Twenty years have elapsed since the discovery of a microRNA (miRNA) gene in Caenorhabditis elegans. Based on growing research progress, we are approaching the nature of this small RNA species, which seemed to be mysterious before. The regulatory activities of miRNAs have been extensively studied through target identification and physiological and phenotypic assays by using bioinformatic, genetic, and biochemical approaches. However, recent evidence points to the fact that the effective levels of miRNAs are determined by transcription, processing, miRNA-induced silencing complex loading, action, turnover use, and decay. Each process is affected by certain factors, such as genomic modifications, RNA editing, miRNA-induced silencing complex loading competition, target abundance and complementarity, and spatiotemporal effects, thus conferring a highly dynamic feature to miRNA activities. To maintain steady-state levels of the functional miRNAs, thus ensuring a normal physiological and biochemical status, plants employ several exquisite strategies, such as feedback regulation and a buffering system, to minimize the influence of external signal fluctuations. In this review, we raise the notion that a more dynamic picture of miRNA activities should be drawn to construct comprehensive miRNA-mediated networks in plants.
MicroRNAs (miRNAs), approximately 21 nucleotides in length, were identified as a small RNA (sRNA) species with essential regulatory roles in various biological processes (Carrington and Ambros, 2003). The transcription of most miRNA genes is guided by RNA polymerase II (Lee et al., 2004; Xie et al., 2005). Following transcription, the single-stranded RNAs with internal stem-loop structures are then recognized by Drosha and Dicer in animals (Kim et al., 2009a) or Dicer-Like1 (DCL1) in plants (Voinnet, 2009) for sequential cleavage, converting the primary microRNAs (pri-miRNAs) to the precursor microRNAs (pre-miRNAs) and finally to the miRNA/miRNA* duplexes. After dissociation from the duplexes, the miRNAs are incorporated into Argonaute (AGO)-associated microRNA-induced silencing complexes (miRISCs; preferentially AGO1-associated miRISCs). Although the sophisticated model of miRNA biogenesis is seemingly settled for each step, there exist many key nodes that influence the final activity of a miRNA gene. The transcription of miRNA genes is under the rigorous surveillance of many cis- and trans-factors, such as chromatin marks and specific transcription factors (TFs). The processing efficiency of the miRNA precursors is basically determined by their own sequences and structures and is regulated in a spatiotemporal manner (Davis and Hata, 2009; Cuperus et al., 2011; Zhu et al., 2011). Furthermore, the sorting of miRNAs into specific AGO complexes should not be oversimplified, since not all the miRNAs are uniformly loaded into AGO1-associated miRISCs. Additionally, loading competition between miRNAs and other sRNAs occurs in planta.
In plants, miRNAs guide the miRISCs to target transcripts containing highly complementary recognition sites to exert their repressive roles on gene expression. This seemingly simple one-to-one regulation occurs with several concomitant events that strongly affect the regulatory intensity. For instance, the degradation rate of a miRNA was reported to be highly dependent on target abundance and complementarity (Chatterjee and Grosshans, 2009; Ameres et al., 2010; Arvey et al., 2010). After one cleavage of a specific target, the miRISC may survive, or the released miRNA could form another miRISC, both of which will involve in another round of targeting. This turnover rate is also determined by the target complementarity. Quite a different situation exists between miRNAs and their low-complementary targets. For miRNAs sequestered by bulge targets, the turnover rate is intensely reduced.
All the evidence points to the conclusion that the regulatory activities of the miRNAs are highly dynamic. From transcription, to precursor processing, to miRISC loading, to target recognition and miRNA-mediated regulation, and finally to miRNA degradation and turnover, numerous crucial factors support the apparently steady-state levels of the mature miRNAs. From another point of view, the miRNA-mediated regulation itself is strictly regulated in many respects, making the miRNA-involved networks more robust. Based on recent research progress, we felt that a dynamic view should be provided to measure the miRNA activities more precisely in plants.
TRANSCRIPTIONAL CONTROL OF MIRNA GENES
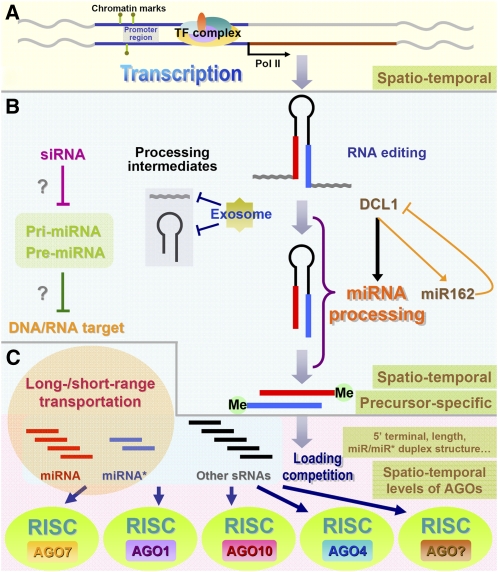
Increasing evidence points to the fact that the accumulation of a specific miRNA is a combinatory effect of its transcription, processing, and degradation (Kai and Pasquinelli, 2010). As the first step of miRNA expression, their transcription is modulated in a highly dynamic manner. Both cis-modifications and trans-acting factors are responsible for the spatiotemporally restricted expression patterns of miRNA genes (Chen, 2009; Davis and Hata, 2009; Winter et al., 2009; Fig. 1A).
Figure 1.
Schematic presentation showing the dynamic nature of miRNA biogenesis in plants. A, RNA polymerase II (Pol II)-dependent transcription of the miRNA genes. Chromatin marks including DNA methylation and histone modifications, and the combinatory regulation of many TFs, together contribute the spatiotemporal expression patterns of the miRNA genes in plants. B, Processing of miRNA precursors and miRNA maturation. RNA editing on the miRNA precursors plays a role in changing the original sequence information encoded by the miRNA gene loci. The exosome was suggested to be implicated in digesting the processing intermediates from the miRNA precursors, ensuring relatively high processing efficiency. The processing efficiency also shows a high precursor sequence-specific dependence. Methylation at the 3′ ends of the miRNA/miRNA* duplex (“Me” here represents the methyl group) is crucial for the stabilization of miRNA and miRNA*. A feedback circuit between DCL1 and miR162 exists within the processing procedure. Moreover, the siRNAs complementary to specific pri-miRNAs and pre-miRNAs exhibit a potential repressive role in miRNA processing. More interestingly, the pri-miRNAs and pre-miRNAs may possess their own targets. C, Sorting into the AGO-associated miRISCs. A drastic loading competition may exist among miRNAs, miRNA*s, and other sRNA species. Not all the miRNAs are incorporated into the AGO1 complex. The 5′ terminal composition and the sequence length of the miRNAs, the structure of the miRNA/miRNA* duplex, and other undetermined factors have a significant influence on the loading patterns of the mature miRNAs.
One major form of cis-regulation is known as chromatin modification, including DNA methylation and histone modification. These epigenetic marks can be present within the upstream and downstream regions or in the bodies of the miRNA genes, which are extraordinarily variable according to cellular contents and environmental stimuli. These marks have shown great potential in influencing the transcriptional status of miRNA genes. As proposed by Rodriguez-Enriquez et al. (2011), somaclonal variation, a featured phenomenon observed in plant tissue culture, is one of the biological consequences caused by miRNA misexpression and the accompanying disordered regulatory pathways. In their hypothesis, the inducers introduced by in vitro tissue culture could result in aberrant miRNA transcription, processing, and miRISC loading, which could have remarkable impacts on the transcriptome and the proteome and further alter the epigenetic status of the genome in cultured cells. In turn, miRNA transcription could be influenced by the altered epigenetic marks surrounding the miRNA genes (Rodriguez-Enriquez et al., 2011). Another example was recently provided by Kim et al. (2009b). A histone acetyltransferase, GCN5 (for general control nonrepressed protein 5), was indicated to interfere with miRNA biogenesis transcriptionally and posttranscriptionally. The GCN5-mediated histone modifications serve as an epigenetic mechanism for modulating miRNA production (Kim et al., 2009b).
The transcription of most miRNA genes is mediated by RNA polymerase II (Lee et al., 2004; Xie et al., 2005). The structures of the miRNA promoters are similar to those of the protein-coding genes (Megraw et al., 2006; Zhou et al., 2007). For example, the distribution patterns of the basic cis-elements for transcriptional control (i.e. the transcription start site, the TATA box, and the CAAT box) on the miRNA promoters were demonstrated to be identical to the protein-coding genes (Meng et al., 2009). In plants, many TFs have been identified as trans-acting factors with roles in the transcriptional modulation of certain miRNA genes. One example is the PHR1 (for PHOSPHATE STARVATION RESPONSE1)-miR399-PHO2 (defined by the mutant pho2) regulatory pathway involved in phosphorous homeostasis (Bari et al., 2006). Upon phosphorous deprivation, miR399 is up-regulated transcriptionally by the activated PHR1, a direct upstream regulator. Then, the repression of PHO2 by miR399 is subsequently reinforced posttranscriptionally. This cascade ensures the expeditious response of the plants under phosphorous-deficient conditions, enabling more efficient use of both environmental and cellular resources of phosphorus. Intriguingly, some TFs were also targeted by the downstream miRNAs, forming feedback circuits in certain signaling pathways. For instance, within the auxin signaling pathway implicated in Arabidopsis (Arabidopsis thaliana) adventitious root development (Gutierrez et al., 2009), miR160 is transcriptionally regulated by AUXIN RESPONSE FACTOR6 (ARF6) and ARF17 and miR167 is regulated by ARF6, ARF8, and ARF17. On the other hand, all three ARF genes are negatively regulated by either of the two miRNAs. These feedback circuits form an interlaced network that could decipher, integrate, and transduce the light and auxin signals to shape normal root system architecture. Another exquisite case was presented by Wu et al. (2009). Both miR156 and miR172 were demonstrated to participate in the regulatory network that was essential for developmental timing in Arabidopsis. These two miRNAs act by repressing the expression of TFs belonging to SQUAMOSA PROMOTER-BINDING PROTEIN LIKE (SPL) and APETALA2-LIKE (AP2-like) gene families, respectively. Certain members of the two TF families were proved to regulate miR156 and miR172 positively, thus forming negative feedback loops that contributed to the normal juvenile-to-adult phase transition. More complicatedly, the transcription of miR172 was directly regulated by SPL9 and SPL10, which were both targeted by miR156. These connections established the miR156-SPL-miR172-AP2 regulatory cascade, which was crucial for vegetative phase change in Arabidopsis (Wu et al., 2009). However, all these findings only uncovered part of the miRNA- and TF-involved networks. The one-to-one and multiple-to-one regulatory relationships between TFs and specific miRNA genes should not be stationary states. Instead, they are highly dynamic due to numerous intrinsic factors, such as the spatiotemporal expression of TF genes and indirect TF-miRNA regulation. Facilitated by these technical advances, a more comprehensive view of such networks could be constructed through genome-wide identification of TF-binding sites (MacQuarrie et al., 2011).
