Abstract
Organization of replicating prokaryotic genomes requires architectural elements that, similarly to eukaryotic systems, induce topological changes such as DNA supercoiling. Bacteriophage ϕ29 protein p6 has been described as a histone-like protein that compacts the viral genome by forming a nucleoprotein complex and plays a key role in the initiation of protein-primed DNA replication. In this work, we analyze the subcellular localization of protein p6 by immunofluorescence microscopy and show that, at early infection stages, it localizes in a peripheral helix-like configuration. Later, at middle infection stages, protein p6 is recruited to the bacterial nucleoid. This migrating process is shown to depend on the synthesis of components of the ϕ29 DNA replication machinery (i.e., terminal protein and DNA polymerase) needed for the replication of viral DNA, which is required to recruit the bulk of protein p6. Importantly, the double-stranded DNA-binding capacity of protein p6 is essential for its relocalization at the nucleoid. Altogether, the results disclose the in vivo organization of a viral histone-like protein in bacteria.
Keywords: DNA topology, protein-priming, architectural proteins, non-sequence-specific DNA binding
The genomes of eukaryotic and prokaryotic organisms are packed and organized by architectural proteins with nonsequence-specific DNA-binding properties that give rise to higher-order nucleoprotein complexes mediating, in addition to control of DNA structure and topology, the regulation of fundamental processes such as DNA replication. Eukaryotic chromosomes are packaged by histones into a periodic nucleoprotein complex, known as chromatin (1). In Eubacteria, the term “histone-like” refers to a number of nucleoid-associated proteins responsible for genome organization, condensation, and DNA supercoiling (2). In Bacillus subtilis, HBsu belongs to a conserved group of bacterial histone-like proteins that distribute along the entire nucleoid and are essential for cell viability (3, 4). HBsu contributes to the existence of discrete chromosomal domains, and its depletion leads to strong chromosome decondensation (4). B. subtilis contains a single circular chromosome (∼4,200 kb) that replicates bidirectionally from a single origin (oriC). The replisome containing two complexes, one for each replication fork, remains at midcell positions whereas the newly synthesized DNA is extruded and moves toward the cell poles (5). A prokaryotic condensin/cohesin-like complex called SMC forms two subcellular centers that organize newly duplicated chromosome regions within each cell half (2). SMC gives rise to DNA loops arranged in rosette-like superstructures and is complexed with two other proteins, ScpA and ScpB, which also contribute to the formation of superhelical domains throughout the B. subtilis-replicated DNA (2).
B. subtilis phage ϕ29 has a linear, double-stranded DNA of 19,285 bp with a terminal protein (TP) (parental TP) covalently linked to the 5′ ends. Initiation of ϕ29 DNA replication (see Fig. S1 for details) takes place by a protein-priming mechanism (6, 7) and starts with the formation of a heterodimer between the ϕ29 DNA polymerase and a free TP molecule (primer TP) that recognizes the origins of replication (containing the parental TP) at both ends of the viral genome. The ϕ29 double-stranded DNA-binding protein p6 (encoded by gene 6) forms a nucleoprotein complex at the replication origins that has been proposed to open the DNA ends, facilitating the initiation step of replication (8, 9). Initiation of viral DNA replication requires the formation of a covalent linkage between the first inserted nucleotide (dAMP) and the hydroxyl group of serine 232 of the priming TP, catalyzed by the ϕ29 DNA polymerase (10, 11). Protein p6 is essential for phage DNA replication in vivo because a ϕ29 suppressor-sensitive (sus) mutant in gene 6 is deficient in viral DNA synthesis (12, 13). Its small size and abundance in infected cells (about 700,000 copies/cell) (14) are features expected for proteins with architectural roles. Because ϕ29 DNA forms a right-handed toroidal superhelix around a multimeric protein p6 core (15, 16), it has been described as a histone-like protein adapted to package and to organize the viral genome (9). In addition, protein p6 has been shown to form dimers that bind cooperatively to DNA every 24 nucleotides (17) and to interact with the viral DNA through the minor groove (15, 18). It possesses higher affinity for both ϕ29 DNA ends, but it is able to bind to most, if not all, the viral genome in vivo (19).
Although the intrinsic mechanisms governing DNA replication of viral genomes in bacteria have been extensively studied in vitro (20), remarkably little is known about the subcellular organization of proteins involved in this fundamental process. Recently, we have provided insights into the in vivo organization of the ϕ29 DNA polymerase and TP (21). After ϕ29 TP-DNA injection takes place, the viral genome associates with the B. subtilis nucleoid by means of the DNA-binding capacity of the parental TP. Once both ϕ29 primer TP and DNA polymerase are synthesized, they form a heterodimeric complex that associates with the bacterial nucleoid through the DNA-binding domain of the priming TP. Hence, the ϕ29 priming TP plays a role in recruiting the DNA polymerase to the specific sites for the initiation of viral DNA replication (20) and recognizes the replication origins at both DNA ends by means of specific interactions with the parental TP (22, 23).
Here, we show that the ϕ29 histone-like protein p6 localizes in a helix-like manner at the beginning of the phage infection cycle. Later, the bulk of protein p6 is recruited to the bacterial nucleoid, a migrating process that requires the accumulation of ϕ29 genomes. The DNA-binding capacity of protein p6 is essential for its subcellular relocalization. A model integrating the results is discussed below.
Results
Subcellular Localization of ϕ29 Protein p6 in B. subtilis.
