Abstract
Decoupling of transcription and translation during postmeiotic germ cell differentiation is critical for successful spermatogenesis. Here we establish that the interaction between microRNAs and actin-associated protein Arpc5 sets the stage for an elaborate translational control mechanism by facilitating the sequestration of germ cell mRNAs into translationally inert ribonucleoprotein particles until they are later translated. Our studies reveal that loss of microRNA-dependent regulation of Arpc5, which controls the distribution of germ cell mRNAs between translationally active and inactive pools, results in abnormal round spermatid differentiation and impaired fertility. Interestingly, Arpc5 functions as a broadly acting translational suppressor, as it inhibits translation initiation by blocking 80S formation and facilitates the transport of mRNAs to chromatoid/P bodies. These findings identify a unique role for actin-associated proteins in translational regulation, and suggest that mRNA-specific and general translational control mechanisms work in tandem to regulate critical germ cell differentiation events and diverse somatic cell functions.
Keywords: spermiogenesis, messenger ribonucleoprotein, Arp2/3, processing body
The spatial and temporal control of protein synthesis in mammalian cells is ensured by checkpoints present at both the transcriptional and translational levels. This is particularly essential during postmeiotic male germ cell maturation, when transcription ceases and newly synthesized mRNAs are stored in translationally inert ribonucleoprotein complexes (mRNPs) (1, 2). The sequestration of germ cell transcripts into mRNPs ensures that necessary mRNAs are synthesized before transcriptional arrest, whereas translation is delayed until protein expression is required. This dynamic translational regulatory mechanism facilitates timely expression of genes essential for proper germ cell differentiation and normal sperm development. For example, early or late expression of protamine in differentiating germ cells results in compromised chromatin integrity, leading to infertility (3, 4). Although the translational regulation of individual genes has been well-documented (4), the mechanisms and factors that play critical roles in incorporating large numbers of mRNAs into mRNPs, and therefore comprehensive understanding of transcriptional/translational uncoupling during haploid germ cell maturation, remain elusive.
MicroRNAs (miRNAs) are small noncoding RNAs that regulate gene expression by targeting mRNAs for translational repression and degradation. Studies in many organisms have established that miRNAs play important roles during normal development and differentiation in various tissues (5). In addition to somatic cells, miRNAs have been reported to play an equally crucial role during germ cell development, as independent targeting of the miRNA-processing enzyme Dicer resulted in impaired germ-line stem cell division and primordial germ cell development (6, 7).
In this study, we examined the role of miRNAs in translational repression during haploid male germ cell differentiation. We found that the loss of miRNA expression resulted in altered translational activation of germ cell transcripts, leading to impaired spermatid differentiation and compromised fertility. This is largely attributed to increased expression of actin-associated protein Arpc5 [a member of the Arp2/3 complex known to facilitate actin nucleation (8)], which functions as a general translational suppressor. Our results indicate that miRNA-dependent expression of Arpc5 is critical for translational regulation during the haploid stages of spermatogenesis. These findings suggest that miRNA-mediated, mRNA-specific, and Arpc5-dependent broadly acting bimodal translational suppression mechanisms act in concert to regulate mRNP assembly and temporal expression of specific proteins in differentiating germ cells. Because a large number of all mRNAs (more than 70%) are associated with mRNP particles in adult male germ cells (9), this study will likely have broader implications, as it will provide important insight into the process of general translational repression/activation during late stages of spermiogenesis.
Results
Dicer Inactivation Results in Abnormal Postmeiotic Male Germ Cell Differentiation.
To understand the mechanism of translational suppression events during germ cell maturation, we inactivated the miRNA-processing enzyme Dicer by mating Dicer-flox mice (10) with protamine 1 (Prm1)-cre mice, which express cre preferentially in haploid spermatids (Fig. 1A) (11). Pups produced from mating Prm1-cre|DicerF/FΔ (Dicer FΔ) males with wild-type control females revealed efficient recombination (albeit not 100%; Fig. S1 A–C). Because expression from the Prm1-cre transgene was observed at postnatal day 18 (Fig. S1D), when the first wave of round spermatids appears (12), we expected to see a discernable effect of Dicer inactivation in round spermatids. In agreement with this, Dicer expression was significantly reduced (∼75%) in Dicer FΔ compared with control mouse testis (Fig. S1 E–H). In addition, the levels of all mature miRNAs examined were reduced in Dicer FΔ purified round spermatids (Fig. 1B).
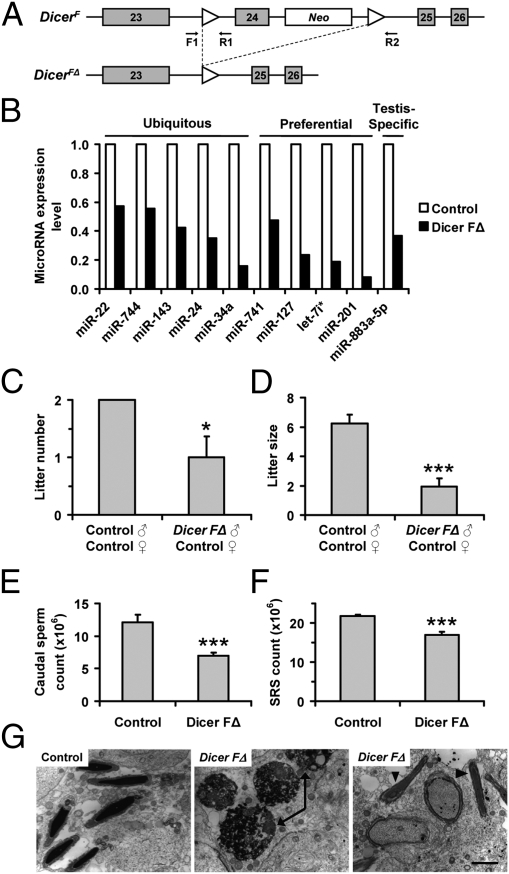
Fig. 1.