Expression pattern analyses of plant miRNAs by high-throughput profiling or fine-scale quantification revealed that numerous miRNAs were expressed in a tissue- or stage-specific manner and that dozens of miRNAs could be induced by external stimuli (Reinhart et al., 2002; Kidner and Martienssen, 2004; Sunkar and Zhu, 2004; Lu et al., 2005; Yao et al., 2007; Liu et al., 2008b; Oh et al., 2008; Hsieh et al., 2009; Johnson et al., 2009; Simon et al., 2009). All these findings indicate that miRNA activities are highly variable at distinct developmental stages, upon diverse treatments, or in different tissues. Conceivably, the strict control of miRNA expression at both the chromatic and transcriptional levels could make a great contribution to this highly dynamic nature.
PROCESSING AND MATURATION, CRUCIAL STEPS FOR MIRNA BIOGENESIS
After transcription, the primary products (i.e. pri-miRNAs) are subjected to DCL1-mediated two-step cleavage in the nucleus in plants (Papp et al., 2003; Kurihara and Watanabe, 2004). Several aspects should be taken into account to assess the dynamic efficiency of the miRNA precursor processing. One of the well-characterized models balancing processing and miRNA activities is the feedback circuit formed between DCL1 and miR162 (Xie et al., 2003). The nucleus-localized DCL1 with RNase III activity is indispensable for the processing of most plant miRNAs (Voinnet, 2009). Thus, the levels of the final products of the miRNA genes (i.e. the mature miRNAs) are highly correlated to the expression of DCL1. On the other hand, DCL1 itself is regulated by miR162 posttranscriptionally to avoid plethoric DCL1 activity (Xie et al., 2003). Moreover, the tissue-specific regulation of miRNA processing by DCLs was proposed recently. The overaccumulated level of DCL3 in specific tissues may result in a shift of the substrates of DCL1 to DCL3s (Vazquez et al., 2008). Similar substrate competition may also occur between DCL1 and other DCLs.
In addition, different miRNA precursors possess different affinities to DCL1 due to distinct sequence characteristics and spatiotemporal distributions. Although the precursor sequences are largely determined by the genomic sequences of the miRNA genes, several pieces of evidence in animals show that RNA editing mediated by ADENOSINE DEAMINASE ACTING ON RNA can occur on the mature miRNAs or the precursors posttranscriptionally, which has remarkable effects on miRNA processing and targeting (Luciano et al., 2004; Blow et al., 2006; Yang et al., 2006b; Kawahara et al., 2007a, 2007b, 2008; Winter et al., 2009; Krol et al., 2010). Different from animals, RNA editing in plants, mostly represented by C-to-U base conversion, is carried out by PENTATRICOPEPTIDE REPEAT (PPR) family proteins (Schmitz-Linneweber and Small, 2008) and is restricted to plant organelles, including mitochondria and plastids, based on available reports (Shikanai, 2006). Recent bioinformatic analyses using huge high-throughput sequencing (HTS) data sets raised the possibility that RNA editing of miRNA gene products could also take place in plants (Ebhardt et al., 2009; Iida et al., 2009; Meng et al., 2010a). Once further experimental validations are available, this kind of sequence modification could be another dynamic layer underlying miRNA processing, maturation, and action.
Depending on the in vivo levels and structures, miRNA precursors, perhaps along with other sRNA precursors, could compete for the machineries functioning in the miRNA biogenesis pathway. One such competition in plants could be raised by different miRNA precursors and other stem-loop-structured precursors for accessibility to DCL1 or other DCLs such as DCL3, mentioned above (Vazquez et al., 2008). In animals, manually introduced short hairpin RNAs could interfere with the expression of endogenous miRNA genes through drastic competition for the nuclear exportation and processing machineries (Grimm et al., 2006; Stewart et al., 2008).
In mouse, posttranscriptional regulation was found to reside within the miRNA processing procedure. During the early developmental stage or in mouse primary tumors, many miRNA precursors were highly accumulated while their processing was intensively blocked (Thomson et al., 2006). This suggests that the expression of miRNA genes could be spatiotemporally modulated at the posttranscriptional level. Although it needs further verification, this strategy is likely to be employed by the plant miRNA biogenesis system. Additionally, several reports in animals point to the fact that AGO proteins participate in miRNA processing (Diederichs and Haber, 2007; O’Carroll et al., 2007; Cheloufi et al., 2010; Cifuentes et al., 2010; Lund et al., 2011). However, to our best knowledge, there has been no such finding in plants yet. The only related study reported that AGO1 was involved in the stabilization of certain miRNAs (Vaucheret et al., 2004, 2006). That is, AGO1 is highly correlated with the degradation rate of the mature miRNAs. If AGO1 indeed acts on miRNA processing in plants, its spatiotemporal distribution pattern could contribute to the tissue- or stage-specific expression of many miRNA genes.
A recent study carried out by Hoffer et al. (2011) discovered that posttranscriptional gene silencing (PTGS) guided by small interfering RNAs (siRNAs) could take place in the nucleus in plants, which has revolutionized the traditional view that the targets of PTGS are mature mRNAs or other transcripts in cytoplasm. On the other hand, Mortensen et al. (2011) showed that in Xenopus laevis oocytes, siRNAs antisense to the miRNA precursors were able to deplete the generation of the mature miRNAs. Since the pri-miRNAs and the pre-miRNAs are both nucleus localized in plant cells, it is reasonable to imagine that certain siRNAs, either intrinsic or extrinsic, may be capable of controlling the cellular levels of specific miRNA precursors. Furthermore, we previously proposed a feedback model between miRNA(*)s and the parental precursors in which the miRNA(*)s could bind to the complementary sites on their precursors to exert a cleavage-based repressive role, thus modulating their own biogenesis. Thus, it provides another regulatory layer of miRNA transcripts (Meng et al., 2010b). It is worth mentioning here that recent evidence in animals showed the great potential of miRNA precursors in target recognition and repression (Trujillo et al., 2010). Also, the loop sequences of the precursors could determine the activities of the corresponding mature miRNAs directly (Liu et al., 2008a). If these findings are also true in plants, an interlaced connection between miRNA processing and action could be established.
The exosome, responsible for 3′-to-5′ RNA processing and degradation, is critical for RNA metabolism in organisms (Mitchell et al., 1997). A transcriptome-wide high-resolution mapping was applied by Chekanova et al. (2007) to exhaustively identify the exosome substrates in Arabidopsis. Intriguingly, miRNA processing intermediates were cloned as one kind of exosome substrate. Since numerous exosome targets have been identified in the nucleus (Bousquet-Antonelli et al., 2000; Torchet et al., 2002; Das et al., 2003; Kadaba et al., 2004, 2006), it is reasonable that the nucleus-localized processing intermediates of plant miRNA precursors are under tight surveillance by the exosomes. Based on this result, the authors proposed that exosome-mediated degradation of these processing intermediates could facilitate efficient recycling of the miRNA-processing machineries (Chekanova et al., 2007). Moreover, the RRP6 exosome subunit in Chlamydomonas reinhardtii possesses a quality-control role in eliminating dysfunctional or damaged sRNA molecules, including mature miRNAs and siRNAs (Ibrahim et al., 2010). From this point of view, the exosome could be another important factor affecting the efficiency of miRNA processing in plants.
After DCL1-mediated two-step cleavage, the miRNA/miRNA* duplexes will be recognized by HUA ENHANCER1 (HEN1) to add methyl groups on the 2′ OH of the 3′-most terminal nucleotides on both strands (Yu et al., 2005). This is another crucial step for miRNA maturation, since terminal methylation could protect them from 3′-end uridylation and adenylation, thus stabilizing the miRNAs and the miRNA*s in vivo (Li et al., 2005; Yang et al., 2006c). Interestingly, HEN1 is not specific to miRNA duplexes. The study by Yang et al. (2006c) showed that both miRNA/miRNA* and siRNA duplexes ranging from 21 to 24 bp could be methylated. Besides, a recent study by Yu et al. (2010) further validated the competition between siRNAs and miRNAs for HEN1-mediated methylation. In this regard, the methylation-based stabilization that relies on both HEN1 activity and substrate levels could contribute to the varying abundances of the effective miRNAs.
Taken together, processing and maturation are two speed-limiting steps for the final miRNA levels (Fig. 1B). In both plants and animals, there are enormous factors and several checkpoints modulating these susceptible steps (Davis and Hata, 2009; Voinnet, 2009; Winter et al., 2009), which confer a highly dynamic nature to the miRNA activities.
RISC SORTING, WITH A DOMINANT BUT NOT COMPLETELY DETERMINED LOADING PATTERN
To exert their regulatory roles, the mature miRNAs released from the miRNA/miRNA* duplexes must be subsequently sorted into AGO-associated miRISCs (Voinnet, 2009). Two components of the sRNA sequence characteristics (i.e. 5′ terminal composition and sequence length) were demonstrated to be the key determinants for their AGO sorting patterns (Kim, 2008; Mi et al., 2008; Montgomery et al., 2008; Ebhardt et al., 2010). By employing HTS technology to investigate sRNA contents within the immunopurified AGO complexes, Mi et al. (2008) observed that the sRNAs that started with 5′ A (adenosine) were preferentially recruited by AGO2 and AGO4 and those that initiated with 5′ C (cytosine) were loaded into the AGO5-associated silencing complex. The miRNAs, most of which favored 5′ U (uridine), were largely incorporated into the AGO1 complex. Additionally, AGO1- and AGO2-associated sRNAs were predominantly 21 nucleotides in length, and AGO4-associated sRNAs tended to be 24 nucleotides, indicating a critical role of sequence length in AGO sorting (Mi et al., 2008). However, these two factors are not sufficient to determine which AGO(s) a specific sRNA should associate with. In a study by Mi et al. (2008), AGO5 was demonstrated to bind the sRNAs belonging to three size classes (i.e. 21, 22, and 24 nucleotides). Moreover, recent results revealed the association between AGO10 and miR166/165, which cannot be explained by the 5′ composition- and sequence length-based rule (Zhu et al., 2011). Another exceptional case, in which miR390 interacts with AGO7 for subsequent ta-siRNA (for trans-acting small interfering RNA) production, suggests that the miRNA species are not always associated with AGO1. The 5′ A of miR390 was suggested to specifically determine the preferential association of miR390 with AGO7 but not AGO1 (Montgomery et al., 2008). In the well-established model of miRNA biogenesis, the strand with a less stably paired 5′ end of a specific duplex is selectively recognized as the guide strand and loaded into AGO1-associated complexes (Jones-Rhoades et al., 2006). From this point of view, other sequence- or structure-based features embedded within the miRNA/miRNA* duplexes could play important roles in determining the destination of the miRNAs.
