unliganded wild-type gp120 core is already in the CD4-bound state, but that after cross-linking, the mobile loops (V1/V2 and V3) occlude access of certain ligands to their sites of recognition; we consider this possibility unlikely because it would require the cross-linked variable loops to impede access of CD4, but not VRC01, b12, b13, and F105 antibodies, to the common gp120 surface targeted by these ligands (19–21).
NBD-556 as a Structure-Specific Probe.
To minimize the contributions of steric factors to ligand binding, we used the small molecule NBD-556 (337.8 Da) as a structure-specific probe (31, 32). NBD-556 mimics CD4, binding gp120 with a large entropic change and fixing a gp120 conformation that functionally resembles the CD4-bound state (32, 33). To determine its mode of binding, we cocrystallized NBD-556 with the clade C strain C1086 version of gp120 coree. The NBD-556-gp120 coree complex crystallized in the same lattice as the unliganded core, and data were collected to 2.7-Å resolution (SI Appendix, Table S2). NBD-556 bound within the Phe-43 cavity of gp120, at the nexus of the inner domain, outer domain, and bridging sheet (Fig. 4 A and B and SI Appendix, Fig. S16). The phenyloxalamide portion of NBD-556 projected deep into the Phe-43 pocket, with only the piperidine ring exposed outside of the pocket (Fig. 4B). The structure confirms NBD-556 as a structure-specific probe for the CD4-bound state. Solution measurements of gp120-NBD-556 were made by surface-plasmon resonance (SPR) binding experiments in which NBD-556 was coupled to an SPR chip through a linker attached to the exposed piperidine ring. The affinity of NBD-556 for coremin was substantially weaker than the NBD-556 affinity to coree, despite both cores retaining all of the gp120 residues that contact NBD-556 (SI Appendix, Fig. S17); moreover, the affinity of NBD-556 for coree was ∼20-fold higher than that for full-length gp120, as evaluated by equilibrium and competition binding assays (Fig. 4C and SI Appendix, Table S8). These results suggest that a higher percentage of the truncated coree molecules sample the CD4-bound conformation, relative to the more truncated coremin or to full-length wild-type gp120.
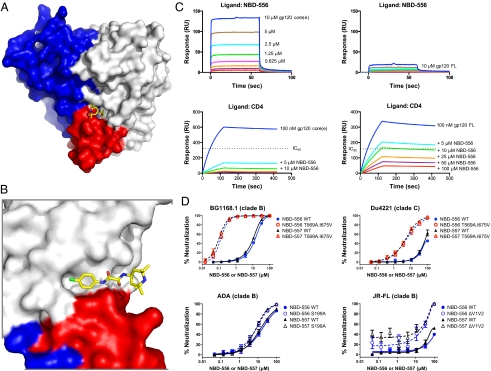
Fig. 4.
Conformational diversity of HIV-1 gp120 as assessed by the small molecule NBD-556 and mechanism of control. The unliganded state of HIV-1 gp120 exists as an equilibrium of conformations, with the coree portion of gp120 displaying a strong intrinsic propensity to snap into the CD4-bound conformation when not restrained by variable loops or by interactions with gp41. (A) Structure of small molecule NBD-556 in complex with HIV-1 gp120 provides atomic-level details of its binding site, and also provides an explanation for its preference for the CD4-bound conformation of gp120. The surface of coree gp120 is colored blue for inner domain, gray for outer domain, and red for bridging sheet, with the small molecule NBD-556 binding at a highly conserved pocket at the nexus of inner domain, outer domain, and bridging sheet minidomain. The NBD-556 is shown in stick representation, colored yellow for carbon, red for oxygen, blue for nitrogen, and green for chlorine. (B) Close-up rotated 90° about a vertical axis from A. (C) Assessment of gp120 conformation in solution. SPR measurements of NBD-556 binding to gp120 in coree (Left) or full-length (Right) gp120 contexts and by direct binding (Upper) or by competition (Lower) indicates that the unliganded coree has a greater propensity to assume the CD4-bound conformation than full-length gp120. (D) Assessment of gp120 conformation in the functional viral spike. Neutralization by NBD-556 or NBD-557 is strongly enhanced by gp41 mutants (Upper) and to a lesser extent by changes in the V1/V2 region (Lower).
