Little is known about the genome of Anthurium other than chromosome observations, which frequently indicate supernumerary (“B”) chromosomes. New genome size estimates for 34 species and nine cultivars presented here provide insights into genome organization and evolution in this very large genus.
Abstract
Background and aims
Anthurium is an important horticultural crop from the family Araceae, order Alismatales, a lineage considered to have diverged from other monocots prior to the cereals. Genome size and its distribution in Anthurium were investigated to gain a basic understanding of genome organization in this large genus and to forge a firm foundation for advancement of molecular approaches for the study of Anthurium. Currently, genome size estimates have been reported for only two Anthurium samples.
Methodology
Bulk nuclear DNA content estimates were obtained by flow cell cytometry using leaf tissue collected from Anthurium species of different subgeneric groups and from commercial cultivars. The most current and well-supported topology of subgeneric, sectional relationships was applied to present genome size estimates in the context of reported chromosome counts, karyotypes, putative phylogenetic relationships, observed phenotypes and pedigree.
Principal results
Genome size estimates based on bulk nuclear DNA content for 77 accessions representing 34 species and 9 cultivars were obtained, including initial estimates for 33 Anthurium species, and both the smallest (Anthurium obtusum; Tetraspermium) and largest (Anthurium roseospadix; Calomystrium) Anthurium genome sizes reported to date. Genome size did not distinguish any subgeneric section, but ranged 5-fold (4.42–20.83 pg/2 C) despite consistent 2N= 30 chromosome counts. Intraspecies genome size variation >20 % is reported for Anthurium ravenii, A. watermaliense and A. gracile.
Conclusions
Genome size estimates for Anthurium species spanning 13 recognized subgeneric sections indicate that genome size does not generally correlate with chromosome count or phylogenetic relationships. Mechanisms of genome expansion and contraction, including amplification and reduction of repetitive elements, polyploidy, chromosome reorganization/loss, may be involved in genome evolution in Anthurium as in other species. The new information on Anthurium genome sizes provides a platform for molecular studies supporting further research on genome evolution as well as cultivar development.
Introduction
Anthurium is the most speciose genus in the Araceae, a monocot family defined by its unique inflorescence composed of a spadix and spathe. The spadix holds hundreds of minute flowers compacted on a spike, which is subtended by a more or less showy sterile leaf-like organ, the spathe. Anthurium is comprised of ∼900 published and 1500 estimated species endemic to the neotropical zones of northern Mexico and south through Central America to southern Brazil, and on the Caribbean Islands (Croat 1988, 1989; Mayo et al. 1997; Govaerts et al. 2011; Boyce and Croat 2012). Anthurium andraeanum, native to Colombia, was first introduced to the island of Oahu in 1889, where it flourished and became widely cultivated by amateur breeders and hobbyists who developed many attractive new varieties throughout the 1940s. Beginning in 1950, an intensive breeding programme at the University of Hawai`i at Manoa yielded many unique and improved cultivars using selective breeding, hybridization and in vitro propagation of Anthurium species. The novel colours and forms, as well as desirable horticultural attributes, generated in these cultivars contributed to the dominance of the anthurium industry by Hawaiian growers for much of the second half of the 20th century (Kamemoto and Kuehnle 1996). We anticipate that future improvements to anthurium cultivars will utilize molecular resources developed to contribute to the basic understanding of this large genus while supporting applied science.
Genome size has implications for molecular biology work, genomics and overall successful implementation as a study organism. Genome size is also correlated with seed mass, cell size, stomatal size, stomatal density, length of cell cycle and a host of derivative phenotypes important for plant success (Beaulieu et al. 2007, 2008; Leitch et al. 2007; Hodgson et al. 2010). Genera with larger genome sizes have limited photosynthetic rates, tend to have limited distribution and tend to be less speciose (Knight et al. 2005). Genome size in angiosperms varies ∼1000-fold (Leitch et al. 2005) with the largest genomes found among the monocots (Leitch et al. 2010; Zonneveld 2010). Variations in angiosperm DNA content (Bennett and Leitch 2005) have been interpreted in a robust phylogenetic context to reconstruct genome size evolution, revealing the ancestral genome size to be relatively small (1.4 pg/1 C) (Soltis et al. 2003). Significant increases in genome size have been attributed to polyploidy and to amplification of repetitive DNA content (SanMiguel et al. 1996; Vicient et al. 1999; Hawkins et al. 2006). Secondary downsizing in lineages embedded within clades having larger genome sizes counters the overall trend towards genome size growth (Leitch et al. 1998; Bennetzen et al. 2005).
In the family Araceae, genome sizes tend to be moderate, including Orontium aquaticum (30 pg/2 C), a species derived from an early-diverging lineage in Araceae (Cabrera et al. 2008; Cusimano et al. 2011). Genome size estimates have been reported for only two Anthurium accessions: A. schlechtendalii (15.27 pg/2 C) and A. grande Hort. (27.00 pg/2 C) (26 July 2011; http://data.kew.org/cvalues). These are also of moderate size with the genome size estimate for A. schlechtendalii 2.8 times that of Zea mays (5.45 pg/2 C) (Bennett and Smith 1976), and the estimate for the accession A. grande Hort. nearly twice as large as that of A. schlechtendalii (Ghosh et al. 2001). However, in Lemnoideae, the sister group to the true Araceae, the group to which Anthurium belongs (Cabrera et al. 2008; Cusimano et al. 2011), the genera Lemna (1.20 pg/2 C) and Spirodela (0.60 pg/2 C, 0.74 pg/2 C) have quite small genome sizes (Geber 1989). The evolutionary relationship of these lineages suggests that the common ancestor of both Lemnoideae and the true Araceae may have had in place the genomic machinery to secondarily generate species with small genomes and that there may also be Anthurium species with small genome sizes.
Understanding the organization and composition of the Anthurium genome is a prerequisite to the development of molecular resources to support improvement of the Anthurium Hort. complex. We set out to document a wider range of Anthurium genome sizes and interpret them in the context of the most recent phylogenetic analysis of Anthurium species (Carlsen 2011), referencing cytological observations and known mechanisms of genome size evolution to identify trends in genome evolution in Anthurium. We sampled most deeply the natural, easily recognized sections Calomystrium and Cardiolonchium from which the Anthurium Hort. complex mainly derives in order to better explore the extent of evolutionary change and gain insight into the events influencing genome evolution in these clades.
Materials and methods
Nuclear genome size estimations were obtained by flow cell cytometry following the protocol described by Arumuganathan and Earle (1991). Genome sizes were obtained for 81 accessions obtained from botanical gardens, anthurium industry growers and cultivar developers. Tissue samples of 50 mg fully expanded, non-senescing leaf tissue were collected and shipped to arrive for analysis within 24 h of collection. Flow cytometry involves chopping of fresh plant material together with an internal standard (Galbraith et al. 1983). Ideal internal standards display minimal variability (Baranyi and Greilhuber 1996), match as closely as possible the configuration of DNA (e.g. chromosome structure) in the nucleus of the sample, and have a genome size larger than that of the sample, but not >4 times larger (Suda and Leitch 2010; Praça-Fontes et al. 2011). Monocots display a greater variability in chromosome organization and amount of DNA in the genome (Leitch et al. 2010), so we provided the monocots wheat (Triticum aestivum cv. ‘Zak’ 30.55 pg/2 C) and barley (Hordeum vulgare line NE86954 9.69 pg/2 C) as internal standards based on the existing 2-fold range of genome sizes for Anthurium found in the plant DNA C values database (26 July 2011; http://data.kew.org/cvalues). We also provided tobacco (Nicotiana tabacum cv. ‘SR-1’ 9.32 pg/2 C). Each nuclear preparation was sampled four times, under the direction of K. Arumuganathan, at the Flow Cell Core Lab, Benaroya Research Institute at Virginia Mason, 1202 Ninth Avenue, Seattle, WA 98101, USA. The mean bulk nuclear DNA content (2 C) of each sample (expressed as picograms) was based on 1000 scanned G0 + G1 nuclei from sample tissue, compared with nuclei of the internal standard.