Notably, several dynamic factors should not be excluded when we try to interrogate the association between a certain AGO protein and a miRNA, considering that unexpectedly drastic loading competition might take place in a specific cellular context (Fig. 1C). Two distinct layers exist in this kind of competition: two or more AGOs compete for one miRNA, and miRNA, miRNA*s, and other sRNAs strive to incorporate into a specific AGO complex. A specific example was provided by Zhu et al. (2011) on the loading balance of miR166/165 between AGO1 and AGO10 in Arabidopsis. miR166/165 occupied the dominant portion of the AGO10-bound miRNAs, and the featured structure of the miR166/miR166* duplex predetermined the preferential association of miR166 with AGO10 (Zhu et al., 2011). This result well supports the structure-based rule for miRNA sorting, as proposed above. More interestingly, loss-of-function mutation of AGO10 significantly enhanced the association of miR166 with AGO1. Considering the elucidated role of AGO10 in maintaining an undifferentiated cell state of the shoot apical meristems (Moussian et al., 1998; Lynn et al., 1999), the authors proposed that AGO10 competed with AGO1 to sequester miR166/165, thus preventing them from targeting HD-ZIP III genes involved in shoot apical meristem maintenance (Prigge et al., 2005; Barton, 2010; Zhu et al., 2011). Considering the partially overlapping roles of some AGO family members within the plant sRNA pathways (Vaucheret, 2008; Mallory et al., 2009; Mallory and Vaucheret, 2010), we suggest that this kind of loading competition (i.e. one miRNA versus multiple AGOs) could be widespread in plants.
In several types of human cells, one investigation carried out by Khan et al. (2009) showed that transfected siRNAs could compete with endogenous miRNAs at several points of the miRNA biogenesis pathway, such as nuclear exportation and miRISC loading. It was observed that many targets of the endogenous miRNAs were significantly up-regulated after siRNA transfection, which exhibited concentration and temporal dependence. The authors proposed that the intracellular machineries for miRNA processing and action could be saturated through the invasion of sRNAs sharing overlapping biogenesis pathways with the miRNAs, and they could also compete with the miRNAs for target binding (Khan et al., 2009). In this regard, it is possible that competition among two or more sRNA species for nuclear exportation, RISC loading, and target binding may occur in plants. This scenario is reasonable considering the notion that several indistinguishable intersections exist within the biogenesis and functioning pathways between the young miRNA genes and the siRNAs in plants (Cuperus et al., 2011). Besides, the miRNA*s were recently reported to possess regulatory roles in both animals (Okamura et al., 2008; Packer et al., 2008; Yang et al., 2011) and plants (Mi et al., 2008; Devers et al., 2011; Meng et al., 2011; Zhang et al., 2011), which points to the possibility that the miRNA*s are likely to participate in the RISC loading competition (Fig. 1C).
In plants, the functional diversification of the AGO proteins is not only attributed to the protein sequences themselves but is also highly dependent on their spatiotemporal expression patterns (Vaucheret, 2008; Mallory and Vaucheret, 2010; Zhu et al., 2011). Thus, the identical subcellular localizations are the prerequisite for the in vivo association between a specific miRNA and an AGO-associated RISC (Havecker et al., 2010).
A HIGHLY DYNAMIC MIRNA-MEDIATED REGULATORY SYSTEM IN PLANTS
miRNA Turnover
Although increasing evidence points to the fact that translational repression could be adopted as one action mode by the plant miRNAs (Chen, 2004; Gandikota et al., 2007; Brodersen et al., 2008; Dugas and Bartel, 2008; Todesco et al., 2010), most highly conserved miRNAs exert their regulatory roles at the posttranscriptional level through cleavage (Jones-Rhoades et al., 2006; Voinnet, 2009). In contrast to the low complementarity between miRNAs and their targets in animals (Carthew and Sontheimer, 2009), the plant miRNA-mediated target slicing largely depends on the near-perfect binding sites within the corresponding transcripts (Mallory et al., 2004).
We must recognize that this kind of regulation is not performed in a steady-state mode but is highly dynamic. The pressing issue that needs to be addressed is how to assess the regulatory activity of a specific miRNA. Besides the checkpoints residing within the transcription, processing, and miRISC loading steps, the spatiotemporal expression patterns of both miRNAs and their targets should be taken into account. However, here, we introduce another notable layer, the turnover use of the mature miRNAs, since less attention has been paid to the fate of the miRNAs subsequent to their first round of target cleavage.
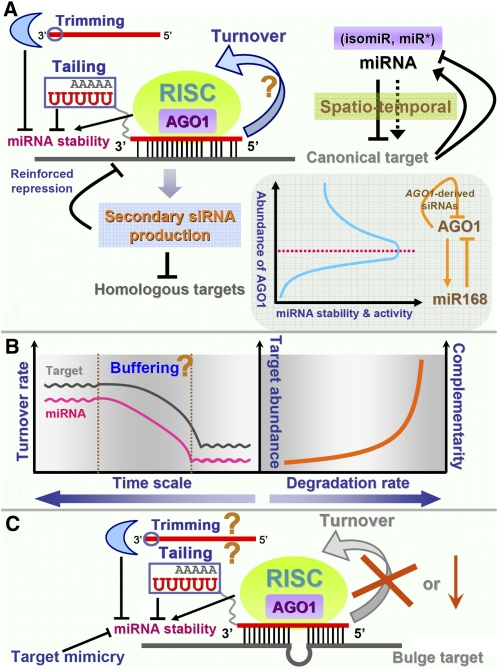
The multiple-turnover model was first raised by Hutvágner and Zamore (2002) during their study of let-7-guided RNA cleavage in human cells. Their results showed that each let-7-programmed RISC was capable of catalyzing approximately 10 rounds of cleavage action on a specific target. Also in mammalian cells, another pioneering work was recently done by Baccarini et al. (2011). Quantitative analysis of miR-223 showed that each miRNA molecule could regulate at least two target transcripts during its life cycle. Then, they demonstrated that the miRNA-mediated nonslicing pathway was multiple turnover. Although this kind of kinetic regulation was unraveled by analyzing only a few miRNAs, and it still needs to be verified whether a similar kinetic model could be applied to most miRNAs in animals (Muers, 2011), it is tempting for the plant biologist to test the possibility that the recycling strategy is also adopted by plant sRNAs for PTGS (Fig. 2A). Once the scenario that miRNAs can be reused is confirmed in plants, our current understanding of miRNA activities will be significantly updated. The turnover use of a specific miRNA molecule could remarkably elevate the targeting efficiency in planta, enabling strict surveillance of a large pool of target transcripts by a relatively small population of miRNAs. However, this does not mean an invariable one-to-multiple regulation mode. The fluctuant turnover rate of a specific miRNA in different cellular contexts or under diverse conditions predetermines the highly dynamic nature of miRNA recycling. From another point of view, the adjustable turnover rate greatly enhances the buffering capacity of the miRNA-involved regulatory system when undergoing the expression fluctuation of certain target genes. This buffering system ensures the physiologically normal expression of the genes (Fig. 2B).
Figure 2.
Schematic summarization of the factors influencing miRNA action and in vivo levels. A, Based on recent reports in animals, the turnover use of the mature miRNAs is proposed in plants, which needs further validation (denoted by the question mark). The 3′ tailing and the 3′-to-5′ trimming greatly affect the stability of mature miRNAs. Certain miRNA targets, such as TF genes, could in turn regulate the miRNAs, thus forming feedback regulatory circuits. The secondary siRNAs amplified from the cleaved target transcripts of a specific miRNA could reinforce miRNA-mediated gene silencing posttranscriptionally. One example is provided by the feedback circuit between AGO1 and miR168 in Arabidopsis. The secondary siRNAs derived from miR168-cleaved AGO1 transcripts could further regulate the expression of AGO1 posttranscriptionally. On the other hand, the abundance of the AGO1 protein significantly affects the activities and the stability of numerous miRNAs. B, The adjustable turnover rate of certain miRNAs may form an elaborate buffering system within the miRNA-mediated regulatory networks, which needs further investigation (denoted by the question mark in the left panel). Based on current hints in plants and animals, a target abundance- and complementarity-dependent model was proposed to be implicated in modulating the miRNA degradation rate (right panel). C, Based on the phenomenon of target mimicry observed by Franco-Zorrilla et al. (2007), the miRNA could be sequestered by a target decoy with a central bulge within the target recognition sites. Thus, miRNA turnover will be inhibited in that case. Although it is still not clear, the 3′ tailing and 3′-to-5′ trimming may also occur on the miRNAs sequestered by the bulged targets (denoted by question marks).
Different from the situation described by Baccarini et al. (2011), that miRNAs were not irreversibly sequestered by their targets in animals, one piece of evidence in Arabidopsis showed that miR399 was sequestered by a spurious target encoded by the noncoding gene INDUCED BY PHOSPHATE STARVATION1 (IPS1; Franco-Zorrilla et al., 2007). Within the miR399-IPS1 binding region, the high complementarity is interrupted by a three-nucleotide bulge at the expected cleavage site. This leads to an inhibitory effect on miR399 by the noncleavable IPS1 transcript, and this phenomenon was termed “target mimicry.” Recently, a large collection of target mimics against dozens of miRNAs were generated to facilitate further functional studies on plant miRNAs (Todesco et al., 2010). The activities of many designated miRNAs were confirmed to be successfully repressed by artificial target mimics, and the abundances of the corresponding targets were observed to be elevated. Hence, target mimicry, or some other similar mechanism, may provide another control layer of the miRNA turnover rate in planta (Fig. 2C).
Silencing Amplification
In addition to the miRNA-guided primary regulation, amplification of the silencing signals through secondary siRNA proliferation should be considered when evaluating plant miRNA activities (Fig. 2A). One piece of evidence supporting such activity of miRNAs is their involvement in ta-siRNA generation (Allen and Howell, 2010). To date, four ta-siRNA gene families, TAS1, TAS2, TAS3, and TAS4, have been discovered to encode ta-siRNAs in Arabidopsis. To enter the RDR6 (for RNA-dependent RNA polymerase 6)-DCL4-dependent pathway for ta-siRNA generation (Peragine et al., 2004; Vazquez et al., 2004; Yoshikawa et al., 2005), the primary TAS transcripts should be cleaved by miR173-, miR390-, or miR828-programmed AGO complexes first (Allen et al., 2005; Axtell et al., 2006; Rajagopalan et al., 2006; Chen et al., 2007; Montgomery et al., 2008). Another specific example was provided by Carrington's group. Certain miRNAs associated with AGO1-containging silencing complexes were found to be competent to trigger secondary siRNAs from the target transcripts. This RDR6-dependent pathway is specifically mediated by the 22-nucleotide miRNA species but not the 21-nucleotide ones (Cuperus et al., 2010). Besides, cleavage of the PPR transcripts by several miRNAs and ta-siRNAs could activate the biogenesis of the PPR-derived secondary siRNAs, which could in turn target the host transcripts in cis or the other homologous genes in trans, reinforcing the miRNA- or ta-siRNA-mediated gene silencing in Arabidopsis (Axtell et al., 2006; Chen et al., 2007; Howell et al., 2007; Addo-Quaye et al., 2008). More recently, the 22-nucleotide-long miR393 was shown to be involved in auxin signal-mediated leaf development through cleavage of the TIR1/AFB2 Auxin Receptor (TAAR) transcripts. Interestingly, the production of the TAAR-derived secondary siRNAs could be initiated by miR393-guided cleavage, which further enhanced the repressive regulation of TAAR-related or other homologous genes (Si-Ammour et al., 2011). Furthermore, the study by Chen et al. (2010) also supports the 22-nucleotide model that the secondary siRNA triggers tend to be the miRNAs and the siRNAs of 22 nucleotides, rather than the 21-nucleotide ones.