We also used NBD-556 to assess the conformation of gp120 in the context of the functional viral spike. We tested the ability of NBD-556 to neutralize diverse strains of HIV-1 in clades B and C (SI Appendix, Fig. S3 and Table S9). Overall, NBD-556 showed weak neutralization of HIV-1. Although NBD-556, soluble CD4 (sCD4), and 17b all require the CD4-bound conformation of gp120 in the viral spike to bind, the IC50 values of NBD-556, sCD4, and 17b exhibited significant correlations only for NBD-556 and 17b (SI Appendix, Fig. S18); in particular, all isolates sensitive to 17b were neutralized by NBD-556, and all isolates sensitive to NBD-556 were neutralized by sCD4 (P value = 0.015). This intermediate level of NBD-556 potency suggested its utility as a specific and sensitive indicator of the CD4-bound conformation on functional envelope glycoproteins.
Assessment of gp120 Conformation in the Functional Viral Spike.
To assess the conformational constraints that portions of the viral spike other than the gp120 core place on the gp120 conformational state, we tested three mutants: two with alterations in V1/V2 (S199A or delta V1/V2) and one with alterations in gp41 (T569A and I675V). All of these changes have been previously shown to make HIV-1 more sensitive to neutralization by sCD4 and CD4-induced antibodies (34–36). In the case of the V1/V2 alterations (S199A or delta V1/V2), no enhancement (S199A) or a small enhancement (delta V1/V2) in NBD-556 neutralization was observed (Fig. 4D). By contrast, the gp41 changes rendered otherwise resistant viruses sensitive to NBD-556 (Fig. 4D). Thus, though the V1/V2 alterations affect the conformational equilibrium for the viral spike, these changes only minimally shift the equilibrium to the CD4-bound state to permit NBD-556 neutralization. The gp41 changes (T569A and I675V) more dramatically shift spike equilibrium, with a resultant large increase in sensitivity to NBD-556 neutralization. Thus, in the context of the functional viral spike, gp41 interactions appear to provide significant constraints on gp120 conformation. These results are consistent with cryoelectron microscopy studies (8, 9), which show V1/V2-deleted viral spikes to be in a closed conformation, more akin to the unliganded state, although perhaps with heightened conformational mobility.
Discussion
Prototypical type I entry machines differ from that used by HIV-1 in several ways: (i) HIV-1 uses an additional cellular receptor; (ii) the unliganded conformation of HIV-1 is highly resistant to antibody-mediated neutralization; and (iii) unlike the receptor-binding domains of most type I fusion machines, which are conformationally fixed, the gp120 component is conformationally mobile. Here we explore the implications of these differences and probe the mechanism of gp120 conformational control. The obtained results (four unliganded HIV-1 gp120 coree structures, a structure of an NBD-556–bound conformation of HIV-1 gp120, isothermal titration calorimetry measurements of gp120 conformational transitions, and cross-linking coupled to ligand selection) reveal the workings of the conformational switch that is normally initiated by HIV-1 binding to CD4.
Three findings: First, although gp120 may exist in a vast ensemble of distinct conformations in the unliganded state, it has evolved to “snap” into the CD4-bound conformation. The propensity to assume the CD4-bound state is preserved across diverse HIV-1 clades. By retaining the ability to refold itself into the functionally critical conformation preferred by CD4, gp120 specifically assists the binding of its receptor, which minimizes the conserved, exposed gp120 contacts with CD4 required to activate HIV-1 entry and thus facilitates antibody evasion. Second, for most primary HIV-1 isolates, the CD4-bound conformation is infrequently sampled in the native unliganded state of the Env spike. Transitions of full-length gp120 into the CD4-bound state are accompanied by large decreases in entropy (22). Cross-linking of cell-surface HIV-1 Env or full-length gp120 results in a specific decrease in recognition by ligands that prefer the CD4-bound state (26). Changes in gp120 core residues that increase the sampling of the CD4-bound conformation result in dramatic increases in HIV-1 cold sensitivity (37). These observations suggest that the functional HIV-1 spike generally avoids the labile, neutralization-sensitive CD4-bound conformation. Third, in the functional viral spike, the propensity of gp120 to assume the CD4-bound conformation is modulated by the gp41 interaction and the V1/V2 and V3 variable loops. These regions reside near the trimer axis and contribute to spike stability (38). Our results explain why many of the diverse biological properties of HIV-1 strains (e.g., CD4 dependence or sensitivity to inhibition) can be determined by the structure of these loops (39–41). Changes in gp41 can also influence the transition of gp120 into an NBD-556-binding conformation. Although NBD-556 binds to a gp120 pocket conserved between clade B and C HIV-1, the relative resistance of clade C HIV-1 to NBD-556 inhibition (SI Appendix, Fig. S19) illustrates that other Env elements significantly influence drug susceptibility, presumably through conformational modulation.