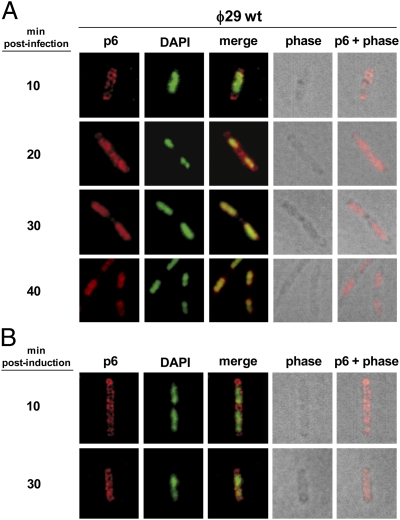
To gain insight into the subcellular distribution of essential components of the ϕ29 DNA replication machinery, we first studied, by immunofluorescence (IF) microscopy, the localization of protein p6 in both infected and noninfected B. subtilis cells. Fig. 1A shows immunofluorescence, DAPI staining, phase contrast, and merged images of cells at different times post infection with wild-type phage ϕ29. Overlay of phase contrast and deconvolved IF images at early times after infection (i.e., 10 min) shows that p6 localized in a peripheral helix-like pattern that spanned the entire length of the infected cell. Interestingly, at 20 min post infection, p6 started to redistribute up to the central region of the cell, spanning the nucleoid area. Overlay of DAPI staining and the p6 fluorescent signal revealed a substantial colocalization. Later, at 30 and 40 min after infection, protein p6 remained localized at the bacterial nucleoid.
Fig. 1.
Subcellular localization of protein p6 in ϕ29-infected and noninfected cells. B. subtilis 110NA and IH-04 (expressing p6) cells were grown at 37 °C in LB medium supplemented with 5 mM MgSO4. At an OD600 of 0.45–0.5, the 110NA culture was infected with wild-type phage ϕ29 at a multiplicity of infection (MOI) of 5 (A), and IPTG was added to a final concentration of 1 mM to the IH-04 culture (B). Samples were withdrawn at the indicated times post infection or after IPTG addition and subjected to IF microscopy using polyclonal antibodies against p6. For clarity, p6 and DAPI fluorescent signals are false-colored red and green, respectively.
To test whether the subcellular localization of protein p6 depends on other phage-encoded proteins, we used a B. subtilis strain (IH-04) harboring episomal plasmid pNDH33-p6, which contains an isopropyl β-d-1-thiogalactopyranoside (IPTG)-inducible ϕ29 gene 6, and analyzed the distribution of protein p6 in noninfected cells. As a control, the amount of protein p6 synthesized was similar to that produced in a phage infection cycle (Fig. S2A), and its expression did not have a significant effect in the transcription of some B. subtilis essential genes such as dnaA, ftsZ, gyrB, hbs, mreB, murB, rpoC, and smc (Fig. S2B). As shown in Fig. 1B, IF microscopy images of IPTG-induced IH-04 cells revealed that the ectopically expressed protein p6 localized in a helix-like configuration at 30 min post induction, indicating that its redistribution to the bacterial nucleoid depends on other ϕ29-encoded protein(s) and/or viral DNA.
Protein p6 Colocalizes with the ϕ29 TP at the Bacterial Nucleoid.
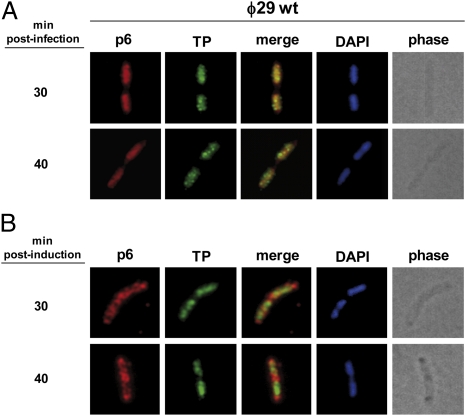
We have recently shown that the ϕ29 TP associates with the host bacterial nucleoid independently of other viral-encoded proteins, recruiting the phage DNA polymerase to the active site of DNA replication (21). Because protein p6 relocalizes to the bacterial nucleoid region at middle infection times (see above), we analyzed simultaneously the subcellular localization of both ϕ29 TP and protein p6 during this stage of the phage infection cycle. For this, B. subtilis 110NA was infected with wild-type phage ϕ29 and subjected to IF microscopy. As shown in Fig. 2A, at 30 min post infection, both protein p6 and TP follow a similar localization pattern in occupying the mass of the nucleoid. Superimposition of the fluorescent signals revealed that protein p6 displays a high degree of colocalization with the viral TP, which is also extended to late times after infection (i.e., 40 min).
Fig. 2.
Protein p6 colocalizes with TP at the cell nucleoid at middle post-infection times. (A) B. subtilis 110NA cells were grown at 37 °C in LB medium containing 5 mM MgSO4. At an OD600 of 0.45–0.5, the culture was infected with wild-type phage ϕ29 at a MOI of 5. Samples were harvested at the indicated times after infection and processed for IF microscopy using polyclonal antibodies against p6 and TP. (B) B. subtilis strain IH-19 (expressing protein p6 constitutively and TP under an inducible IPTG promoter) was grown at 37 °C in LB medium containing 5 mM MgSO4. At an OD600 of 0.4, 10 μM IPTG was added to induce expression of TP. Samples were harvested at 30 and 40 min post induction.
To test whether the ϕ29 TP directly recruits protein p6 to the chromosomal region, we constructed a B. subtilis strain (IH-19) able to express simultaneously ϕ29 TP and protein p6. Results in noninfected cells show that, as expected, the TP localized at the bacterial nucleoid region (21), whereas protein p6 remained organized in a helix-like configuration (Fig. 2B). Hence, and differently from the phage DNA polymerase (21), protein p6 is not directly recruited by the viral TP to the nucleoid area.
Redistribution of Protein p6 to the Bacterial Nucleoid Requires ϕ29 DNA Replication.