Results
Genome sizing
The terms ‘C value’ and ‘genome size’ have specific meaning independent of the number of chromosomes or base pairs in the cell. The term ‘C value’ originally referred to a constant value observed across all different tissue types in animals, whereas the term ‘genome size’ is used to describe the bulk nuclear DNA content of cells, both the more easily obtained somatic (holoploid) cells and also gametic (monoploid) cells (Bennett and Leitch 2005; Greilhuber et al. 2005). The terms 2 C and 1 C have been proposed to distinguish between the DNA content of holoploid somatic cells and monoploid gametes, respectively, and we follow this convention (Greilhuber et al. 2005). Measurements were derived from somatic (i.e. leaf) tissue, therefore values for 1 C are obtained by dividing the measured 2 C value in half and are useful for estimating and comparing the DNA content of the monoploid genome. After excluding four accessions having uncertain provenance, we report the arithmetic mean of four instrument readings of nuclei, ±standard deviation, for 77 accessions, increasing reported genome size estimates for Anthurium spp. by 33 species and 9 cultivars (Table 1).
Table 1.
Genome sizes of accessions sampled, listed alphabetically by Anthurium species, followed by cultivars
| Species name | Accession | Genome sizingstandard | Genome sizea (pg/2 C) ± S.D. (n=4) | DNA content(Mbp/1 C)b |
|---|---|---|---|---|
| A. amnicola Dressler | ABG 19911372 | W | 10.53 ± 0.11 | 5147 |
| MBG 84952 | W | 10.81 ± 0.57 | 5287 | |
| MSBG 1976-0053-002A | W | 9.74 ± 0.21 | 4765 | |
| A. andraeanum Linden | ABG 19911368 | W | 9.59 ± 1.20 | 4688 |
| A. andraeanum sp. aff. (presumed Hort.) | USBG s.n. | W | 8.92 ± 0.05 | 4631 |
| A. antioquiense Engler | MBG 81407a/b | W | 10.35 ± 0.07 | 5059 |
| MSBG 1996-0276A | W | 9.91 ± 0.06 | 4845 | |
| NYBG 1383/78*A*C | W | 9.23 ± 0.14 | 4512 | |
| A. armeniense Croat | MBG 63434e | W | 11.38 ± 0.15 | 5563 |
| MSBG 1979-1055A | W | 12.64 ± 0.35 | 6180 | |
| A. bakeri Hooker f. | MBG 78747c | W | 9.89 ± 0.08 | 4835 |
| NYBG 897/63*A*B*C | W | 8.71 ± 0.35 | 4260 | |
| USBG 77-0090 | W | 9.24 ± 0.13 | 4519 | |
| A. cerrocampanense Croat | MBG 76663/b | W | 11.73 ± 0.18 | 5734 |
| MSBG 1980-0429A | W | 11.16 ± 0.09 | 5455 | |
| A. clavigerum Poepp.& Endl. | MSBG 1991-0174A | W | 13.27 ± 0.17 | 6490 |
| ABG 20011433 | T | 15.26+/-0.28 | 7462 | |
| A. clidemioides Standl. | JBVL 850002 | W | 9.47 ± 0.17 | 4632 |
| MBG 79567 | W | 8.86 ± 0.65 | 4333 | |
| A. coriaceum G. Don | NYBG 574/65*A | W | 14.32 ± 0.17 | 7003 |
| A. esmeraldense Sodiro | ABG 20072399 | W | 8.79 ± 0.17 | 4297 |
| A. flexile ssp muelleri (J.F. Macbr.) Croat & Baker | MBG 100348 | W | 9.46 ± 0.19 | 4628 |
| A. formosum Schott | MSBG 1991-0158A | W | 8.77 ± 0.28 | 4286 |
| A. fragrantissimum Croat | MBG 76860 | W | 6.21 ± 0.39 | 3036 |
| A. gracile (Rudge) Lindl. | ABG 19980680 | W | 9.66 ± 0.20 | 4721 |
| MBG 95664 | W | 13.51 ± 0.22 | 6607 | |
| MSBG 2001-0232A | W | 13.98 ± 0.06 | 6835 | |
| A. gymnopus Griseb. | CJBN 2009.3.213 | A. coriaceum NYBG 574/65*A | 11.21 ± 0.14 | 5482 |
| A. hoffmanni Schott | MBG 66203 | W | 9.56 ± 0.38 | 4674 |
| MSBG 1993-0176A | W | 9.89 ± 0.09 | 4835 | |
| A. kamemotoanum Croat | UH s.n. | W | 9.47 ± 0.07 | 4629 |
| A. lentii Croat & Baker | MBG 35902 | W | 13.96 ± 0.06 | 6824 |
| A. leuconeurum Lemaire | MSBG 1980-1683B | W | 14.35 ± 0.07 | 7016 |
| A. lucens Standl. ex Yuncker | MBG 78702 | W | 13.24 ± 0.50 | 6473 |
| MSBG 1980-1619A | W | 12.71 ± 0.38 | 6216 | |
| NYBG 103/81*A*B | W | 13.16 ± 0.68 | 6435 | |
| A. microspadix Schott | MBG 100186 | W | 12.25 ± 0.13 | 5992 |
| A. nymphaefolium C. Koch & Bouché | MBG 55262 | W | 9.45 ± 0.27 | 4623 |
| A. obtusum (Engl.) Grayum | ABG 19970478 | T | 5.80 ± 0.08 | 2835 |
| MBG 82905 | T | 4.42 ± 0.09 | 2160 | |
| A. ochranthum K. Koch | MBG 69861a | W | 10.87 ± 0.12 | 5317 |
| MBG 75190a | W | 10.64 ± 0.10 | 5201 | |
| A. pittieri Engl. | JBM 84-2010 | T | 5.61 ± 0.03 | 2745 |
| A. radicans K. Koch & A. Haage | ABG 19911495 | W | 15.10 ± 0.32 | 7103 |
| MSBG 1975-0053-0003A | W | 14.52 ± 0.11 | 7103 | |
| USBG 98-2591 | W | 13.46 ± 0.33 | 6581 | |
| A. ravenii Croat & Baker | MBG 74778 | W | 13.32 ± 0.29 | 6515 |
| MSBG 1980-0425A | W | 7.54 ± 0.15 | 3689 | |
| A. roseospadix Croat | MBG 74076 | W | 20.83 ± 0.36 | 10187 |
| A. scandens (Aubl.) ssp. pusillum Engler | USBG 98-1900 | W | 5.12 ± 0.14 | 2506 |
| A. scandens (Aubl.) ssp. scandens Scheffer | ABG 19911433 | W | 9.98 ± 0.57 | 4882 |
| ABG 19980667 | W | 9.67 ± 0.15 | 4726 | |
| MBG 47671 | W | 9.28 ± 0.16 | 4537 | |
| A. schlechtendalii ssp. schlechtendalii Kunth | MBG 78640 | W | 11.54 ± 0.13 | 5643 |
| MSBG 1977-3108A | W | 14.33 ± 0.15 | 7008 | |
| NYBG 933/79*A | W | 12.25 ± 0.18 | 5991 | |
| NYBG 993/93*A-C | W | 12.07 ± 0.10 | 5903 | |
| A. solitarium Schott | JBM 2083-2000 | W | 14.69 ± 0.23 | 7181 |
| MBG 53699 | W | 16.03 ± 0.21 | 7840 | |
| A. warocqueanum J. Moore | ABG 19930478 | W | 8.65 ± 0.14 | 4232 |
| MBG 101538 | W | 8.97 ± 0.09 | 4386 | |
| MSBG 2006-0001A | W | 8.95 ± 0.12 | 4374 | |
| A. watermaliense Hort ex. L.H. Bailey & Nash | MBG 78766 | W | 9.47 ± 0.32 | 4632 |
| MSBG 1977-2832A | T | 7.57 ± 0.02 | 3701 | |
| A. wendlingeri Barroso | ABG 20072507 | W | 7.87 ± 0.20 | 3846 |
| MBG 95418 | W | 7.04 ± 0.15 | 3441 | |
| MSBG 1977-1989A | T | 6.59 ± 0.31 | 3221 | |
| USBG 01-1412 | W | 8.31 ± 0.22 | 4066 | |
| cv. ‘Marian Seefurth’ | HAIA-MS1 | W | 9.22 ± 0.07 | 4509 |
| cv. ‘Midori’ | HAIA-Md1 | W | 9.48 ± 0.25 | 4636 |
| cv. ‘Miss June Purple’ | HAIA-MJP | W | 14.05 ± 0.32 | 6870 |
| cv. ‘New Pahoa Red’ | HAIA-NPR2 | W | 8.93 ± 0.08 | 4367 |
| cv. ‘Princess Aiko’ | NG-10-02-Aiko | W | 9.39 ± 0.27 | 4592 |
| cv. ‘Puanani’ | NG-10-Pu | W | 9.44 ± 0.10 | 4616 |
| cv. ‘Purple Passion’ | NG-10-PP | W | 10.82 ± 0.14 | 5291 |
| cv. ‘Regina’ | NG-10-1-Reg | W | 9.80 ± 0.22 | 4792 |
| cv. ‘Shibori’ | NG-10-Shi | W | 9.73 ± 0.20 | 4758 |
Mbp, million base pairs; W, wheat (Triticum aestivum cv. ‘Zak,’ 30.55 pg/2 C); T, tobacco (Nicotiana tabacum cv. ‘SR-1’ 9.32 pg/2 C); ABG, Atlanta Botanical Garden; CJBN, Conservatoire et Jardins Botaniques de Nancy; JBM, Jardin Botanique de Montréal; JBVL, Jardin Botanique de la Ville de Lyon; MBG, Missouri Botanical Garden; MSBG, Marie Selby Botanical Gardens; NYBG, New York Botanical Garden; UH, University of Hawai`i College of Tropical Agriculture and Human Resources; USBG, United States Botanic Garden; NG, Novelty Greens; HAIA, Hawaiian Anthurium Industry Association.