Action Mode Conversion
Besides the two action modes (i.e. transcript cleavage and translational repression), which are employed by plant miRNAs for target regulation, a miRNA-mediated pathway has been shown to be implicated in DNA methylation (Chellappan et al., 2010; Wu et al., 2010; Chen et al., 2011). Interestingly, in Physcomitrella patens, the miRNA-guided mRNA slicing could be converted to another action mode, DNA methylation, which was conditionally dependent on the ratio of a miRNA to its target (Khraiwesh et al., 2010). Thus, the dosage-dependent action mode conversion displays another dynamic layer of miRNA-mediated regulation in plants.
Subcellular Localization
The canonical model of miRNA-mediated regulation indicates that miRNAs recognize their targets mostly in the cytoplasm after their nuclear exportation and maturation (Carthew and Sontheimer, 2009; Voinnet, 2009). However, current evidence uncovered the novel nucleus-localized expression patterns of mature miRNAs in both plants and animals (Politz et al., 2006, 2009; Wong et al., 2011). More specifically, in human beings, Hwang et al. (2007) discovered a hexanucleotide cis-element residing within miR-29b, which could direct nuclear import of the examined miRNAs and siRNAs. Based on this result, the authors proposed that the seemingly redundant miRNAs with identical 5′ seed regions could be functionally diversified under the influence of certain cis-motifs, such as the 3′ transferable nuclear localization motif (Hwang et al., 2007). To date, only a few cases of nucleus-localized miRNAs have been discovered, but the variable subcellular localizations point to the possibility that miRNAs could target nuclear transcripts such as the primary gene transcripts and the miRNA precursors in plants.
miRNA Diffusion
The dynamic nature of miRNA-mediated regulation in plants is also strongly reflected by their diffusion effect (Fig. 1C). The sRNA molecules, including certain miRNAs and siRNAs, could not only perform cell-to-cell movement but also are implicated in long-distance transport through the phloem (Yoo et al., 2004; Kehr and Buhtz, 2008; Brosnan and Voinnet, 2011), which were demonstrated to serve as systemic signals for leaf development (Juarez et al., 2004) and phosphate homeostasis (Pant et al., 2008). From this point of view, in many cases, the plant miRNAs could exert non-cell-autonomous control over plant growth and development through cell-to-cell, tissue-to-tissue, or even organ-to-organ communication. Henceforth, we should not restrict the miRNA-mediated regulation to a limited context, since the diffusion effect must be treated as an important factor when assessing miRNA activities.
Regulatory Network
The tissue- or stage-specific expression patterns and the dynamic subcellular localizations of the miRNAs emphasize the importance of spatiotemporal colocalization of miRNA regulators and their targets for regulatory effectiveness. However, another regulatory layer, indirect targeting, should not be ignored, especially when attempting to construct miRNA-mediated regulatory networks (Rubio-Somoza et al., 2009). Within the regulatory module miR390-TAS3-ARF2/3/4, ARF2, ARF3, and ARF4 are the indirect targets of miR390 (Marin et al., 2010). Besides, the ARF transcripts targeted by miR160, miR167, and miR390 (Mallory et al., 2005; Wang et al., 2005; Wu et al., 2006; Yang et al., 2006a; Marin et al., 2010) encode ARF TFs that could interact with other ARFs and specific auxin/indole-3-acetic acid (Aux/IAA) repressors (another family of TFs) at the protein level, modulating the auxin signaling pathway in plants. Furthermore, ARFs could in turn regulate the expression of miR160, miR167 (Gutierrez et al., 2009), or other sRNA genes transcriptionally. Thus, certain ARFs, Aux/IAAs, and the sRNA genes regulated by the ARFs become the indirect targets of miR160, miR167, and miR390, contributing to the complexity of the miRNA-mediated networks. Besides, as mentioned above, the secondary siRNAs generated from the miR393-cleaved TAAR transcripts could further amplify the silencing signal involved in auxin-mediated leaf development (Si-Ammour et al., 2011).
In summary, miRNA recycling, amplification of silencing signals, dosage- and localization-dependent regulation, feedback regulation between certain miRNAs and their targets, and indirect targeting together orchestrate a fascinating, highly dynamic regulatory network with a buffering system in plants (Fig. 2).
THE FATE OF A MIRNA: TAILING/TRIMMING-INDUCED DECAY OR STABILIZATION
In addition to transcription, maturation, miRISC loading, and recycling, in vivo stability greatly influences the levels of the active miRNAs. Recent evidence gained from HTS and other methods showed that a large portion of miRNAs were tailed with one to several nontemplated 3′ U or A (adenine) nucleotides in both plants and animals, which served as a signal modulating miRNA stability (Li et al., 2005; Ramachandran and Chen, 2008; Katoh et al., 2009; Lu et al., 2009; Ameres et al., 2010; Ibrahim et al., 2010; Baccarini et al., 2011). In both kingdoms, several studies reached the consensus that adenylation increased the miRNA stability whereas uridylation promoted miRNA degradation (Katoh et al., 2009; Lu et al., 2009; Ibrahim et al., 2010; Baccarini et al., 2011). However, some exceptional cases were reported at the same time. In Drosophila, highly complementary targets triggered tailing of the small silencing RNAs, including the miRNA species, and both 3′ uridylation and 3′ adenylation were suggested to promote miRNA degradation, which was also conserved in human cells (Ameres et al., 2010). In Arabidopsis, in vitro analysis demonstrated that miRNAs could be uridylated at the 3′ ends that were not methylated, and the uridylation protected those miRNAs from SMALL RNA-DEGRADING NUCLEASE1-mediated degradation (Ramachandran and Chen, 2008). This seems quite complicated according to the above observations. However, the roles of terminal modifications on miRNA stability should become more clear with continuing research on this topic. On the other hand, the 5′-to-3′ and 3′-to-5′ trimming of miRNAs, as one means of degradation, was demonstrated to have a regulatory role in controlling the abundance of miRNAs in plants and mammalian cells (Lu et al., 2009; Ameres et al., 2010; Baccarini et al., 2011).
However, we should note that miRNA stability cannot be calculated by a simple linear equation and is affected by numerous dynamic factors. For example, adenylation of the miRNAs in Populus trichocarpa showed a tissue-specific dependence (Lu et al., 2009). More interestingly, emerging evidence in animals pointed to the fact that miRNA fate was greatly influenced by their targets. As shown in Figure 2B, the degradation rate of a miRNA was primarily affected by two factors, the complementarity and the abundance of its target. A recent study carried out by Ameres et al. (2010) proposed that extensive complementarity between a target transcript and a miRNA triggered tailing and 3′-to-5′ trimming of the miRNA. Considering the fact that most targets in plants are highly complementary to the miRNA regulators (Jones-Rhoades et al., 2006; Voinnet, 2009), whether the sequence complementarity has a great influence on the stability of plant miRNAs needs further investigation. One intriguing hint was obtained by Todesco et al. (2010). In that study, the authors examined the levels of targeted miRNAs in all the target-mimic lines of Arabidopsis. Their results showed that the abundances of nearly all the targeted miRNAs were significantly decreased, leading to the conclusion that interactions between the target decoys and the miRNAs could reduce the miRNA stability in plants (Todesco et al., 2010). Coincidentally, a repressive regulatory role of partially complementary transcripts in controlling sRNA activities was also reported in bacteria (Figueroa-Bossi et al., 2009; Overgaard et al., 2009). Another important factor, the target abundance, was reported to have a dilution effect on miRNA activities after an expression-based examination in dozens of miRNA- and siRNA-transfected HeLa S3 cell lines (Arvey et al., 2010). Considering the evidence above, we propose that the high abundances of the targets with high complementarity to the miRNAs in animals (maybe the bulge targets in plants) could not only dilute the miRNA activities through sequestration but also promote miRNA degradation more efficiently. However, even though the complementarity- and abundance-dependent model of target-induced miRNA degradation is established (Fig. 2B), some exceptions cannot be excluded. In Caenorhabditis elegans, both in vitro and in vivo analyses showed the unexpected result that miRNA degradation could be blocked by the addition of target RNAs, which was then defined as “target-mediated miRNA protection” (TMMP; Chatterjee and Grosshans, 2009; Chatterjee et al., 2011).
In addition to the negative factors promoting miRNA decay, the miRNAs could be protected and stabilized by AGO-associated miRISCs (Vaucheret et al., 2004; Kai and Pasquinelli, 2010). As reviewed by Kai and Pasquinelli (2010), in both animals and plants, AGO proteins may function in both the biogenesis and stabilization of the mature miRNAs (Kai and Pasquinelli, 2010). Considering the novel phenomenon TMMP discovered in Caenorhabditis elegans (Chatterjee and Grosshans, 2009; Chatterjee et al., 2011), we reasoned that the target-bound miRISCs could efficiently stabilize the incorporated miRNAs, and this protection may be partially dependent on the target structures (Ameres et al., 2007). As suggested by Chatterjee et al. (2011), TMMP adds another mutual regulatory layer between miRNAs and their targets, enabling dynamic expression and functional evolution of the miRNA genes. The early pioneering work by Vaucheret et al. (2004) showed that complete depletion of AGO1 led to a drop in the abundances of some miRNAs in Arabidopsis, indicating the involvement of AGO1 in miRNA biogenesis and/or stabilization (Vaucheret et al., 2004). However, things will develop in the opposite direction when they become extreme. A large excess of AGO1 protein in transgenic plants resulted in a decrease in miRNA accumulation. The authors determined that excessive AGO1 could interfere with the normal function of miRISCs and might sequester the mature miRNAs, thus inhibiting their activities (Vaucheret et al., 2004). In that study, the authors also provided the first evidence for the feedback regulation between AGO1 and miR168 and further illustrated the importance of miR168-mediated regulation of AGO1 mRNA for proper plant development (Vaucheret et al., 2004; Fig. 2A). The follow-up experiments presented more detailed insights into this feedback regulatory module (Vaucheret et al., 2006; Mallory and Vaucheret, 2009; Vaucheret, 2009). As mentioned above, AGO proteins have great potential in participating in the biogenesis and stabilization of miRNAs in various organisms (Vaucheret et al., 2004, 2006; Diederichs and Haber, 2007; O’Carroll et al., 2007; Cheloufi et al., 2010; Cifuentes et al., 2010; Kai and Pasquinelli, 2010; Lund et al., 2011).