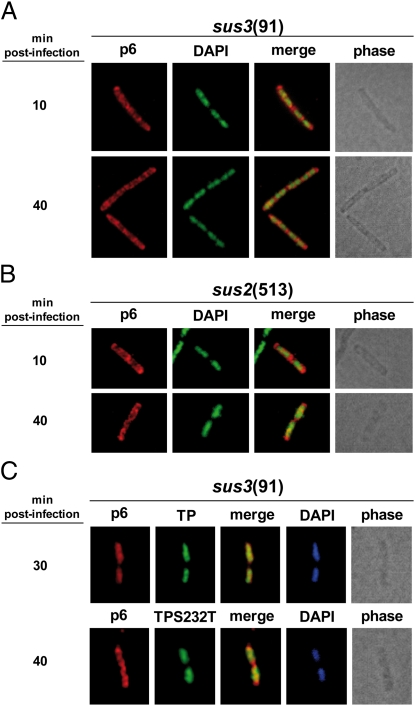
To investigate whether the nucleoid association of protein p6 depends on ϕ29 DNA replication, we infected the nonsuppressor B. subtilis strain 110NA with the mutant phage sus3(91), which is unable to synthesize primer TP and hence to replicate its genome, and analyzed by IF microscopy the subcellular distribution of p6 at different post infection times. Fig. 3A shows that, at 10 min post infection, protein p6 localized in helix-like structures. Importantly, at late post infection times (i.e., 40 min), p6 was not redistributed to the chromosomal site, showing that its association with the bacterial nucleoid depends on the synthesis of ϕ29 TP and supporting the possibility that the accumulation of ϕ29 DNA genomes may be required for the recruitment of the bulk of p6. Accordingly, when nonsuppresor B. subtilis cells were infected with phage sus2(513), unable to produce ϕ29 DNA polymerase, protein p6 did not relocalize to the bacterial nucleoid (Fig. 3B).
Fig. 3.
The nucleoidal localization of protein p6 depends on viral DNA replication. (A and B) B. subtilis 110NA cells were grown at 37 °C in LB medium supplemented with 5 mM MgSO4. At an OD600 of 0.45–0.5, the cultures were infected with sus3(91) or sus2(513) mutant phages at a MOI of 5. Samples were harvested at the indicated times post infection and processed for IF microscopy using polyclonal antibodies against p6. For clarity, p6 and DAPI fluorescent signals are false-colored red and green, respectively. (C) Strains IH-16 (expressing TP wild type) and IH-18 (expressing TP-S232T mutant) were grown at 37 °C in LB medium supplemented with 5 mM MgSO4. At an OD600 of 0.45–0.5, the cultures were infected with sus3(91) mutant phage at a MOI of 5, and 10 μM IPTG was added to the medium 10 min later. Samples were withdrawn 30 or 40 min post infection and subjected to IF microscopy using polyclonal antibodies against both proteins, p6 and TP.
It has been shown that the TP residue Ser232 establishes a covalent phosphoester bond between its −OH group and the first dAMP incorporated at the initiation step of ϕ29 DNA replication (10). A single mutation of Ser232 to Thr completely abolished the TP-priming capacity, retaining its ability to interact with the ϕ29 DNA polymerase and viral DNA (24). To further unravel the mechanism that gives rise to the p6 nucleoid association, we studied the subcellular localization of protein p6 in cells expressing a S232T mutant TP. In this scenario, the DNA polymerase/TP heterodimer would efficiently recognize the origins of replication of the phage TP-DNA at the bacterial nucleoid; however, ϕ29 DNA replication cannot be initiated. We therefore engineered B. subtilis strains expressing ectopically wild-type TP (IH-16) or S232T mutant TP (IH-18) in an IPTG-dependent way, infected them with mutant phage sus3(91), and subjected them to IF microscopy assays. As shown in Fig. 3C, in cells producing S232T mutant TP, protein p6 was not recruited to the bacterial nucleoid and displayed a helix-like configuration even at 40 min after infection. In a different way, protein p6 relocalized to the bacterial nucleoid when wild-type TP was ectopically provided. As an internal control, both wild-type TP and S232T mutant TP were shown to be associated with the bacterial nucleoid (Fig. 3C). Altogether, these results show that ϕ29 DNA replication is required for the recruitment of the bulk of protein p6 to the bacterial nucleoid.
Colocalization Experiments of Protein p6 and ϕ29-Replicated DNA.
To gain further insight into the subcellular distribution of the ϕ29 DNA replication machinery, we analyzed simultaneously the localization of both ϕ29-replicated DNA and protein p6 in B. subtilis-infected cells. Localization of ϕ29 DNA in infected cells has been previously studied by IF (25) using incorporation of the thymine analog 5-bromo-2′-deoxyuridine (BrdU) and simultaneous inhibition of the B. subtilis DNA polymerase III holoenzyme by 6-(p-hydroxyphenylazo)-uracil (HpUra) (26). Fig. S3 shows that, on initiation of phage DNA replication (at 10 min post infection), ϕ29 DNA appeared to localize to a single focus within the cell. At this time, almost all of the p6 fluorescent signal was visualized as a peripheral helix-like structure. Later, during the middle stage of infection, as the replicated ϕ29 DNA redistributed and occupied the bulk of the nucleoid, protein p6 was recruited to the central part of the cell and displayed a substantial degree of colocalization with the replicated viral genomes.
Nucleoid-Associated Localization of Protein p6 Depends on Its Double-Stranded DNA-Binding Capacity.