aGenome size and standard deviation have been rounded to two decimal places.
bMbp/1 C DNA for plant species is based on 1 pg=978 Mbp according to Doležel and Greilhuber (2010) and was calculated prior to rounding genome size (pg/2 C) to two decimal places.
Wheat and barley, both monocots, were chosen as internal standards to better reflect the DNA configuration in our Anthurium samples. However, when barley was used, sample peaks frequently overlapped those of the internal standard, so most genome sizes are reported using wheat as the internal standard. When overlapping peaks prevented interpretation of results with wheat, or if the genome size was closer to that of the eudicot tobacco, results are reported using that species as an internal standard. In one case (A. gymnopus) the sample was processed using a previously evaluated Anthurium species as the internal standard (Table 1) because standards were not available at the time of sampling. Genome sizes for 26 accessions obtained using both tobacco and wheat as internal standards were generally within 10 % of the mean, confirming that the use of either standard produces essentially the same results [see Additional Information].
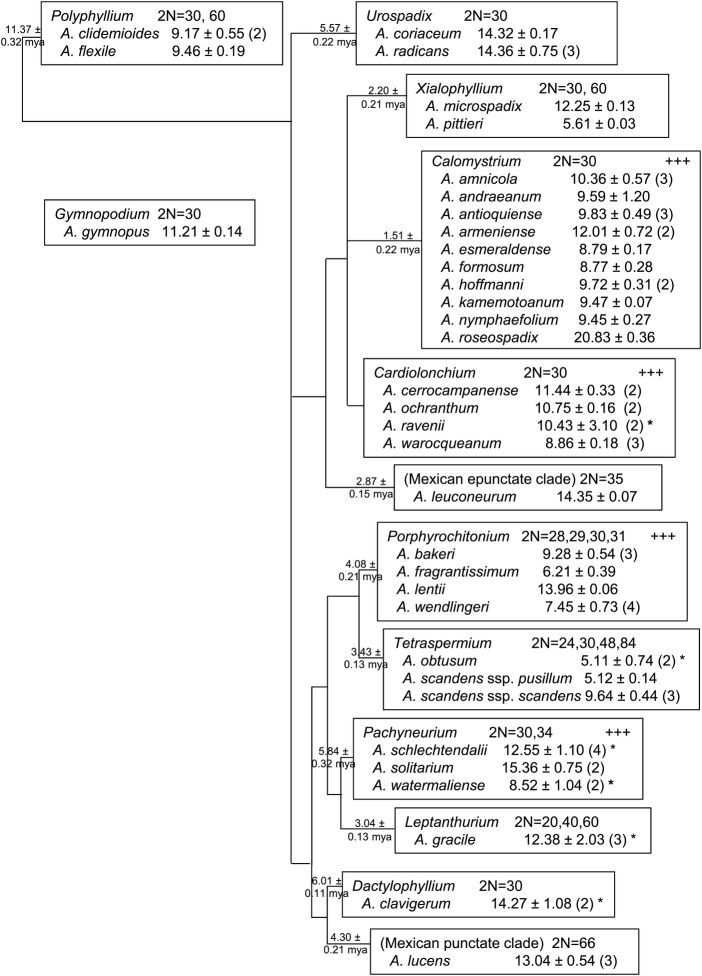
The mean pg/2 C genome size of all accessions sampled for each species is presented (Fig. 1) along with previously published chromosome counts [see Additional Information], organized according to accepted sectional assignments based on traditional characters of morphology, habit, flower/inflorescence, secondary metabolites, karyotype and, most recently, molecular data (Croat 1983, 1991; Croat and Sheffer 1983; Carlsen and Croat 2007; Carlsen 2011). The most recent phylogenetic analysis of 102 samples broadly retains the composition and identity of natural sections, those found least controversial by traditional systematics, and proposes relative relationships among them, which can be extended to other species assigned to those sections. Carlsen sampled 84 species that we did not include, and we sampled 16 species that Carlsen did not include (Carlsen 2011). These are placed according to their existing sectional assignment (Carlsen 2011). The relationship of the monotypic section Gymnopus to other sections has not been determined (Fig. 1).
Fig. 1.
Genome size and chromosome counts of Anthurium species presented by subgeneric sections. Clades of punctate and epunctate Mexican species are proposed new sections (Carlsen 2011). Relative relationships of sections and estimated dates of crown group divergence are indicated at nodes, as per Carlsen (2011). Dates expressed in millions of years ago (mya). Chromosome counts for each section represent values reported for species in that section. ‘+++’ indicates supernumerary chromosomes have been observed in that section. Genome size is reported as the mean of all accessions sampled, in pg/2 C ± S.D., followed by number of accessions sampled in parentheses. Superscript asterisk indicates between-sample (accession) variance >10 % of the species mean.
Published chromosome counts report 2N= 30 (N= monoploid chromosome number) for most Anthurium species (Fig. 1), with frequent reports of supernumerary chromosomes (‘B’, chromosomes, satellites or fragments) (Petersen 1989), distinguished mainly by size, dispensability and behaviour at meioisis (Jones and Rees 1982) [see Additional Information]. Of the species we sampled, recent cytogenetic analyses report supernumerary chromosomes exclusively in species assigned to sections Calomystrium, Cardiolonchium, Porphyrochitonium and Pachyneurium (Fig. 1) (Sharma and Bhattacharyya 1961; Sheffer and Kamemoto 1976; Sheffer and Croat 1983; Marutani et al. 1993; Cotias-de-Oliveira et al. 1999).
Genome sizes in a phylogenetic framework
The organization of Anthurium species chromosome counts and genome sizes according to the accepted subgeneric grouping of species into sections suggests all clades derived from a progenitor lineage having 2N= 30 chromosomes, with occasional polyploidy and cytological variation in most sections. The genome sizes associated with 2N= 30 vary >4-fold (Table 1, Fig. 1). The genome sizes of the two species sampled in section Polyphyllium (9.17–9.46 pg/2 C) are within 10 % of each other, a variance not greater than within-sample variance (K. Arumuganathan, Flow Cell Core Lab, Benaroya Research Institute, Seattle, WA, USA, pers. commun.). There is evidence for polyploidization in section Polyphyllium, with twice as many chromosomes reported for A. flexile (2N= 60) as for A. clidemioides (2N= 30) (Table 1, Fig. 1). The near equivalence of genome size despite the doubled number of chromosomes may reflect an early polyploidization in the Polyphyllium crown group which arose over 11 million years ago (mya) (Fig. 1). The genome sizes of the two species sampled in section Urospadix (14.32–14.36 pg/2 C) are also within 10 % of each other, but are reported to share the same chromosome count (Table 1, Fig. 1) [see Additional Information].
The phylogenetic relationships among the traditionally described sections Xialophyllium, Calomystrium and Cardiolonchium are unclear, although it is certain that section Cardiolonchium as traditionally defined is not monophyletic (Carlsen 2011). The genome sizes of two species sampled in section Xialophyllium are 5.61–12.25 pg/2 C, displaying an approximately proportional relationship between genome size and chromosome count between the two species, suggesting a polyploid event occurring no more than 2.20 mya, the estimated crown group divergence date (Fig. 1).
