A novel mechanism is found by which Drosophila male germline stem cells (GSCs) slow their cell cycle under limited nutrient conditions. Upon culturing in poor media, GSCs misorient their centrosomes with respect to the stem cell niche, activating the centrosome orientation checkpoint and leading to slowdown of the cell cycle.
Abstract
Drosophila male germline stem cells (GSCs) divide asymmetrically, balancing self-renewal and differentiation. Although asymmetric stem cell division balances between self-renewal and differentiation, it does not dictate how frequently differentiating cells must be produced. In male GSCs, asymmetric GSC division is achieved by stereotyped positioning of the centrosome with respect to the stem cell niche. Recently we showed that the centrosome orientation checkpoint monitors the correct centrosome orientation to ensure an asymmetric outcome of the GSC division. When GSC centrosomes are not correctly oriented with respect to the niche, GSC cell cycle is arrested/delayed until the correct centrosome orientation is reacquired. Here we show that induction of centrosome misorientation upon culture in poor nutrient conditions mediates slowing of GSC cell proliferation via activation of the centrosome orientation checkpoint. Consistently, inactivation of the centrosome orientation checkpoint leads to lack of cell cycle slowdown even under poor nutrient conditions. We propose that centrosome misorientation serves as a mediator that transduces nutrient information into stem cell proliferation, providing a previously unappreciated mechanism of stem cell regulation in response to nutrient conditions.
INTRODUCTION
Many adult stem cells use asymmetric stem cell division to maintain the critical balance between self-renewal and differentiation (Morrison and Kimble, 2006). Although asymmetric stem cell division balances stem cell self-renewal and differentiation, it does not govern the rate at which new differentiated cells are produced; instead, the stem cell division rate must be modulated in response to environmental stimuli (Drummond-Barbosa, 2008). As is true for essentially all cell types, nutrient conditions have been shown to control the division rate of many stem cells, including Drosophila and Caenorhabditis elegans germline stem cells (GSCs), Drosophila follicle stem cells, and intestinal stem cells, via insulin signaling (Drummond-Barbosa and Spradling, 2001; LaFever and Drummond-Barbosa, 2005; Narbonne and Roy, 2006; Ueishi et al., 2009; McLeod et al., 2010; Michaelson et al., 2010; Choi et al., 2011; O'Brien et al., 2011). Stem cells are characterized by their high proliferation activity, yet how nutrient conditions specifically affect these cell types compared with other, more slowly cycling cell types is not known. Despite intensive studies on the mechanisms by which cells respond to nutrient conditions in general (Saltiel and Kahn, 2001; Taguchi and White, 2008), it is poorly understood how nutrients regulate the rate of stem cell division, and whether this regulation is distinct from that of other cell types. Although it is clear that stem cells are influenced by nutrients (LaFever and Drummond-Barbosa, 2005; Hsu et al., 2008; Hsu and Drummond-Barbosa, 2009; Michaelson et al., 2010; Choi et al., 2011; O'Brien et al., 2011), whether stem cell behavior is regulated in response to nutrient conditions in a stem cell–specific manner and, if so, the nature of the underlying cellular mechanisms that allow stem cells to respond differently compared with other cell types remain unknown.
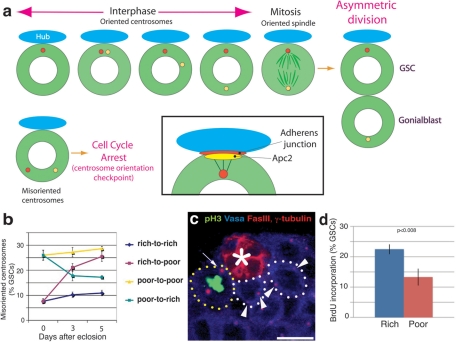
Drosophila male germline stem cells (GSCs) always divide asymmetrically by orienting their mitotic spindle perpendicular to hub cells, a major component of the stem cell niche (Figure 1a; Yamashita et al., 2003; Fuller and Spradling, 2007). The stereotypical movement of centrosomes during interphase prepares spindle orientation in GSCs (Yamashita et al., 2003, 2007). The centrosome is believed to be anchored to the hub-GSC interface through astral microtubules and Apc2, a homologue of the adenomatous polyposis coli tumor suppressor (Figure 1a, inset). Previously we demonstrated that misoriented GSCs (i.e., GSCs in which neither of the two centrosomes is juxtaposed to the hub-GSC interface) do not undergo mitosis (Figure 1a). Instead, these GSCs are arrested in the cell cycle until the centrosome orientation is corrected (Cheng et al., 2008; Inaba et al., 2010; Yuan et al., 2012) due to a checkpoint (the centrosome orientation checkpoint) involving a functional centrosome as well as DE-cadherin (Inaba et al., 2010).