Dicer FΔ mice exhibit reproductive defects. (A) Schematic of the Dicer-flox allele before (DicerF) and after (DicerFΔ) recombination. Neo, neomycin resistance cassette. F1, R1, and R2 represent genotyping primers. (B) QRT-PCR on total RNA from purified round spermatids using primers listed in Table S3. Expression normalized to Snora74a. (C and D) Litter number (n = 12; *P < 0.05) and size (n = 12; ***P < 0.0005) from 8-wk timed matings. (E and F) Caudal epididymal sperm count (n = 11; ***P < 0.0005) and sonication-resistant spermatid (SRS) count (n = 6; ***P < 0.0005). (G) TEM of testis sections showing round spermatid apoptosis (Center, arrows) and failure of sperm release (Right, arrowheads) in Dicer FΔ mice. (Scale bar, 0.5 μm.)
Next, we assessed whether Dicer inactivation in round spermatids produced any reproductive defects. Interestingly, Dicer FΔ males were subfertile, as they sired not only fewer litters but also reduced litter sizes (Fig. 1 C and D). The decrease in litter size was only seen in pairings between Dicer FΔ males and control females but not between control males and Dicer FΔ females, indicating the fertility defect was present in males. Dicer FΔ mice also exhibited significantly reduced epididymal sperm counts (Fig. 1E), likely due to a testicular defect, as they also possessed significantly fewer sonication-resistant spermatids (the most differentiated spermatids; Fig. 1F). Transmission electron microscopy (TEM) revealed additional defects in Dicer FΔ testes, including apoptotic round spermatids (Fig. 1G, Center) and the presence of mature spermatids with step 10 spermatids at stage X of the seminiferous epithelial cycle, indicating failure of sperm release (Fig. 1G, Right). Furthermore, sperm from Dicer FΔ mice exhibited various morphogenetic abnormalities including sperm with rounded (globozoospermia) or bent heads (10.8% ± 1.5% versus 5.2% ± 0.6%) and double heads (1.0% ± 0.2% versus 0.1% ± 0.1%; Fig. 2A and Table S1).
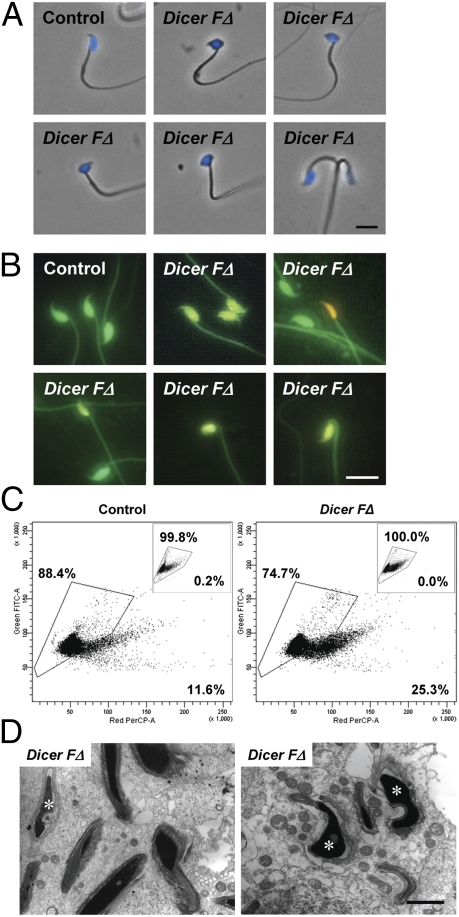
Fig. 2.
Abnormal head morphology and defective chromatin compaction in Dicer FΔ sperm. (A) Phase–contrast microscopy of DAPI-stained sperm. Representative abnormal head morphologies are shown (Upper Center and Right and Lower Left, bent head; Lower Center, rounded head and kinked tail; Lower Right, two-headed). (Scale bar, 10 μm.) (B) AO fluorescence of acid-denaturized sperm. (Scale bar, 10 μm.) (C–E) FACS showing increased number of sperm with chromatin defects in Dicer FΔ mice. (C) Bivariate histograms of green versus red fluorescence for untreated (Insets) and sonicated acid-denatured AO-stained sperm heads (Fig. S2A). Percentage inside the gated area represents sperm with normal chromatin compaction. (D) TEM of testis sections showing abnormal chromatin condensation (Left, asterisk) and acrosomes (Right, asterisks) in Dicer FΔ mice compared with control mice (Fig. 1G). (Scale bar, 0.5 μm.)
