Abstract
Protein phosphorylation, catalyzed by the opposing actions of protein kinases and phosphatases, is a cornerstone of cellular signaling and regulation. Since their discovery, protein phosphatases have emerged as highly regulated enzymes with specificity that rivals their counteracting kinase partners. However, despite years of focused characterization in mammalian and yeast systems, many protein phosphatases in plants remain poorly or incompletely characterized. Here, we describe a bioinformatic, biochemical, and cellular examination of an ancient, Bacterial-like subclass of the phosphoprotein phosphatase (PPP) family designated the Shewanella-like protein phosphatases (SLP phosphatases). The SLP phosphatase subcluster is highly conserved in all plants, mosses, and green algae, with members also found in select fungi, protists, and bacteria. As in other plant species, the nucleus-encoded Arabidopsis (Arabidopsis thaliana) SLP phosphatases (AtSLP1 and AtSLP2) lack genetic redundancy and phylogenetically cluster into two distinct groups that maintain different subcellular localizations, with SLP1 being chloroplastic and SLP2 being cytosolic. Using heterologously expressed and purified protein, the enzymatic properties of both AtSLP1 and AtSLP2 were examined, revealing unique metal cation preferences in addition to a complete insensitivity to the classic serine/threonine PPP protein phosphatase inhibitors okadaic acid and microcystin. The unique properties and high conservation of the plant SLP phosphatases, coupled to their exclusion from animals, red algae, cyanobacteria, archaea, and most bacteria, render understanding the function(s) of this new subclass of PPP family protein phosphatases of particular interest.
Since their discovery, protein phosphatases have emerged as key components of cellular regulation, with roles in essentially all aspects of biology. Numerous phosphatases exist in nature, ranging from phosphate-scavenging acid phosphatases (Bozzo et al., 2002) to highly specific protein phosphatases, which remove covalently attached phosphate groups from amino acids phosphorylated by a specific protein kinase (DeLong, 2006; Moorhead et al., 2007). Reversible protein phosphorylation has been documented to occur on nine amino acids in eukaryotes, however, most phosphorylation events utilize Ser, Thr, and Tyr residues (Olsen et al., 2006; Sugiyama et al., 2008). From mass spectrometry data, it has been estimated that 70% of all proteins are regulated by reversible phosphorylation, emphasizing the prevalence of regulatory phosphorylation events in biological systems (Olsen et al., 2010) and rendering the functional characterization of the Ser/Thr protein phosphatases of continued interest to most areas of cell biology.
There are four major families of protein phosphatases, the phosphoprotein phosphatases (PPP), phosphoprotein metallophosphatases (PPM), phosphotyrosine phosphatases, and Asp-based protein phosphatases (Kerk et al., 2008). Each family contains at least one member capable of dephosphorylating phospho-Ser/Thr residues; however, the vast majority of Ser/Thr phosphatases are found in the PPP and PPM families (Kerk et al., 2008). In higher plants, such as Arabidopsis (Arabidopsis thaliana), these two families comprise 102 of the approximately 150 protein phosphatase catalytic subunits, compared with only 31 of the approximately 148 protein phosphatases in humans (Kerk et al., 2008). This likely indicates an increased diversity of functions for Ser/Thr (de)phosphorylation in plant cell function.
Despite having similar catalytic mechanisms and targeting the same phosphorylated residues, the PPP and PPM phosphatases differ significantly. Unlike the PPP enzymes, PPMs are Mg2+/Mn2+ dependent, lack associated regulatory subunits, and are insensitive to small molecule inhibitors, such as okadaic acid (OA) and microcystin (MC; Shi, 2009). Most PPP family phosphatase catalytic subunits lack accessory domains but typically associate with additional proteins (regulatory subunits) to direct their cellular functions (Moorhead et al., 2009). In plants, the PPP family consists of PP1, PP2A (PP2), PP4, PP5, PP6, and PP7 mammalian protein homologs but lack PP2B (also known as PP3)-related phosphatases (Moorhead et al., 2009). In addition to these highly conserved PPP subclasses, Arabidopsis also possesses six conserved PPP family phosphatases that are absent in mammals (Kerk et al., 2008; Moorhead et al., 2009). Four of these phosphatases are Kelch-repeat domain-containing protein phosphatases shown to be involved in brassinosteroid signaling (Mora-García et al., 2004), while the other two phosphatases are distantly related to several bacterial phosphatases first noted in Shewanella and as such were termed Shewanella-like phosphatases, or SLPs (Andreeva and Kutuzov, 2004). Due to their likely ancient prokaryotic origin and a number of unique structural elements within their amino acid sequences, characterization of the SLP protein phosphatases from Arabidopsis was undertaken to elucidate the biological and biochemical properties of this unique PPP protein phosphatase family subclass.
RESULTS
Prevalence of SLPs across the Domains of Life
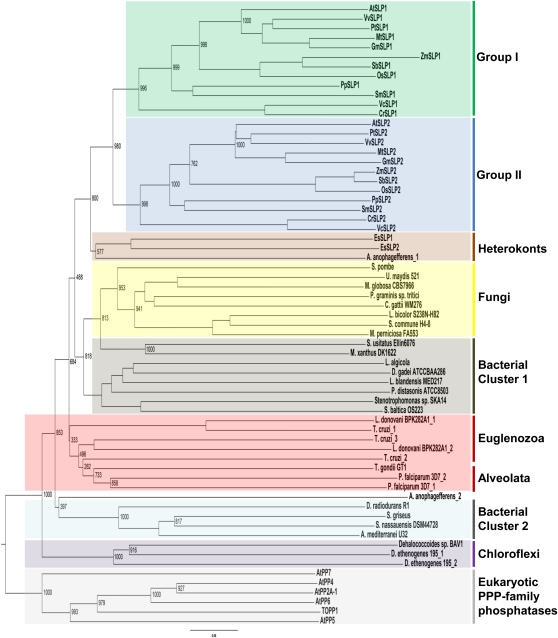
Since the initial documentation of genes encoding AtSLP1 and AtSLP2 (Andreeva and Kutuzov, 2004), numerous genomes from a variety of higher and lower plants, as well as other organisms, have been sequenced. Preliminary BLASTP analyses of the Arabidopsis genome located at the National Center for Biotechnology Information (NCBI) with either full-length AtSLP1 or AtSLP2 retrieved multiple PPP family phosphatases, with PP1 and PP2A catalytic subunits offering the highest level of identity other than themselves or each other. AtSLP1 and AtSLP2 were found to be 10% to 14% and 9% to 13% identical to PP1 and PP2A catalytic subunits, respectively, suggesting that these enzymes are PPP family protein phosphatases, consistent with previous observations (Andreeva and Kutuzov 2004; Moorhead et al., 2009). Further supporting this observation, no other protein phosphatase family members outside of the PPP family were retrieved in this search, and the catalytic signatures of each of these families is absent in the AtSLPs (Kerk et al., 2008; Moorhead et al., 2009). Additional SLP phosphatases were identified by screening the protein data sets of organisms across the domains of life, significantly expanding on previous bioinformatic surveys (Andreeva and Kutuzov, 2004; Kutuzov and Andreeva, 2008; Moorhead et al., 2009). Homologs of each AtSLP phosphatase were identified for every plant, moss, and green algal genome examined. These identified SLP1 and SLP2 phosphatases consistently clustered into separate groups (groups I and II) yet remained distinct from other PPP family phosphatases (Fig. 1; Supplemental Fig. S1; Supplemental Table S1). SLP homologs uncovered in eukaryotes outside of Plantae included those found in stramenopiles (heterokonts; in this case, brown algae and diatoms), alveolates (collectively the chromalveolates), fungi, and euglenozoa. SLPs were also identified in several bacterial species, and phylogenetic inference of representative SLPs indicates an ancestral prokaryotic origin for the SLPs (Fig. 1; Supplemental Fig. S1), consistent with previous studies (Andreeva and Kutuzov, 2004). Interestingly, the bacterial SLPs cluster into two branches while the alveolata and euglenozoa SLPs group with bacterial cluster 1. As well, no SLP phosphatase homologs were documented in any of the sequenced red algal or cyanobacterial genomes by BLASTP analysis, in addition to their absence in amoebozoa, animalia, and archaea.
Figure 1.
Phylogenetic analysis of select SLP phosphatases from prokaryotic ancestors to conserved eukaryotic homologs. A rooted tree was generated through the comparison of homologous full-length SLP sequences with the exception of SmSLP2 (partial sequence), obtained by BLASTP analyses. Select eukaryotic Arabidopsis PPPs were used as an outgroup to anchor the tree. Homologous SLPs were aligned using ClustalX, and the bootstrap tree was generated using the neighbor-joining function of ClustalX. The group I and group II designations were assigned based on prediction algorithms that identified either a chloroplast or cytosol subcellular localization (Supplemental Fig. S1). Numbers represent bootstrap values for that node.
Each group of the plant SLPs not only corresponds to the two distinct SLP1 and SLP2 phosphatases but also signifies distinct subcellular localization predictions. Group I denotes SLP1 phosphatases putatively targeted to the chloroplast, while group II SLP2 phosphatases are predicted to be cytosolic (Supplemental Table S1). The predicted subcellular localization designations of groups I and II SLP phosphatases were formulated based upon the overall consensus of four prediction algorithms and also provided a means to locate the putative chloroplast transit peptide cleavage site of AtSLP1. This was defined between N-terminal amino acids Ser-53 and Ala-54.
AtSLP1 and AtSLP2 Localize to Different Cellular Compartments
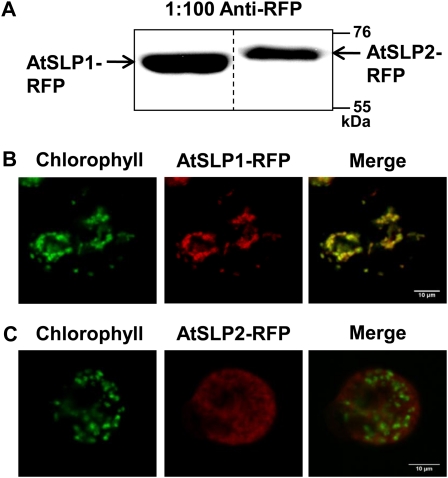
Full-length AtSLP1 and AtSLP2 were fused to a C-terminal red fluorescent protein (RFP) and transfected into Arabidopsis cell culture to create protoplasts constitutively expressing either AtSLP1-RFP or AtSLP2-RFP for live-cell imaging. Stable transfected cell culture was extracted, resolved by SDS-PAGE, and probed with anti-RFP to verify expression of the fusion protein (Fig. 2A). Positive AtSLP1-RFP cell culture demonstrated chloroplastic localization (Fig. 2B; Supplemental Fig. S2A), while positive AtSLP2-RFP cell culture maintained a nonchloroplastic localization (Fig. 2C; Supplemental Fig. S2B).