These findings suggest a coherent model for gp120 in the unliganded and CD4-bound states (Fig. 5). Unlike most other type 1 viral fusion machines, the HIV-1 viral spike requires two major conformational transitions: an unliganded to CD4-bound transition followed by a CD4-bound to coreceptor-triggered transition. Our results indicate that the default conformation for unliganded coree gp120 is the CD4-bound one. The transition from the unliganded conformation of gp120 in the functional viral spike to the CD4-bound conformation can thus occur by merely following an energetic gradient from a deformed higher-energy state to a default-ground state. Because there are many ways to deform a particular conformation, such a mechanism would intrinsically allow for a diversity in potential unliganded conformations and would also be consistent with prior suggestions (42, 43) that the unliganded state is an evolutionary addition to a primordial “one receptor-triggered” entry mechanism. We note in this regard that an ability to assume many potential unliganded conformations provides advantages for immune evasion, with our results providing insight into how the transition between immune-evading unliganded conformations and the CD4-bound conformation are influenced by variable loops and, in the viral spike, by interactions with gp41 and other subunits. Thus, in addition to facilitating an understanding of HIV-1 Env states, their conformational diversity, and mechanisms of control, our results should expedite progress on inhibition or premature activation of conformational transitions for prophylactic or therapeutic aims.
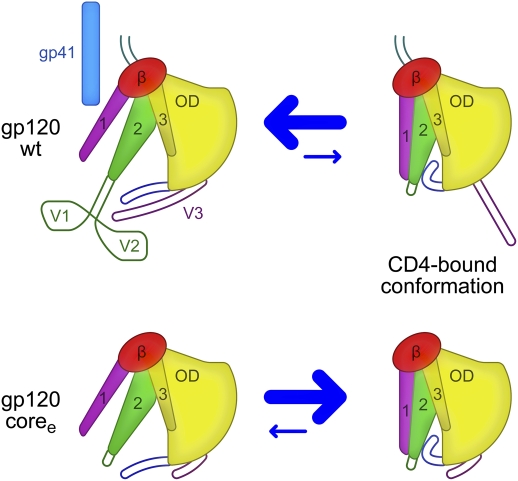
Fig. 5.
Mechanism of gp120 conformation control. In full-length WT gp120 (Upper), interactions with gp41 and between the variable loops V1/V2 and V3 shift the conformational equilibrium of the gp120 core away from the CD4-bound conformation (equilibrium is denoted by size of blue arrows). The intrinsic propensity of coree gp120 (Lower) to assume the CD4-bound conformation is revealed when the gp41-interactive region and variable loops are removed.
Materials and Methods
Supernatants of transiently transfected 293F or HEK293S GnTi− cells expressing gp120 variants were passed through a 17b-conjugated protein A column, washed with PBS, and eluted with elution buffer (Pierce). The 17b column-purified gp120s were deglycosylated with endoglycosidase H and purified with Con A-Sepharose (Sigma) and Superdex 200 (GE Healthcare) chromatography using protocols described previously (18). Crystallization conditions were identified by small-volume robotic screening (0.2 μL protein-containing solution plus 0.2 μL of the crystallization reservoir with hanging-drop vapor diffusion at 20 °C) and optimized manually. Diffraction data were collected at the Advanced Photon Source (insertion device 22) and indexed, integrated, and scaled with HKL2000 (44). Structures were solved by molecular replacement starting with either core or outer domain version of gp120 and refined with PHENIX (45). Isothermal titration calorimetry experiments were performed on an ITC200 Microcalorimeter from MicroCal Inc. Glutaraldehyde cross-linking followed by ligand selection was performed following procedures described previously (26). SPR experiments were performed on a Biacore T100 (GE Healthcare) at 25 °C. Kinetic data were extracted by fitting the responses globally with a 1:1 interaction model. HIV-1 Env pseudoviruses were prepared by transfecting 293T cells with 10 μg of rev/env expression plasmid and 30 μg of an env-deficient HIV-1 backbone vector (pSG3Δenv) and by using Fugene 6 transfection reagents (Invitrogen). Pseudovirus-containing culture supernatants were harvested 2 d after transfection, filtered (0.45 μm), and stored at –80 °C or in the vapor phase of liquid nitrogen. Neutralization capacities were measured by using HIV-1 Env pseudoviruses to infect TZM-bl cells as described previously (46). Additional methodological details are presented in SI Appendix, Materials and Methods.