As mice with even one-tenth the typical sperm number can sire litters, we surmised that other defects were present in the normal-appearing sperm of Dicer FΔ mice that contributed to the compromised fertility. Because sperm chromatin compaction state is an important independent prognostic characteristic associated with fertility, we examined whether loss of Dicer activity can affect this process. To analyze chromatin compaction, sperm were acid-treated followed by acridine orange (AO) staining. Acid treatment denatures loosely compacted DNA, causing intercalated AO to fluoresce yellow/orange, whereas tightly compacted DNA will fluoresce green (13). Interestingly, Dicer FΔ mice had increased numbers of sperm heads with yellow/orange fluorescence, many of which also exhibited abnormal morphology, indicating compromised chromatin integrity (Fig. 2B). FACS of AO- and propidium iodide (PI)-stained sperm heads further confirmed impaired chromatin integrity in Dicer FΔ mice (Fig. 2C and Fig. S2 B and C). Moreover, sperm head defects and abnormal chromatin compaction in Dicer FΔ mice were also recapitulated in TEM studies (Fig. 2D). Taken together, the increased apoptosis, acrosomal defects, improper chromatin compaction, and failure in spermiation may account for the subfertile phenotype observed in Dicer FΔ males. The lack of total sterility in Dicer FΔ mice is possibly due to incomplete cre-lox recombination, resulting in residual Dicer expression (Fig. S1 D–H). Dicer expression from spermatids that escaped recombination may partially compensate for Dicer loss (through cytoplasmic bridges). The residual level of Dicer (Fig. S1 E–H) may be sufficient for Dicer FΔ males to maintain some fertility.
Impaired Translational Activation of Germ Cell Transcripts in Dicer FΔ Mice.
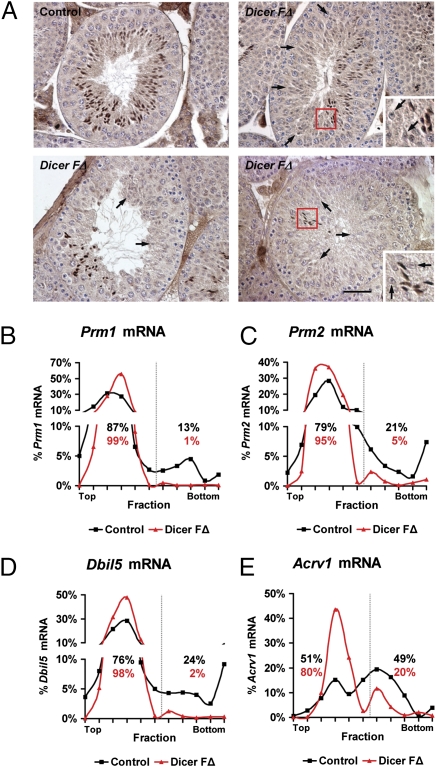
To explore the mechanism underlying compromised spermatid differentiation and chromatin integrity observed in Dicer FΔ mice, we examined the expression levels and patterns of testicular basic nuclear proteins: transition proteins and protamines. These proteins ensure proper chromatin condensation in differentiating spermatids (14), the disruption of which leads to compromised fertility (3, 15). Surprisingly, although expression levels and processing were unaffected (Fig. S3 A and B), Dicer FΔ testis sections showed mosaic expression patterns of Prm1 in elongating spermatids (Fig. 3A). This is interesting, as mosaic expression indicating impaired protamine translational activation was also reported in Dicer-interacting protein TRBP knockout mice (4). Because many postmeiotic germ cell transcripts are stored for up to a week after transcription in translationally inert mRNPs, we hypothesized that impaired protamine translational activation may be due to altered cytoplasmic distribution of protamine mRNAs. We used sucrose-gradient centrifugation to examine the levels of protamine mRNAs in RNP, monosome, and polysome subcellular fractions, identified based on the presence of ribosomal protein S6, a component of the 40S ribosomal subunit (Fig. S3C) (16). Quantitative (Q)RT-PCR analyses of fractionated Dicer FΔ total testis lysates revealed a shift of Prm1 and Prm2 mRNA away from polysome into RNP/40S fractions (Fig. 3 B and C). These findings suggest that altered translational activation of protamine in Dicer FΔ mice may be due to increased sequestration of mRNA into translationally inert complexes, resulting in their inaccessibility to the translational machinery. To our surprise, in addition to protamine, other haploid germ cell-specific mRNAs (Dbil5 and Acrv1), but not pachytene- (Sycp3) or Sertoli-specific (Rhox5) mRNAs, showed increased sequestration away from polysomes into RNP/40S in Dicer FΔ mice (Fig. 3 D and E and Fig. S3 D and E), suggesting miRNA-dependent regulation is a part of a more general control mechanism during postmeiotic germ cell differentiation.
Fig. 3.
Increased sequestration of haploid germ cell-specific transcripts into mRNPs in Dicer FΔ mice. (A) Immunohistochemistry on testis sections using anti-Prm1. Arrows indicate spermatids lacking Prm1 expression in Dicer FΔ testis. Areas in red boxes are magnified (Insets). (Scale bar, 50 μm.) (B–E) QRT-PCR of fractionated lysates was performed using the indicated primers. Dotted line indicates separation between complexes. Percentages indicate the amount of the specified mRNA in the fraction compared with the total amount of the specified mRNA in all fractions.
Loss of miRNA-Dependent Regulation of Actin-Associated Protein Arpc5 Impairs Protamine Translational Activation.