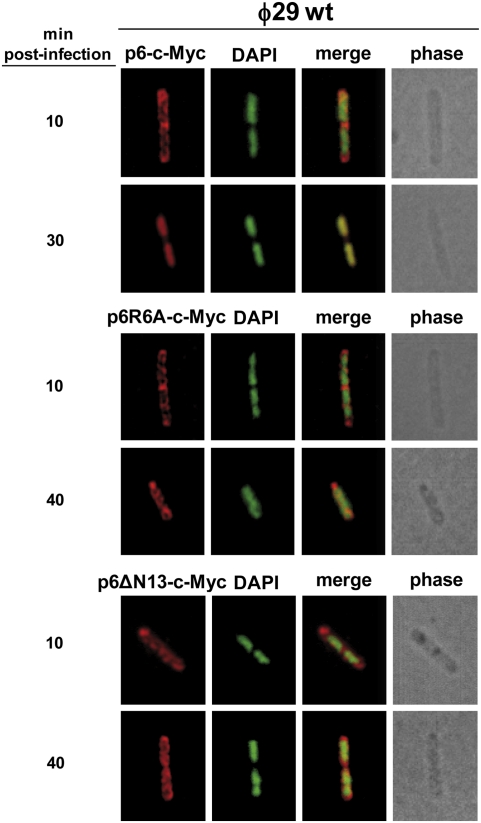
As described before, protein p6 binds to most, if not all, the viral genome in vivo, although with higher affinity for both DNA ends (19). Previous studies with protein p6 N-terminal deletion mutants showed that the first 13 amino acids are essential for the p6 DNA-binding capacity in vitro (27). In fact, a p6R6A single-mutant protein failed to bind DNA and did not show any activation of initiation of ϕ29 DNA replication in vitro (18). To determine whether the DNA-binding capacity of protein p6 is important for its recruitment at the bacterial nucleoid, we engineered B. subtilis strains expressing ectopically wild-type p6, p6ΔN13, or p6R6A under an IPTG-inducible promoter; they were infected with the replication-deficient mutant phage sus6(626) and subjected to IF microscopy. Complementation experiments by agarose gel electrophoresis and real-time PCR confirmed that, differently from the wild-type p6, when the p6ΔN13 and p6R6A variants were expressed, ϕ29 DNA replication was impaired (Fig. S4 A and B). Fig. S5 shows that, when wild-type p6 was ectopically induced, it localized in a helix-like pattern at 10 min after infection with the sus6(626) mutant phage. Later on, protein p6 was seen relocalizing to the bacterial nucleoid. In contrast, ectopically induced p6R6A and p6ΔN13 mutants, even at late stages of infection with the sus6(626) mutant, displayed a peripheral helix-like configuration. These latter results were consistent with the view that the p6 DNA-binding capacity might play a role in allowing its nucleoid recruitment. However, because ϕ29 DNA replication was impaired under p6R6A or p6ΔN13 expression, this possibility could not be assessed. Therefore, we engineered B. subtilis strains producing C-terminal c-Myc fusions of wild-type p6 (IH-10), p6R6A (IH-12), or p6ΔN13 (IH-14) in an IPTG-dependent way. Induced cultures were infected with wild-type phage ϕ29 (able to produce wild-type p6) and subjected to IF experiments by using monoclonal antibodies against c-Myc to recognize the p6-c-Myc fusion. Agarose gel electrophoresis and real-time PCR complementation experiments using mutant phage sus6(626) to infect strains IH-10, IH-12, and IH-14 confirmed that p6-c-Myc was functional, but differently from the p6R6A-c-Myc and p6ΔN13-c-Myc variants (Fig. S6 A and B). In agreement with this, p6-c-Myc, but not the p6R6A-c-Myc and p6ΔN13-c-Myc variants, was efficiently redistributed at the bacterial nucleoid in a sus6(626) infection (Fig. S7), similarly to the results obtained with the wild-type protein p6 (Fig. S5). Under an active ϕ29 DNA replication background (i.e., when cells were infected with a wild-type phage ϕ29), the fluorescent signal corresponding to the p6-c-Myc fusion was also observed to relocalize to the bacterial nucleoid (Fig. 4). Importantly, neither the p6R6A-c-Myc nor the p6ΔN13-c-Myc fusions relocalized to the host chromosomal site but displayed a peripheral helix-like pattern throughout the infection cycle. This result shows that the DNA-binding capacity of protein p6 is essential for its migration toward the nucleoid and suggests that the accumulation of replicated ϕ29 TP-DNA is required to capture the bulk of protein p6.
Fig. 4.
Protein p6 mutants in DNA binding are not distributed at the bacterial nucleoid. B. subtilis strains IH-10 (expressing p6-c-Myc), IH-12 (expressing p6R6A-c-Myc), and IH-14 (expressing p6ΔN13-c-Myc) were grown at 37 °C in LB medium containing 5 mM MgSO4. At an OD600 of 0.45–0.5, cells were infected with wild-type phage ϕ29 at a MOI of 5, and 50 μM IPTG was added to the medium 5 min later. Samples were withdrawn at the indicated times post infection and subjected to IF microscopy using polyclonal antibodies against p6. For clarity, p6 and DAPI fluorescent signals are false-colored red and green, respectively.