FIGURE 1:
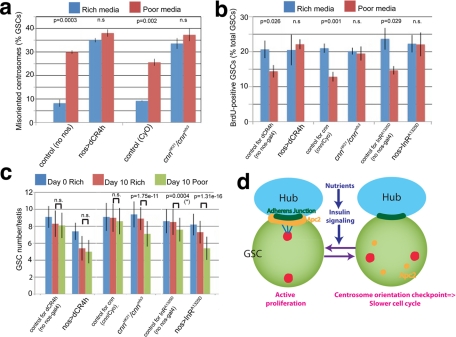
Germline stem cell centrosome orientation is modulated by nutrient availability. (a) Schematic diagram of centrosome movement during the GSC cell cycle. (b) Centrosome misorientation frequencies in GSCs from flies cultured in rich or poor media. Numerical data are presented as the mean ± SD in all figures. n > 300/data point. Flies were cultured in the indicated media until the adult stage and then kept in/transferred to the indicated media at day 0. (c) An example of an apical testis tip from a fly cultured in poor media. Although many interphase GSCs (white circles) are misoriented, the mitotic spindle is correctly oriented (yellow circle). Blue, Vasa (germ cells); green, phosphorylated histone H3 (pH3, mitotic chromosomes); red, γ-tubulin (centrosomes). The hub is marked by an asterisk. Scale bar, 10 μm. (d) Bromodeoxyuridine incorporation in GSCs under poor or rich media conditions. n > 100/data point.
Here we show that, in Drosophila male GSCs, modulation of the division rate in response to nutrient conditions involves regulation of centrosome orientation, mediated via the insulin receptor pathway. Poor nutrient conditions or low insulin signaling leads to centrosome misorientation as a result of delocalization of Apc2, a cortical anchor for the GSC centrosome. The centrosome misorientation ultimately results in activation of the centrosome orientation checkpoint, thus slowing GSC proliferation. Mutant GSCs defective in the centrosome orientation checkpoint do not slow their cell cycle even in poor nutrient conditions, suggesting that the centrosome orientation checkpoint may mediate the slowing of GSC proliferation in response to poor nutrient conditions. We further show that slowdown of GSC proliferation is required to maintain the GSC number under poor nutrient conditions. Together, our results reveal a previously unappreciated cellular mechanism by which nutrients and insulin signaling modulate stem cell proliferation.
RESULTS
Drosophila male GSCs show increased centrosome misorientation under poor nutrient conditions
We noticed that young wild-type flies, which we previously reported to have stereotypically oriented centrosomes (Yamashita et al., 2003), sometimes have a high frequency of misoriented centrosomes. While investigating the cause, we realized that this high frequency of centrosome misorientation was associated with an overcrowded culture, leading us to hypothesize that GSC centrosome misorientation might reflect a response to limited nutrient availability. To test this possibility, we compared GSC centrosome orientation in flies cultured in nutrient-rich versus nutrient-poor media (see Supplemental Table S1 for media recipe). The poor media, which contained a lower amount of yeast (protein source) and sugar (carbon source) than rich media, supported normal development and reproduction, albeit with a somewhat reduced fecundity (David et al., 1971; Mair et al., 2004). The centrosomes were scored as misoriented when neither of the two centrosomes is located near the hub-GSC junction (see Cheng et al., 2008, for the definition of centrosome misorientation). As reported previously, in flies grown in rich media ∼7% of the GSCs had misoriented centrosomes. This percentage increased to ∼25% in the flies grown in poor media (Figure 1, b and c). However, we rarely observed misoriented spindles (Figure 1c and Supplemental Table S2), suggesting that the centrosome orientation checkpoint was intact. This increase in centrosome misorientation in nutrient-poor media, combined with the intact centrosome orientation checkpoint, indicates that the GSCs have a slower proliferation rate. Indeed, slowing of the cell cycle was confirmed by a lower S-phase index of GSCs in poor media (Figure 1d; see Materials and Methods). Considering that the majority of misoriented GSCs contain two centrosomes, we believe that they are arrested in the G2 phase of the cell cycle (or at least post G1/S transition), although we cannot exclude the possibility that a minor fraction of GSCs are arrested in G1.