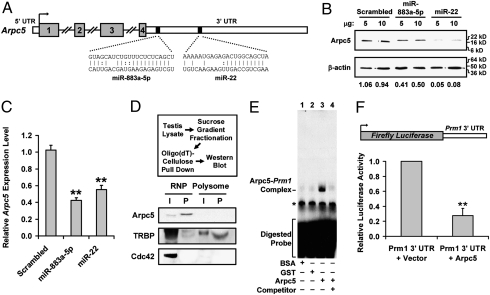
To address the underlying mechanism of altered haploid germ cell mRNA distribution, we performed gene expression analysis on testes from control and Dicer FΔ mice. Among the genes with altered expression in Dicer FΔ mice, we were primarily interested in those that showed up-regulation and contained predicted binding sites for miRNAs with testis-preferential or -specific expression, as they may be direct targets of these miRNAs in postmeiotic germ cells. We focused on one such gene: actin-related protein Arpc5, a subunit of the actin-nucleating Arp2/3 complex (8), which showed significantly higher expression in Dicer FΔ germ cells (Table S2). Actin-associated proteins are known to be a part of mRNPs and have well-documented roles in mRNA transport and localized translation (17). Furthermore, the Arpc5 3′ UTR is predicted to contain binding sites for miR-22/883a-5p (Fig. 4A), two miRNAs preferentially expressed in testicular germ cells (18) that show highly reduced expression in Dicer FΔ haploid germ cells (Fig. 1B). To determine whether miR-22/883a-5p are bona fide regulators of Arpc5, we cotransfected luciferase-Arpc5 3′ UTR reporters with miR-22/883a-5p mimics. Luciferase activity was significantly reduced by the miRNA mimics (Fig. S4A), suggesting that Arpc5 expression is regulated by binding sequences within its 3′ UTR. We further substantiated these findings by determining the effect of the miRNA mimics on endogenous Arpc5. Western blot analyses revealed that exogenous expression of the miRNA mimics resulted in reduced levels of Arpc5 protein (Fig. 4B). In addition, Arpc5 mRNA levels also decreased in mimic-transfected cells (Fig. 4C), indicating that these miRNAs regulate Arpc5 expression at both the mRNA and protein levels.
Fig. 4.
miR-22/883a-5p–targeted Arpc5 interacts with mRNA and mediates translational suppression. (A) Schematic of putative miR-22/883a-5p binding sites. (B) NIH 3T3 cells transfected with the indicated mimic analyzed by Western blot. Gel photograph is representative of three independent experiments. (C) QRT-PCR of RNA isolated from cells transfected as in C. Expression is normalized to Rpl19 (n = 3; **P < 0.005). (D) Arpc5 associates with mRNA in RNP fractions. Pooled testis RNP and polysome fractions were subjected to RNP capture assay (Upper). Bound proteins were analyzed by Western blot (Lower). TRBP is known to interact with Prm1 mRNA (positive control), whereas Cdc42 does not interact with mRNA (negative control). I, input; P, pull down. (E) RNA-EMSA using in vitro transcribed, radiolabeled Prm1 3′ UTR incubated with BSA (lane 1), GST (lane 2), or recombinant Arpc5 (lanes 3 and 4). Lane 4 was preincubated with a 10-fold excess of cold competitor. Arpc5–Prm1 complex is indicated; a star indicates a nonspecific complex. (F) Arpc5 overexpression inhibits reporter activity. HeLa cells were cotransfected with the luciferase-Prm1 3′ UTR construct (Upper), Renilla luciferase, and either vector or FLAG-Arpc5. Values are normalized against Renilla luciferase activity (n = 3; **P < 0.005).
Next, to examine whether Arpc5 has a role in regulating protamine mRNA compartmentalization, we identified the subcellular localization of Arpc5 protein in testicular cells. RNP capture assay revealed that Arpc5 was not only exclusively enriched in RNP fractions but was also associated with poly(A) RNA (Fig. 4D). To determine whether Arpc5 is part of the protamine–mRNP complex, we performed RNA electrophoretic mobility shift assay (RNA-EMSA) using Prm1 3′ UTR 69–152 as the template, as several proteins bind to regulatory sites in this region (4, 19). In vitro transcribed, radiolabeled probe produced a protein–RNA complex with recombinant Arpc5 whereas cold competition with excess unlabeled probe competed out this complex (Fig. 4E, lanes 3 and 4), suggesting the Arpc5–Prm1 interaction is specific. RNA-EMSA supershift against Arpc5 further confirmed the Arpc5–Prm1 interaction (Fig. S4B, lane 4, arrowheads). To determine whether this interaction had any regulatory significance, we performed reporter assay on cells expressing luciferase-Prm1 3′ UTR in the presence of Arpc5. Interestingly, Arpc5 overexpression resulted in significantly reduced luciferase activity from the reporter construct (Fig. 4F), suggesting that Arpc5 suppressed protamine translation by sequestering protamine mRNA away from polysomes. Importantly, reporter mRNA levels were not affected, suggesting that Arpc5-dependent regulation does not occur at the level of mRNA turnover.
Arpc5 Is a Broadly Acting Translational Suppressor.