Discussion
DNA-binding proteins that display the essential function of organizing and compacting chromosomes have been described from higher eukaryotes to prokaryotes. In Eubacteria, these architectural factors are commonly known as “histone-like” proteins (28), which share some basic features such as high levels of expression, nonsequence-specific DNA-binding capacity, and the induction of pleiotropic effects. Phage ϕ29 protein p6 is one of the most abundant viral proteins in B. subtilis-infected cells, constituting about 4% of the total bacterial proteins. It has been estimated that protein p6 reaches intracellular concentrations around 1 mM, which would give rise to the formation of oligomers (14). Protein p6 has a small size (103 amino acids) and reaches abundant levels in B. subtilis (about 700,000 copies/cell), which represents 1.4 times the amount necessary to cover all of the ϕ29 TP-DNA molecules present at middle-infection times (14). In comparison, HBsu reaches about 50,000 copies per cell in B. subtilis, and the Escherichia coli histone-like proteins Dps and HU reach 180,000 and 60,000 copies/cell, respectively (29, 30). Hence, protein p6 possesses the expected features of a histone-like protein and resembles these architectural proteins in bacteria, which have been found to be uniformly distributed within the entire nucleoid contributing to the maintenance of superhelical density (2).
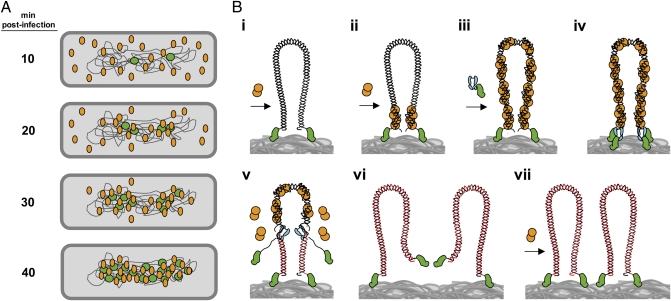
The results described in this paper, together with those obtained previously (19, 21, 31), allow us to propose a comprehensive model about the in vivo organization of protein p6 and its role in ϕ29 DNA replication, which is illustrated schematically in Fig. 5. By using IF microscopy, we here provide direct evidence that the bulk of protein p6 displays a peripheral helix-like localization at 10 min after infection, whereas at this post infection time the replicated-ϕ29 DNA was observed as a single focus in the central part of the cell. Due to the fact that protein p6 is essential to initiate ϕ29 DNA replication, a small amount of protein p6 must be recruited early at the bacterial nucleoid, establishing the appropriate conditions at the phage DNA ends to achieve the first rounds of replication. As more de novo-synthesized ϕ29 genomes are generated during the phage DNA replication cycle, the bulk of protein p6 is recruited to the bacterial nucleoid (Fig. 5A). This result fits well with the view that high numbers of replicated ϕ29 genomes are required as a target for the high levels of protein p6 that are synthesized. Accordingly, this migrating process requires synthesis of ϕ29 TP and DNA polymerase, essential components of the phage DNA replication machinery. When initiation of ϕ29 DNA replication was abolished by using a S232T mutant TP, protein p6 did not migrate to the bacterial nucleoid and displayed a peripheral helix-like localization.
Fig. 5.
Model of nucleoid-associated ϕ29 DNA replication and recruitment of protein p6 to the viral genome. (A) Once ϕ29 infection takes place, the TP-DNA genomes (green circles) are recruited to the bacterial nucleoid. Soon after the injection process, early promoters mediate a rapid expression of protein p6 (orange ovals). At 10 min post infection, p6 is shown to distribute following a peripheral helix-like pattern. Later, and simultaneously with the generation of new replicated ϕ29 genomes, the bulk of protein p6 is recruited to the bacterial nucleoid. (B, i) A ϕ29 TP-DNA molecule (linear dsDNA drawn as a double helix) is shown attached to the bacterial nucleoid surface (gray mass at bottom) by the parental TPs (green). Protein p6 (orange circles) is recruited to the bacterial nucleoid by binding to the viral DNA. (B, ii) Protein p6 binds the origins of replication of the ϕ29 genome through its DNA-binding domain, forming a nucleoprotein complex that helps to open the DNA ends, facilitating the initiation step of replication. (B, iii) The p6-bound origins act as a nucleation site from which more p6 molecules bind to the rest of the viral genome. (B, iv) The heterodimer formed by TP and DNA polymerase (cyan) recognizes the p6-bound replication origins and initiates DNA replication. (B, v) After a transition step, the DNA polymerase dissociates and continues processive elongation of the nascent DNA strands (red lines) coupled to strand displacement, removing p6 molecules from the viral genome during the polymerization process. (B, vi and vii) Once DNA replication is completed, two ϕ29 TP-DNA molecules are ready for another round of replication. Proteins, viral DNA, and bacterial nucleoid are not drawn to scale.
In a more detailed view, after ϕ29 TP-DNA injection takes place, the ϕ29 genome associates with the B. subtilis nucleoid by the nonsequence-specific DNA-binding capacity of the parental TPs covalently linked to the phage 5′ DNA ends (21) (Fig. 5B, i). In this way, and despite its linear conformation, the ϕ29 TP-DNA may be topologically constrained in vivo. Binding of protein p6 to the viral DNA ends forms a nucleoprotein complex by which a right-handed superhelix of DNA wraps tightly around a multimeric p6 core that was proposed to open the DNA ends facilitating initiation of ϕ29 DNA replication (15, 16, 32) (Fig. 5B, ii). This multimeric nucleoprotein complex, in which the protein p6 repeated motif is formed by protein dimers bound to 24-bp DNA segments, originates a nucleation-dependent oligomerization process to cover most, if not all, of the ϕ29 genome (19), thereby displaying a key role as an architectural protein in organizing the viral DNA (Fig. 5B, iii). The ϕ29 primer TP and DNA polymerase form a heterodimeric complex that associates with the bacterial nucleoid and recognizes the viral replication origins through specific contacts with the parental TP (22, 23) (Fig. 5B, iv). Enabled by the p6 nucleoprotein complex, the DNA polymerase then catalyzes the addition of the first dAMP and, after a transition step, it dissociates from the primer TP and continues processive elongation coupled to strand displacement and the simultaneous removal of protein p6 from the viral DNA (Fig. 5B, v). Continuous elongation by the DNA polymerase completes replication of the parental strand (Fig. 5B, vi), and the generated full-length ϕ29 DNA molecules then would be ready for successive rounds of replication by recruiting protein p6 at the DNA ends (Fig. 5B, vii). In the course of the late stages of the phage DNA replication cycle, the viral DNA accumulated is packaged inside the viral particle with a left-to-right polarity (33). The ϕ29 assembled particles are distributed mainly throughout the bacterial nucleoid as shown by electron microscopy (34).