When flies were transferred from poor to rich media, proper centrosome orientation was significantly restored within 3–5 d (Figure 1b), demonstrating the reversible nature of centrosome orientation in response to nutrient availability. We also found that amino acids are the essential component of the rich media required for correct centrosome orientation (Supplemental Figure S1), similar to a report in female GSCs and follicle cells (Drummond-Barbosa and Spradling, 2001). We further found that essential amino acids are the major determinants of centrosome orientation, with a minor (but statistically significant) contribution from nonessential amino acids. Methionine, which was reported to be a major nutrient determinant of longevity (Grandison et al., 2009), had a moderate but statistically significant effect on centrosome orientation (Supplemental Figure S1). Together, these results show that centrosome orientation is under tight control that correlates with the nutrient conditions, in particular the presence of amino acids.
Induction of centrosome misorientation under poor nutrient conditions is not due to increased dedifferentiation
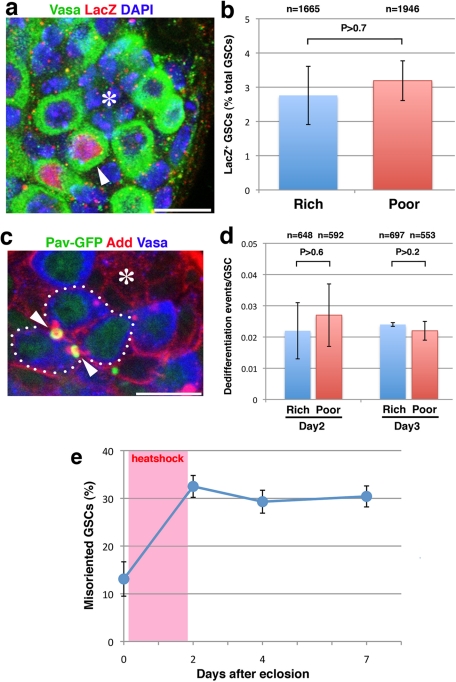
Previously we showed that GSCs dedifferentiated from spermatogonia have a high frequency of centrosome misorientation (Cheng et al., 2008). Thus it is possible that poor nutrient conditions induce GSC loss and subsequent dedifferentiation of spermatogonia, resulting in an increase in GSC centrosome misorientation. However, it is unlikely that dedifferentiation underlies the increase in centrosome misorientation under poor nutrient conditions for the following two reasons, First, using two previously described independent criteria—1) LacZ marking of cells that have once undergone differentiation (Figure 2a) and 2) multiple germ cells, while attaching the hub, being connected by ring canals and disintegrating fusomes (Figure 2c; Brawley and Matunis, 2004; Cheng et al., 2008)—we did not detect any increase in dedifferentiation upon culturing in poor media (Figure 2, b and d). Second, dedifferentiated GSCs, induced by transient expression of Bam (Sheng et al., 2009), never recovered their interphase centrosome orientation (Figure 2e), in contrast to GSCs that are shifted from poor to rich media (Figure 1b). These results suggest that distinct mechanisms underlie centrosome misorientation induced by poor nutrient conditions or dedifferentiation.
FIGURE 2:
Nutrient-poor media do not increase dedifferentiation of germline cells. (a) An example of an apical testis tip with a dedifferentiated GSC, as observed by LacZ expression. LacZ expression was induced by Bam-gal4/UAS-FLP–mediated activation of the LacZ gene, thus marking cells that had once committed to differentiation (Cheng et al., 2008). Green, Vasa (germ cells); red, LacZ; blue, 4′,6-diamidino-2-phenylindole. Hub, asterisk. Scale bar, 10 μm. (b) Frequency of LacZ-positive GSCs in rich and poor media (mean ± SD). (c) An example of apical testes tip containing dedifferentiating GSCs and spermatogonia that are still connected to one another. Green, Pavarotti-GFP (contractile ring and ring canal); red, adducin-like (spectrosome/fusome); blue, Vasa. (d) Frequency of dedifferentiation in rich and poor media (mean ± SD). Flies raised in rich media were transferred to new vials containing either rich or poor media, and testes were examined 2 or 3 d later. (e) Dedifferentiated GSCs do not recover proper centrosome orientation after a prolonged time period. Young adult hs-Bam flies (day 0) were subjected to five heat shocks (30 min each) over the course of 2 d (Sheng et al., 2009) and then cultured at 25°C. GSC centrosome orientation was scored at the indicated time.