Figure 2.
In vivo subcellular localization of AtSLP1 and AtSLP2 using stably transfected Arabidopsis cell culture. A, Western-blot verification of stably transfected cell culture constitutively expressing AtSLP1-RFP or AtSLP2-RFP. Each lane contains 30 μg of crude cell culture lysate probed with 1:100 anti-RFP IgG (Chromotek). B and C, Fluorescence images of protoplasts derived from stably transfected Arabidopsis cell culture expressing AtSLP1-RFP or AtSLP2-RFP (red), respectively. Chlorophyll autofluorescence is also shown (green), and images were merged to reveal localization. All images are single slices obtained using a confocal laser scanning microscope (Leica). Bars = 10 μm.
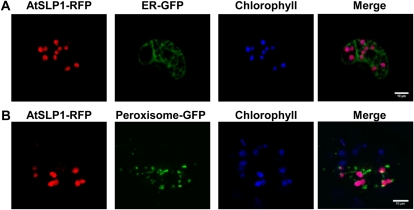
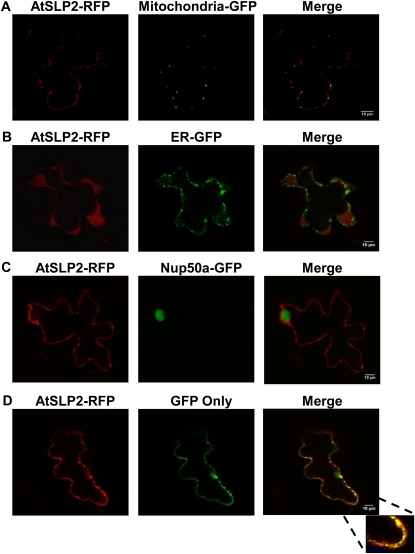
Transient coexpression of RFP-tagged AtSLPs in fava bean (Vicia faba) epidermal leaf cells with various fluorescence marker proteins was also performed to further resolve the specific subcellular localization of AtSLP1 and AtSLP2. AtSLP1-RFP coexpressed with a GFP construct specifically targeted to the endoplasmic reticulum (ER) or peroxisome confirmed the chloroplast subcellular localization of AtSLP1 by the colocalization of AtSLP1-RFP with chlorophyll autofluorescence and not with either GFP construct (Fig. 3). The localization of AtSLP2 was further resolved through the transient coexpression of AtSLP2-RFP with nontargeted GFP (GFP alone resides in both the cytosol and nucleus; Fig. 4D) and GFP targeted specifically to either the ER or mitochondria (Fig. 4, A and B). AtSLP2-RFP demonstrated specific colocalization with only the nontargeted (cytosolic) GFP, indicating a cytosolic subcellular localization (Fig. 4D). AtSLP2-RFP was also coexpressed with GFP-tagged nucleoporin protein 50a (Nup50a), a known nuclear protein (Tamura et al., 2010), to account for the dual subcellular localization of nontargeted GFP to the cytosol and nucleus (Seibel et al., 2007). AtSLP2-RFP did not colocalize with Nup50a-GFP, indicating that AtSLP2 is only cytosolic (Fig. 4C).
Figure 3.
Colocalization of AtSLP1 with fluorescent marker constructs in fava bean leaves. A and B show representative images of guard cells cobombarded with AtSLP1-RFP (red) and either ER- or peroxisome-targeted GFP (green), respectively. Chlorophyll autofluorescence (blue) is shown along with the merged image (right). The specific organelle targeting of GFP was achieved though fusion with conserved targeting motifs (Nelson et al., 2007). All images are single slices obtained using a confocal laser scanning microscope (Leica). Bars = 10 μm.
Figure 4.
Colocalization of AtSLP2 with fluorescent marker constructs in fava bean leaves. A and B, Images of pavement cells cobombarded with AtSLP2-RFP and either mitochondria- or ER-targeted GFP, respectively. The specific organelle targeting of GFP was achieved as in Figure 3. C, AtSLP2-RFP cobombarded with Nup50a-GFP (Tamura et al., 2010). D, AtSLP2 colocalized with GFP only (no targeting motif). GFP only localized to both the nuclear and cytosolic compartments (Seibel et al., 2007) but colocalizes with AtSLP2-RFP only in the cytosolic fraction. All images are single slices obtained using a confocal laser scanning microscope (Leica). Bars = 10 μm.
Spatial and Temporal Expression of AtSLP1 and AtSLP2
Initial characterization of endogenous AtSLP phosphatases consisted of an analysis of transcriptional expression using Genevestigator (https://www.genevestigator.com). The Genevestigator search engine houses a number of Arabidopsis microarray experiments based on the commercially available Arabidopsis ATH1 22k array (http://www.affymetrix.com) to explore gene expression under various experimental conditions. This screen indicates that the genes encoding the AtSLPs (At1g07010 and At1g18480) are differentially expressed among Arabidopsis plant tissues and throughout plant development (Supplemental Fig. S3). AtSLP1 had highest expression in cauline leaves, sepals, and photosynthetic tissues but was absent in roots and other nonphotosynthetic tissues (i.e. seeds), while AtSLP2 demonstrated elevated transcriptional expression in protoplasts, roots, and the endosperm of seeds (Supplemental Fig. S3A). Both AtSLP1 and AtSLP2 also exhibited different transcriptional expression patterns across plant development (Supplemental Fig. S3B) and in response to light/dark diurnal cycling. AtSLP1 transcript demonstrated a sharp increase upon germination followed by a subsequent decrease toward plant maturity (Supplemental Fig. S3B) as well as a clear diurnal fluctuation in expression pattern (Supplemental Fig. S4A). AtSLP2, however, did not demonstrate a major transcriptional fluctuation over the course of plant development (Supplemental Fig. S3B) or during the diurnal cycle (Supplemental Fig. S4B).
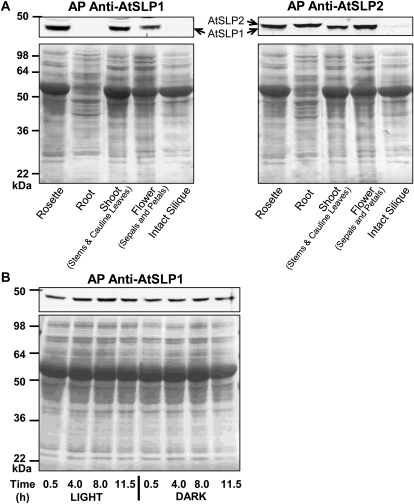
To examine if AtSLP gene expression was paralleled at the protein level, antibodies were raised using heterologously expressed His6-AtSLP1 and His6-AtSLP2 protein purified by nickel-nitrilotriacetic acid agarose (Ni-NTA; Supplemental Fig. S5). Affinity purification of each antibody was performed to enhance the specific detection of each phosphatase (Supplemental Fig. S6). The affinity purification of anti-AtSLP1 IgG provided monospecific detection of AtSLP1, while affinity purification of anti-AtSLP2 IgG resulted in an antibody that detects both AtSLP1 and AtSLP2, but with greater affinity for AtSLP2 (Supplemental Fig. S6, A and B). The documented transcriptional expression trend of AtSLP1 and AtSLP2 uncovered using Genevestigator was largely paralleled at the protein level (Fig. 5), with AtSLP1 protein observed in all photosynthetic tissues with the exception of siliques. Conversely, AtSLP2 was only definitively detectable in roots and intact siliques (containing seeds). Blotting was less conclusive for other tissues due to the cross-reactivity of the affinity-purified anti-AtSLP2 IgG with AtSLP1 and an irresolvable 2-kD mass difference between endogenous AtSLP1 (approximately 41 kD) and AtSLP2 (approximately 43 kD). Interestingly, AtSLP2 was barely detectable in intact siliques despite comparatively high levels of transcriptional expression in seeds relative to other plant tissues (Fig. 5A; Supplemental Fig. S3).
Figure 5.
Spatial and temporal western-blot analysis of AtSLP1 and AtSLP2 protein expression. A, Tissue-specific expression of AtSLP1 and AtSLP2 from different Arabidopsis Columbia tissues. Each lane contains 30 μg of clarified cell lysate. B, Time-course analysis of AtSLP1 protein expression over a 24-h light/dark cycle. Each lane contains 30 μg of clarified lysate from rosette leaf tissue. All tissues were harvested from 21-d-old plants. The bottom panels of both A and B represent Ponceau S (0.1%, w/v)-stained membranes demonstrating equal protein loading prior to incubation with either affinity-purified (AP) anti-AtSLP1 or anti-AtSLP2 IgG.
AtSLP1 protein expression was also examined over a light/dark cycle consisting of 12 h of light and 12 h of dark. Initial examination of online-accessible transcriptional expression data from the NCBI Gene Expression Omnibus (GEO) Profiles (http://www.ncbi.nlm.nih.gov/geoprofiles) demonstrated a diurnal cycling of AtSLP1 transcript but no such pattern for AtSLP2 expression (Supplemental Fig. S4). Despite the noted changes in AtSLP1 transcript levels, AtSLP1 protein was basally detectable over the entire diurnal cycle, with protein expression beginning to increase at 4 h of light and peaking at the 8-h light time point before decreasing back to basal expression levels (Fig. 5B).
To further explore the biological relevance of SLP phosphatases, numerous single T-DNA promoter and exon insertional mutant lines were obtained from both the RIKEN Experimental Plant Division (http://www.brc.riken.jp/lab/epd/Eng/) and the Arabidopsis Biological Resource Center (http://www.arabidopsis.org/). Homozygous lines for each atslp insertional mutant line were created and propagated for phenotypic analysis. As a result of each AtSLP phosphatase expression pattern, shoot and root growth were targeted for examination. The results of these preliminary analyses failed to produce any obviously discernible phenotypic differences between the insertional mutant lines and their respective Columbia and Nossen wild-type backgrounds (data not shown).