The results obtained in this work illustrate that the DNA-binding capacity of protein p6 is essential to its recruitment at the nucleoid environment. Indeed, we exclude the possibility of a direct recruitment by the ϕ29 TP or by any other B. subtilis protein. Taking into account that protein p6 does not recognize any specific DNA sequence, it could be expected to recognize bacterial DNA with the same affinity as phage DNA. However, in the absence of ϕ29 DNA replication (i.e., no ϕ29 replicated genomes are available), protein p6 is not recruited to the bacterial nucleoid, and hence it is able to discriminate the bacterial DNA. It has been proposed that the negative supercoiling of bacterial DNA would strongly impair protein p6 complex formation and that the presumably lower negative superhelicity of ϕ29 DNA likely makes the viral genome an appropriate target for the binding of p6 (9, 19). In fact, protein p6 is able to restrain positive supercoiling of the DNA in vitro (16, 17) and binds all along ϕ29 DNA in vivo with a much higher affinity than for plasmid DNA (19), which, similarly to the bacterial chromosome, consists of a circular DNA molecule. In addition, protein p6 binding to plasmid DNA is enhanced by decreasing its negative supercoiling with novobiocin, a DNA gyrase inhibitor (19). On the other hand, the p6 binding to plasmid DNA was not enhanced by adding nalidixic acid, which also inhibits the DNA gyrase but has no topological effects. On the basis of these results, the DNA supercoiling is proposed to play an important role in the recognition of phage DNA by protein p6. A topologic-dependent viral DNA binding of protein p6 may constitute a sophisticated strategy to avoid its sequestration by the higher volume of the bacterial DNA.
In conclusion, the results presented in this paper provide further insights into the dynamic organization of the ϕ29 DNA replication machinery and lead us to disclose, both spatially and temporally, the subcellular localization of a viral histone-like protein in bacteria.
Materials and Methods
General Methods and DNA Techniques.
Because phage ϕ29 DNA replication is inhibited by Spo0A (35), spo0A deletion strains were used when indicated. All DNA manipulations were carried out according to standard methods (36).
Phages, Bacterial Strains, and Growth Conditions.
Phages and bacterial strains used are listed in Tables S1 and S2. E. coli strain XL1-Blue, used for cloning, was grown in Luria–Bertani (LB) medium containing ampicillin (100 μg/mL). B. subtilis strains were grown at 37 °C in LB medium containing 5 mM MgSO4 and supplemented with chloramphenicol (5 μg/mL) and/or kanamycin (5 μg/mL), as indicated. Generally, overnight cultures were diluted 1/100 in fresh medium and incubated at 37 °C for 2–3 h to re-establish exponential growth before manipulation. Ectopically expressed proteins were induced by addition of IPTG.
Bacterial Transformation and DNA Labeling.
B. subtilis cells were transformed by standard procedures (37, 38). Selection for B. subtilis transformants was carried out in LB-agar plates, supplemented with chloramphenicol (5 μg/mL) and/or kanamycin (5 μg/mL).
In DNA-labeling experiments, the thymine analog BrdU (Sigma) was added to the growth medium at a final concentration of 150 μM. Incorporation of BrdU into chromosomal DNA of B. subtilis was inhibited by the addition of 75 μM of HpUra (26) to the growth medium 3 min before the BrdU addition.
Analysis of Viral DNA Synthesis by Gel Electrophoresis.
Analysis of viral DNA synthesis in vivo was carried out as described (39). Basically, total intracellular DNA was isolated at different times post infection and analyzed in 0.6% agarose gels.
Real-Time PCR.
Aliquots of 1 mL of B. subtilis cultures were withdrawn at the indicated times after infection, processed, and analyzed by real-time PCR essentially as described (40). The primer sets R-25 and R-OUT-SUPER were used to amplify a 297-bp fragment corresponding to the right end of the ϕ29 genome (Tables S3 and S4). The data obtained were interpolated to standard curves constructed with known amounts of phage DNA.
Immunofluorescence Microscopy.
For IF microscopy, overnight cultures were diluted in LB medium containing 5 mM MgSO4 and grown at 37 °C to early exponential phase. At an OD600 of 0.45–0.5, the culture was infected with the indicated ϕ29 phage at a multiplicity of infection (MOI) of 5. Samples were fixed after the indicated times of infection and processed essentially as described (41). See SI Materials and Methods for details.
Supplementary Material
Acknowledgments
We thank Dr. Mario Mencía for helpful discussions, Dr. Antonio Maraver for critical reading of the manuscript, and the Genomics Core Facility at the Centro de Biología Molecular “Severo Ochoa” (Consejo Superior de Investigaciones Científicas-Universidad Autónoma de Madrid) for RT-quantitative PCR and data analysis. This investigation was supported by Grant BFU2008-00215 and Consolider-Ingenio Grant 2010 24717 from the Spanish Ministry of Science and Innovation to M.S. and by an institutional grant from Fundación Ramón Areces to the Centro de Biología Molecular “Severo Ochoa.” D.M.-E. was a holder of a Consolider-Ingenio contract. I.H. and D.B.-P. are holders of a Formación de Profesorado Universitario and a Formación de Personal Investigador fellowship, respectively, from the Spanish Ministries of Education, and Science and Innovation.