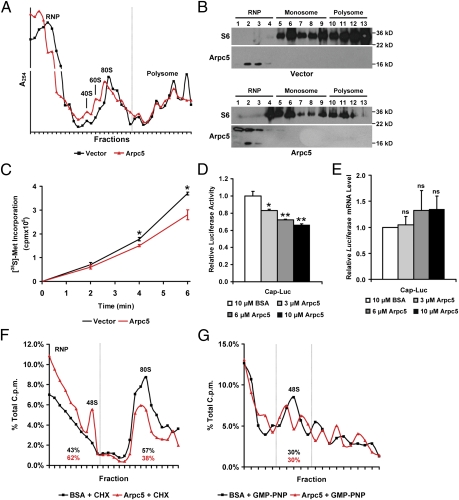
Because germ cell transcripts were sequestered away from polysome fractions in Dicer FΔ mice and Arpc5 is highly expressed in somatic cells (20), we hypothesized that Arpc5 may be a broadly acting translational suppressor. Interestingly, similar to germ cells, Arpc5 overexpression in somatic cells also resulted in increased sequestration of RNA away from polysomes into RNP/40S fractions (Fig. 5A). Consistent with this, S6 shifted from polysomes into RNP/monosome fractions in cells overexpressing Arpc5 (Fig. 5B). Additional evidence that Arpc5 may function to suppress translation is provided by decreased rates of [35S]methionine incorporation in cells overexpressing Arpc5 (Fig. 5C). To further establish that Arpc5 directly functions in translational suppression, we asked whether it repressed translation in vitro. Addition of increasing concentrations of recombinant Arpc5 to reporter mRNA-programmed rabbit reticulocyte lysate (RRL) significantly reduced luciferase activity in a dose-dependent manner (Fig. 5D). Importantly, the translational suppression activity was not due to destabilization of luciferase mRNA (Fig. 5E). These results suggest that Arpc5 may be involved in rate-limiting steps to determine the fate of localizing mRNA.
Fig. 5.
Arpc5 is a broadly acting translational suppressor. (A) Polysome profile of HeLa cells transfected with vector or Arpc5 as in Fig. S3C. Results are representative of four independent experiments. RNP, 40S/60S, 80S, and polysome fractions are indicated. Dotted line indicates separation between complexes. (B) Arpc5 overexpression leads to shift of S6 from polysome to RNP/80S fractions. Protein isolated from pooled fractions of vector or Arpc5-transfected cells was analyzed by Western blot. The lower Arpc5 band in Arpc5-transfected cells is endogenous protein, whereas the upper band is expressed from transfected FLAG-Arpc5. Gel photograph is representative of three independent experiments. (C) Incorporation of [35S]methionine in HeLa cells transfected with vector or Arpc5. The time after label addition is indicated (n = 4; *P < 0.05). (D) Relative luciferase activity from in vitro transcribed luciferase mRNA assayed in RRL. The indicated amount of recombinant Arpc5 or BSA was added. Luciferase activity is normalized to translation in the presence of BSA (n = 2; *P < 0.05, **P < 0.01). (E) QRT-PCR of RNA from D. Expression is normalized to Rpl19 [n = 2; no significant change (ns)]. (F and G) Arpc5 inhibited 80S but not 48S formation in vitro. RRL programmed with radiolabeled luciferase reporter and CHX (F) or GMP-PNP (G) in the presence of recombinant Arpc5 or BSA. Lysates were fractionated and cpm was measured for each fraction. RNP, 48S, and 80S complexes (F) or 48S complex (G) are indicated. Percentages indicate cpm of the fraction compared with the total cpm of all fractions.
Arpc5 Inhibits 80S Formation.
Translation initiation can be broadly divided into three steps. The first step requires association of eukaryotic translation initiation factor (eIF)4F with the cap structure. Second, eIF4F recruits the 43S preinitiation complex (which includes 40S), resulting in the 48S preinitiation complex. Finally, the 48S preinitiation complex scans for the AUG start codon, and upon binding recruits the 60S ribosomal subunit to form the 80S ribosome. To gain insight into the specific step at which Arpc5 may be affecting translation, RRL (with the addition of Arpc5) was programmed with radiolabeled luciferase reporter mRNA in the presence of two translational inhibitors: cycloheximide (CHX), which allows for the assembly of the 80S but blocks translation elongation (21), and GMP-PNP, which blocks recruitment of the 60S and causes accumulation of the 48S on mRNAs (22). Interestingly, addition of recombinant Arpc5 reduced CHX-mediated 80S accumulation and instead caused accumulation of 48S (Fig. 5F). Consistent with this, Arpc5 exhibited no effect on GMP-PNP–induced 48S accumulation (Fig. 5G). These results suggested that Arpc5 represses translation in vitro by inhibiting the ability of 80S to form on mRNA.
Arpc5 Colocalizes with Chromatoid/P-Body Components.