AtSLP Primary Sequences and Enzymatic Properties
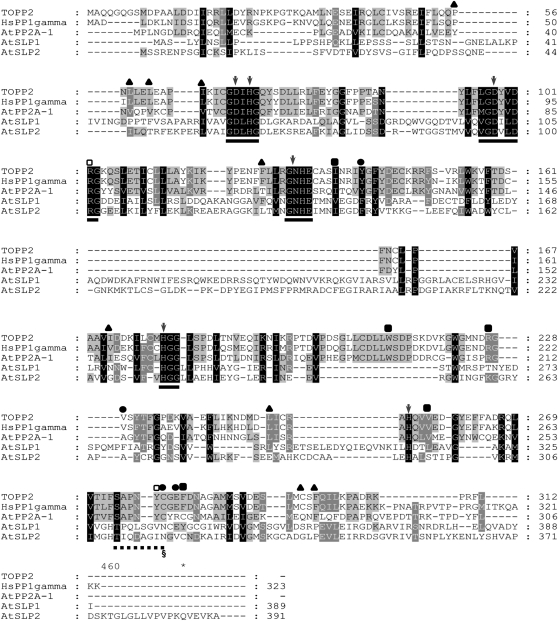
Alignment of the AtSLPs with the other Arabidopsis PPP family phosphatases and representative human PPP family phosphatases uncovered many amino acid substitutions and unique regions within each AtSLP primary amino acid sequence. A condensed alignment containing representative PPP family phosphatases is shown along with AtSLP1 and AtSLP2 (Fig. 6). Human PP1γ (HsPP1γ) and an Arabidopsis PP1 (TOPP2) were found to be 76% identical to each other but only possessed approximately 10% identity (24%–25% similarity) to either of the AtSLP phosphatases. Regions of conserved identity between HsPP1γ, TOPP2, and the AtSLP phosphatases were found to largely reside in the key amino acid clusters comprising the active sites of PPP family phosphatases (Fig. 6; Shi, 2009). AtSLP1 and AtSLP2 however, maintain 31% identity (43% overall similarity) to each other and possess many overlapping regions of identity beyond those comprising the PPP family phosphatase active sites (Fig. 6). SLP phosphatases were also found to lack seven of the eight key amino acids involved in the docking of mammalian and yeast PP1 regulatory subunits through the RVXF sequence motif found in essentially all PP1 interactors (Egloff et al., 1997; Moorhead et al., 2007; Dancheck et al., 2011). Key amino acid stretches shown to be involved in the formation of the PP2A trimeric complex were also absent (Xu et al., 2006).
Figure 6.
Primary amino acid alignment of AtSLP1 and AtSLP2 full-length sequences with representative PP1 sequences from Arabidopsis (TOPP2 [At5g59160] and AtPP2A-1 [At1g59830]) and human (HsPP1γ; NP_002701). Alignment was performed using ClustalX followed by import into GeneDoc for image assembly. Solid lines represent highly conserved motifs comprising the active site of canonical Ser/Thr phosphatases. Dashed lines represent a highly conserved region in PPPs that conveys sensitivity to MC-LR through its covalent attachment to Cys-273 (HsPP1γ) and an equivalent residue in a plant PP1, Cys-279 (TOPP2; section mark). The amino acids marked by black circles represent those specifically important to MC-LR binding, while those marked by black squares are specific to OA binding but are also key in coordinating MC-LR. The arrows denote amino acids found to play a role in metal ion coordination, while the black triangles denote amino acids found to dock the PP1 I-2 protein. Amino acids denoted with white squares have been implicated in the binding of MC-LR, OA, and I-2.
Of the many differences revealed at the primary amino acid level between the SLPs and the eukaryotic PPP family phosphatases, the absence of the canonical SAPNYC motif was of particular interest (Fig. 6). Although not directly involved in catalysis or metal ion coordination, the Cys within this motif forms a covalent bond with the potent PPP inhibitor MC-Leu and Arg (LR) (MacKintosh et al., 1995). SLP phosphatases were also found to lack other conserved amino acids within the SAPNYC motif and throughout their full-length sequence that coordinate OA (Maynes et al., 2001) and the PP1-specific inhibitor-2 (I-2) protein (Hurley et al., 2007; Fig. 6). This would suggest that the SLPs are resistant to the toxins and proteins that have evolved to inhibit the PPP family phosphatases by docking these regions. To explore this idea, enzyme assays were performed using a selection of phosphatase inhibitors.
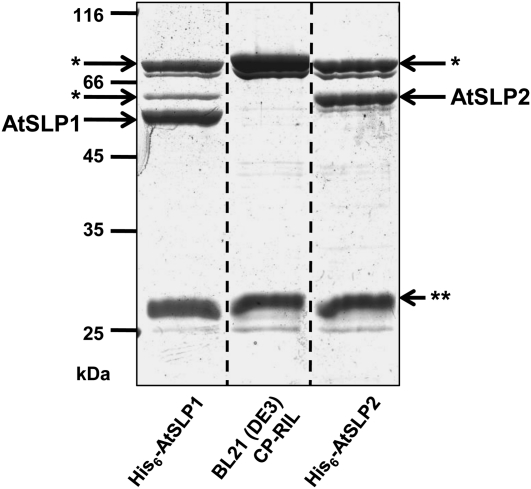
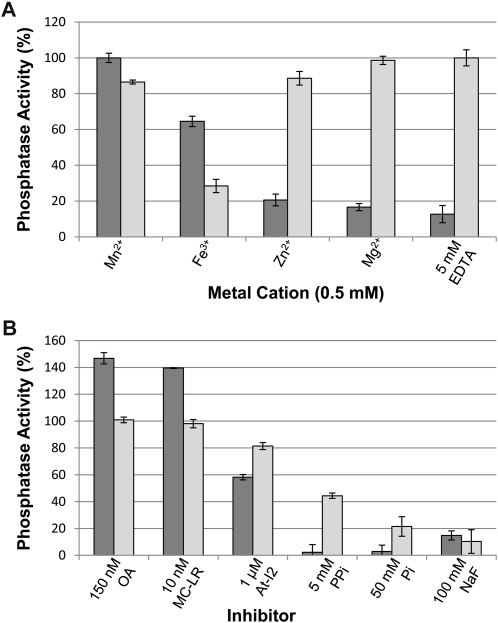
To assess the enzymatic properties of the AtSLP phosphatases, each protein was expressed as a His6 fusion protein in Escherichia coli and purified to near homogeneity by Ni-NTA (Fig. 7). Both His6-AtSLP1 and His6-AtSLP2 Ni-NTA eluates had additional nonspecific copurifying (NSCP) proteins, rendering a control Ni-NTA eluate from the same E. coli expression strain necessary to control for potential NSCP phosphatase activity during enzymatic analysis of each AtSLP (Fig. 7). The main contaminant protein was identified by mass spectrometry (data not shown) as glucosamine-fructose-6-phosphate aminotransferase, a well-documented contaminant of Ni-NTA protein purification that has no documented phosphatase activity (Badet et al., 1987; Trinkle-Mulcahy et al., 2008). Enzymatic assays were conducted using the artificial phosphatase substrate paranitrophenyl phosphate (pNPP) to assess the activity of each AtSLP phosphatase. His6-AtSLP1 was most active in the presence of Mn2+ but could also use Fe3+ to generate 65% of its Mn2+ activity (Fig. 8A). Contrary to His6-AtSLP1, His6-AtSLP2 was seemingly most active in the absence of additional metal ions (i.e. 5 mm EDTA); however, application of Student’s t test found no significant difference between the activity achieved with 5 mm EDTA and that achieved with either 0.5 mm Mg2+ or 0.5 mm Zn2+. A significant inhibitory effect was observed in the presence of Fe3+, which reduced His6-AtSLP2 activity by 70% (Fig. 8A).
Figure 7.
Twelve percent SDS-PAGE of Ni-NTA-purified His6-AtSLP1, His6-AtSLP2, and uninduced bacterial cell line control eluates stained with colloidal blue. His6-AtSLP1 and His6-AtSLP2 were partially purified by Ni-NTA along with NSCP Ni-NTA-binding proteins isolated in parallel from untransformed, uninduced BL21 (DE3) Codon(+)-RIL E. coli grown under identical conditions. The BL21 (DE3) Codon(+)-RIL E. coli Ni-NTA eluate controlled for nonspecific cleavage of the phosphatase substrate pNPP by NSCP proteins during enzymatic analysis of Ni-NTA-purified His6-AtSLP1 and His6-AtSLP2. The NSCP protein glucosamine-fructose-6-phosphate aminotransferase (asterisks) was identified by mass spectrometry, while double asterisks represent another NSCP protein. Each lane contains 5 μg of concentrated Ni-NTA eluate.
Figure 8.
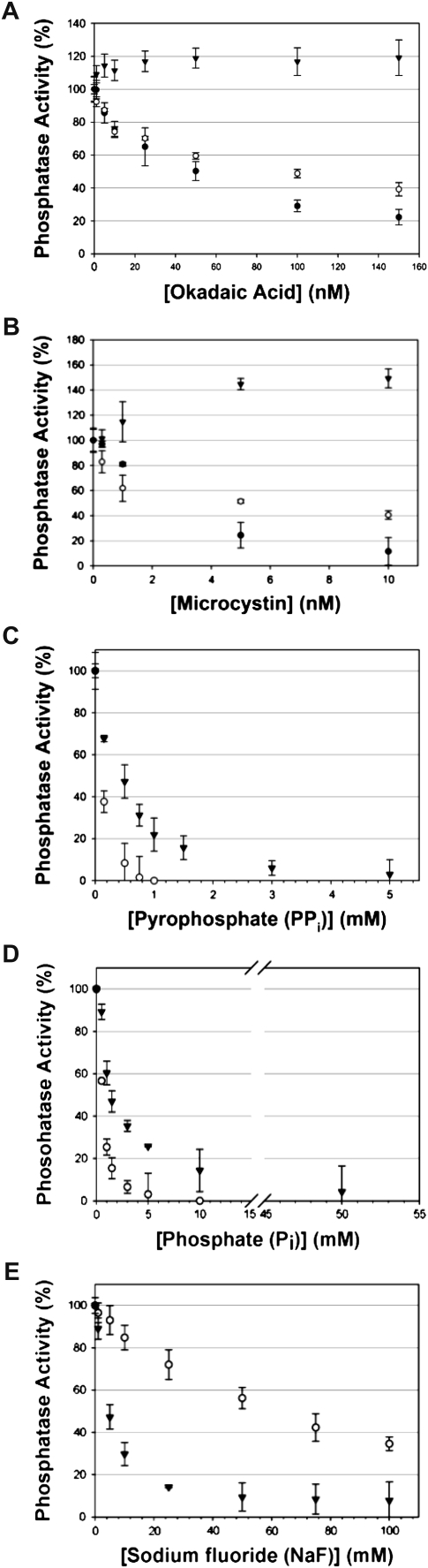
Enzymatic analysis of His6-AtSLP1 and His6-AtSLP2. A, Metal dependency assessment of each SLP phosphatase was performed using a variety of metal cations as well as 5 mm EDTA. B, Analysis of known Ser/Thr phosphatase inhibitors employed at concentrations known to fully inhibit bacterially expressed and purified human and Arabidopsis PP1 protein phosphatases. Dark gray bars represent His6-AtSLP1, while light gray bars represent His6-AtSLP2. Assays were conducted as outlined in “Materials and Methods” and were performed with or without OA, MC-LR, At-I2 (At5g52200), PPi, Pi, and NaF. One hundred percent enzyme activity in A and B is defined by the presence of 0.5 mm Mn2+ (AtSLP1 assays) or 5 mm EDTA (AtSLP2 assays). Each bar represents n = 3, and error bars indicate se.