Footnotes
The authors declare no conflict of interest.
This article contains supporting information online at www.pnas.org/lookup/suppl/doi:10.1073/pnas.1203824109/-/DCSupplemental.
References
- 1.Fischle W, Wang Y, Allis CD. Histone and chromatin cross-talk. Curr Opin Cell Biol. 2003;15(2):172–183. doi: 10.1016/s0955-0674(03)00013-9. [DOI] [PubMed] [Google Scholar]
- 2.Graumann PL. Bacillus, Cellular and Molecular Biology. Norfolk, UK: Caister Academic Press; 2007. [Google Scholar]
- 3.Micka B, Groch N, Heinemann U, Marahiel MA. Molecular cloning, nucleotide sequence, and characterization of the Bacillus subtilis gene encoding the DNA-binding protein HBsu. J Bacteriol. 1991;173:3191–3198. doi: 10.1128/jb.173.10.3191-3198.1991. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Köhler P, Marahiel MA. Association of the histone-like protein HBsu with the nucleoid of Bacillus subtilis. J Bacteriol. 1997;179:2060–2064. doi: 10.1128/jb.179.6.2060-2064.1997. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Webb CD, et al. Bipolar localization of the replication origin regions of chromosomes in vegetative and sporulating cells of B. subtilis. Cell. 1997;88:667–674. doi: 10.1016/s0092-8674(00)81909-1. [DOI] [PubMed] [Google Scholar]
- 6.Salas M. Protein-priming of DNA replication. Annu Rev Biochem. 1991;60:39–71. doi: 10.1146/annurev.bi.60.070191.000351. [DOI] [PubMed] [Google Scholar]
- 7.Salas M. Mechanisms of initiation of linear DNA replication in prokaryotes. Genet Eng (N Y) 1999;21:159–171. doi: 10.1007/978-1-4615-4707-5_8. [DOI] [PubMed] [Google Scholar]
- 8.Blanco L, Gutiérrez J, Lázaro JM, Bernad A, Salas M. Replication of phage ϕ29 DNA in vitro: Role of the viral protein p6 in initiation and elongation. Nucleic Acids Res. 1986;14:4923–4937. doi: 10.1093/nar/14.12.4923. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Serrano M, et al. Phage ϕ29 protein p6: A viral histone-like protein. Biochimie. 1994;76:981–991. doi: 10.1016/0300-9084(94)90023-x. [DOI] [PubMed] [Google Scholar]
- 10.Hermoso JM, Méndez E, Soriano F, Salas M. Location of the serine residue involved in the linkage between the terminal protein and the DNA of phage ϕ29. Nucleic Acids Res. 1985;13:7715–7728. doi: 10.1093/nar/13.21.7715. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Blanco L, Salas M. Characterization and purification of a phage ϕ29-encoded DNA polymerase required for the initiation of replication. Proc Natl Acad Sci USA. 1984;81:5325–5329. doi: 10.1073/pnas.81.17.5325. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Carrascosa JL, et al. Bacillus subtilis phage ϕ29. Characterization of gene products and functions. Eur J Biochem. 1976;66:229–241. doi: 10.1111/j.1432-1033.1976.tb10512.x. [DOI] [PubMed] [Google Scholar]
- 13.Escarmís C, Guirao D, Salas M. Replication of recombinant ϕ29 DNA molecules in Bacillus subtilis protoplasts. Virology. 1989;169(1):152–160. doi: 10.1016/0042-6822(89)90051-2. [DOI] [PubMed] [Google Scholar]
- 14.Abril AM, Salas M, Andreu JM, Hermoso JM, Rivas G. Phage ϕ29 protein p6 is in a monomer-dimer equilibrium that shifts to higher association states at the millimolar concentrations found in vivo. Biochemistry. 1997;36:11901–11908. doi: 10.1021/bi970994e. [DOI] [PubMed] [Google Scholar]
- 15.Serrano M, Salas M, Hermoso JM. A novel nucleoprotein complex at a replication origin. Science. 1990;248:1012–1016. doi: 10.1126/science.2111580. [DOI] [PubMed] [Google Scholar]
- 16.Serrano M, Gutiérrez C, Salas M, Hermoso JM. Superhelical path of the DNA in the nucleoprotein complex that activates the initiation of phage ϕ29 DNA replication. J Mol Biol. 1993;230:248–259. doi: 10.1006/jmbi.1993.1140. [DOI] [PubMed] [Google Scholar]
- 17.Prieto I, Serrano M, Lázaro JM, Salas M, Hermoso JM. Interaction of the bacteriophage ϕ29 protein p6 with double-stranded DNA. Proc Natl Acad Sci USA. 1988;85:314–318. doi: 10.1073/pnas.85.2.314. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Freire R, Salas M, Hermoso JM. A new protein domain for binding to DNA through the minor groove. EMBO J. 1994;13:4353–4360. doi: 10.1002/j.1460-2075.1994.tb06755.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.González-Huici V, Salas M, Hermoso JM. Genome wide, supercoiling-dependent in vivo binding of a viral protein involved in DNA replication and transcriptional control. Nucleic Acids Res. 2004;32:2306–2314. doi: 10.1093/nar/gkh565. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Calendar R. The Bacteriophages. New York, Oxford: Oxford University Press; 2006. pp. 1–746. [Google Scholar]
- 21.Muñoz-Espín D, Holguera I, Ballesteros-Plaza D, Carballido-López R, Salas M. Viral terminal protein directs early organization of phage DNA replication at the bacterial nucleoid. Proc Natl Acad Sci USA. 2010;107:16548–16553. doi: 10.1073/pnas.1010530107. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.González-Huici V, Lázaro JM, Salas M, Hermoso JM. Specific recognition of parental terminal protein by DNA polymerase for initiation of protein-primed DNA replication. J Biol Chem. 2000;275:14678–14683. doi: 10.1074/jbc.m910058199. [DOI] [PubMed] [Google Scholar]