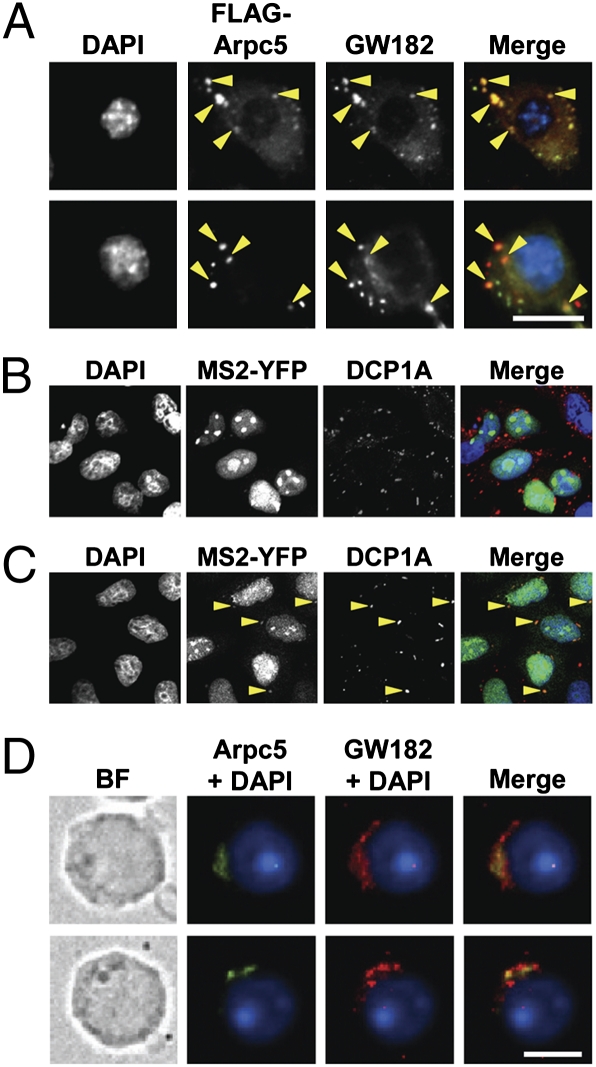
Nontranslating mRNAs have been reported to assemble into mRNPs that accumulate within P bodies. Because Arpc5 binds to mRNA in RNP fractions and suppresses translation, we reasoned that Arpc5-associated mRNA may accumulate in P bodies. Supporting this notion, endogenous (Fig. S5) as well as ectopically expressed Arpc5 (Fig. 6A) colocalized with P-body component GW182. To further substantiate these results, we performed colocalization studies with P-body component DCP1A in cells ectopically expressing MS2-YFP fusion protein and MS2 transcript in the presence of Arpc5. Whereas no colocalization was observed in vector-transfected cells, Arpc5 overexpression resulted in MS2-YPF/DCP1A colocalization, suggesting that Arpc5 mediates the transport of MS2-YFP–tagged MS2 transcripts to P bodies (Fig. 6 B and C). Similar to P bodies, Arpc5 also colocalized with GW182 in chromatoid bodies, germ cell cytoplasmic foci that are compositionally and functionally analogous to P bodies (Fig. 6D). Taken together, these results suggest that Arpc5 is a translational regulator that facilitates movement of translationally inert mRNAs into chromatoid/P bodies.
Fig. 6.
Arpc5 associates with chromatoid/P bodies in haploid germ cells/somatic cells. (A) GW182 colocalizes with Arpc5. HeLa cells transfected with FLAG-Arpc5 (red), endogenous GW182 (green), and nuclei (blue), shown in black and white for clarity. Arrowheads indicate merge of green and red signals (yellow). (Scale bar, 10 μm.) (B and C) Arpc5 facilitates transport of MS2 transcripts to P bodies. HeLa cells were cotransfected with constructs expressing MS2-YFP fusion protein, MS2 transcript, and either vector (B) or Arpc5 (C). MS2 mRNA tagged with MS2-YFP protein (green), endogenous DCP1A (red), and nuclei (blue), shown in black and white for clarity. Arrowheads indicate the merge of green and red signals (yellow; C). Note the lack of colocalization in vector-transfected cells (B). (D) Localization of Arpc5 to chromatoid bodies. Arpc5 (green), GW182 (red), and nuclei (blue) in purified round spermatids. BF, bright field. (Scale bar, 5 μm.)
Discussion
Subcellular localization of mRNAs coupled with localized translation is an important posttranscriptional mechanism that controls faithful spatial and temporal protein synthesis. This is largely accomplished by the packaging of mRNAs into mRNP particles that function to transiently silence translation during transport (17). This regulatory step is especially critical during meiotic and postmeiotic germ cell maturation, when transient translational suppression of mRNAs ensures proper development of differentiating germ cells (1, 2). Although individual components implicated in the localization and translation of specific proteins have been identified (17), a broadly acting control mechanism coordinating translational suppression and mRNP assembly with mRNA transport remains unclear. In this study, we established that the interaction between miRNAs and actin-associated protein Arpc5 sets the stage for the uncoupling of transcription and translation during postmeiotic male germ cell maturation. Our studies reveal that miRNA-dependent regulation of Arpc5, which acts as a translational suppressor, is crucial in this process. We show that Arpc5 colocalizes with chromatoid/P-body component GW182 and inhibits translation initiation by blocking 80S formation, leading to sequestration of mRNAs into translationally inert complexes.
Our results suggest that the miRNA-mediated, mRNA-specific, and Arpc5-dependent broadly acting translational control mechanisms create a robust translational regulation system essential for proper germ cell differentiation. Because most translationally suppressed mRNAs in germ cells are overrepresented, it is conceivable that global translational suppression may be necessary to prevent the deleterious effects of protein overproduction (2), whereas miRNA-specific translational suppression may provide an additional layer of safeguard control by fine-tuning the expression of individual overrepresented mRNAs. In addition, translational fine-tuning may facilitate the turnover of specific key molecules required for regulating a wide range of downstream events. Consistent with this, we show that miR-22/883a-5p–mediated regulation of Arpc5 is critical for controlling the expression of genes known to be involved in chromatin remodeling during haploid germ cell differentiation (3, 15). Another possibility is that mRNA-specific translational control can regulate a cell's ability to respond to extracellular cues, such as Sertoli cell- or androgen-dependent signaling during postmeiotic germ cell differentiation (23, 24). In addition, our results describing the generality of Arpc5 translational regulation suggest that similar to germ cells, the miRNA/Arpc5-mediated bimodal translational suppression mechanism may also be critical for restricting the precocious translation of proteins in somatic cells (17).