Along with examining metal cation dependency, the AtSLPs were tested for sensitivity to classic protein phosphatase inhibitors (Fig. 8B). Both His6-AtSLP1 and His6-AtSLP2 demonstrated a complete lack of inhibition in the presence of the PPP inhibitors OA (150 nm) and MC-LR (10 nm). Control reactions using purified His6-TOPP2 and HsPP1γ (Supplemental Fig. S7) demonstrated complete inhibition at the concentrations of PPP inhibitor employed above (data not shown). Surprisingly, His6-AtSLP1 phosphatase activity was activated in the presence of OA (approximately 140%) and MC-LR (approximately 145%), while these exhibited no overall effect on His6-AtSLP2 activity (Fig. 8B). His6-AtSLP1 also showed enhanced inhibition by 5 mm pyrophosphate (PPi) and 50 mm phosphate (Pi) relative to His6-AtSLP2, while both His6-AtSLP1 and His6-AtSLP2 showed similar inhibition by 100 mm sodium fluoride (NaF; Fig. 8B). Both AtSLPs were also tested for their sensitivity to the well-characterized and specific PP1 protein inhibitor I-2 from Arabidopsis (AtI-2; Templeton et al., 2011). Both AtSLP1 and AtSLP2 exhibited minimal sensitivity to AtI-2, being inhibited approximately 40% and approximately 20%, respectively, at 1 μm concentration (Fig. 8B).
Characterization of Purified Untagged AtSLP1
Due to the inability to identify conditions permitting the stable purification and storage of AtSLP2 past initial Ni-NTA elution, only the chloroplast-localized AtSLP1 was purified to homogeneity. The initial His6-AtSLP1 Ni-NTA eluate was concentrated and subjected to Superdex 200 size-exclusion chromatography (Supplemental Fig. S8A) followed by subsequent Mono-Q anion-exchange separation of the Superdex 200 peak fractions (Supplemental Fig. S8B). Pooled and concentrated Mono-Q peak fractions were used for enzymatic analyses of untagged AtSLP1 (Supplemental Figs. S8B and S9A). Some degradation of full-length AtSLP1 was observed, as both a full-length protein and a 20-kD fragment were confirmed as AtSLP1 by matrix-assisted laser-desorption ionization time of flight mass spectrometry (Supplemental Table S2) and immunoblot analysis (Supplemental Fig. S9B).
Enzymatic characterization of pure, His6 tag-free AtSLP1 was conducted to elucidate the impact of the His6 affinity tag on both small molecule and protein inhibitor sensitivity as well as its affinity for a protein substrate. For comparison, control assays containing the same metal cations and inhibitors at the same experimental concentrations were performed in parallel using purified His6-TOPP2 and HsPP1γ. AtSLP1 metal cation dependency was reexamined and found to be very similar to that observed with His6-AtSLP1 (Supplemental Fig. S10). As observed with His6-AtSLP1, untagged AtSLP1 was again activated by both OA and MC-LR (Fig. 9, A and B). Activation by OA was moderately reduced in the absence of the His6 tag, while MC-LR elicited the same 40% increase in AtSLP1 phosphatase activity. PPi, Pi, and NaF all had inhibitory effects identical to those observed with His6-AtSLP1 (Fig. 9, C–E). Lastly, purified AtSLP1 demonstrated a specific activity of 1.03 units mg−1 when assayed using the model protein substrate glycogen phosphorylase a in place of pNPP, an activity comparable to rabbit skeletal muscle PP2A (Cohen et al., 1988). The activity of AtSLP1 toward glycogen phosphorylase a further suggests that the SLP phosphatases are in fact protein phosphatases.
Figure 9.
Assessment of purified, untagged AtSLP1 sensitivity to small molecule inhibitors. A and B, Inhibition curves comparing the phosphatase inhibitor sensitivity of AtSLP1 (black triangles) with TOPP2 (white circles) and HsPP1γ (black circles) using the potent PPP inhibitors OA and MC. C to E, Inhibition curves comparing the PPi, Pi, and NaF sensitivity of AtSLP1 with TOPP2. All phosphatase assays were conducted using the small molecule substrate pNPP as outlined in “Materials and Methods.”
DISCUSSION
SLP Phosphatases Are a Novel Subclass of PPP Family Phosphatases
The SLP enzymes were designated Shewanella like after their initial recognition as being related to Shewanella phosphatases (Andreeva and Kutuzov, 2004). Our bioinformatic search for the presence of these enzymes across the domains of life revealed a distribution in bacteria, fungi, heterokonts, and euglenozoa. Both SLP1 and SLP2 were present in plants, mosses, and green algae but absent in the sequenced genomes of all red algae, cyanobacteria, animals, and archaea. This distribution indicates a likely complex evolutionary history for SLP phosphatases. The absence of an SLP phosphatase in modern cyanobacteria casts doubt on the chloroplast version of SLP arising from the original endosymbiotic event. What this suggests is a gene duplication event in early photosynthetic eukaryotes, leading to one isoform being targeted to the chloroplast (Miyagishima, 2011), although a secondary gene loss in all cyanobacteria cannot be discounted. Further adding to the evolutionary complexity of SLPs is the presence of orthologs in brown algae. Analysis of the brown algae sequences indicates cytosolic and plastid localization for the two SLPs. These organisms are thought to have obtained plastids from a red algal ancestor (Gould et al., 2008; Keeling, 2010; Miyagishima, 2011), yet no sequenced genome of modern red algae is documented to possess an SLP phosphatase, suggesting selective gene loss in red algae and a possible gene duplication event in brown algae, again leading to one isoform being plastid targeted. Dissecting the intricate evolutionary history of these enzymes will require the sequencing of additional genomes and analysis using more sophisticated bioinformatic methods; however, among this incomplete evolutionary history is one clear outcome: predicted chloroplast and cytosolic isoforms of SLP phosphatases in plant genomes are conserved. These compartmental predictions were confirmed in Arabidopsis, supporting the notion that these enzymes play important, compartment-specific, likely nonoverlapping roles in plants.
Subcellular Targeting of the AtSLP Phosphatases
Previous bioinformatic and proteomic studies suggested that SLP phosphatases occupy a number of subcellular locations. In particular, SLP1-like phosphatases were documented as peroxisomal (Fukao et al., 2002) and speculated to be either chloroplast (Andreeva and Kutuzov, 2004) and/or ER targeted (Kutuzov and Andreeva, 2008), while SLP2-like phosphatases were similarly suggested to reside in the ER (Kutuzov and Andreeva, 2008). Using a combination of fluorescent fusion constructs stably transfected into cell culture and transiently coexpressed fava bean leaves (Fig. 2-4), a clear subcellular localization was resolved for AtSLP1 (chloroplast) and AtSLP2 (cytosol). These AtSLP findings substantiated the in silico subcellular localization prediction consensus for groups I and II SLP phosphatases (Supplemental Table S1), emphasizing the likelihood of a conserved chloroplastic and cytosolic location for the respective enzymes in all plants.
Manual and prediction algorithm-assisted inspection of AtSLP1 and other group I SLP phosphatase amino acid sequences using the peroxisomal targeting sequence (PTS1) predictor (http://mendel.imp.ac.at/mendeljsp/sat/pts1/PTS1predictor.jsp) as well as Predotar and WoLF pSORT found no evidence of a canonical C-terminal PTS1 signaling motif (data not shown). The possibility of an N-terminal PTS2 signal or a noncanonical PTS signaling motif was also considered (Lazarow, 2006; Girzalsky et al., 2009), but with the C-terminal AtSLP1-RFP fusion protein lacking observable peroxisome localization in either the stably transfected cell culture or transiently expressed conditions, it is unlikely that AtSLP1 resides in the peroxisome (Figs. 2 and 4). Previous accounts of AtSLP1 peroxisome targeting were based on the proteomic analysis of isolated plant peroxisomes (Fukao et al., 2002), rendering contamination by plastid species (i.e. chloroplasts) during purification a possibility. A previous bioinformatic analysis of the SLPs has suggested that AtSLP1 was also ER targeted (Kutuzov and Andreeva, 2008). These predictions were based on an in silico subcellular prediction result from the online resource Predotar (Kutuzov and Andreeva, 2008). Despite these findings, the consensus of the four subcellular localization prediction algorithms employed in this study, combined with experimentally verified subcellular localization results of AtSLP1, does not support ER localization for group I SLP phosphatases. One of the four in silico targeting programs used here in the subcellular localization prediction analysis was Predotar, and as in the previous study, it also predicted several group I SLP phosphatases to be ER targeted (Supplemental Table S1). ER localization, however, was not suggested by any other program, with the majority of programs predicting the same SLP phosphatases to be chloroplast targeted (Supplemental Table S1). Given the overall predicted chloroplast localization consensus of group I SLP phosphatases, the experimentally verified chloroplast localization of RFP-tagged AtSLP1 (Figs. 2B and 3), and the high level of conserved identity (approximately 50%–70%) across group I SLP phosphatase homologs (Fig. 1), all group I SLP phosphatases likely reside solely in the chloroplast.