In section Calomystrium, the most recently established clade (1.5 mya), genome sizes of the 10 species sampled range from 8.77 to 20.83 pg/2 C, a 2.4-fold difference (Figs 1 and 2), despite a consistent number of chromosomes (Fig. 1). Among the Calomystrium species, the genome sizes of A. formosum and A. esmeraldense are very similar (8.77–8.79 pg/2 C), as are the genome sizes of A. nymphaefolium, A. kamemotoanum, A. andraeanum, A. hoffmanni and A. antioquiense (9.45–9.83 pg/2 C) (Table 1, Fig. 1). The genome sizes of A. amnicola and A. armeniense are larger, but the genome size of A. roseospadix (20.83 pg/2 C), the largest Anthurium genome size to date, is two or more times that of most other species sampled in section Calomystrium (Table 1, Fig. 1). Although B chromosomes have been occasionally reported in section Calomystrium, there have been no reports of polyploidy as in other Anthurium sections having such large inter- and intraspecies differences in genome size (Fig. 1).
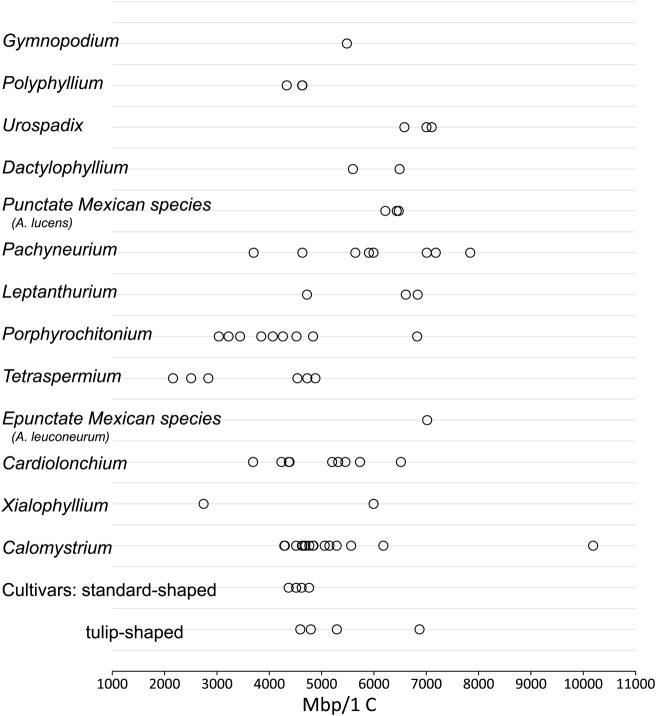
Fig. 2.
Genome sizes for Anthurium accessions sampled. Estimated genome sizes are shown as million base pairs (Mbp) per 1 C.
The mean genome size estimates for the four species represented in section Cardiolonchium range from 8.86 to 11.44 pg/2 C, with consistent 2N= 30 and reported occurrence of B chromosomes (Figs 1 and 2). The range of the mean genome sizes for these species is less than the range of genome size estimates reported for the two accessions sampled for the species A. ravenii (Table 1). The intraspecies variation reported for A. ravenii (7.54–13.32 pg/2 C) reflects ∼43 % difference in genome sizes between the two accessions sampled, each of which had reliable provenance information and resembled no species other than A. ravenii. The larger genome size reported for this species is 1.76 times that of the smaller one, suggesting some extent of autopolyploidy in A. ravenii and that we may have sampled an accession with a different cytotype. Other species sampled in this section display unremarkable intraspecies variance.
The genome size of A. leuconeurum, a member of a newly described (Carlsen 2011) and yet unnamed clade of epunctate Mexican species, is much larger than all those in its sister clade (consisting of Cardiolonchium, Calomystrium, Xialophyllium) with the exception of A. roseospadix (Fig. 1). The 2N= 35 chromosome count report for A. leuconeurum has not been confirmed in over 50 years (Mookerjea 1955), although it is speculated to have possibly included observation of 1–5 B chromosomes (Sheffer and Kamemoto 1976).
The four species sampled in section Porphyrochitonium, a section arising ∼ 4.08 mya, display a more than two-fold range in genome sizes (6.21–13.96 pg/2 C). Supernumerary chromosomes have been reported in A. bakeri, the only species sampled from section Porphyrochitonium varying from 2N = 30. In section Tetraspermium, sister clade to Porphyrochitonium, A. obtusum and A. scandens ssp. pusillum have similar genome sizes, with one accession of A. obtusum having the smallest genome size (4.42 pg/2 C) reported to date in Anthurium (Fig. 1, Table 1). Anthurium obtusum has been reported as having 2N= 30, and also as 2N= 24, suggesting a ready loss of six chromosomes, or a 20 % decrease. The genome sizes of the two accessions for this species differ by ∼25 % (Table 1, Fig. 1), suggesting that our accessions may have had the different numbers of chromosomes reported for this species. Anthurium scandens ssp. pusillum has been reported as having 2N= 24. A polyploid event in A. scandens may be responsible for the 2N= 48 chromosomes found in A. scandens ssp. scandens. A different loss of six chromosomes appears evident in the 2N= 84 variant of A. scandens ssp. scandens, which could arise by the A. scandens ssp. scandens 2N= 48 cytotype losing six chromosomes to 2N= 42, followed by a polyploidization event to yield the observed 2N= 84.
The mean genome sizes of the three species in section Pachyneurium (estimated divergence 5.8 mya) range from 8.52 to 15.36 pg/2 C (Fig. 1). Relatively wide intraspecies variance is observed between accessions in A. watermaliense, which varied ∼20 % from the mean, approaching the intraspecies genome size changes that correlate with chromosome count changes in A. scandens of section Tetraspermium. However, supernumerary chromosome are the only cytotypic changes reported to occur in section Pachyneurium, suggesting that the genome size differences between different accessions observed here might be related to the presence of extra-chromosomal DNA, or that somatic changes in the accessions sampled may have been extensive.
The species sampled of section Leptanthurium, section Dactylophyllium, and A. lucens, representing a newly described (Carlsen 2011) and yet unnamed clade of punctate Mexican species, had relatively larger genome sizes (Table 1, Figs 1 and 2). In section Leptanthurium, one of the three different accessions sampled of A. gracile has a genome size nearly 30 % smaller than the other two, which are very similar, suggesting that we sampled two accessions having the same cytotype and a third accession having a different one.
Genome size and phenotype
Species contributing to the pedigree of cultivars were expected to be reflected in the genome sizes of those cultivars. Anthurium andraeanum hybridizes most easily with A. amnicola, A. antioquiense, A. armeniense, A. formosum, A. hoffmanni, A. kamemotoanum, A. lindenianum, A. nymphaefolium and A. roseospadix, all members of section Calomystrium (Kamemoto and Kuehnle 1996). Of these, A. lindenianum was not available for sampling. The cultivars ‘Marian Seefurth’, ‘Midori’, ‘New Pahoa Red’, ‘Puanani’ and ‘Shibori’ all share the ‘standard’ blistered, heart-shaped spathe of A. andraeanum (Fig. 3A–F) and lack any documented contribution by species other than A. andraeanum. Genome sizes for those five cultivars (Table 1, Fig. 2) are similar to those of most members of the Calomystrium series (Figs 2 and 4), but only the species A. andraeanum (Fig. 3A) has a heart-shaped spathe.
Fig. 3.
Images of A. andraeanum and related cultivars. (A) A. andraeanum Linden (used with permission of Michael Wenzel), (B) A. andraeanum Hort. cv. ‘New Pahoa Red’, (C) A. andraeanum Hort. cv. ‘Marian Seefurth’, (D) A. andraeanum Hort. cv. ‘Midori’, (E) A. andraeanum Hort. cv. ‘Puanani’, (F) A. andraeanum Hort. cv. ‘Shibori’, (G) Anthurium Hort. ‘Princess Aiko’, (H) Anthurium Hort. ‘Regina’ and (I) Anthurium Hort. ‘Miss June Purple.’
Fig. 4.
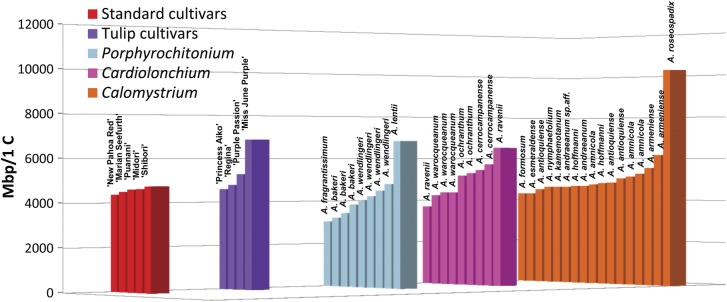
Genome size estimates of accessions of cultivars and species from Calomystrium, Cardiolonchium and Porphyrochitonium, subgeneric sections with known contributors to commercial anthurium hybrids. Individual accessions within each series are ordered by increasing genome size.