In plants, to prevent the excessive expression of AGO1, miR168 is employed as a critical regulator controlling the abundance of AGO1 mRNAs. Interestingly, the two nodes of the feedback circuit have a common expression pattern, which was demonstrated to be coregulated transcriptionally (Vaucheret et al., 2006). Besides miR168, other regulators, both positive and negative, were discovered to modulate AGO1 activities in plants. Loss-of-function mutations of SQUINT (SQN), an ortholog of CYCLOPHILIN40 (CyP40) in Arabidopsis, caused a reduction in AGO1 activity. This supports the notion that CyP40 maintains the proper function of AGO1 or the AGO1-associated silencing complexes and that it is required for the regulatory activities of plant miRNAs (Smith et al., 2009). Additionally, HEAT SHOCK PROTEIN90 (HSP90), functioning as a molecular chaperone, was demonstrated to facilitate the in vitro assembly of AGO1-associated RISCs. The ATP-dependent, HSP90-bound, AGO1 complex-mediated process could ensure correct incorporation of the functional strands of the siRNA duplexes into the designated RISCs (Iki et al., 2010). There are also several negative regulators modulating the activities of AGO1. During a screening for the mutations suppressing the sqn phenotype, Earley et al. (2010) identified the F box gene F-BOX WITH WD-40-2, which was further shown to negatively regulate the abundance of AGO1 at the protein level in Arabidopsis. In aged fly brain, the Drosophila AGO1 protein level is negatively regulated by LEUCINE-RICH REPEAT KINASE2 (Gehrke et al., 2010). More interestingly, the siRNAs derived from the transcripts of AGO1 itself could trigger AGO1 cosuppression through the RDR6-, SUPPRESSOR OF GENE SILENCING3-, SILENCING DEFECTIVE5-, and DCL2/4-dependent silencing pathways in Arabidopsis (Mallory and Vaucheret, 2009; Fig. 2A). Furthermore, the authors showed that the siRNA-mediated AGO1 silencing depended on the correct cleavage of AGO1 transcripts by miR168, pointing to the coordinated regulatory actions of the miRNA and the siRNA pathways for maintaining AGO1 homeostasis.
Taken together, nontemplated 3′ tailing has a significant impact on the stabilization of the miRNAs. The miRNA degradation rate is highly dependent on the abundances and the complementarity of its targets. The AGO-associated miRISCs have a protective role for the mature miRNAs. More complicatedly, the activities of AGO1 and its associated silencing complex are also under strict surveillance. All these factors make the in vivo activities of the plant miRNAs more variable and more tolerant to external fluctuations.
MIRNA DYNAMICS: AN EVOLUTIONARY PERSPECTIVE
As described above, the spatiotemporal transcription and processing, the loading competition, the feedback regulation, the miRNA recycling, the buffering system, and the target-dependent decay together contribute to the dynamic activities of plant miRNAs. Apart from these aspects, the evolution of miRNA processing and functional diversification also underscores the dynamic nature of miRNA-based regulation in complex regulatory networks (Allen et al., 2004; Cuperus et al., 2011). The miRNA genes were suggested to originate from inverted repeats that could form self-complementary regions, such as the nonautonomous transposons containing flanking terminal inverted repeats (Allen et al., 2004; Vazquez et al., 2010; Cuperus et al., 2011). During evolutionary history, new miRNA families were spawned, and some might have been lost at a high frequency. The transitional loci of the newly born miRNA genes may be difficult to identify, since some indivisible intersections exist between the miRNA and siRNA pathways (Cuperus et al., 2011). From the proto-miRNAs, to the inverted repeat-miRNAs, to the young miRNAs, and finally to the highly conserved miRNA genes (Vazquez et al., 2010), the evolution of the miRNA genes was indicated to be a neutral process (Cuperus et al., 2011). However, we could not completely exclude the selective evolutionary process that the miRNAs with essential biological roles might be preferentially retained and functionally diversified in different plant species while the functionally inert ones tend to be lost more frequently.
This dynamic evolution may also lead to the discrepancy of miRNA processing and action between the plant and animal systems. The established miRNA-mediated regulatory modes tell us that the plant miRNAs recognize and cleave the target transcripts with high complementarity, whereas in animals, the imperfect interactions between specific miRNAs and their targets lead to translational repression in most cases (Carthew and Sontheimer, 2009; Voinnet, 2009). Considering the notion mentioned above that high target complementarity might result in an elevated degradation rate of mature miRNAs (Fig. 2B), we suspect that the evolved HEN1-mediated 3′ methylation of miRNA/miRNA* duplexes in plants could protect the miRNAs from tailing and trimming when they interact with highly complementary targets (Li et al., 2005; Yu et al., 2005). On the other hand, the relatively low complementarity between the targets and the animal miRNAs might be able to compensate for the lack of the methylation machinery for miRNA maturation. In support of this notion, the maturation of Piwi (for P element-induced wimpy testis)-interacting RNAs in animals, which function in transposon silencing in germline cells, requires 3′ trimming and methylation. Similar to most miRNA-target interactions in plants, the targets are highly complementary to the Piwi-interacting RNAs (Vagin et al., 2006; Horwich et al., 2007; Saito et al., 2007; Kurth and Mochizuki, 2009; Senti and Brennecke, 2010). Additional evidence provided by Ameres et al. (2010) showed that the Drosophila AGO2-bound siRNAs targeting viral and transposon RNAs with high complementarity possessed a 2′-O-methyl group at their 3′ ends, while the AGO1-associated miRNAs did not. More interestingly, increasing the complementarity between a target transcript and the regulatory miRNA could significantly reduce its stability by triggering tailing and 3′-to-5′ trimming of this unmethylated miRNA (Ameres et al., 2010). Whether these observations indicate the nexus of the coevolved miRNA-mediated regulatory systems between plants and animals needs clarification.
TOWARD THE GENUINE ACTIVITIES OF MIRNAS
In this review, we summarized all the major processes (i.e. transcriptional control, processing and maturation, miRISC loading, and miRNA action and fate) influencing the in vivo levels of plant miRNAs. Based on current reports in both plants and animals, numerous important factors embedded within each process were presented, which made great contributions to the dynamic nature of the regulatory activities of miRNAs. In addition to the spatiotemporal expression of the miRNA-target pairs and the machineries responsible for miRNA processing and action and miRNA-involved loading competition, several novel actions, such as recycling, buffering, feedback regulation, secondary amplification, and target-dependent stabilization, converge to a complex equation for miRNA activity calculation. Furthermore, both the birth and death and the functional diversification of miRNA genes represent another dynamic aspect of plant miRNA genes. All these dynamic features of miRNAs infuse more energy to the buffering systems of the gene regulatory networks in plants.
Finally, we want to emphasize the importance of taking all the variable elements into account when assessing the real activities of miRNAs in planta. At the same time, more and more such factors are being uncovered. It is foreseeable that once the comprehensive equation is reached, miRNA activities could be precisely quantified by drawing an elaborate curve showing their dynamic variation in a specific cellular context.
Acknowledgments
We sincerely apologize to those whose original works could not be cited in this review due to space constraints.
References
- Addo-Quaye C, Eshoo TW, Bartel DP, Axtell MJ. (2008) Endogenous siRNA and miRNA targets identified by sequencing of the Arabidopsis degradome. Curr Biol 18: 758–762 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Allen E, Howell MD. (2010) miRNAs in the biogenesis of trans-acting siRNAs in higher plants. Semin Cell Dev Biol 21: 798–804 [DOI] [PubMed] [Google Scholar]
- Allen E, Xie Z, Gustafson AM, Carrington JC. (2005) MicroRNA-directed phasing during trans-acting siRNA biogenesis in plants. Cell 121: 207–221 [DOI] [PubMed] [Google Scholar]
- Allen E, Xie Z, Gustafson AM, Sung GH, Spatafora JW, Carrington JC. (2004) Evolution of microRNA genes by inverted duplication of target gene sequences in Arabidopsis thaliana. Nat Genet 36: 1282–1290 [DOI] [PubMed] [Google Scholar]
- Ameres SL, Horwich MD, Hung JH, Xu J, Ghildiyal M, Weng Z, Zamore PD. (2010) Target RNA-directed trimming and tailing of small silencing RNAs. Science 328: 1534–1539 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ameres SL, Martinez J, Schroeder R. (2007) Molecular basis for target RNA recognition and cleavage by human RISC. Cell 130: 101–112 [DOI] [PubMed] [Google Scholar]
- Arvey A, Larsson E, Sander C, Leslie CS, Marks DS. (2010) Target mRNA abundance dilutes microRNA and siRNA activity. Mol Syst Biol 6: 363. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Axtell MJ, Jan C, Rajagopalan R, Bartel DP. (2006) A two-hit trigger for siRNA biogenesis in plants. Cell 127: 565–577 [DOI] [PubMed] [Google Scholar]
- Baccarini A, Chauhan H, Gardner TJ, Jayaprakash AD, Sachidanandam R, Brown BD. (2011) Kinetic analysis reveals the fate of a microRNA following target regulation in mammalian cells. Curr Biol 21: 369–376 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bari R, Datt Pant B, Stitt M, Scheible WR. (2006) PHO2, microRNA399, and PHR1 define a phosphate-signaling pathway in plants. Plant Physiol 141: 988–999 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Barton MK. (2010) Twenty years on: the inner workings of the shoot apical meristem, a developmental dynamo. Dev Biol 341: 95–113 [DOI] [PubMed] [Google Scholar]
- Blow MJ, Grocock RJ, van Dongen S, Enright AJ, Dicks E, Futreal PA, Wooster R, Stratton MR. (2006) RNA editing of human microRNAs. Genome Biol 7: R27. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bousquet-Antonelli C, Presutti C, Tollervey D. (2000) Identification of a regulated pathway for nuclear pre-mRNA turnover. Cell 102: 765–775 [DOI] [PubMed] [Google Scholar]