Previous bioinformatic work also predicted AtSLP2 to be an ER-localized protein (Kutuzov and Andreeva, 2008). Our results, however, found a consistent cytosolic localization consensus for group II SLP phosphatases, with none of the in silico targeting programs predicting the group II SLP phosphatases to be ER localized (Supplemental Table S1). Moreover, AtSLP2 expressed as an RFP fusion protein in both stably transfected cell culture or transiently expressed in fava bean leaves lacked detectable ER localization (Figs. 2 and 4) but rather had a consistent cytosolic localization under both experimental conditions. Compared with group I SLP phosphatases, group II SLP phosphatases exhibit an even higher level of sequence identity (approximately 50%–80%), rendering it unlikely that an alternative localization in other plants would be observed.
Temporal and Spatial Differences in AtSLP Protein Phosphatase Expression
Both AtSLP1 and AtSLP2 were found to be expressed in different Arabidopsis plant tissues, indicating that they may differ in biological function. Combined with differing subcellular locations, AtSLP1 protein expression was only detected in photosynthetic tissues, with the exception of intact siliques, while maintaining complete absence from roots (Fig. 5A; Supplemental Fig. S3). Conversely, AtSLP2 demonstrated clear expression in roots, with marginally detectable expression in intact siliques (Fig. 5A; Supplemental Fig. S3). When compared, AtSLP1 and AtSLP2 tissue-specific protein expression largely correlates with transcriptional expression (Supplemental Fig. S3). However, the presence of AtSLP2 protein in photosynthetic tissues cannot be discounted, as data suggest that at least some AtSLP2 transcriptional expression is occurring in rosettes, shoots, and flowers (Supplemental Fig. S3). Unfortunately, the clear detection of AtSLP2 protein in these tissues via western blot was hindered by the cross-reactivity of affinity-purified anti-AtSLP2 IgG with AtSLP1, coupled with the small size differences between AtSLP1 and AtSLP2 (Supplemental Fig. S6). Interestingly, parallel temporal expression of AtSLP phosphatase transcripts and proteins did not hold true for the light/dark cycle of Arabidopsis leaves. AtSLP1 presented a clear diurnal cycle in transcript abundance (Supplemental Fig. S4), which did not completely translate to parallel AtSLP1 protein expression (Fig. 5B). AtSLP1 transcript abundance was shown to increase in the dark, peaking at the 24-h-dark/0-h-light interface, followed by a reduction in transcriptional abundance as the light period progressed (Supplemental Fig. S4A). Comparatively, AtSLP1 protein abundance seemed to peak at 8 h of light, followed by a reduction back to a basal lower level of abundance for the remainder of the 24-h light/dark cycle. This finding may indicate that AtSLP1 is not only transcriptionally regulated but also regulated posttranslationally and is reminiscent of other chloroplast phosphatases, such as the dual-specificity phosphatase starch excess 4, which demonstrates diurnal gene expression while maintaining constant protein levels (Lu et al., 2005).
Collectively, the AtSLP1 expression findings indicate two things: (1) AtSLP1 transcript levels seem to peak prior to AtSLP1 protein production, analogous to other chloroplast-targeted proteins that exhibit a preemptive transcript accumulation prior to translation as part of a diurnal cycle, to anticipate the coming metabolic and cellular changes accompanying illumination transition (Smith et al., 2004; Lu et al., 2005); and (2) AtSLP1 transcripts and AtSLP1 protein lack complete correlative expression, indicating that AtSLP1 may possess more than just a light-regulated role in the chloroplast and/or a complex cellular regulation mechanism involving a combination of transcriptional and posttranslational control factors. AtSLP2, on the other hand, was not investigated for a diurnal fluctuation in protein expression, since it was shown to be cytosolic (Fig. 4), expressed mainly in nonphotosynthetic root tissue (Fig. 5A; Supplemental Fig. S3), and exhibited no preexisting indication of diurnally regulated transcript fluctuations (Supplemental Fig. S4B).
Conservation of Essential PPP Family Phosphatase Motifs in AtSLP1 and AtSLP2
The large number of sequenced plant genomes available online enabled the large-scale alignment of group I and group II SLP phosphatases (Fig. 6). All SLPs possess the core signature motifs of PPP family phosphatases (GDxHG, GDxVDRG, and GNHE) constituting the active site (Egloff et al., 1995; Xu et al., 2006) and lack any of the defining motifs for other protein phosphatase families (Kerk et al., 2008). These motifs also served as an initial indicator that the SLP phosphatases are Ser/Thr protein phosphatases. As shown in Figure 6, the SLP phosphatases all have the canonical amino acids involved in coordinating active site metal ions required for catalysis in PPP family phosphatases (Egloff et al., 1995; Zhang et al., 1996; Xu et al., 2006). These six amino acids are Asp-64 (Asp-54), His-66 (His-56), Asp-98 (Asp-88), Asn-124 (Asn-114), His-173 (His-164), and His-248 (His-238) of the mammalian HsPP1γ and PP2A (in parentheses), respectively (Fig. 6).
Also important are the regulatory protein interactor coordination motifs of PP1 and PP2A catalytic subunits. Our alignments revealed that AtSLPs possess almost none of the amino acids involved in coordinating the canonical RVXF motif of PP1 regulatory subunits (Egloff et al., 1997). Nonetheless, AtI-2, an ancient PP1 interactor with an RVXF motif, did weakly inhibit the SLPs. Human I-2 contacts PP1 at multiple sites, and we propose that the contribution of several interaction sites (Fig. 6) allows a weak association in vitro. This is consistent with data showing that human PP2A can be inhibited by I-2 at high concentrations (Brautigan et al., 1986). Since both AtSLP phosphatases have higher sequence identity to PP2Ac, the presence of amino acid stretches involved in mediating PP2Ac-regulatory subunit interactions was also examined. Neither AtSLP phosphatase was found to possess regions of amino acids resembling these stretches (Xu et al., 2006). Together, this suggests that SLP phosphatases either have unique targeting subunits or no targeting subunits at all.
Metal Cation Preferences of AtSLP Phosphatases
The atomic structures of PP1 and PP2A revealed the presence of Mn2+/Fe3+ and Mn2+/Mn2+ dimetal cation arrangements in their active sites, respectively (Egloff et al., 1995; Cho and Xu, 2007). These metal cations were suggested to function in coordinating water in the active site to initiate nucleophilic attack on a protein-coupled phosphoryl group and mediate its release (Egloff et al., 1995; Xu et al., 2006; Cho and Xu, 2007). Our results revealed PP1-like metal-dependent phosphatase activity for AtSLP1 with Mn2+ and also Fe3+ (Fig. 8A; Supplemental Fig. S10). Zn2+ and Mg2+ were also examined, as both these cations have documented importance in the catalytic mechanisms of nonprotein phosphatases (Kim and Wyckoff, 1991) and PP2C protein phosphatases, respectively (Ingebritsen and Cohen, 1983; Cohen, 1997; Klumpp et al., 2006). Neither Zn2+ nor Mg2+ could recover the activity of AtSLP1 beyond the basal activity observed in the presence of 5 mm EDTA-chelating agent (Fig. 8A; Supplemental Fig. S10), further indicating that AtSLP1 is a PPP family protein phosphatase.
Conversely, AtSLP2 displayed high activity whether additional metal ions were present (Mg2+ or Zn2+) or not (EDTA), but it did appear to be inhibited by Fe3+ (Fig. 8A). The lack of discernible metal-dependent activity suggests that AtSLP2 likely maintains a conformation that renders its bound metal ions unable to be quenched by 5 mm EDTA. Despite this, AtSLP2 activity was easily quenched by 200 mm excess EDTA, indicating that active site metal ions are catalytically important but not easily displaced in vitro (data not shown).
Inhibition by Classic PPP Family Protein Phosphatase Inhibitors
As AtSLPs have the greatest identity to eukaryotic PPP family protein phosphatases PP1 and PP2A, each AtSLP phosphatase was examined for sensitivity to known small molecule and protein inhibitors of PPPs. Both PP1- and PP2A-type phosphatases have well-documented sensitivity to MC-LR and OA, with PP1 possessing greater sensitivity to MC and PP2A to OA (Bialojan and Takai, 1988; MacKintosh et al., 1995). As well, these compounds have also been shown to inhibit the closely related PPP family protein phosphatases PP4, PP5, and PP6 (Heidari et al., 2011). Neither AtSLP1 nor AtSLP2 was sensitive to MC-LR or OA (Figs. 8B and 9). In fact, AtSLP1 demonstrated an unexpected activation by both MC-LR and OA (Figs. 8B and 9). Other Ser/Thr protein phosphatases have been documented to lack sensitivity to these compounds, including AtPP7 (Kutuzov et al., 1998) and PP2C protein phosphatases (Bialojan and Takai, 1988), but to our knowledge, no other PPP family protein phosphatase has demonstrated activation by either MC-LR or OA. This enzymatic activation phenomenon was exhibited by both tagged and untagged AtSLP1 protein, eliminating the possibility of His6 tag influence over AtSLP1 conformation and sensitivity to inhibitors (Fig. 9). Lack of MC-LR and OA inhibition was likely caused by the absence of a canonical inhibitor-binding motif (SAPNYC) that comprises a hydrophobic binding pocket that both covalently (MC-LR) and noncovalently (OA) accommodates these inhibitors (Zhang et al., 1996; Maynes et al., 2001). Furthermore, site-directed mutagenesis of amino acid residues Arg-221 and Phe-276 of HsPP1γ, found outside the SAPNYC motif, conveyed substantive decreases in OA inhibition (Maynes et al., 2001). AtSLP phosphatases also lack Arg-221 and Phe-276 residues at equivalent positions (Fig. 6), supporting the lack of observable SLP phosphatase inhibition.
Additional PPP family phosphatase inhibitors tested here included PPi, Pi, and NaF, which have all been shown to inhibit Arabidopsis type 1 phosphatases (Stubbs et al., 2001). Previous work affinity purified six of the nine AtPP1 (TOPP) phosphatase isoforms (TOPP1 to -6) from Arabidopsis cell culture using a MC-conjugated Sepharose matrix (Stubbs et al., 2001). TOPP2 was among these six phosphatases and was used here in its heterologously expressed and affinity-purified form as an enzyme assay control (Fig. 9). Stubbs and colleagues (2001) found that the affinity-purified TOPPs were almost completely inhibited by 100, 1, and 10 mm Pi, PPi, and NaF, respectively. Comparatively, the AtSLP phosphatases demonstrated a different pattern of inhibition by these compounds, possibly emphasizing differences in their cellular roles relative to the TOPP phosphatases (Takemiya et al., 2009). Both AtSLP1 and AtSLP2 are relatively insensitive to PPi inhibition, while AtSLP1 exhibits enhanced sensitivity to Pi, with complete inhibition at a 50 mm concentration (Fig. 9). An inability to completely inhibit either AtSLP phosphatase at 100 mm NaF signifies a notable enzymatic difference from the affinity of purified TOPPs, which were almost entirely inhibited at 20 mm NaF (Stubbs et al., 2001).