The cultivars with tulip-shaped spathes (Fig. 3G–I), ‘Princess Aiko’, 'Regina’, ‘Purple Passion’ (photograph not available) and ‘Miss June Purple’, may have been derived from tulip-shaped species in section Calomystrium, or from species in other sections (Cardiolonchium, Porphyrochitonium) known to contribute to hybrids derived from A. andraeanum (e.g. A. antrophyoides, A. ochranthum, A. cerrocampanense and A. lentii) (Kamemoto and Kuehnle 1996). Alternatively, cultivars with tulip-shaped spathes may be derived entirely from species reproductively incompatible with A. andraeanum (e.g. Anthurium wendlingeri, A. bakeri, A. scherzerianum, A. lancifolium, A. caperatum and A. garagaranum). The pedigrees of the cultivars ‘Aiko’ and ‘Regina’ are in fact known, as they were developed in the University of Hawai`i, Manoa, anthurium breeding programme. The genome size of the ‘Princess Aiko’ cultivar (Table 1, Fig. 3G) is similar to that of the cultivars with standard-shaped spathes (Figs 2 and 4) and other accessions sampled in Calomystrium, reflecting its derivation from the standard cultivar ‘Tatsuta Pink Obake’ and the tulip-shaped section Calomystrium species, A. antioquiense (Kuehnle et al. 2004). The cultivar ‘Regina’ (Table 1, Fig. 3H) is derived from earlier cultivars composed of contributions from the smaller-genome-sized Calomystrium species (i.e. A. amnicola, A. formosum, A. andraeanum and A. kamemotoanum) (Kamemoto and Kuehnle 1996; Kuehnle et al. 2004), and the genome size of ‘Regina’ is consistent with that pedigree (Figs 2 and 4). The cultivar ‘Purple Passion’ resembles (phenotypically) no species other than A. amnicola, and the genome size estimate is consistent with that (Table 1, Figs 2 and 4). The cultivar ‘Miss June Purple’ phenotypically resembles ‘Regina’ in many aspects (Fig. 3H and I), but the genome size is ∼43 % larger (Table 1, Fig. 4), suggesting a species with a larger genome size in its pedigree (perhaps A. roseospadix or A. lentii), or possibly an endogenous genome size change occurred in this cultivar.
Discussion
Genome sequencing and comparative mapping have revealed the ancient polyploid nature of angiosperms and provided an insight into the effects of polyploidy on genome evolution in plants (The Arabidopsis Genome Initiative 2000; Vision et al. 2000; Blanc and Wolfe 2004; Adams and Wendel 2005; Cui et al. 2006; Fawcett et al. 2009), while studies in Oryza (Wang et al. 2005), Arabidopsis (Lagercrantz 1998; Yogeeswaran et al. 2005) and others (Wendel 2000; Leitch and Bennett 2004; Leitch et al. 2008) reveal downsizing in genomes occurring after polyploidization events. We have used genome size to gain insight into some aspects of genome evolution in Anthurium, a genus for which the angiosperm genome size database previously contained data only for one species and one cultivar. In this study, apparent correlation between chromosome count and genome size is only clearly evident in species found in sections Xialophyllium and Tetraspermium. Although the correlations may be incidental, their importance as clues for the various ongoing processes involved in genome evolution in Anthurium should not be excluded.
Genome sizes and reported chromosome counts of two species are suggestive of polyploidy events in section Xialophyllium. In section Tetraspermium, with the smallest genome sizes reported to date, genome sizes reflect reported intraspecies variations in chromosome counts in A. scandens (Sheffer and Kamemoto 1976; Sheffer and Croat 1983), including polyploidy, and suggest additional, subsequent genome changes occurred. The age of genome changes such as polyploidy influences the confidence with which they can be identified since subsequent mutations tend to obscure the original event (Doyle and Egan 2009). Although the ages of polyploidy events discussed here are not known, they are maximally the ages of the relatively young crown groups, Xialophyllium and Tetraspermium, to which they belong, which are estimated to have arisen 2.2 and 3.43 mya, respectively. In sections other than Xialophyllium and Tetraspermium, our data display incongruity between interspecific and intraspecific genome size and chromosome counts reported by others for Anthurium and other genera.
In sections Calomystrium, Cardiolonchium, Porphyrochitoniumand Pachyneurium, we report interspecies genome size variation without any apparent relationship between genome size and base chromosome number, suggesting that the size difference may be unrelated to a polyploidization event. Therefore, interspecies genome size variation in these sections, particularly in the youngest crown group, Calomystrium, suggests a mechanism of genome size change capable of producing large differences in a short span of time. Transposable elements are capable of producing such changes. Up to 80 % of the current Z. mays genome is composed of retroelements, most inserted in the last 1–3 million years (Rabinowicz and Bennetzen 2006). Transposable elements may be deleted after initial amplification (Shirasu et al. 2000), or may persist and play a part in local adaptation, as exemplified by the intraspecific expansion of BARE-1 retroelements in barley in response to elevation and aridity (Kalendar et al. 2000). In Arabidopsis and Oryza, genome size variations are associated with changes in repetitive DNA content occurring in the last 3 million years (Bennetzen et al. 2005). Considering Calomystrium is estimated to have arisen ∼1.5 mya, genome changes due to rapid invasion and evolution of repetitive elements may play a role in genome size differences. However, the timeframe for comparable changes to occur in Anthurium, a long-lived tropical perennial, may be different than that of the annuals Zea spp., Arabidopsis spp. and Oryza spp.
Some genome size changes in these sections may be attributable to DNA changes associated with chromosomal reorganization (Kalendar et al. 2000). Chromosome reorganization in Anthurium was reported by Marutani et al. (1993), who detected differences in the karyotypes of A. nymphaefolium (Calomystrium) and A. ochranthum (Cardiolonchium), which she proposed to have resulted from chromosomal rearrangement. She also observed very similar karyotypes among closely related species in section Calomystrium, noting that the A. roseospadix karyotype resembled those of A. kamemotoanum and A. formosum (Marutani et al. 1993). This is particularly interesting given that the genome size estimate for A. roseospadix is more than twice that of each of the other two, suggesting that this may be an example where similar karyotypes among related species may be composed of chromosomes of different structure or DNA mass. However, the cytotype of the A. roseospadix accession we sampled would have to be determined before further inferences can be made.
In Polyphyllium, the oldest crown group in Anthurium, we have a case of extreme differences between reported chromosome count and expected potential genome size, with reported ploidy difference between two species with measured genome sizes that were essentially the same. As ploidy differences within other Anthurium species do exist, it is possible that we may have sampled a previously unreported cytotype, as is suspected for A. ravenii in section Cardiolonchium. However, if verified, this apparent incongruity between chromosome count and measured genome size allows us to consider a loss or gain of bulk nuclear DNA and permits inferences based on evidence from mechanisms of genome evolution elucidated by studies in other species. For example, in rice, Wang et al. (2005) estimate that 35–60 % of duplicated genes were lost shortly after genome size expansion as recently as 5 mya. In Arabidopsis, Lagercrantz (1998) estimated ∼90 chromosomal rearrangements since Arabidopsis and Brassica diverged ∼14–24 mya, and Yogeeswaran et al. (2005) estimated ∼10 chromosomal rearrangements occurred in the divergence of Arabidopsis thaliana and Arabidopsis lyrata ∼5 mya. Carlsen (2011) estimated the crown group Polyphyllium arose ∼11 mya, well within the time required to accomplish the scope of chromosomal changes as observed in the genus Arabidopsis.
The polyploid origin of A. clidemioides is unknown. However, polyploids arising by interspecies hybridization (allopolyploidy) are subject to mismatch repair during recombination of homeologous chromosomes which may generate large-scale deletions contributing to chromosome loss and reorganization (Leitch and Bennett 2004). The more broadly applicable mechanisms of ongoing unequal recombination and illegitimate recombination of homologous chromosomes also contribute to genome size reduction (Shirasu et al. 2000; Devos et al. 2002), in part by double-strand break repair, an essential but error-prone housekeeping function causing increases, decreases and chromosomal reorganizations, which can lead to chromosome loss (Gorbunova and Levy 1999; Kirik et al. 2000). Although the suppositions are intriguing, the disparity between measured genome size and reported chromosome count of A. flexile compared with A. clidemioides warrants further investigation. It was not possible to evaluate cytotypes for the accessions included in this study. Most samples were contributed by botanical gardens, and thus we did not have the plants available locally for fresh root tip sampling and cytotype determination.