Supplementary Material
Acknowledgments
We thank J. Hoxie, R. Sandier, I. A. Wilson, and members of the Structural Biology Section and Structural Bioinformatics Core, Vaccine Research Center, for discussions and comments on the manuscript. Support for this work was provided by the Intramural Research Program of the National Institutes of Health (NIH) and by grants from the NIH and from the International AIDS Vaccine Initiative's Neutralizing Antibody Consortium. Use of sector 22 (Southeast Region Collaborative Access Team) at the Advanced Photon Source was supported by the US Department of Energy, Basic Energy Sciences, Office of Science Contract W-31-109-Eng-38.
Footnotes
The authors declare no conflict of interest.
*This Direct Submission article had a prearranged editor.
Data deposition: The coordinates and structure factors for the four unliganded HIV-1 gp120 and the NBD-556 HIV-1 gp120 structures reported in this paper have been deposited with the Protein Data Bank, www.pdb.org (PDB ID codes 3TGQ, 3TGR, 3TGS, 3TGT, and 3TIH).
This article contains supporting information online at www.pnas.org/lookup/suppl/doi:10.1073/pnas.1112391109/-/DCSupplemental.
References
- 1.Eckert DM, Kim PS. Mechanisms of viral membrane fusion and its inhibition. Annu Rev Biochem. 2001;70:777–810. doi: 10.1146/annurev.biochem.70.1.777. [DOI] [PubMed] [Google Scholar]
- 2.Colman PM, Lawrence MC. The structural biology of type I viral membrane fusion. Nat Rev Mol Cell Biol. 2003;4:309–319. doi: 10.1038/nrm1076. [DOI] [PubMed] [Google Scholar]
- 3.Wyatt R, Sodroski J. The HIV-1 envelope glycoproteins: Fusogens, antigens, and immunogens. Science. 1998;280:1884–1888. doi: 10.1126/science.280.5371.1884. [DOI] [PubMed] [Google Scholar]
- 4.Sattentau QJ. HIV gp120: Double lock strategy foils host defences. Structure. 1998;6:945–949. doi: 10.1016/s0969-2126(98)00096-3. [DOI] [PubMed] [Google Scholar]
- 5.Berger EA, Murphy PM, Farber JM. Chemokine receptors as HIV-1 coreceptors: Roles in viral entry, tropism, and disease. Annu Rev Immunol. 1999;17:657–700. doi: 10.1146/annurev.immunol.17.1.657. [DOI] [PubMed] [Google Scholar]
- 6.Liu J, Bartesaghi A, Borgnia MJ, Sapiro G, Subramaniam S. Molecular architecture of native HIV-1 gp120 trimers. Nature. 2008;455:109–113. doi: 10.1038/nature07159. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Wu SR, et al. Single-particle cryoelectron microscopy analysis reveals the HIV-1 spike as a tripod structure. Proc Natl Acad Sci USA. 2010;107:18844–18849. doi: 10.1073/pnas.1007227107. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.White TA, et al. Molecular architectures of trimeric SIV and HIV-1 envelope glycoproteins on intact viruses: Strain-dependent variation in quaternary structure. PLoS Pathog. 2010;6:e1001249. doi: 10.1371/journal.ppat.1001249. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Hu G, Liu J, Taylor KA, Roux KH. Structural comparison of HIV-1 envelope spikes with and without the V1/V2 loop. J Virol. 2011;85:2741–2750. doi: 10.1128/JVI.01612-10. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Chan DC, Fass D, Berger JM, Kim PS. Core structure of gp41 from the HIV envelope glycoprotein. Cell. 1997;89:263–273. doi: 10.1016/s0092-8674(00)80205-6. [DOI] [PubMed] [Google Scholar]
- 11.Weissenhorn W, Dessen A, Harrison SC, Skehel JJ, Wiley DC. Atomic structure of the ectodomain from HIV-1 gp41. Nature. 1997;387:426–430. doi: 10.1038/387426a0. [DOI] [PubMed] [Google Scholar]
- 12.Kwong PD, et al. Structure of an HIV gp120 envelope glycoprotein in complex with the CD4 receptor and a neutralizing human antibody. Nature. 1998;393:648–659. doi: 10.1038/31405. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Kwong PD, et al. Structures of HIV-1 gp120 envelope glycoproteins from laboratory-adapted and primary isolates. Structure. 2000;8:1329–1339. doi: 10.1016/s0969-2126(00)00547-5. [DOI] [PubMed] [Google Scholar]