A considerable body of evidence shows that actin-associated proteins are intimately involved in mRNA transport and localized protein synthesis. Because localizing mRNAs must be transported as translationally silenced mRNPs (17), our results showing Arpc5 interaction with mRNAs in mRNPs and its role as a translational suppressor suggest that in addition to being the regulators of mRNA transport, actin-associated proteins may be actively engaged in blocking premature translation during transport. One possible mechanism for Arpc5-mediated translational suppression is that Arpc5 facilitates the assembly of a repressor complex that sequesters mRNAs into translationally inert mRNPs and therefore precludes accessibility to translational machinery (Fig. S6). Consistent with this, our results show that Arpc5 colocalized with GW182 in chromatoid/P bodies. It is likely that association of Arpc5-containing mRNPs with chromatoid/P bodies may further facilitate the aggregation of repressor complexes, resulting in more quiescent and translationally inert mRNPs.
An alternative mechanism could be that Arpc5 competitively binds with factors that promote 60S joining or recruit corepressors that inhibit 80S formation. Examples of such repressors include GW182, which interacts with poly(A)-binding protein PABC1 and interferes with the ability of PABC1 to stimulate 80S formation (25). Furthermore, translational repressors (such as ZBP1 and Puff) have been shown to block 60S joining by competitively binding to eIF5B (Fig. S6) (26, 27). Future studies aimed at identifying Arpc5-interacting proteins in germ/somatic cells will likely reveal the molecular composition and specific physiological cues required for promoting Arpc5-mediated translational suppression.
In summary, our findings suggest that miRNA-dependent regulation of Arpc5 plays an important role in ensuring that protein synthesis occurs in a timely manner and is restricted to target destinations in mammalian cells. The role of Arpc5 as a regulator of mRNA distribution into translationally active and inactive pools suggests that subtle alterations in the regulation (i.e., miRNA-dependent) of key molecules (such as Arpc5) have significant consequences in deciding the fate of mRNAs. Our study highlights a unique posttranscriptional role for actin-associated proteins known to be intimately involved at various levels of gene regulation, including chromatin remodeling, transcription, mRNA transport, and localized translation (17).
Materials and Methods
Animals, Genotyping, and Reproductive Phenotype Analyses.
Animal experiments were performed in accordance with the National Institutes of Health Guide for the Care and Use of Laboratory Animals and with the approval of the Institutional Animal Care and Use Committee at the University of Texas Health Science Center at San Antonio. Eight-week timed matings, caudal sperm count, and sonication-resistant spermatid count were conducted as previously described (28).
Elutriation and RNA/Protein Analyses.
Elutriation and RNA/protein analyses have been previously described (28, 29).
General Methods.
Other general methods are described in detail in SI Materials and Methods.
Statistical Analyses.
All values and error bars in graphs are means ± SEM; respective n values are indicated in the figure legends; P values were determined by two-tailed Student's t tests.
Supplementary Material
Acknowledgments
We thank Yidong Chen, Krystle Harris, Jennifer Rebeles, and Jennifer Martindale for assistance with experiments and evaluation of data. We also thank Daniel Schoenberg, Rod Balhorn, Marvin Fritzler, and Peter Sarnow for generously providing us with reagents. Thanks to Alexander Pertsemlidis for critical reading of the manuscript. M.K.R. was supported by National Institute of Child Health and Human Development Grant HD057118, National Institutes of Health (NIH); and S.S.S. and M.G. were supported by the Intramural Research Program, National Institute on Aging, NIH.
Footnotes
The authors declare no conflict of interest.
This article is a PNAS Direct Submission.
This article contains supporting information online at www.pnas.org/lookup/suppl/doi:10.1073/pnas.1117837109/-/DCSupplemental.
References
- 1.Hecht NB. Post-meiotic gene expression during spermatogenesis. Prog Clin Biol Res. 1988;267:291–313. [PubMed] [Google Scholar]
- 2.Kleene KC. A possible meiotic function of the peculiar patterns of gene expression in mammalian spermatogenic cells. Mech Dev. 2001;106(1-2):3–23. doi: 10.1016/s0925-4773(01)00413-0. [DOI] [PubMed] [Google Scholar]
- 3.Lee K, Haugen HS, Clegg CH, Braun RE. Premature translation of protamine 1 mRNA causes precocious nuclear condensation and arrests spermatid differentiation in mice. Proc Natl Acad Sci USA. 1995;92:12451–12455. doi: 10.1073/pnas.92.26.12451. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Zhong J, Peters AH, Lee K, Braun RE. A double-stranded RNA binding protein required for activation of repressed messages in mammalian germ cells. Nat Genet. 1999;22(2):171–174. doi: 10.1038/9684. [DOI] [PubMed] [Google Scholar]
- 5.Bernstein E, et al. Dicer is essential for mouse development. Nat Genet. 2003;35:215–217. doi: 10.1038/ng1253. [DOI] [PubMed] [Google Scholar]