The genus Anthurium displays considerable flexibility of nuclear DNA quantity and organization, even within species. We report here different genome sizes for different accessions of A. ravenii, A. watermaliense and A. gracile varying >20 % from the mean for the species. Conceptually, intraspecies variation can be viewed as incipient interspecies genome size variation (Greilhuber 1998). Once, genome size seemed to offer promise for delineating species (Ohri 1998; Ghosh et al. 2001), so reports of intraspecies genome size variations have been scrutinized to identify and eliminate systematic sources of variation, leading to standardization of methods, attention to detail in sample handling and careful selection of internal standards (Greilhuber 1998, 2005). Still, intraspecies variations persist: approximately 10 % of Curcuma species sampled displayed intraspecies variation in genome size estimates (2007), while both genome sizes and ploidy levels varied widely in a survey of 244 Dianthus broteri individuals collected from 25 populations (Balao et al. 2009), similar to results reported here for A. scandens. While variant cytotypes may explain the largest differences observed, a lesser amount of intraspecific variation in bulk nuclear DNA content may be attributed to the presence of extrachromosomal material, which can only be convincingly excluded from genome size estimates by determining the cytotype of each accession sampled (Teoh and Rees 1976). In particular, the origin and evolution of B chromosomes seems to be associated with amplification of tandem repeats on A chromosomes, and can be generated spontaneously following allopolyploidization (Jones and Houben 2003). It may be that activity of extra-chromosomal DNA in sections Calomystrium, Cardiolonchium, Porphyrochitonium and Pachyneurium has contributed to the range of genome sizes among accessions of the same species in those sections.
Furthermore, Anthurium cultural practices, including in vitro cultivation, clonal propagation and selection for sports, impose extreme selective pressures, capable of activating transposable elements causing intraspecies genome size variations without imposing a reproductive barrier (Peschke and Phillips 1991; Hirochika et al. 1996). Indeed, individual cultivars may be selected for phenotypes associated with transposable element activity which has affected genome size, but has more noticeably affected phenotype. For example, variegated cultivars of maize (McClintock 1965–1966), Antirrhinum (snapdragon) (Coen and Carpenter 1986), Convolvulus (morning glory) (Hoshino et al. 1995), Dahlia (Ohno et al. 2011) and Sorghum carry transposable elements associated with variegation (Chopra et al. 1999), and it may be that the mottled ‘Shibori’ cultivar (Figs 3I and 4), with a slightly larger genome than that of the other standard cultivars, is accomplishing its variegation by similar means.
Conclusions and forward look
Genome sizes in Anthurium display variation suggestive of repeated polyploidy, with evidence for possible re-diploidization in the oldest crown group Polyphyllium, and ongoing expansion in the youngest crown group, Calomystrium. Anthurium genome size distribution was not distinctly demarcated by ploidy level, as Leong-Škorničková et al. (2007) similarly reported in an analysis of nearly half the Curcuma (Zingiberales) species found on the Indian continent. Also, as in Curcuma, we found genome size to be useful, together with phenotypic similarities, for insight into the pedigree of cultivars (Leong-Škorničková et al. 2007). The new information on genome sizes in Anthurium will serve as a useful framework from which to launch molecular investigations including map- and sequence-based studies, which may provide further insight into the processes resulting in genome size variation observed in Anthurium, which may be similar to those described in other monocots.
Additional information
The following additional information is available in the online version of this article –
File 1. Genome size estimates for 26 Anthurium accessions evaluated with two internal standards.
File 2. References for cytological observations summarized for Anthurium species sampled.
Sources of funding
This work was funded by the United States Department of Agriculture, Agriculture Research Service.
Contributions by the authors
B.J.B. and J.Y.S. designed the study. B.J.B. coordinated sampling and genome sizing, analysed data, organized figures and authored the manuscript. J.Y.S. supported the research and contributed to the development and revision of the manuscript.
Conflict of interest statement
None declared.
Acknowledgements
Colleagues at public and private botanical gardens provided plant tissues: Michael Wenzel of Atlanta Botanical Garden (ABG), Jon Peter of New York Botanical Garden (NYBG), Bruce Holst of the Marie Selby Botanical Gardens (MSBG), Kyle Wallick of United States Botanic Garden, David Scherberich of Jardin Botanique de la Ville de Lyon (JBVL), Renée Gaudette of Jardin Botanique de Montréal (JBM), Geneviève Ferry of Conservatoire et Jardins Botaniques de Nancy (CJBN), Thomas Croat of Missouri Botanical Garden (MBG). Monica Carlsen, University of Missouri St Louis (UMSL), contributed immensely to our understanding of Anthurium systematics and current phylogenetic relationships. Claudia Henriquez, Washington University in St Louis, selected, collected, packaged and shipped plant material from Missouri Botanical Garden. The Hawaiian Anthurium Industry Association (HAIA) donated cultivars. Teresita Amore and Joanne Lichty from University of Hawai`i, Manoa (UH) provided plant tissues, propagules and historical perspective. Special thanks to Thomas Croat, who verified determinations of some accessions, as well as Monica Carlsen, University of Missouri St Louis (UMSL), who shared pre-publication findings to provide us with the most current hypothesis of phylogenetic relationships in Anthurium.
References
- Adams KL, Wendel JF. Polyploidy and genome evolution in plants. Current Opinion in Plant Biology. 2005;8:135–141. doi: 10.1016/j.pbi.2005.01.001. [DOI] [PubMed] [Google Scholar]
- The Arabidopsis Genome Initiative. Analysis of the genome sequence of the flowering plant Arabidopsis thaliana. Nature. 2000;408:796–815. doi: 10.1038/35048692. [DOI] [PubMed] [Google Scholar]
- Arumuganathan K, Earle E. Estimation of nuclear DNA content of plants by cell-flow cytometry. Plant Molecular Biology Reporter. 1991;9:229–233. [Google Scholar]
- Balao F, Casimiro-Soriguer R, Talavera M, Herrera J, Talavera S. Distribution and diversity of cytotypes in Dianthus broteri as evidenced by genome size variations. Annals of Botany. 2009;104:965–973. doi: 10.1093/aob/mcp182. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Baranyi M, Greilhuber J. Flow cytometric and Feulgen densitometric analysis of genome size variation in Pisum. TAG Theoretical and Applied Genetics. 1996;92:297–307. doi: 10.1007/BF00223672. [DOI] [PubMed] [Google Scholar]
- Beaulieu JM, Moles AT, Leitch IJ, Bennett MD, Dickie JB, Knight CA. Correlated evolution of genome size and seed mass. New Phytologist. 2007;173:422–437. doi: 10.1111/j.1469-8137.2006.01919.x. [DOI] [PubMed] [Google Scholar]
- Beaulieu JM, Leitch IJ, Patel S, Pendharkar A, Knight CA. Genome size is a strong predictor of cell size and stomatal density in angiosperms. New Phytologist. 2008;179:975–986. doi: 10.1111/j.1469-8137.2008.02528.x. [DOI] [PubMed] [Google Scholar]
- Bennett MD, Leitch IJ. Nuclear DNA amounts in angiosperms: progress, problems and prospects. Annals of Botany. 2005;95:45–90. doi: 10.1093/aob/mci003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bennett MD, Smith JB. Nuclear-DNA amounts in angiosperms. Philosophical Transactions of the Royal Society of London Series B: Biological Sciences. 1976;274:227–274. doi: 10.1098/rstb.1976.0044. [DOI] [PubMed] [Google Scholar]
- Bennetzen JL, Ma J, Devos KM. Mechanisms of recent genome size variation in flowering plants. Annals of Botany. 2005;95:127–132. doi: 10.1093/aob/mci008. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Blanc G, Wolfe KH. Widespread paleopolyploidy in model plant species inferred from age distributions of duplicate genes. The Plant Cell Online. 2004;16:1667–1678. doi: 10.1105/tpc.021345. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Boyce PC, Croat TB. 2012. The Überlist of Araceae: Totals for published and estimated number of species in aroid generahttp://www.aroid.org/genera/120110uberlist.pdf. 12 January 2012.