- 14.Huang CC, et al. Structure of a V3-containing HIV-1 gp120 core. Science. 2005;310:1025–1028. doi: 10.1126/science.1118398. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Huang CC, et al. Structures of the CCR5 N terminus and of a tyrosine-sulfated antibody with HIV-1 gp120 and CD4. Science. 2007;317:1930–1934. doi: 10.1126/science.1145373. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Pancera M, et al. Structure of HIV-1 gp120 with gp41-interactive region reveals layered envelope architecture and basis of conformational mobility. Proc Natl Acad Sci USA. 2010;107:1166–1171. doi: 10.1073/pnas.0911004107. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Chen B, et al. Structure of an unliganded simian immunodeficiency virus gp120 core. Nature. 2005;433:834–841. doi: 10.1038/nature03327. [DOI] [PubMed] [Google Scholar]
- 18.Kwong PD, et al. Probability analysis of variational crystallization and its application to gp120, the exterior envelope glycoprotein of type 1 human immunodeficiency virus (HIV-1) J Biol Chem. 1999;274:4115–4123. doi: 10.1074/jbc.274.7.4115. [DOI] [PubMed] [Google Scholar]
- 19.Zhou T, et al. Structural definition of a conserved neutralization epitope on HIV-1 gp120. Nature. 2007;445:732–737. doi: 10.1038/nature05580. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Chen L, et al. Structural basis of immune evasion at the site of CD4 attachment on HIV-1 gp120. Science. 2009;326:1123–1127. doi: 10.1126/science.1175868. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Zhou T, et al. Structural basis for broad and potent neutralization of HIV-1 by antibody VRC01. Science. 2010;329:811–817. doi: 10.1126/science.1192819. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Myszka DG, et al. Energetics of the HIV gp120-CD4 binding reaction. Proc Natl Acad Sci USA. 2000;97:9026–9031. doi: 10.1073/pnas.97.16.9026. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Walker LM, et al. Broad and potent neutralizing antibodies from an African donor reveal a new HIV-1 vaccine target. Science. 2009;326:285–289. doi: 10.1126/science.1178746. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Dey B, et al. Structure-based stabilization of HIV-1 gp120 enhances humoral immune responses to the induced co-receptor binding site. PLoS Pathog. 2009;5:e1000445. doi: 10.1371/journal.ppat.1000445. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Chen B, et al. Determining the structure of an unliganded and fully glycosylated SIV gp120 envelope glycoprotein. Structure. 2005;13:197–211. doi: 10.1016/j.str.2004.12.004. [DOI] [PubMed] [Google Scholar]
- 26.Yuan W, Bazick J, Sodroski J. Characterization of the multiple conformational states of free monomeric and trimeric human immunodeficiency virus envelope glycoproteins after fixation by cross-linker. J Virol. 2006;80:6725–6737. doi: 10.1128/JVI.00118-06. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Kong L, et al. Local conformational stability of HIV-1 gp120 in unliganded and CD4-bound states as defined by amide hydrogen/deuterium exchange. J Virol. 2010;84:10311–10321. doi: 10.1128/JVI.00688-10. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Kwong PD, et al. HIV-1 evades antibody-mediated neutralization through conformational masking of receptor-binding sites. Nature. 2002;420:678–682. doi: 10.1038/nature01188. [DOI] [PubMed] [Google Scholar]
- 29.Wu X, et al. Rational design of envelope identifies broadly neutralizing human monoclonal antibodies to HIV-1. Science. 2010;329:856–861. doi: 10.1126/science.1187659. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Xiang SH, et al. Mutagenic stabilization and/or disruption of a CD4-bound state reveals distinct conformations of the human immunodeficiency virus type 1 gp120 envelope glycoprotein. J Virol. 2002;76:9888–9899. doi: 10.1128/JVI.76.19.9888-9899.2002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Zhao Q, et al. Identification of N-phenyl-N′-(2,2,6,6-tetramethyl-piperidin-4-yl)-oxalamides as a new class of HIV-1 entry inhibitors that prevent gp120 binding to CD4. Virology. 2005;339:213–225. doi: 10.1016/j.virol.2005.06.008. [DOI] [PubMed] [Google Scholar]