- Brodersen P, Sakvarelidze-Achard L, Bruun-Rasmussen M, Dunoyer P, Yamamoto YY, Sieburth L, Voinnet O. (2008) Widespread translational inhibition by plant miRNAs and siRNAs. Science 320: 1185–1190 [DOI] [PubMed] [Google Scholar]
- Brosnan CA, Voinnet O. (2011) Cell-to-cell and long-distance siRNA movement in plants: mechanisms and biological implications. Curr Opin Plant Biol 14: 580–587 [DOI] [PubMed] [Google Scholar]
- Carrington JC, Ambros V. (2003) Role of microRNAs in plant and animal development. Science 301: 336–338 [DOI] [PubMed] [Google Scholar]
- Carthew RW, Sontheimer EJ. (2009) Origins and mechanisms of miRNAs and siRNAs. Cell 136: 642–655 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chatterjee S, Fasler M, Büssing I, Grosshans H. (2011) Target-mediated protection of endogenous microRNAs in C. elegans. Dev Cell 20: 388–396 [DOI] [PubMed] [Google Scholar]
- Chatterjee S, Grosshans H. (2009) Active turnover modulates mature microRNA activity in Caenorhabditis elegans. Nature 461: 546–549 [DOI] [PubMed] [Google Scholar]
- Chekanova JA, Gregory BD, Reverdatto SV, Chen H, Kumar R, Hooker T, Yazaki J, Li P, Skiba N, Peng Q, et al. (2007) Genome-wide high-resolution mapping of exosome substrates reveals hidden features in the Arabidopsis transcriptome. Cell 131: 1340–1353 [DOI] [PubMed] [Google Scholar]
- Chellappan P, Xia J, Zhou X, Gao S, Zhang X, Coutino G, Vazquez F, Zhang W, Jin H. (2010) siRNAs from miRNA sites mediate DNA methylation of target genes. Nucleic Acids Res 38: 6883–6894 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cheloufi S, Dos Santos CO, Chong MM, Hannon GJ. (2010) A dicer-independent miRNA biogenesis pathway that requires Ago catalysis. Nature 465: 584–589 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chen D, Meng Y, Yuan C, Bai L, Huang D, Lv S, Wu P, Chen LL, Chen M. (2011) Plant siRNAs from introns mediate DNA methylation of host genes. RNA 17: 1012–1024 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chen HM, Chen LT, Patel K, Li YH, Baulcombe DC, Wu SH. (2010) 22-nucleotide RNAs trigger secondary siRNA biogenesis in plants. Proc Natl Acad Sci USA 107: 15269–15274 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chen HM, Li YH, Wu SH. (2007) Bioinformatic prediction and experimental validation of a microRNA-directed tandem trans-acting siRNA cascade in Arabidopsis. Proc Natl Acad Sci USA 104: 3318–3323 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chen X. (2004) A microRNA as a translational repressor of APETALA2 in Arabidopsis flower development. Science 303: 2022–2025 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chen X. (2009) Small RNAs and their roles in plant development. Annu Rev Cell Dev Biol 25: 21–44 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cifuentes D, Xue H, Taylor DW, Patnode H, Mishima Y, Cheloufi S, Ma E, Mane S, Hannon GJ, Lawson ND, et al. (2010) A novel miRNA processing pathway independent of Dicer requires Argonaute2 catalytic activity. Science 328: 1694–1698 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cuperus JT, Carbonell A, Fahlgren N, Garcia-Ruiz H, Burke RT, Takeda A, Sullivan CM, Gilbert SD, Montgomery TA, Carrington JC. (2010) Unique functionality of 22-nt miRNAs in triggering RDR6-dependent siRNA biogenesis from target transcripts in Arabidopsis. Nat Struct Mol Biol 17: 997–1003 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cuperus JT, Fahlgren N, Carrington JC. (2011) Evolution and functional diversification of MIRNA genes. Plant Cell 23: 431–442 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Das B, Butler JS, Sherman F. (2003) Degradation of normal mRNA in the nucleus of Saccharomyces cerevisiae. Mol Cell Biol 23: 5502–5515 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Davis BN, Hata A. (2009) Regulation of microRNA biogenesis: a miRiad of mechanisms. Cell Commun Signal 7: 18. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Devers EA, Branscheid A, May P, Krajinski F. (2011) Stars and symbiosis: microRNA- and microRNA*-mediated transcript cleavage involved in arbuscular mycorrhizal symbiosis. Plant Physiol 156: 1990–2010 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Diederichs S, Haber DA. (2007) Dual role for argonautes in microRNA processing and posttranscriptional regulation of microRNA expression. Cell 131: 1097–1108 [DOI] [PubMed] [Google Scholar]
- Dugas DV, Bartel B. (2008) Sucrose induction of Arabidopsis miR398 represses two Cu/Zn superoxide dismutases. Plant Mol Biol 67: 403–417 [DOI] [PubMed] [Google Scholar]
- Earley K, Smith M, Weber R, Gregory B, Poethig R. (2010) An endogenous F-box protein regulates ARGONAUTE1 in Arabidopsis thaliana. Silence 1: 15. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ebhardt HA, Fedynak A, Fahlman RP. (2010) Naturally occurring variations in sequence length creates microRNA isoforms that differ in argonaute effector complex specificity. Silence 1: 12. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ebhardt HA, Tsang HH, Dai DC, Liu Y, Bostan B, Fahlman RP. (2009) Meta-analysis of small RNA-sequencing errors reveals ubiquitous post-transcriptional RNA modifications. Nucleic Acids Res 37: 2461–2470 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Figueroa-Bossi N, Valentini M, Malleret L, Fiorini F, Bossi L. (2009) Caught at its own game: regulatory small RNA inactivated by an inducible transcript mimicking its target. Genes Dev 23: 2004–2015 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Franco-Zorrilla JM, Valli A, Todesco M, Mateos I, Puga MI, Rubio-Somoza I, Leyva A, Weigel D, García JA, Paz-Ares J. (2007) Target mimicry provides a new mechanism for regulation of microRNA activity. Nat Genet 39: 1033–1037 [DOI] [PubMed] [Google Scholar]
- Gandikota M, Birkenbihl RP, Höhmann S, Cardon GH, Saedler H, Huijser P. (2007) The miRNA156/157 recognition element in the 3′ UTR of the Arabidopsis SBP box gene SPL3 prevents early flowering by translational inhibition in seedlings. Plant J 49: 683–693 [DOI] [PubMed] [Google Scholar]
- Gehrke S, Imai Y, Sokol N, Lu B. (2010) Pathogenic LRRK2 negatively regulates microRNA-mediated translational repression. Nature 466: 637–641 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Grimm D, Streetz KL, Jopling CL, Storm TA, Pandey K, Davis CR, Marion P, Salazar F, Kay MA. (2006) Fatality in mice due to oversaturation of cellular microRNA/short hairpin RNA pathways. Nature 441: 537–541 [DOI] [PubMed] [Google Scholar]
- Gutierrez L, Bussell JD, Pacurar DI, Schwambach J, Pacurar M, Bellini C. (2009) Phenotypic plasticity of adventitious rooting in Arabidopsis is controlled by complex regulation of AUXIN RESPONSE FACTOR transcripts and microRNA abundance. Plant Cell 21: 3119–3132 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Havecker ER, Wallbridge LM, Hardcastle TJ, Bush MS, Kelly KA, Dunn RM, Schwach F, Doonan JH, Baulcombe DC. (2010) The Arabidopsis RNA-directed DNA methylation argonautes functionally diverge based on their expression and interaction with target loci. Plant Cell 22: 321–334 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hoffer P, Ivashuta S, Pontes O, Vitins A, Pikaard C, Mroczka A, Wagner N, Voelker T. (2011) Posttranscriptional gene silencing in nuclei. Proc Natl Acad Sci USA 108: 409–414 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Horwich MD, Li C, Matranga C, Vagin V, Farley G, Wang P, Zamore PD. (2007) The Drosophila RNA methyltransferase, DmHen1, modifies germline piRNAs and single-stranded siRNAs in RISC. Curr Biol 17: 1265–1272 [DOI] [PubMed] [Google Scholar]
- Howell MD, Fahlgren N, Chapman EJ, Cumbie JS, Sullivan CM, Givan SA, Kasschau KD, Carrington JC. (2007) Genome-wide analysis of the RNA-DEPENDENT RNA POLYMERASE6/DICER-LIKE4 pathway in Arabidopsis reveals dependency on miRNA- and tasiRNA-directed targeting. Plant Cell 19: 926–942 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hsieh LC, Lin SI, Shih AC, Chen JW, Lin WY, Tseng CY, Li WH, Chiou TJ. (2009) Uncovering small RNA-mediated responses to phosphate deficiency in Arabidopsis by deep sequencing. Plant Physiol 151: 2120–2132 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hutvágner G, Zamore PD. (2002) A microRNA in a multiple-turnover RNAi enzyme complex. Science 297: 2056–2060 [DOI] [PubMed] [Google Scholar]
- Hwang HW, Wentzel EA, Mendell JT. (2007) A hexanucleotide element directs microRNA nuclear import. Science 315: 97–100 [DOI] [PubMed] [Google Scholar]
- Ibrahim F, Rymarquis LA, Kim EJ, Becker J, Balassa E, Green PJ, Cerutti H. (2010) Uridylation of mature miRNAs and siRNAs by the MUT68 nucleotidyltransferase promotes their degradation in Chlamydomonas. Proc Natl Acad Sci USA 107: 3906–3911 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Iida K, Jin H, Zhu JK. (2009) Bioinformatics analysis suggests base modifications of tRNAs and miRNAs in Arabidopsis thaliana. BMC Genomics 10: 155. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Iki T, Yoshikawa M, Nishikiori M, Jaudal MC, Matsumoto-Yokoyama E, Mitsuhara I, Meshi T, Ishikawa M. (2010) In vitro assembly of plant RNA-induced silencing complexes facilitated by molecular chaperone HSP90. Mol Cell 39: 282–291 [DOI] [PubMed] [Google Scholar]
- Johnson C, Kasprzewska A, Tennessen K, Fernandes J, Nan GL, Walbot V, Sundaresan V, Vance V, Bowman LH. (2009) Clusters and superclusters of phased small RNAs in the developing inflorescence of rice. Genome Res 19: 1429–1440 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jones-Rhoades MW, Bartel DP, Bartel B. (2006) MicroRNAs and their regulatory roles in plants. Annu Rev Plant Biol 57: 19–53 [DOI] [PubMed] [Google Scholar]
- Juarez MT, Kui JS, Thomas J, Heller BA, Timmermans MC. (2004) MicroRNA-mediated repression of rolled leaf1 specifies maize leaf polarity. Nature 428: 84–88 [DOI] [PubMed] [Google Scholar]