- Cabrera LI, Salazar GA, Chase MW, Mayo SJ, Bogner J, Dávila P. Phylogenetic relationships of aroids and duckweeds (Araceae) inferred from coding and noncoding plastid DNA. American Journal of Botany. 2008;95:1153–1165. doi: 10.3732/ajb.0800073. [DOI] [PubMed] [Google Scholar]
- Carlsen M. St Louis, MO, USA: University of Missouri; 2011. Understanding the origin and rapid diversification of the genus Anthurium Schott (Araceae), integrating molecular phylogenetics, morphology and fossils. PhD Dissertation. [Google Scholar]
- Carlsen MM, Croat TB. Taxonomic revision of Anthurium section Semaeophyllium Schott (Araceae) Harvard Papers in Botany. 2007;12:173–234. [Google Scholar]
- Chopra S, Brendel V, Zhang J, Axtell JD, Peterson T. Molecular characterization of a mutable pigmentation phenotype and isolation of the first active transposable element from Sorghum bicolor. Proceedings of the National Academy of Sciences of the USA. 1999;96:15330–15335. doi: 10.1073/pnas.96.26.15330. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Coen ES, Carpenter R. Transposable elements in Antirrhinum majus: generators of genetic diversity. Trends in Genetics. 1986;2:292–296. [Google Scholar]
- Cotias-de-Oliveira ALP, Guedes MLS, Barreto EC. Chromosome numbers for Anthurium and Philodendron spp. (Araceae) occurring in Bahia, Brazil. Genetics and Molecular Biology. 1999;22:237–242. [Google Scholar]
- Croat TB. A revision of the genus Anthurium (Araceae) of Mexico and Central America. Part I. Mexico and Middle America. Annals of the Missouri Botanical Garden. 1983;70:211–420. [Google Scholar]
- Croat TB. Ecology and life forms of Araceae. Aroideana. 1988;11:4–56. [Google Scholar]
- Croat TB. Ecology and life forms of Araceae: a follow-up. Aroideana. 1989;12:6–8. [Google Scholar]
- Croat TB. A revision of Anthurium section Pachyneurium (Araceae) Annals of the Missouri Botanical Garden. 1991;78:539–855. [Google Scholar]
- Croat TB, Sheffer RD. The sectional groupings of Anthurium (Araceae) Aroideana. 1983;6:85–123. [Google Scholar]
- Cui L, Wall PK, Leebens-Mack JH, Lindsay BG, Soltis DE, Doyle JJ, Soltis PS, Carlson JE, Arumuganathan K, Barakat A, Albert VA, Ma H, dePamphilis CW. Widespread genome duplications throughout the history of flowering plants. Genome Research. 2006;16:738–749. doi: 10.1101/gr.4825606. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cusimano N, Bogner J, Mayo SJ, Boyce PC, Wong SY, Hesse M, Hetterscheid WLA, Keating RC, French JC. Relationships within the Araceae: comparison of morphological patterns with molecular phylogenies. American Journal of Botany. 2011;98:654–668. doi: 10.3732/ajb.1000158. [DOI] [PubMed] [Google Scholar]
- Devos KM, Brown JKM, Bennetzen JL. Genome size reduction through illegitimate recombination counteracts genome expansion in Arabidopsis. Genome Research. 2002;12:1075–1079. doi: 10.1101/gr.132102. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Doležel J, Greilhuber J. Nuclear genome size: are we getting closer? Cytometry Part A. 2010;77A:635–642. doi: 10.1002/cyto.a.20915. [DOI] [PubMed] [Google Scholar]
- Doyle JJ, Egan AN. Dating the origins of polyploidy events. New Phytologist. 2009;186:73–85. doi: 10.1111/j.1469-8137.2009.03118.x. [DOI] [PubMed] [Google Scholar]
- Fawcett JA, Maere S, Van de Peer Y. Plants with double genomes might have had a better chance to survive the Cretaceous–Tertiary extinction event. Proceedings of the National Academy of Sciences of the USA. 2009;106:5737–5742. doi: 10.1073/pnas.0900906106. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Galbraith D, Harkins K, Maddox J, Ayres N, Sharma D, Firoozabady E. Rapid flow cytometric analysis of the cell cycle in intact plant tissues. Science. 1983;220:1049–1051. doi: 10.1126/science.220.4601.1049. [DOI] [PubMed] [Google Scholar]
- Geber G. Vienna: University of Vienna; 1989. Zur Karyosystematik der Lemnaceae. PhD Dissertation. [Google Scholar]
- Ghosh P, Mukherjee S, Sharma A. Cytophotometric estimation of in situ DNA content in several species of Araceae. Cytobios. 2001;105:177–183. [PubMed] [Google Scholar]
- Gorbunova V, Levy AA. How plants make ends meet: DNA double-strand break repair. Trends in Plant Science. 1999;4:263–269. doi: 10.1016/s1360-1385(99)01430-2. [DOI] [PubMed] [Google Scholar]
- Govaerts R, Bogner J, Boos J, Boyce P, Cosgriff B, Croat T, Gonçalves E, Grayum M, Hay A, Hetterscheid W, Ittenbach S, Landolt E, Mayo S, Murata J, Nguyen VD, Sakuragui CM, Singh Y, Thompson S, Zhu G. 2011. World checklist of Araceaehttp://www.kew.org/wcsp/ 16 September 2011.
- Greilhuber J. Intraspecific variation in genome size: a critical reassessment. Annals of Botany. 1998;82:27–35. [Google Scholar]
- Greilhuber J. Intraspecific variation in genome size in angiosperms: identifying its existence. Annals of Botany. 2005;95:91–98. doi: 10.1093/aob/mci004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Greilhuber J, Doležel J, Lysák MA, Bennett MD. The origin, evolution and proposed stabilization of the terms ‘genome size’ and ‘C-value’ to describe nuclear DNA contents. Annals of Botany. 2005;95:255–260. doi: 10.1093/aob/mci019. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hawkins JS, Kim H, Nason JD, Wing RA, Wendel JF. Differential lineage-specific amplification of transposable elements is responsible for genome size variation in Gossypium. Genome Research. 2006;16:1252–1261. doi: 10.1101/gr.5282906. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hirochika H, Sugimoto K, Otsuki Y, Tsugawa H, Kanda M. Retrotransposons of rice involved in mutations induced by tissue culture. Proceedings of the National Academy of Sciences of the USA. 1996;93:7783–7788. doi: 10.1073/pnas.93.15.7783. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hodgson JG, Sharafi M, Jalili A, Díaz S, Montserrat-Martí G, Palmer C, Cerabolini B, Pierce S, Hamzehee B, Asri Y, Jamzad Z, Wilson P, Raven JA, Band SR, Basconcelo S, Bogard A, Carter G, Charles M, Castro-Díez P, Cornelissen JHC, Funes G, Jones G, Khoshnevis M, Pérez-Harguindeguy N, Pérez-Rontomé MC, Shirvany FA, Vendramini F, Yazdani S, Abbas-Azimi R, Boustani S, Dehghan M, Guerrero-Campo J, Hynd A, Kowsary E, Kazemi-Saeed F, Siavash B, Villar-Salvador P, Craigie R, Naqinezhad A, Romo-Díez A, de Torres Espuny L, Simmons E. Stomatal vs. genome size in angiosperms: the somatic tail wagging the genomic dog? Annals of Botany. 2010;105:573–584. doi: 10.1093/aob/mcq011. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hoshino A, Inagaki Y, Iida S. Structural analysis of Tpn1, a transposable element isolated from Japanese morning glory bearing variegated flowers. Molecular and General Genetics. 1995;247:114–117. doi: 10.1007/BF00425828. [DOI] [PubMed] [Google Scholar]
- Jones N, Houben A. B chromosomes in plants: escapees from the A chromosome genome? Trends in Plant Science. 2003;8:417–423. doi: 10.1016/S1360-1385(03)00187-0. [DOI] [PubMed] [Google Scholar]
- Jones RN, Rees H. B chromosomes. London: Academic Press; 1982. [Google Scholar]
- Kalendar R, Tanskanen J, Immonen S, Nevo E, Schulman AH. Genome evolution of wild barley (Hordeum spontaneum) by BARE-1 retrotransposon dynamics in response to sharp microclimatic divergence. Proceedings of the National Academy of Sciences of the USA. 2000;97:6603–6607. doi: 10.1073/pnas.110587497. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kamemoto H, Kuehnle AR. Breeding Anthuriums in Hawai`i. Honolulu: University of Hawai`i Press; 1996. [Google Scholar]
- Kirik A, Salomon S, Puchta H. Species-specific double-strand break repair and genome evolution in plants. EMBO J. 2000;19:5562–5566. doi: 10.1093/emboj/19.20.5562. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Knight CA, Molinari NA, Petrov DA. The large genome constraint hypothesis: evolution, ecology and phenotype. Annals of Botany. 2005;95:177–190. doi: 10.1093/aob/mci011. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kuehnle AR, Amore TD, Kamemoto H, Kunisaki JT, Lichty JS, Uchida JY. 2004. New plants for Hawai‘i anthurium cultivar release: ‘Princess Aiko’ (‘Imperial’) and ‘Regina’, two novelty anthuriums. Research Extension Series 152, Publication NPH-A-7. 2 pp.