- 23.Serna-Rico A, Illana B, Salas M, Meijer WJJ. The putative coiled coil domain of the ϕ29 terminal protein is a major determinant involved in recognition of the origin of replication. J Biol Chem. 2000;275:40529–40538. doi: 10.1074/jbc.M007855200. [DOI] [PubMed] [Google Scholar]
- 24.Garmendia C, Salas M, Hermoso JM. Site-directed mutagenesis in the DNA linking site of bacteriophage ϕ29 terminal protein: Isolation and characterization of a Ser232 Thr mutant. Nucleic Acids Res. 1988;16:5727–5740. doi: 10.1093/nar/16.13.5727. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Meijer WJJ, Lewis PJ, Errington J, Salas M. Dynamic relocalization of phage ϕ29 DNA during replication and the role of the viral protein p16.7. EMBO J. 2000;19:4182–4190. doi: 10.1093/emboj/19.15.4182. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Brown NC. 6-(p-hydroxyphenylazo)-uracil: A selective inhibitor of host DNA replication in phage-infected Bacillus subtilis. Proc Natl Acad Sci USA. 1970;67:1454–1461. doi: 10.1073/pnas.67.3.1454. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Otero MJ, Lázaro JM, Salas M. Deletions at the N terminus of bacteriophage ϕ29 protein p6: DNA binding and activity in ϕ29 DNA replication. Gene. 1990;95(1):25–30. doi: 10.1016/0378-1119(90)90409-k. [DOI] [PubMed] [Google Scholar]
- 28.Drlica K, Rouviere-Yaniv J. Histonelike proteins of bacteria. Microbiol Rev. 1987;51:301–319. doi: 10.1128/mr.51.3.301-319.1987. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Azam TA, Hiraga S, Ishihama A. Two types of localization of the DNA-binding proteins within the Escherichia coli nucleoid. Genes Cells. 2000;5:613–626. doi: 10.1046/j.1365-2443.2000.00350.x. [DOI] [PubMed] [Google Scholar]
- 30.Ross MA, Setlow P. The Bacillus subtilis HBsu protein modifies the effects of alpha/beta-type, small acid-soluble spore proteins on DNA. J Bacteriol. 2000;182:5556–5562. doi: 10.1128/jb.182.7.1942-1948.2000. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.González-Huici V, Alcorlo M, Salas M, Hermoso JM. Binding of phage ϕ29 architectural protein p6 to the viral genome: Evidence for topological restriction of the phage linear DNA. Nucleic Acids Res. 2004;32:3493–3502. doi: 10.1093/nar/gkh668. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Salas M, et al. Protein-primed replication of bacteriophage ϕ29 DNA. Biochim Biophys Acta. 1988;951:419–424. doi: 10.1016/0167-4781(88)90115-7. [DOI] [PubMed] [Google Scholar]
- 33.Bjornsti MA, Reilly BE, Anderson DL. Morphogenesis of bacteriophage ϕ29 of Bacillus subtilis: Oriented and quantized in vitro packaging of DNA protein gp3. J Virol. 1983;45:383–396. doi: 10.1128/jvi.45.1.383-396.1983. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Jiménez F, Camacho A, De La Torre J, Viñuela E, Salas M. Assembly of Bacillus subtilis phage ϕ29. 2. Mutants in the cistrons coding for the non-structural proteins. Eur J Biochem. 1977;73(1):57–72. doi: 10.1111/j.1432-1033.1977.tb11291.x. [DOI] [PubMed] [Google Scholar]
- 35.Castilla-Llorente V, Muñoz-Espín D, Villar L, Salas M, Meijer WJJ. Spo0A, the key transcriptional regulator for entrance into sporulation, is an inhibitor of DNA replication. EMBO J. 2006;25:3890–3899. doi: 10.1038/sj.emboj.7601266. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Sambrook J, Fritsch EF, Maniatis T. Molecular Cloning: A Laboratory Manual. Cold Spring Harbor, NY: Cold Spring Harbor Laboratory Press; 1989. [Google Scholar]
- 37.Ward JB, Jr, Zahler SA. Genetic studies of leucine biosynthesis in Bacillus subtilis. J Bacteriol. 1973;116:719–726. doi: 10.1128/jb.116.2.719-726.1973. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Formstone A, Errington J. A magnesium-dependent mreB null mutant: Implications for the role of mreB in Bacillus subtilis. Mol Microbiol. 2005;55:1646–1657. doi: 10.1111/j.1365-2958.2005.04506.x. [DOI] [PubMed] [Google Scholar]
- 39.Bravo A, Hermoso JM, Salas M. A genetic approach to the identification of functional amino acids in protein p6 of Bacillus subtilis phage ϕ29. Mol Gen Genet. 1994;245:529–536. doi: 10.1007/BF00282215. [DOI] [PubMed] [Google Scholar]
- 40.González-Huici V, Salas M, Hermoso JM. The push-pull mechanism of bacteriophage ϕ29 DNA injection. Mol Microbiol. 2004;52:529–540. doi: 10.1111/j.1365-2958.2004.03993.x. [DOI] [PubMed] [Google Scholar]
- 41.Muñoz-Espín D, et al. The actin-like MreB cytoskeleton organizes viral DNA replication in bacteria. Proc Natl Acad Sci USA. 2009;106:13347–13352. doi: 10.1073/pnas.0906465106. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.