- Kadaba S, Krueger A, Trice T, Krecic AM, Hinnebusch AG, Anderson J. (2004) Nuclear surveillance and degradation of hypomodified initiator tRNAMet in S. cerevisiae. Genes Dev 18: 1227–1240 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kadaba S, Wang X, Anderson JT. (2006) Nuclear RNA surveillance in Saccharomyces cerevisiae: Trf4p-dependent polyadenylation of nascent hypomethylated tRNA and an aberrant form of 5S rRNA. RNA 12: 508–521 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kai ZS, Pasquinelli AE. (2010) MicroRNA assassins: factors that regulate the disappearance of miRNAs. Nat Struct Mol Biol 17: 5–10 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Katoh T, Sakaguchi Y, Miyauchi K, Suzuki T, Kashiwabara S, Baba T, Suzuki T. (2009) Selective stabilization of mammalian microRNAs by 3′ adenylation mediated by the cytoplasmic poly(A) polymerase GLD-2. Genes Dev 23: 433–438 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kawahara Y, Megraw M, Kreider E, Iizasa H, Valente L, Hatzigeorgiou AG, Nishikura K. (2008) Frequency and fate of microRNA editing in human brain. Nucleic Acids Res 36: 5270–5280 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kawahara Y, Zinshteyn B, Chendrimada TP, Shiekhattar R, Nishikura K. (2007a) RNA editing of the microRNA-151 precursor blocks cleavage by the Dicer-TRBP complex. EMBO Rep 8: 763–769 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kawahara Y, Zinshteyn B, Sethupathy P, Iizasa H, Hatzigeorgiou AG, Nishikura K. (2007b) Redirection of silencing targets by adenosine-to-inosine editing of miRNAs. Science 315: 1137–1140 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kehr J, Buhtz A. (2008) Long distance transport and movement of RNA through the phloem. J Exp Bot 59: 85–92 [DOI] [PubMed] [Google Scholar]
- Khan AA, Betel D, Miller ML, Sander C, Leslie CS, Marks DS. (2009) Transfection of small RNAs globally perturbs gene regulation by endogenous microRNAs. Nat Biotechnol 27: 549–555 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Khraiwesh B, Arif MA, Seumel GI, Ossowski S, Weigel D, Reski R, Frank W. (2010) Transcriptional control of gene expression by microRNAs. Cell 140: 111–122 [DOI] [PubMed] [Google Scholar]
- Kidner CA, Martienssen RA. (2004) Spatially restricted microRNA directs leaf polarity through ARGONAUTE1. Nature 428: 81–84 [DOI] [PubMed] [Google Scholar]
- Kim VN. (2008) Sorting out small RNAs. Cell 133: 25–26 [DOI] [PubMed] [Google Scholar]
- Kim VN, Han J, Siomi MC. (2009a) Biogenesis of small RNAs in animals. Nat Rev Mol Cell Biol 10: 126–139 [DOI] [PubMed] [Google Scholar]
- Kim W, Benhamed M, Servet C, Latrasse D, Zhang W, Delarue M, Zhou DX. (2009b) Histone acetyltransferase GCN5 interferes with the miRNA pathway in Arabidopsis. Cell Res 19: 899–909 [DOI] [PubMed] [Google Scholar]
- Krol J, Loedige I, Filipowicz W. (2010) The widespread regulation of microRNA biogenesis, function and decay. Nat Rev Genet 11: 597–610 [DOI] [PubMed] [Google Scholar]
- Kurihara Y, Watanabe Y. (2004) Arabidopsis micro-RNA biogenesis through Dicer-like 1 protein functions. Proc Natl Acad Sci USA 101: 12753–12758 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kurth HM, Mochizuki K. (2009) 2′-O-Methylation stabilizes Piwi-associated small RNAs and ensures DNA elimination in Tetrahymena. RNA 15: 675–685 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lee Y, Kim M, Han J, Yeom KH, Lee S, Baek SH, Kim VN. (2004) MicroRNA genes are transcribed by RNA polymerase II. EMBO J 23: 4051–4060 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Li J, Yang Z, Yu B, Liu J, Chen X. (2005) Methylation protects miRNAs and siRNAs from a 3′-end uridylation activity in Arabidopsis. Curr Biol 15: 1501–1507 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Liu G, Min H, Yue S, Chen CZ. (2008a) Pre-miRNA loop nucleotides control the distinct activities of mir-181a-1 and mir-181c in early T cell development. PLoS ONE 3: e3592. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Liu HH, Tian X, Li YJ, Wu CA, Zheng CC. (2008b) Microarray-based analysis of stress-regulated microRNAs in Arabidopsis thaliana. RNA 14: 836–843 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lu S, Sun YH, Chiang VL. (2009) Adenylation of plant miRNAs. Nucleic Acids Res 37: 1878–1885 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lu S, Sun YH, Shi R, Clark C, Li L, Chiang VL. (2005) Novel and mechanical stress-responsive microRNAs in Populus trichocarpa that are absent from Arabidopsis. Plant Cell 17: 2186–2203 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Luciano DJ, Mirsky H, Vendetti NJ, Maas S. (2004) RNA editing of a miRNA precursor. RNA 10: 1174–1177 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lund E, Sheets MD, Imboden SB, Dahlberg JE. (2011) Limiting Ago protein restricts RNAi and microRNA biogenesis during early development in Xenopus laevis. Genes Dev 25: 1121–1131 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lynn K, Fernandez A, Aida M, Sedbrook J, Tasaka M, Masson P, Barton MK. (1999) The PINHEAD/ZWILLE gene acts pleiotropically in Arabidopsis development and has overlapping functions with the ARGONAUTE1 gene. Development 126: 469–481 [DOI] [PubMed] [Google Scholar]
- MacQuarrie KL, Fong AP, Morse RH, Tapscott SJ. (2011) Genome-wide transcription factor binding: beyond direct target regulation. Trends Genet 27: 141–148 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mallory A, Vaucheret H. (2010) Form, function, and regulation of ARGONAUTE proteins. Plant Cell 22: 3879–3889 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mallory AC, Bartel DP, Bartel B. (2005) MicroRNA-directed regulation of Arabidopsis AUXIN RESPONSE FACTOR17 is essential for proper development and modulates expression of early auxin response genes. Plant Cell 17: 1360–1375 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mallory AC, Hinze A, Tucker MR, Bouché N, Gasciolli V, Elmayan T, Lauressergues D, Jauvion V, Vaucheret H, Laux T. (2009) Redundant and specific roles of the ARGONAUTE proteins AGO1 and ZLL in development and small RNA-directed gene silencing. PLoS Genet 5: e1000646. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mallory AC, Reinhart BJ, Jones-Rhoades MW, Tang G, Zamore PD, Barton MK, Bartel DP. (2004) MicroRNA control of PHABULOSA in leaf development: importance of pairing to the microRNA 5′ region. EMBO J 23: 3356–3364 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mallory AC, Vaucheret H. (2009) ARGONAUTE 1 homeostasis invokes the coordinate action of the microRNA and siRNA pathways. EMBO Rep 10: 521–526 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Marin E, Jouannet V, Herz A, Lokerse AS, Weijers D, Vaucheret H, Nussaume L, Crespi MD, Maizel A. (2010) miR390, Arabidopsis TAS3 tasiRNAs, and their AUXIN RESPONSE FACTOR targets define an autoregulatory network quantitatively regulating lateral root growth. Plant Cell 22: 1104–1117 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Megraw M, Baev V, Rusinov V, Jensen ST, Kalantidis K, Hatzigeorgiou AG. (2006) MicroRNA promoter element discovery in Arabidopsis. RNA 12: 1612–1619 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Meng Y, Chen D, Jin Y, Mao C, Wu P, Chen M. (2010a) RNA editing of nuclear transcripts in Arabidopsis thaliana. BMC Genomics (Suppl 4) 11: S12. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Meng Y, Gou L, Chen D, Wu P, Chen M. (2010b) High-throughput degradome sequencing can be used to gain insights into microRNA precursor metabolism. J Exp Bot 61: 3833–3837 [DOI] [PubMed] [Google Scholar]
- Meng Y, Huang F, Shi Q, Cao J, Chen D, Zhang J, Ni J, Wu P, Chen M. (2009) Genome-wide survey of rice microRNAs and microRNA-target pairs in the root of a novel auxin-resistant mutant. Planta 230: 883–898 [DOI] [PubMed] [Google Scholar]
- Meng Y, Shao C, Gou L, Jin Y, Chen M. (2011) Construction of microRNA- and microRNA*-mediated regulatory networks in plants. RNA Biol 8: 1–25 [DOI] [PubMed] [Google Scholar]
- Mi S, Cai T, Hu Y, Chen Y, Hodges E, Ni F, Wu L, Li S, Zhou H, Long C, et al. (2008) Sorting of small RNAs into Arabidopsis argonaute complexes is directed by the 5′ terminal nucleotide. Cell 133: 116–127 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mitchell P, Petfalski E, Shevchenko A, Mann M, Tollervey D. (1997) The exosome: a conserved eukaryotic RNA processing complex containing multiple 3′→5′ exoribonucleases. Cell 91: 457–466 [DOI] [PubMed] [Google Scholar]
- Montgomery TA, Howell MD, Cuperus JT, Li D, Hansen JE, Alexander AL, Chapman EJ, Fahlgren N, Allen E, Carrington JC. (2008) Specificity of ARGONAUTE7-miR390 interaction and dual functionality in TAS3 trans-acting siRNA formation. Cell 133: 128–141 [DOI] [PubMed] [Google Scholar]
- Mortensen RD, Serra M, Steitz JA, Vasudevan S. (2011) Posttranscriptional activation of gene expression in Xenopus laevis oocytes by microRNA-protein complexes (microRNPs). Proc Natl Acad Sci USA 108: 8281–8286 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Moussian B, Schoof H, Haecker A, Jürgens G, Laux T. (1998) Role of the ZWILLE gene in the regulation of central shoot meristem cell fate during Arabidopsis embryogenesis. EMBO J 17: 1799–1809 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Muers M. (2011) Small RNAs: recycling for silencing. Nat Rev Genet 12: 227. [DOI] [PubMed] [Google Scholar]
- O’Carroll D, Mecklenbrauker I, Das PP, Santana A, Koenig U, Enright AJ, Miska EA, Tarakhovsky A. (2007) A Slicer-independent role for Argonaute 2 in hematopoiesis and the microRNA pathway. Genes Dev 21: 1999–2004 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Oh TJ, Wartell RM, Cairney J, Pullman GS. (2008) Evidence for stage-specific modulation of specific microRNAs (miRNAs) and miRNA processing components in zygotic embryo and female gametophyte of loblolly pine (Pinus taeda). New Phytol 179: 67–80 [DOI] [PubMed] [Google Scholar]
- Okamura K, Phillips MD, Tyler DM, Duan H, Chou YT, Lai EC. (2008) The regulatory activity of microRNA* species has substantial influence on microRNA and 3′ UTR evolution. Nat Struct Mol Biol 15: 354–363 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Overgaard M, Johansen J, Møller-Jensen J, Valentin-Hansen P. (2009) Switching off small RNA regulation with trap-mRNA. Mol Microbiol 73: 790–800 [DOI] [PubMed] [Google Scholar]