- Lagercrantz U. Comparative mapping between Arabidopsis thaliana and Brassica nigra indicates that Brassica genomes have evolved through extensive genome replication accompanied by chromosome fusions and frequent rearrangements. Genetics. 1998;150:1217–1228. doi: 10.1093/genetics/150.3.1217. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Leitch IJ, Bennett MD. Genome downsizing in polyploid plants. Biological Journal of the Linnean Society. 2004;82:651–663. [Google Scholar]
- Leitch IJ, Chase MW, Bennett MD. Phylogenetic analysis of DNA C-values provides evidence for a small ancestral genome size in flowering plants. Annals of Botany. 1998;82:85–94. [Google Scholar]
- Leitch IJ, Soltis DE, Soltis PS, Bennett MD. Evolution of DNA amounts across land plants (Embryophyta) Annals of Botany. 2005;95:207–217. doi: 10.1093/aob/mci014. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Leitch IJ, Beaulieu JM, Cheung K, Hanson L, Lysak MA, Fay MF. Punctuated genome size evolution in Liliaceae. Journal of Evolutionary Biology. 2007;20:2296–2308. doi: 10.1111/j.1420-9101.2007.01416.x. [DOI] [PubMed] [Google Scholar]
- Leitch IJ, Hanson L, Lim KY, Kovarik A, Chase MW, Clarkson JJ, Leitch AR. The ups and downs of genome size evolution in polyploid species of Nicotiana (Solanaceae) Annals of Botany. 2008;101:805–814. doi: 10.1093/aob/mcm326. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Leitch IJ, Beaulieu JM, Chase MW, Leitch AR, Fay MF. Genome size dynamics and evolution in monocots. Journal of Botany. 2010;2010 Available at http://dx.doi.org/10.1155/2010/862516. [Google Scholar]
- Leong-Škorničková J, Šída O, Jarolímová V, Sabu M, Fér T, Trávníček P, Suda J. Chromosome numbers and genome size variation in Indian species of Curcuma (Zingiberaceae) Annals of Botany. 2007;100:505–526. doi: 10.1093/aob/mcm144. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Marutani M, Sheffer RD, Kamemoto H. Cytological analysis of Anthurium andraeanum (Araceae), its related taxa and their hybrids. American Journal of Botany. 1993;80:93–103. [Google Scholar]
- Mayo SJ, Bogner J, Boyce P, Catherine E Royal Botanic Gardens Kew. The genera of Araceae. London: Royal Botanic Gardens, Kew; 1997. [Google Scholar]
- McClintock B. Regulation of pattern of gene expression by controlling elements in maize. Washington, DC: Yearbook of The Carnegie Institution of Washington; 1965–1966. [Google Scholar]
- Mookerjea A. Cytology of different species of aroids with a view to trace the basis of their evolution. Caryologia. 1955;7:221–291. [Google Scholar]
- Ohno S, Hosokawa M, Hoshino A, Kitamura Y, Morita Y, Park K-I, Nakashima A, Deguchi A, Tatsuzawa F, Doi M, Iida S, Yazawa S. A bHLH transcription factor, DvIVS, is involved in regulation of anthocyanin synthesis in dahlia (Dahlia variabilis) Journal of Experimental Botany. 2011;62:5105–5116. doi: 10.1093/jxb/err216. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ohri D. Genome size variation and plant systematics. Annals of Botany. 1998;82:75–83. [Google Scholar]
- Peschke VM, Phillips RL. Activation of the maize transposable element Suppressor-mutator (Spm) in tissue culture. Theoretical and Applied Genetics. 1991;81:90–97. doi: 10.1007/BF00226117. [DOI] [PubMed] [Google Scholar]
- Petersen G. Cytology and systematics of Araceae. Nordic Journal of Botany. 1989;9:119–166. [Google Scholar]
- Praça-Fontes M, Carvalho C, Clarindo W, Cruz C. Revisiting the DNA C-values of the genome size-standards used in plant flow cytometry to choose the ‘best primary standards. Plant Cell Reports. 2011;30:1183–1191. doi: 10.1007/s00299-011-1026-x. [DOI] [PubMed] [Google Scholar]
- Rabinowicz PD, Bennetzen JL. The maize genome as a model for efficient sequence analysis of large plant genomes. Current Opinion in Plant Biology. 2006;9:149–56. doi: 10.1016/j.pbi.2006.01.015. [DOI] [PubMed] [Google Scholar]
- SanMiguel P, Tikhonov A, Jin Y-K, Motchoulskaia N, Zakharov D, Melake-Berhan A, Springer PS, Edwards KJ, Lee M, Avramova Z, Bennetzen JL. Nested retrotransposons in the intergenic regions of the maize genome. Science. 1996;274:765–768. doi: 10.1126/science.274.5288.765. [DOI] [PubMed] [Google Scholar]
- Sharma AK, Bhattacharyya UC. Structure and behavior of chromosomes in species of Anthurium with special reference to the accessory chromosomes. Proceedings of the National Institute of India, Part B, Biological Science. 1961;27:317–328. [Google Scholar]
- Sheffer RD, Croat TB. Chromosome numbers in the genus Anthurium (Araceae) II. American Journal of Botany. 1983;70:858–871. [Google Scholar]
- Sheffer RD, Kamemoto H. Chromosome numbers in genus Anthurium. American Journal of Botany. 1976;63:74–81. [Google Scholar]
- Shirasu K, Schulman AH, Lahaye T, Schulze-Lefert P. A contiguous 66-kb barley DNA sequence provides evidence for reversible genome expansion. Genome Research. 2000;10:908–915. doi: 10.1101/gr.10.7.908. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Soltis DE, Soltis PS, Bennett MD, Leitch IJ. Evolution of genome size in the angiosperms. American Journal of Botany. 2003;90:1596–1603. doi: 10.3732/ajb.90.11.1596. [DOI] [PubMed] [Google Scholar]
- Suda J, Leitch IJ. The quest for suitable reference standards in genome size research. Cytometry Part A. 2010;77A:717–720. doi: 10.1002/cyto.a.20907. [DOI] [PubMed] [Google Scholar]
- Teoh SB, Rees H. Nuclear DNA amounts in populations of Picea and Pinus species. Heredity. 1976;36:123–137. [Google Scholar]
- Vicient CM, Kalendar R, Anamthawat-Jonsson K, Schulman AH. Structure, functionality, and evolution of the BARE-1 retrotransposon of barley. Genetica. 1999;107:53–63. [PubMed] [Google Scholar]
- Vision TJ, Brown DG, Tanksley SD. The origins of genomic duplications in Arabidopsis. Science. 2000;290:2114–7. doi: 10.1126/science.290.5499.2114. [DOI] [PubMed] [Google Scholar]
- Wang X, Shi X, Hao B, Ge S, Luo J. Duplication and DNA segmental loss in the rice genome: implications for diploidization. New Phytologist. 2005;165:937–946. doi: 10.1111/j.1469-8137.2004.01293.x. [DOI] [PubMed] [Google Scholar]
- Wendel JF. Genome evolution in polyploids. Plant Molecular Biology. 2000;42:225–249. [PubMed] [Google Scholar]
- Yogeeswaran K, Frary A, York TL, Amenta A, Lesser AH, Nasrallah JB, Tanksley SD, Nasrallah ME. Comparative genome analyses of Arabidopsis spp.: inferring chromosomal rearrangement events in the evolutionary history of A. thaliana. Genome Research. 2005;15:505–515. doi: 10.1101/gr.3436305. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zonneveld BJM. New record holders for maximum genome size in eudicots and monocots. Journal of Botany. 2010;2010 Available at http://dx.doi.org/10.1155/2010/527357. [Google Scholar]