- 6.Hayashi K, et al. MicroRNA biogenesis is required for mouse primordial germ cell development and spermatogenesis. PLoS One. 2008;3:e1738. doi: 10.1371/journal.pone.0001738. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Maatouk DM, Loveland KL, McManus MT, Moore K, Harfe BD. Dicer1 is required for differentiation of the mouse male germline. Biol Reprod. 2008;79:696–703. doi: 10.1095/biolreprod.108.067827. [DOI] [PubMed] [Google Scholar]
- 8.Goley ED, Welch MD. The ARP2/3 complex: An actin nucleator comes of age. Nat Rev Mol Cell Biol. 2006;7:713–726. doi: 10.1038/nrm2026. [DOI] [PubMed] [Google Scholar]
- 9.Kleene KC. Multiple controls over the efficiency of translation of the mRNAs encoding transition proteins, protamines, and the mitochondrial capsule selenoprotein in late spermatids in mice. Dev Biol. 1993;159:720–731. doi: 10.1006/dbio.1993.1277. [DOI] [PubMed] [Google Scholar]
- 10.Harfe BD, McManus MT, Mansfield JH, Hornstein E, Tabin CJ. The RNaseIII enzyme Dicer is required for morphogenesis but not patterning of the vertebrate limb. Proc Natl Acad Sci USA. 2005;102:10898–10903. doi: 10.1073/pnas.0504834102. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.O'Gorman S, Dagenais NA, Qian M, Marchuk Y. Protamine-Cre recombinase transgenes efficiently recombine target sequences in the male germ line of mice, but not in embryonic stem cells. Proc Natl Acad Sci USA. 1997;94:14602–14607. doi: 10.1073/pnas.94.26.14602. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Bellvé AR, et al. Spermatogenic cells of the prepuberal mouse. Isolation and morphological characterization. J Cell Biol. 1977;74(1):68–85. doi: 10.1083/jcb.74.1.68. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Kosower NS, Katayose H, Yanagimachi R. Thiol-disulfide status and acridine orange fluorescence of mammalian sperm nuclei. J Androl. 1992;13:342–348. [PubMed] [Google Scholar]
- 14.Balhorn R, Weston S, Thomas C, Wyrobek AJ. DNA packaging in mouse spermatids. Synthesis of protamine variants and four transition proteins. Exp Cell Res. 1984;150:298–308. doi: 10.1016/0014-4827(84)90572-x. [DOI] [PubMed] [Google Scholar]
- 15.Shirley CR, Hayashi S, Mounsey S, Yanagimachi R, Meistrich ML. Abnormalities and reduced reproductive potential of sperm from Tnp1- and Tnp2-null double mutant mice. Biol Reprod. 2004;71:1220–1229. doi: 10.1095/biolreprod.104.029363. [DOI] [PubMed] [Google Scholar]
- 16.Wool IG. The structure and function of eukaryotic ribosomes. Annu Rev Biochem. 1979;48:719–754. doi: 10.1146/annurev.bi.48.070179.003443. [DOI] [PubMed] [Google Scholar]
- 17.Besse F, Ephrussi A. Translational control of localized mRNAs: Restricting protein synthesis in space and time. Nat Rev Mol Cell Biol. 2008;9:971–980. doi: 10.1038/nrm2548. [DOI] [PubMed] [Google Scholar]
- 18.Song R, et al. Many X-linked microRNAs escape meiotic sex chromosome inactivation. Nat Genet. 2009;41:488–493. doi: 10.1038/ng.338. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Schumacher JM, Lee K, Edelhoff S, Braun RE. Spnr, a murine RNA-binding protein that is localized to cytoplasmic microtubules. J Cell Biol. 1995;129:1023–1032. doi: 10.1083/jcb.129.4.1023. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Millard TH, Behrendt B, Launay S, Fütterer K, Machesky LM. Identification and characterisation of a novel human isoform of Arp2/3 complex subunit p16-ARC/ARPC5. Cell Motil Cytoskeleton. 2003;54(1):81–90. doi: 10.1002/cm.10087. [DOI] [PubMed] [Google Scholar]
- 21.Schneider-Poetsch T, et al. Inhibition of eukaryotic translation elongation by cycloheximide and lactimidomycin. Nat Chem Biol. 2010;6:209–217. doi: 10.1038/nchembio.304. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Peterson DT, Safer B, Merrick WC. Role of eukaryotic initiation factor 5 in the formation of 80 S initiation complexes. J Biol Chem. 1979;254:7730–7735. [PubMed] [Google Scholar]
- 23.He WW, Kumar MV, Tindall DJ. A frame-shift mutation in the androgen receptor gene causes complete androgen insensitivity in the testicular-feminized mouse. Nucleic Acids Res. 1991;19:2373–2378. doi: 10.1093/nar/19.9.2373. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Mruk DD, Cheng CY. Sertoli-Sertoli and Sertoli-germ cell interactions and their significance in germ cell movement in the seminiferous epithelium during spermatogenesis. Endocr Rev. 2004;25:747–806. doi: 10.1210/er.2003-0022. [DOI] [PubMed] [Google Scholar]
- 25.Zekri L, Huntzinger E, Heimstädt S, Izaurralde E. The silencing domain of GW182 interacts with PABPC1 to promote translational repression and degradation of microRNA targets and is required for target release. Mol Cell Biol. 2009;29:6220–6231. doi: 10.1128/MCB.01081-09. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Deng Y, Singer RH, Gu W. Translation of ASH1 mRNA is repressed by Puf6p-Fun12p/eIF5B interaction and released by CK2 phosphorylation. Genes Dev. 2008;22:1037–1050. doi: 10.1101/gad.1611308. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Hüttelmaier S, et al. Spatial regulation of β-actin translation by Src-dependent phosphorylation of ZBP1. Nature. 2005;438:512–515. doi: 10.1038/nature04115. [DOI] [PubMed] [Google Scholar]
- 28.Chang YF, Lee-Chang JS, Harris KY, Sinha-Hikim AP, Rao MK. Role of β-catenin in post-meiotic male germ cell differentiation. PLoS One. 2011;6:e28039. doi: 10.1371/journal.pone.0028039. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Chang YF, Lee-Chang JS, Panneerdoss S, MacLean JA, II, Rao MK. Isolation of Sertoli, Leydig, and spermatogenic cells from the mouse testis. Biotechniques. 2011;51:341–344. doi: 10.2144/000113764. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.