- Packer AN, Xing Y, Harper SQ, Jones L, Davidson BL. (2008) The bifunctional microRNA miR-9/miR-9* regulates REST and CoREST and is downregulated in Huntington’s disease. J Neurosci 28: 14341–14346 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pant BD, Buhtz A, Kehr J, Scheible WR. (2008) MicroRNA399 is a long-distance signal for the regulation of plant phosphate homeostasis. Plant J 53: 731–738 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Papp I, Mette MF, Aufsatz W, Daxinger L, Schauer SE, Ray A, van der Winden J, Matzke M, Matzke AJ. (2003) Evidence for nuclear processing of plant microRNA and short interfering RNA precursors. Plant Physiol 132: 1382–1390 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Peragine A, Yoshikawa M, Wu G, Albrecht HL, Poethig RS. (2004) SGS3 and SGS2/SDE1/RDR6 are required for juvenile development and the production of trans-acting siRNAs in Arabidopsis. Genes Dev 18: 2368–2379 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Politz JC, Hogan EM, Pederson T. (2009) MicroRNAs with a nucleolar location. RNA 15: 1705–1715 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Politz JC, Zhang F, Pederson T. (2006) MicroRNA-206 colocalizes with ribosome-rich regions in both the nucleolus and cytoplasm of rat myogenic cells. Proc Natl Acad Sci USA 103: 18957–18962 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Prigge MJ, Otsuga D, Alonso JM, Ecker JR, Drews GN, Clark SE. (2005) Class III homeodomain-leucine zipper gene family members have overlapping, antagonistic, and distinct roles in Arabidopsis development. Plant Cell 17: 61–76 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rajagopalan R, Vaucheret H, Trejo J, Bartel DP. (2006) A diverse and evolutionarily fluid set of microRNAs in Arabidopsis thaliana. Genes Dev 20: 3407–3425 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ramachandran V, Chen X. (2008) Degradation of microRNAs by a family of exoribonucleases in Arabidopsis. Science 321: 1490–1492 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Reinhart BJ, Weinstein EG, Rhoades MW, Bartel B, Bartel DP. (2002) MicroRNAs in plants. Genes Dev 16: 1616–1626 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rodriguez-Enriquez J, Dickinson HG, Grant-Downton RT. (2011) MicroRNA misregulation: an overlooked factor generating somaclonal variation? Trends Plant Sci 16: 242–248 [DOI] [PubMed] [Google Scholar]
- Rubio-Somoza I, Cuperus JT, Weigel D, Carrington JC. (2009) Regulation and functional specialization of small RNA-target nodes during plant development. Curr Opin Plant Biol 12: 622–627 [DOI] [PubMed] [Google Scholar]
- Saito K, Sakaguchi Y, Suzuki T, Suzuki T, Siomi H, Siomi MC. (2007) Pimet, the Drosophila homolog of HEN1, mediates 2′-O-methylation of Piwi-interacting RNAs at their 3′ ends. Genes Dev 21: 1603–1608 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Schmitz-Linneweber C, Small I. (2008) Pentatricopeptide repeat proteins: a socket set for organelle gene expression. Trends Plant Sci 13: 663–670 [DOI] [PubMed] [Google Scholar]
- Senti KA, Brennecke J. (2010) The piRNA pathway: a fly’s perspective on the guardian of the genome. Trends Genet 26: 499–509 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Shikanai T. (2006) RNA editing in plant organelles: machinery, physiological function and evolution. Cell Mol Life Sci 63: 698–708 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Si-Ammour A, Windels D, Arn-Bouldoires E, Kutter C, Ailhas J, Meins F, Jr, Vazquez F. (2011) miR393 and secondary siRNAs regulate expression of the TIR1/AFB2 auxin receptor clade and auxin-related development of Arabidopsis leaves. Plant Physiol 157: 683–691 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Simon SA, Meyers BC, Sherrier DJ. (2009) MicroRNAs in the rhizobia legume symbiosis. Plant Physiol 151: 1002–1008 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Smith MR, Willmann MR, Wu G, Berardini TZ, Möller B, Weijers D, Poethig RS. (2009) Cyclophilin 40 is required for microRNA activity in Arabidopsis. Proc Natl Acad Sci USA 106: 5424–5429 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Stewart CK, Li J, Golovan SP. (2008) Adverse effects induced by short hairpin RNA expression in porcine fetal fibroblasts. Biochem Biophys Res Commun 370: 113–117 [DOI] [PubMed] [Google Scholar]
- Sunkar R, Zhu JK. (2004) Novel and stress-regulated microRNAs and other small RNAs from Arabidopsis. Plant Cell 16: 2001–2019 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Thomson JM, Newman M, Parker JS, Morin-Kensicki EM, Wright T, Hammond SM. (2006) Extensive post-transcriptional regulation of microRNAs and its implications for cancer. Genes Dev 20: 2202–2207 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Todesco M, Rubio-Somoza I, Paz-Ares J, Weigel D. (2010) A collection of target mimics for comprehensive analysis of microRNA function in Arabidopsis thaliana. PLoS Genet 6: e1001031. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Torchet C, Bousquet-Antonelli C, Milligan L, Thompson E, Kufel J, Tollervey D. (2002) Processing of 3′-extended read-through transcripts by the exosome can generate functional mRNAs. Mol Cell 9: 1285–1296 [DOI] [PubMed] [Google Scholar]
- Trujillo RD, Yue SB, Tang Y, O’Gorman WE, Chen CZ. (2010) The potential functions of primary microRNAs in target recognition and repression. EMBO J 29: 3272–3285 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Vagin VV, Sigova A, Li C, Seitz H, Gvozdev V, Zamore PD. (2006) A distinct small RNA pathway silences selfish genetic elements in the germline. Science 313: 320–324 [DOI] [PubMed] [Google Scholar]
- Vaucheret H. (2008) Plant ARGONAUTES. Trends Plant Sci 13: 350–358 [DOI] [PubMed] [Google Scholar]
- Vaucheret H. (2009) AGO1 homeostasis involves differential production of 21-nt and 22-nt miR168 species by MIR168a and MIR168b. PLoS ONE 4: e6442. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Vaucheret H, Mallory AC, Bartel DP. (2006) AGO1 homeostasis entails coexpression of MIR168 and AGO1 and preferential stabilization of miR168 by AGO1. Mol Cell 22: 129–136 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Vaucheret H, Vazquez F, Crété P, Bartel DP. (2004) The action of ARGONAUTE1 in the miRNA pathway and its regulation by the miRNA pathway are crucial for plant development. Genes Dev 18: 1187–1197 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Vazquez F, Blevins T, Ailhas J, Boller T, Meins F., Jr (2008) Evolution of Arabidopsis MIR genes generates novel microRNA classes. Nucleic Acids Res 36: 6429–6438 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Vazquez F, Legrand S, Windels D. (2010) The biosynthetic pathways and biological scopes of plant small RNAs. Trends Plant Sci 15: 337–345 [DOI] [PubMed] [Google Scholar]
- Vazquez F, Vaucheret H, Rajagopalan R, Lepers C, Gasciolli V, Mallory AC, Hilbert JL, Bartel DP, Crété P. (2004) Endogenous trans-acting siRNAs regulate the accumulation of Arabidopsis mRNAs. Mol Cell 16: 69–79 [DOI] [PubMed] [Google Scholar]
- Voinnet O. (2009) Origin, biogenesis, and activity of plant microRNAs. Cell 136: 669–687 [DOI] [PubMed] [Google Scholar]
- Wang JW, Wang LJ, Mao YB, Cai WJ, Xue HW, Chen XY. (2005) Control of root cap formation by microRNA-targeted auxin response factors in Arabidopsis. Plant Cell 17: 2204–2216 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Winter J, Jung S, Keller S, Gregory RI, Diederichs S. (2009) Many roads to maturity: microRNA biogenesis pathways and their regulation. Nat Cell Biol 11: 228–234 [DOI] [PubMed] [Google Scholar]
- Wong CE, Zhao YT, Wang XJ, Croft L, Wang ZH, Haerizadeh F, Mattick JS, Singh MB, Carroll BJ, Bhalla PL. (2011) MicroRNAs in the shoot apical meristem of soybean. J Exp Bot 62: 2495–2506 [DOI] [PubMed] [Google Scholar]
- Wu G, Park MY, Conway SR, Wang JW, Weigel D, Poethig RS. (2009) The sequential action of miR156 and miR172 regulates developmental timing in Arabidopsis. Cell 138: 750–759 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wu L, Zhou H, Zhang Q, Zhang J, Ni F, Liu C, Qi Y. (2010) DNA methylation mediated by a microRNA pathway. Mol Cell 38: 465–475 [DOI] [PubMed] [Google Scholar]
- Wu MF, Tian Q, Reed JW. (2006) Arabidopsis microRNA167 controls patterns of ARF6 and ARF8 expression, and regulates both female and male reproduction. Development 133: 4211–4218 [DOI] [PubMed] [Google Scholar]
- Xie Z, Allen E, Fahlgren N, Calamar A, Givan SA, Carrington JC. (2005) Expression of Arabidopsis MIRNA genes. Plant Physiol 138: 2145–2154 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Xie Z, Kasschau KD, Carrington JC. (2003) Negative feedback regulation of Dicer-Like1 in Arabidopsis by microRNA-guided mRNA degradation. Curr Biol 13: 784–789 [DOI] [PubMed] [Google Scholar]
- Yang JH, Han SJ, Yoon EK, Lee WS. (2006a) Evidence of an auxin signal pathway, microRNA167-ARF8-GH3, and its response to exogenous auxin in cultured rice cells. Nucleic Acids Res 34: 1892–1899 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yang JS, Phillips MD, Betel D, Mu P, Ventura A, Siepel AC, Chen KC, Lai EC. (2011) Widespread regulatory activity of vertebrate microRNA* species. RNA 17: 312–326 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yang W, Chendrimada TP, Wang Q, Higuchi M, Seeburg PH, Shiekhattar R, Nishikura K. (2006b) Modulation of microRNA processing and expression through RNA editing by ADAR deaminases. Nat Struct Mol Biol 13: 13–21 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yang Z, Ebright YW, Yu B, Chen X. (2006c) HEN1 recognizes 21-24 nt small RNA duplexes and deposits a methyl group onto the 2′ OH of the 3′ terminal nucleotide. Nucleic Acids Res 34: 667–675 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yao Y, Guo G, Ni Z, Sunkar R, Du J, Zhu JK, Sun Q. (2007) Cloning and characterization of microRNAs from wheat (Triticum aestivum L.). Genome Biol 8: R96. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yoo BC, Kragler F, Varkonyi-Gasic E, Haywood V, Archer-Evans S, Lee YM, Lough TJ, Lucas WJ. (2004) A systemic small RNA signaling system in plants. Plant Cell 16: 1979–2000 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yoshikawa M, Peragine A, Park MY, Poethig RS. (2005) A pathway for the biogenesis of trans-acting siRNAs in Arabidopsis. Genes Dev 19: 2164–2175 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yu B, Bi L, Zhai J, Agarwal M, Li S, Wu Q, Ding SW, Meyers BC, Vaucheret H, Chen X. (2010) siRNAs compete with miRNAs for methylation by HEN1 in Arabidopsis. Nucleic Acids Res 38: 5844–5850 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yu B, Yang Z, Li J, Minakhina S, Yang M, Padgett RW, Steward R, Chen X. (2005) Methylation as a crucial step in plant microRNA biogenesis. Science 307: 932–935 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhang X, Zhao H, Gao S, Wang WC, Katiyar-Agarwal S, Huang HD, Raikhel N, Jin H. (2011) Arabidopsis Argonaute 2 regulates innate immunity via miRNA393(∗)-mediated silencing of a Golgi-localized SNARE gene, MEMB12. Mol Cell 42: 356–366 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhou X, Ruan J, Wang G, Zhang W. (2007) Characterization and identification of microRNA core promoters in four model species. PLoS Comput Biol 3: e37. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhu H, Hu F, Wang R, Zhou X, Sze SH, Liou LW, Barefoot A, Dickman M, Zhang X. (2011) Arabidopsis Argonaute10 specifically sequesters miR166/165 to regulate shoot apical meristem development. Cell 145: 242–256 [DOI] [PMC free article] [PubMed] [Google Scholar]