- 32.Madani N, et al. Small-molecule CD4 mimics interact with a highly conserved pocket on HIV-1 gp120. Structure. 2008;16:1689–1701. doi: 10.1016/j.str.2008.09.005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Schön A, et al. Thermodynamics of binding of a low-molecular-weight CD4 mimetic to HIV-1 gp120. Biochemistry. 2006;45:10973–10980. doi: 10.1021/bi061193r. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Kolchinsky P, Kiprilov E, Sodroski J. Increased neutralization sensitivity of CD4-independent human immunodeficiency virus variants. J Virol. 2001;75:2041–2050. doi: 10.1128/JVI.75.5.2041-2050.2001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Wyatt R, et al. Functional and immunologic characterization of human immunodeficiency virus type 1 envelope glycoproteins containing deletions of the major variable regions. J Virol. 1993;67:4557–4565. doi: 10.1128/jvi.67.8.4557-4565.1993. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Cao J, et al. Replication and neutralization of human immunodeficiency virus type 1 lacking the V1 and V2 variable loops of the gp120 envelope glycoprotein. J Virol. 1997;71:9808–9812. doi: 10.1128/jvi.71.12.9808-9812.1997. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Kassa A, et al. Transitions to and from the CD4-bound conformation are modulated by a single-residue change in the human immunodeficiency virus type 1 gp120 inner domain. J Virol. 2009;83:8364–8378. doi: 10.1128/JVI.00594-09. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Xiang SH, et al. A V3 loop-dependent gp120 element disrupted by CD4 binding stabilizes the human immunodeficiency virus envelope glycoprotein trimer. J Virol. 2010;84:3147–3161. doi: 10.1128/JVI.02587-09. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Sullivan N, et al. Determinants of human immunodeficiency virus type 1 envelope glycoprotein activation by soluble CD4 and monoclonal antibodies. J Virol. 1998;72:6332–6338. doi: 10.1128/jvi.72.8.6332-6338.1998. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.O'Rourke SM, et al. Mutation at a single position in the V2 domain of the HIV-1 envelope protein confers neutralization sensitivity to a highly neutralization-resistant virus. J Virol. 2010;84:11200–11209. doi: 10.1128/JVI.00790-10. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Musich T, et al. A conserved determinant in the V1 loop of HIV-1 modulates the V3 loop to prime low CD4 use and macrophage infection. J Virol. 2011;85:2397–2405. doi: 10.1128/JVI.02187-10. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.de Parseval A, Grant CK, Sastry KJ, Elder JH. Sequential CD134-CXCR4 interactions in feline immunodeficiency virus (FIV): Soluble CD134 activates FIV Env for CXCR4-dependent entry and reveals a cryptic neutralization epitope. J Virol. 2006;80:3088–3091. doi: 10.1128/JVI.80.6.3088-3091.2006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Endres MJ, et al. CD4-independent infection by HIV-2 is mediated by fusin/CXCR4. Cell. 1996;87:745–756. doi: 10.1016/s0092-8674(00)81393-8. [DOI] [PubMed] [Google Scholar]
- 44.Otwinowski Z, Minor W. Processing of X-ray diffraction data collected in oscillation mode. Methods Enzymol. 1997;276:307–326. doi: 10.1016/S0076-6879(97)76066-X. [DOI] [PubMed] [Google Scholar]
- 45.Adams PD, et al. Recent developments in the PHENIX software for automated crystallographic structure determination. J Synchrotron Radiat. 2004;11:53–55. doi: 10.1107/s0909049503024130. [DOI] [PubMed] [Google Scholar]
- 46.Seaman MS, et al. Tiered categorization of a diverse panel of HIV-1 Env pseudoviruses for assessment of neutralizing antibodies. J Virol. 2010;84:1439–1452. doi: 10.1128/JVI.02108-09. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.