Insulin signaling mediates GSC centrosome orientation in response to nutrient conditions
Next we addressed whether centrosome misorientation induced under poor nutrient conditions is physiologically relevant in the regulation of stem cell behavior. Although the poor nutrient conditions used in this study fully support the growth of flies as described earlier, it is formally possible that malnutrition leads to multiple downstream phenomena that compromise “normal” stem cell behavior. To address this question, we tested whether insulin signaling, the major regulator of nutrient responses (Drummond-Barbosa, 2008; Taguchi and White, 2008), is involved in the regulation of GSC centrosome orientation. Expression of a dominant-negative form of insulin receptor (InR; nos-gal4>UAS-InRK1409A; Wu et al., 2005) resulted in a high centrosome misorientation frequency, even in rich media (Figure 3a), without increasing dedifferentiation (Supplemental Figure S2). Conversely, and more important, expression of an active form of InR (nos-gal4>UAS-InRA1325D, UAS-InRDel, or UAS-InRR418P; Wu et al., 2005; Werz et al., 2009) led to a significantly lower frequency of centrosome misorientation, even in poor media (Figure 3a). The hypomorphic InR mutant (inrE19/inr05515) also showed a high centrosome misorientation frequency irrespective of the media conditions (Figure 3a), confirming the results obtained from overexpression of the mutant form of InR. These data suggest that centrosome orientation is downstream of insulin signaling regulated by nutrient availability. The fact that correct centrosome orientation is acquired by activating InR even in poor media also strongly argues against a nonspecific influence of malnutrition on GSC centrosome misorientation.
FIGURE 3:
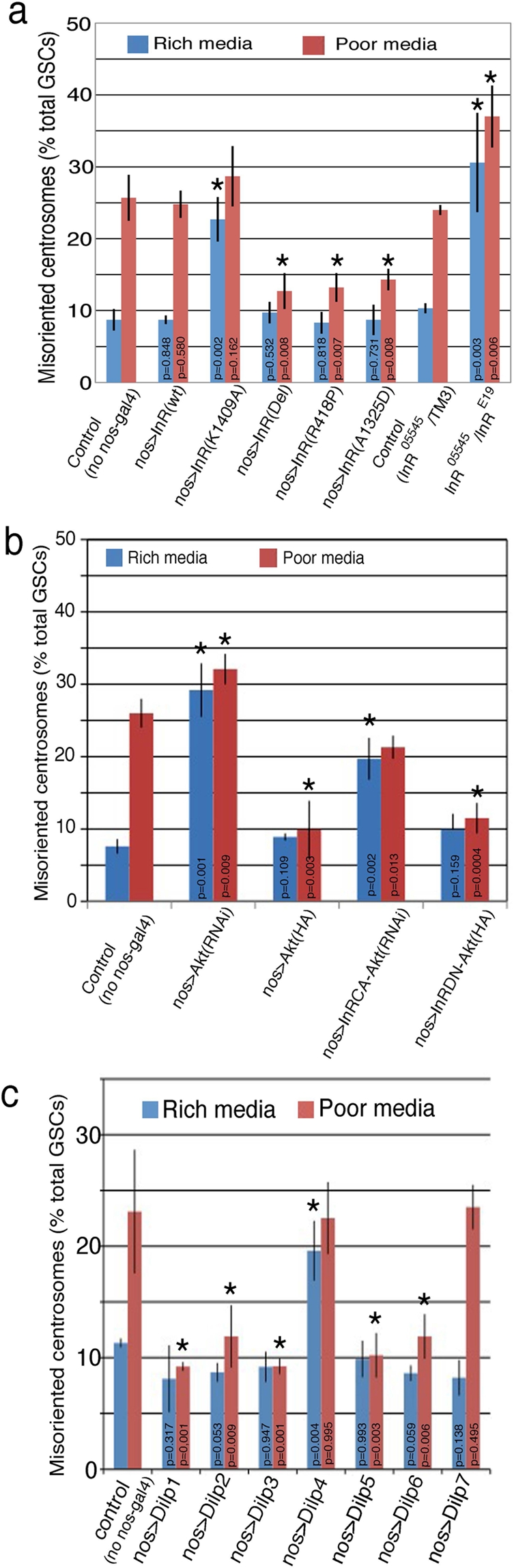
Insulin signaling regulates GSC centrosome orientation. (a) Centrosome misorientation frequency upon overexpression of a dominant-negative (K1409A) or a constitutively active form (Del, R418P, or A1325D) of InR using nos-gal4. n > 300/data point. Siblings from the same cross without the nos-gal4 driver served as controls. p values (Student's t test, two-tailed) are provided on each column compared with its control. Statistically significant values (p < 0.01) are highlighted with asterisks. (b) Akt functions downstream of InR in regulating centrosome orientation. n > 300/data point. (c) Local expression of dilp1, 2, 3, 5, and 6 in the testis reduces centrosome misorientation in poor media. Each dilp was expressed in the testis (nos-gal4>UAS-dilp).