The sensitivity of AtSLP1 to Pi inhibition (concentration giving 50% inhibition of initial activity approximately 1.5 mm) coupled to its chloroplast localization may indicate a chloroplast energy-sensing function, as the chloroplast stroma has been documented to maintain Pi levels of 12 mm or greater to support the production of ATP upon the onset of light (Bligny et al., 1990). Intriguingly, a light-induced decrease in stromal Pi levels to produce ATP would parallel the corresponding increase in detectable AtSLP1 in Arabidopsis rosette leaves, suggesting that decreased stromal Pi levels could activate AtSLP1 to dephosphorylate its substrates (Fig. 3B). Conversely, the relative insensitivity of AtSLP2 to Pi coupled with a 50% lower Pi concentration in the cytosol (6 mm) would seem to indicate a function unrelated to cellular energy levels (Bligny et al., 1990).
CONCLUSION
This work has characterized, to our knowledge for the first time, two novel, highly conserved plant phosphatases. The remarkable degree of conservation of both the cytosolic and chloroplastic isoforms of the SLPs in plants indicates an ancient and nonoverlapping plant function. The enzymatic analysis presented here coincides with bioinformatic analyses suggesting that SLPs are a subcluster of the PPP family protein phosphatases. Future endeavors will need to employ additional cell biological and biochemical approaches to identify possible regulatory subunits as well as the substrates of these unique enzymes. As well, despite the lack of information uncovered through an initial phenotypic analysis of single atslp insertional mutant lines, the creation of a double atslp1/atslp2 knockout in conjunction with more intensive phenotype screening methodologies will hopefully elucidate the physiological role(s) of the SLP phosphatases.
MATERIALS AND METHODS
Bioinformatics
BLASTP searches (Altschul et al., 1997) were conducted using the NCBInr database (http://blast.ncbi.nlm.nih.gov/Blast.cgi) along with targeted BLASTP searches using Phytozome version 6.0 (http://www.phytozome.net/). Entire AtSLP1 (At1g07010) and AtSLP2 (At1g18480) sequences were used as initial BLASTP bait to uncover homolog proteins in other species. Identified homologous proteins were aligned using ClustalX2 (http://www.clustal.org/) and subsequently used to create a bootstrap neighbor-joining phylogenetic tree. Bootstrap values were produced from 1,000 iterations. In silico subcellular localization predictions were conducted using freely accessible online programs that employed plant-specific prediction algorithms: Predotar (http://urgi.versailles.inra.fr/predotar/predotar.html), TargetP (http://www.cbs.dtu.dk/services/TargetP/), WoLF pSORT (http://wolfpsort.org/), and ChloroP (http://www.cbs.dtu.dk/services/ChloroP/). In conjunction with their relatedness based on sequence identity, the consensus of subcellular prediction determined the group I and group II designations for SLPs. The online Genevestigator search engine (https://www.genevestigator.com/gv/index.jsp) containing the pooled ATH1 22k microarray data and NCBI GEO profiles (http://www.ncbi.nlm.nih.gov/geoprofiles) were used to assess the transcriptional expression of At1g07010 (AtSLP1) and At1g18480 (AtSLP2) in various tissue types and across plant development. The Bio-Array Resource for Plant Biology (http://esc4037-shemp.csb.utoronto.ca/welcome.htm) was also utilized as a source of direction for subsequent experimentation.
Molecular Cloning
Fluorescent construct creation utilized full-length clones obtained from The Arabidopsis Information Resource (http://www.arabidopsis.org/) propagated by PCR with Gateway-compatible primers and inserted into pDONR201 (Invitrogen). Each construct was subcloned into the plant expression vector pB7RWG2 (http://gateway.psb.ugent.be/) to create C-terminal RFP fusion constructs. All cloning done in conjunction with fluorescent construct creation was accomplished using Escherichia coli DH5α. Agrobacterium tumefaciens strain GV3101 was transformed with each fluorescent construct for wild-type Arabidopsis (Arabidopsis thaliana) cell culture transfection purposes. Heterologous protein expression employed full-length At1g07010 (minus the predicted chloroplast transit peptide; amino acids 1–53) and At1g18480 PCR products containing EcoRI and NotI restriction sites. Each construct was initially cloned into pGEM-T (Promega) and subsequently subcloned into pET47b(+) for antibody production and into pET48b(+) for purification and enzymatic characterization (EMD Chemicals).
Cell Culture Transfection and Protoplast Creation
Wild-type Arabidopsis cell culture transfection was modeled from previous studies (Forreiter et al., 1997). Empirical testing of the cell culture transfection process used 3-d-old Arabidopsis liquid cell culture grown under 24 h of light in 1× Murashige and Skoog (MS) medium along with 3% (w/v) Suc, 0.5 mg mL−1 naphthalic acid anhydride, and 0.05 mg mL−1 kinetin (1× growth medium) combined with transformed Agrobacterium grown to an optical density at 600 nm = 0.5 at 28°C and 150 rpm resuspended in 10 mL of 1× growth medium, as ideal for successful Arabidopsis cell transfection. Incubation of Agrobacterium and Arabidopsis liquid cell culture was performed for 2 d at room temperature under 24 h of light shaking at 150 rpm. The Agrobacterium-Arabidopsis cell culture mixture was then pelleted at 300g for 1 min and resuspended in 1× growth medium containing 100 μg mL−1 ampicillin (repeated three times). Cells were then spread on 0.8% (w/v) agar plates containing 1× growth medium and 10 μg mL−1 Basta and grown under 24 h of light for 2 weeks to select for positively transfected Arabidopsis cells. Positive transfectants grown as callus cells were visually identified (green papules) and replated on 1× selective medium, then they were subcultured to liquid 1× growth medium and grown as above under 24 h of light. Protoplasts were subsequently derived from 10 mL of positively transfected Arabidopsis cell culture as described previously (Lingard et al., 2008). Protein expression was confirmed by western blotting with anti-RFP IgG (Chromotek).
Transient AtSLP Expression in Fava Bean Epidermal Leaf Cells
Transient coexpression of AtSLP1-RFP and AtSLP2-RFP with marker constructs was conducted using equimolar amounts of gold coupled with DNA and particle bombardment. Plasmid DNA (10 μg) was coated onto 1-mg gold microcarriers, washed, spotted on a macrocarrier, and accelerated at the epidermal layer of fava bean (Vicia faba) leaves using a PDS-1000/He System (Bio-Rad) according to Russell et al. (1992). AtSLP1-RFP was cobombarded with GFP constructs specifically designed to target the ER, Golgi, and peroxisome (Nelson et al., 2007), while AtSLP2-RFP was cobombarded with established mitochondria, ER, Golgi (Nelson et al., 2007), and cytosol/nucleus-targeted GFP (vector p2FGW7; GFP with no targeting motif). AtSLP2-RFP was also cobombarded with GFP-tagged Nup50a to illuminate the nucleus (Tamura et al., 2010).
Microscopy
All imaging was conducted using a Leica DMIRE2 spectral confocal and multiphoton microscope with a Leica TCS SP2 acoustic optical beam splitter (Leica Microsystems), with all cells and leaves visualized using the 63× water-immersion lens. The excitation/emission wavelengths (nm) employed were as follows: GFP, 488/505-515; RFP, 594/610-650; chlorophyll autofluorescence, 488 and 594/685 to 715. Subsequent image processing was performed using the freely accessible MacBiophotonics ImageJ (http://www.macbiophotonics.ca/downloads.htm).
Plant Growth Conditions
All plants were sterilized and stratified at 4°C for 3 d in the dark on 0.5× MS agar plates supplemented with 1% (w/v) Suc. Plants were germinated under a regular light cycle consisting of 12 h of light and 12 h of dark at 22°C with a light level of 125 μmol m−2 s−1 for 7 d before being sown to soil supplemented with 0.5 g L−1 20:20:20 fertilizer. Once sown to soil, plants were grown using the above conditions. Plant (shoot) growth was qualitatively assessed relative to corresponding wild-type plants over a 21-d period (data not shown). Root growth was measured quantitatively over a 7-d period using vertical 0.5× MS agar plates supplemented with 1% (w/v) Suc (data not shown).
Heterologous Protein Expression and Antibody Production
AtSLP1- and AtSLP2-pET48 were expressed in BL21 (DE3) Codon(+)-RIL E. coli at 8°C and 200 rpm by induction with 0.1 mm isopropyl β-d-1-thiogalactopyranoside for 24 h. Bacteria were pelleted at 4,000g for 15 min and resuspended in 1× extraction buffer (50 mm HEPES-NaOH, pH 7.5, 150 mm NaCl, 5% [v/v] glycerol, and 10 mm imidazole) followed by snap freezing and storage at −80°C prior to use. The day of purification saw the addition of 1% (v/v) Tween 20, 20 mm imidazole, 1 mm phenylmethanesulfonyl fluoride, 1 mm benzamidine, 2 μg mL−1 leupeptin, and 5 μg mL−1 pepstatin upon thawing. Extraction involved mechanical lysis via French press at 3 × 16,000 p.s.i. (Sim-Aminco Spectronic Instruments). Crude lysates were clarified at 20,000g for 30 min at 4°C. Supernatants were removed, and 1 mm phenylmethanesulfonyl fluoride and 1 mm benzamidine were added prior to incubation with Ni-NTA agarose matrix (Qiagen) end-over-end mixing at 4°C for 1 h. Matrix was poured into a column and washed with 500 column volumes (cv) of wash buffer A (1× extraction buffer with 1 m NaCl, 1% [v/v] Tween 20, and 20 mm imidazole minus protease inhibitors) by gravity followed by 100 cv of wash buffer B (1× extraction buffer minus protease inhibitors). Ni-NTA-bound proteins were eluted using 1× extraction buffer containing 500 mm imidazole, pH 7.5. Protein eluates were concentrated using a 30,000 molecular weight cutoff Amicon concentrator (Millipore).
Purification of AtSLP1 to homogeneity involved four steps: (1) Ni-NTA purification; (2) size-exclusion chromatography; (3) cleavage of the affinity tag by HRV3c protease (Novagen); and (4) anion-exchange chromatography. Heterologous His6-AtSLP1 expression and subsequent purification by Ni-NTA was conducted as outlined above from 4 × 1 L of bacterial cell culture. The Ni-NTA His6-AtSLP1 eluate was concentrated to 500 μL and subjected to size-exclusion chromatography using a HiLoad 16/60 Superdex 200 (Amersham) column. The sample was run at 0.5 mL min−1, collecting 1-mL fractions in buffer A (50 mm HEPES-NaOH, pH 7.5, 50 mm NaCl, and 5% [v/v] glycerol). Column fractions were resolved by SDS-PAGE (12%) and visualized by colloidal blue. Peak fractions were pooled and concentrated to 500 μL in a 30,000 molecular weight cutoff Amicon concentrator. Cleavage of the His6 tag was performed on 500 to 700 μg of total protein from the concentrated size-exclusion chromatography eluate using 10 units of Novagen HRV3c protease (EMD Chemicals) in the presence of 1 mm dithiothreitol (DTT) overnight at 4°C, rotating gently end over end. Anion-exchange chromatography was performed on a Mono-Q 5/50 GL (Amersham) column employing buffer A (as above) and buffer B consisting of 50 mm HEPES-NaOH, pH 7.5, 1 m NaCl, 5% (v/v) glycerol, and 1 mm DTT. The concentrated Superdex 200 eluate was loaded at 0.5 mL min−1, the column was washed at 1 mL min−1 for 10 cv, and protein was eluted using a gradient of 50 to 400 mm NaCl over 35 cv, collecting 1-mL fractions. Peak fractions were resolved by SDS-PAGE, pooled, concentrated as above, aliquoted, snap frozen in liquid nitrogen, and stored at −80°C for subsequent use.
AtSLP1- and AtSLP2-pET47b were expressed in BL21 (DE3) Codon(+)-RIL E. coli at 37°C and induced with 0.5 mm isopropyl β-d-1-thiogalactopyranoside for 4 h and purified from inclusion bodies using Ni-NTA according to the manufacturer’s instructions (Qiagen). Purified protein was dialyzed against water, freeze dried, and used for polyclonal antibody production in a New Zealand White rabbit as described (Tran et al., 2004). Both the AtSLP1 and AtSLP2 antibodies were affinity purified as described previously, but in the absence of bovine serum albumin (Plaxton, 1989).
Enzymatic Analysis
Enzymatic assessment of AtSLP1 and AtSLP2 was conducted using the small molecule phosphatase substrate pNPP (Sigma). Enzymatic assays comparing AtSLP1 and AtSLP2 directly were conducted using day-of purified and concentrated Ni-NTA eluates. All assays were done in parallel with Ni-NTA eluates generated from uninduced BL21 (DE3) Codon(+)-RIL E. coli to account for potential background pNPP cleavage from NSCP proteins. His6-AtSLP1 and His6-AtSLP2 percentage activities were calculated relative to assays conducted with 0.5 mm Mn2+ and 5 mm EDTA, respectively. Due to its stability, pure, untagged AtSLP1 was used to perform more thorough enzymatic analyses in conjunction with completely purified TOPP2 (At5g59160) and partially purified (approximately 50%) HsPP1γ (NP_002701) as assay benchmarks. Both TOPP2 and HsPP1γ were cloned, expressed, and purified from E. coli as described previously (Douglas et al., 2001; Templeton et al., 2011). Enzymatic assay strategy was adapted from the SensoLyte pNPP Protein Phosphatase Assay Kit (http://www.anaspec.com). Base buffer consisted of 100 mm HEPES, pH 7.5, and 150 mm NaCl. Dilution buffer(s) consisted of base buffer plus 4 mm DTT and 0.2 mm EDTA with either 0.5 mm metal cation (metal-dependent activity) or 5 mm EDTA (metal-independent activity). Assay buffer consisted of dilution buffer plus 5 mm pNPP. All protein samples were premixed under their experimental conditions, brought to 20 μL with the respective dilution buffer, and preincubated at 30°C for 10 min prior to enzymatic analysis. Enzyme assays were initiated by the addition of 180 μL of assay buffer and incubation at 30°C for 30 min (Ni-NTA AtSLP eluates and controls) or 20 min (pure AtSLP1 and controls). For the Ni-NTA AtSLP and control eluates, 750 ng of total protein was used per assay. Assays involving pure AtSLP1 employed 100 ng of AtSLP1, 200 ng of TOPP2, and 400 ng of total HsPP1γ protein (200 ng of pure HsPP1γ). Assays were quenched with 200 μL of 0.5 m EDTA, and pNPP cleavage was assessed using an Ultrospec 2000 spectrophotometer set to 405 nm. When stated, [32P]glycogen phosphorylase a was also examined as substrate. Phosphorylase a was prepared and used as described (Moorhead et al., 1995).
Sequence data from this article can be found in the GenBank/EMBL data libraries under accession numbers NP_172182.2 and NP_564053.1.
Supplemental Data
The following materials are available in the online version of this article.
Supplemental Figure S1. Complete phylogenetic analysis of SLP phosphatases from prokaryotic ancestors to conserved eukaryotic homologs.
Supplemental Figure S2. Subcellular localization of AtSLP1 and AtSLP2.
Supplemental Figure S3. Genevestigator expression data (www.genevestigator.com).
Supplemental Figure S4. NCBI GEO profiles of diurnal transcript cycling (www.ncbi.nlm.nih.gov/geoprofiles).
Supplemental Figure S5. Colloidal blue-stained 12% SDS-PAGE gels of the His6-AtSLP1 and His6-AtSLP2 Ni-NTA eluates employed in producing anti-AtSLP1 and anti-AtSLP2 polyclonal antibodies.
Supplemental Figure S6. Western-blot assessment of affinity-purified anti-AtSLP1 and anti-AtSLP2 IgG detection limits and specificity.
Supplemental Figure S7. Colloidal blue-stained 12% SDS-PAGE of purified HsPP1γ and TOPP2.
Supplemental Figure S8. Size-exclusion and anion-exchange chromatography steps of AtSLP1 purification.
Supplemental Figure S9. Analysis of the major end-point purification steps employed to completely purify AtSLP1.
Supplemental Figure S10. Metal cation dependence of purified untagged AtSLP1.
Supplemental Table S1. Consensus in silico computational subcellular localization data used in grouping the SLPs after phylogenetic tree construction.
Supplemental Table S2. Matrix-assisted laser-desorption ionization time of flight mass spectrometry identification of lower Mr polypeptides in the Superdex 200 pool postdigest (Supplemental Fig. S6).
Acknowledgments
We thank Dr. Doug Muench and Dr. Howard Ceri (University of Calgary) for assistance with aspects of the localization work and for use of the confocal microscopy facilities, respectively, and Dr. Ikuko Hara-Nishimura (Department of Botany, Kyoto University) for use of his Nup50a-GFP construct. As well, thanks to George Templeton, Habir Gill, and Bhavneet Kahlon (University of Calgary) for helpful conversations and support throughout this work.
References
- Altschul SF, Madden TL, Schäffer AA, Zhang J, Zhang Z, Miller W, Lipman DJ. (1997) Gapped BLAST and PSI-BLAST: a new generation of protein database search programs. Nucleic Acids Res 25: 3389–3402 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Andreeva AV, Kutuzov MA. (2004) Widespread presence of “bacterial-like” PPP phosphatases in eukaryotes. BMC Evol Biol 4: 47–61 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Badet B, Vermoote P, Haumont PY, Lederer F, LeGoffic F. (1987) Glucosamine synthetase from Escherichia coli: purification, properties, and glutamine-utilizing site location. Biochemistry 26: 1940–1948 [DOI] [PubMed] [Google Scholar]
- Bialojan C, Takai A. (1988) Inhibitory effect of a marine-sponge toxin, okadaic acid, on protein phosphatases: specificity and kinetics. Biochem J 256: 283–290 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bligny R, Gardestrom P, Roby C, Douce R. (1990) 31P NMR studies of spinach leaves and their chloroplasts. J Biol Chem 265: 1319–1326 [PubMed] [Google Scholar]
- Bozzo GG, Raghothama KG, Plaxton WC. (2002) Purification and characterization of two secreted purple acid phosphatase isozymes from phosphate-starved tomato (Lycopersicon esculentum) cell cultures. Eur J Biochem 269: 6278–6286 [DOI] [PubMed] [Google Scholar]
- Brautigan DL, Gruppuso PA, Mumby M. (1986) Protein phosphatase type-1 and type-2 catalytic subunits both bind inhibitor-2 and monoclonal immunoglobulins. J Biol Chem 261: 14924–14928 [PubMed] [Google Scholar]
- Cho US, Xu W. (2007) Crystal structure of a protein phosphatase 2A heterotrimeric holoenzyme. Nature 445: 53–57 [DOI] [PubMed] [Google Scholar]
- Cohen P, Alemany S, Hemmings BA, Resink TJ, Strålfors P, Tung HY. (1988) Protein phosphatase-1 and protein phosphatase-2A from rabbit skeletal muscle. Methods Enzymol 159: 390–408 [DOI] [PubMed] [Google Scholar]
- Cohen PT. (1997) Novel protein serine/threonine phosphatases: variety is the spice of life. Trends Biochem Sci 22: 245–251 [DOI] [PubMed] [Google Scholar]
- Dancheck B, Ragusa MJ, Allaire M, Nairn AC, Page R, Peti W. (2011) Molecular investigations of the structure and function of the protein phosphatase 1-spinophilin-inhibitor 2 heterotrimeric complex. Biochemistry 50: 1238–1246 [DOI] [PMC free article] [PubMed] [Google Scholar]
- DeLong A. (2006) Switching the flip: protein phosphatase roles in signaling pathways. Curr Opin Plant Biol 9: 470–477 [DOI] [PubMed] [Google Scholar]
- Douglas P, Moorhead GB, Ye R, Lees-Miller SP. (2001) Protein phosphatases regulate DNA-dependent protein kinase activity. J Biol Chem 276: 18992–18998 [DOI] [PubMed] [Google Scholar]
- Egloff MP, Cohen PT, Reinemer P, Barford D. (1995) Crystal structure of the catalytic subunit of human protein phosphatase 1 and its complex with tungstate. J Mol Biol 254: 942–959 [DOI] [PubMed] [Google Scholar]
- Egloff MP, Johnson DF, Moorhead G, Cohen PT, Cohen P, Barford D. (1997) Structural basis for the recognition of regulatory subunits by the catalytic subunit of protein phosphatase 1. EMBO J 16: 1876–1887 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Forreiter C, Kirschner M, Nover L. (1997) Stable transformation of an Arabidopsis cell suspension culture with firefly luciferase providing a cellular system for analysis of chaperone activity in vivo. Plant Cell 9: 2171–2181 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fukao Y, Hayashi M, Nishimura M. (2002) Proteomic analysis of leaf peroxisomal proteins in greening cotyledons of Arabidopsis thaliana. Plant Cell Physiol 43: 689–696 [DOI] [PubMed] [Google Scholar]
- Girzalsky W, Platta HW, Erdmann R. (2009) Protein transport across the peroxisomal membrane. Biol Chem 390: 745–751 [DOI] [PubMed] [Google Scholar]
- Gould SB, Waller RF, McFadden GI. (2008) Plastid evolution. Annu Rev Plant Biol 59: 491–517 [DOI] [PubMed] [Google Scholar]
- Heidari B, Matre P, Nemie-Feyissa D, Meyer C, Rognli OA, Møller SG, Lillo C. (2011) Protein phosphatase 2A B55 and A regulatory subunits interact with nitrate reductase and are essential for nitrate reductase activation. Plant Physiol 156: 165–172 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hurley TD, Yang J, Zhang L, Goodwin KD, Zou Q, Cortese M, Dunker AK, DePaoli-Roach AA. (2007) Structural basis for regulation of protein phosphatase 1 by inhibitor-2. J Biol Chem 282: 28874–28883 [DOI] [PubMed] [Google Scholar]
- Ingebritsen TS, Cohen P. (1983) Protein phosphatases: properties and role in cellular regulation. Science 221: 331–338 [DOI] [PubMed] [Google Scholar]
- Keeling PJ. (2010) The endosymbiotic origin, diversification and fate of plastids. Philos Trans R Soc Lond B Biol Sci 365: 729–748 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kerk D, Templeton G, Moorhead GB. (2008) Evolutionary radiation pattern of novel protein phosphatases revealed by analysis of protein data from the completely sequenced genomes of humans, green algae, and higher plants. Plant Physiol 146: 351–367 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kim EE, Wyckoff HW. (1991) Reaction mechanism of alkaline phosphatase based on crystal structures: two-metal ion catalysis. J Mol Biol 218: 449–464 [DOI] [PubMed] [Google Scholar]
- Klumpp S, Thissen MC, Krieglstein J. (2006) Protein phosphatases types 2Calpha and 2Cbeta in apoptosis. Biochem Soc Trans 34: 1370–1375 [DOI] [PubMed] [Google Scholar]
- Kutuzov MA, Andreeva AV. (2008) Protein Ser/Thr phosphatases of parasitic protozoa. Mol Biochem Parasitol 161: 81–90 [DOI] [PubMed] [Google Scholar]
- Kutuzov MA, Evans DE, Andreeva AV. (1998) Expression and characterization of PP7, a novel plant protein Ser/Thr phosphatase distantly related to RdgC/PPEF and PP5. FEBS Lett 440: 147–152 [DOI] [PubMed] [Google Scholar]
- Lazarow PB. (2006) The import receptor Pex7p and the PTS2 targeting sequence. Biochim Biophys Acta 1763: 1599–1604 [DOI] [PubMed] [Google Scholar]
- Lingard MJ, Gidda SK, Bingham S, Rothstein SJ, Mullen RT, Trelease RN. (2008) Arabidopsis PEROXIN11c-e, FISSION1b, and DYNAMIN-RELATED PROTEIN3A cooperate in cell cycle-associated replication of peroxisomes. Plant Cell 20: 1567–1585 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lu Y, Gehan JP, Sharkey TD. (2005) Daylength and circadian effects on starch degradation and maltose metabolism. Plant Physiol 138: 2280–2291 [DOI] [PMC free article] [PubMed] [Google Scholar]
- MacKintosh RW, Dalby KN, Campbell DG, Cohen PT, Cohen P, MacKintosh C. (1995) The cyanobacterial toxin microcystin binds covalently to cysteine-273 on protein phosphatase 1. FEBS Lett 371: 236–240 [DOI] [PubMed] [Google Scholar]
- Maynes JT, Bateman KS, Cherney MM, Das AK, Luu HA, Holmes CF, James MN. (2001) Crystal structure of the tumor-promoter okadaic acid bound to protein phosphatase-1. J Biol Chem 276: 44078–44082 [DOI] [PubMed] [Google Scholar]
- Miyagishima SY. (2011) Mechanism of plastid division: from a bacterium to an organelle. Plant Physiol 155: 1533–1544 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Moorhead G, MacKintosh C, Morrice N, Cohen P. (1995) Purification of the hepatic glycogen-associated form of protein phosphatase-1 by microcystin-Sepharose affinity chromatography. FEBS Lett 362: 101–105 [DOI] [PubMed] [Google Scholar]
- Moorhead GB, De Wever V, Templeton G, Kerk D. (2009) Evolution of protein phosphatases in plants and animals. Biochem J 417: 401–409 [DOI] [PubMed] [Google Scholar]
- Moorhead GB, Trinkle-Mulcahy L, Ulke-Lemée A. (2007) Emerging roles of nuclear protein phosphatases. Nat Rev Mol Cell Biol 8: 234–244 [DOI] [PubMed] [Google Scholar]
- Mora-García S, Vert G, Yin Y, Caño-Delgado A, Cheong H, Chory J. (2004) Nuclear protein phosphatases with Kelch-repeat domains modulate the response to brassinosteroids in Arabidopsis. Genes Dev 18: 448–460 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nelson BK, Cai X, Nebenführ A. (2007) A multicolored set of in vivo organelle markers for co-localization studies in Arabidopsis and other plants. Plant J 51: 1126–1136 [DOI] [PubMed] [Google Scholar]
- Olsen JV, Blagoev B, Gnad F, Macek B, Kumar C, Mortensen P, Mann M. (2006) Global, in vivo, and site-specific phosphorylation dynamics in signaling networks. Cell 127: 635–648 [DOI] [PubMed] [Google Scholar]
- Olsen JV, Vermeulen M, Santamaria A, Kumar C, Miller ML, Jensen LJ, Gnad F, Cox J, Jensen TS, Nigg EA, et al. (2010) Quantitative phosphoproteomics reveals widespread full phosphorylation site occupancy during mitosis. Sci Signal 3: ra3. [DOI] [PubMed] [Google Scholar]
- Plaxton WC. (1989) Molecular and immunological characterization of plastid and cytosolic pyruvate kinase isozymes from castor-oil-plant endosperm and leaf. Eur J Biochem 181: 443–451 [DOI] [PubMed] [Google Scholar]
- Russell JA, Roy MK, Sanford JC. (1992) Physical trauma and tungsten toxicity reduce the efficiency of biolistic transformation. Plant Physiol 98: 1050–1056 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Seibel NM, Eljouni J, Nalaskowski MM, Hampe W. (2007) Nuclear localization of enhanced green fluorescent protein homomultimers. Anal Biochem 368: 95–99 [DOI] [PubMed] [Google Scholar]
- Shi Y. (2009) Serine/threonine phosphatases: mechanism through structure. Cell 139: 468–484 [DOI] [PubMed] [Google Scholar]
- Smith SM, Fulton DC, Chia T, Thorneycroft D, Chapple A, Dunstan H, Hylton C, Zeeman SC, Smith AM. (2004) Diurnal changes in the transcriptome encoding enzymes of starch metabolism provide evidence for both transcriptional and posttranscriptional regulation of starch metabolism in Arabidopsis leaves. Plant Physiol 136: 2687–2699 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Stubbs MD, Tran HT, Atwell AJ, Smith CS, Olson D, Moorhead GB. (2001) Purification and properties of Arabidopsis thaliana type 1 protein phosphatase (PP1). Biochim Biophys Acta 1550: 52–63 [DOI] [PubMed] [Google Scholar]
- Sugiyama N, Nakagami H, Mochida K, Daudi A, Tomita M, Shirasu K, Ishihama Y. (2008) Large-scale phosphorylation mapping reveals the extent of tyrosine phosphorylation in Arabidopsis. Mol Syst Biol 4: 193–200 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Takemiya A, Ariyoshi C, Shimazaki K. (2009) Identification and functional characterization of inhibitor-3, a regulatory subunit of protein phosphatase 1 in plants. Plant Physiol 150: 144–156 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tamura K, Fukao Y, Iwamoto M, Haraguchi T, Hara-Nishimura I. (2010) Identification and characterization of nuclear pore complex components in Arabidopsis thaliana. Plant Cell 22: 4084–4097 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Templeton GW, Nimick M, Morrice N, Campbell D, Goudreault M, Gingras AC, Takemiya A, Shimazaki K, Moorhead GB. (2011) Identification and characterization of AtI-2, an Arabidopsis homologue of an ancient protein phosphatase 1 (PP1) regulatory subunit. Biochem J 435: 73–83 [DOI] [PubMed] [Google Scholar]
- Tran HT, Ulke A, Morrice N, Johannes CJ, Moorhead GB. (2004) Proteomic characterization of protein phosphatase complexes of the mammalian nucleus. Mol Cell Proteomics 3: 257–265 [DOI] [PubMed] [Google Scholar]
- Trinkle-Mulcahy L, Boulon S, Lam YW, Urcia R, Boisvert FM, Vandermoere F, Morrice NA, Swift S, Rothbauer U, Leonhardt H, et al. (2008) Identifying specific protein interaction partners using quantitative mass spectrometry and bead proteomes. J Cell Biol 183: 223–239 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Xu Y, Xing Y, Chen Y, Chao Y, Lin Z, Fan E, Yu JW, Strack S, Jeffrey PD, Shi Y. (2006) Structure of the protein phosphatase 2A holoenzyme. Cell 127: 1239–1251 [DOI] [PubMed] [Google Scholar]
- Zhang J, Zhang Z, Brew K, Lee EY. (1996) Mutational analysis of the catalytic subunit of muscle protein phosphatase-1. Biochemistry 35: 6276–6282 [DOI] [PubMed] [Google Scholar]