We further investigated the involvement of insulin signaling by testing other components of the pathway. Akt, a major downstream effector of insulin signaling (Taguchi and White, 2008), was required for regulation of GSC centrosome orientation. RNA interference (RNAi)–mediated knockdown of Akt (Parrish et al., 2009) resulted in high centrosome misorientation, even in rich media, whereas overexpression of hemagglutinin (HA)-tagged Akt (Akt-HA; Verdu et al., 1999) resulted in low centrosome misorientation, even in poor media (Figure 3b). Coexpression of Akt-RNAi with constitutively active InR (InRA1325D) led to higher centrosome misorientation frequency, whereas coexpression of Akt-HA with dominant-negative InR (InRK1409A) resulted in low centrosome misorientation frequency. These results are consistent with Akt being downstream of InR (Figure 3b).
The involvement of insulin signaling was further suggested by the fact that increasing the local concentration of Drosophila insulin-like peptides (dilp1, 2, 3, 5, and 6, but not dilp4 and 7) in the testes (Brogiolo et al., 2001; Hsu and Drummond-Barbosa, 2009) leads to a lower centrosome misorientation frequency, even in poor media (Figure 3c). Although these results show the potential of dilps to modulate GSC centrosome orientation, the identity of the physiologically relevant dilp(s) and their tissue of origin remain to be determined. In addition, it should be noted that the expression of dilp4 caused constitutively high centrosome misorientation irrespective of the media conditions, although the underlying mechanism remains to be elucidated in future studies. Together, these data indicate the involvement of insulin signaling in the regulation of GSC centrosome orientation and thus GSC proliferation.
Changes in the localization of Apc2 mediate GSC centrosome orientation in response to the nutrient conditions
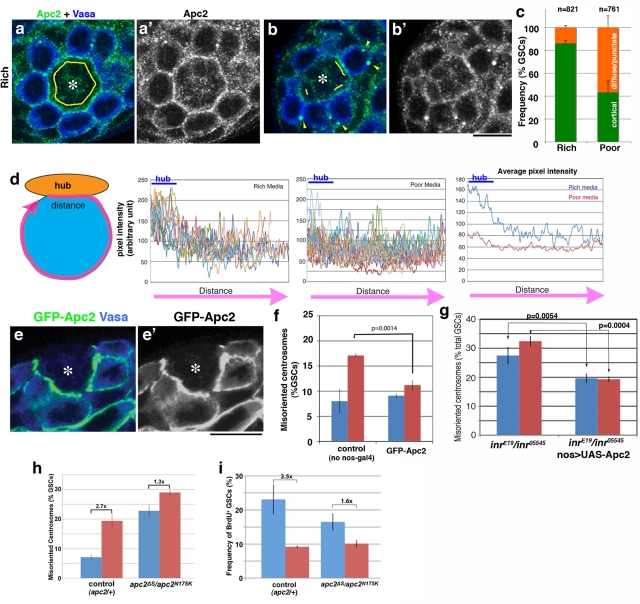
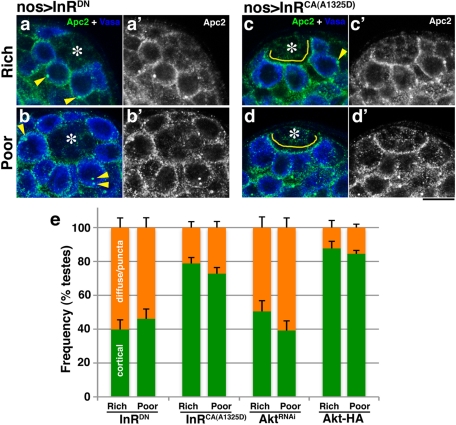
We next investigated the cellular basis for the regulation of centrosome orientation under the influence of nutrient signaling. Previously Akt was reported to regulate spindle orientation in Drosophila embryos via regulation of the cortical localization of Apc2 (Buttrick et al., 2008). Because Apc2 is believed to function as a cortical anchor of centrosomes in male GSCs (Yamashita et al., 2003), we tested the possibility that insulin signaling and Akt might regulate GSC centrosome orientation via regulation of the Apc2 protein. Indeed, we found that localization of Apc2 dynamically changes in response to nutrient conditions. In rich media, Apc2 was predominantly associated with the cell cortex at the hub-GSC interface (Figure 4a), as reported previously (Yamashita et al., 2003). In contrast, in poor media Apc2 was localized to the hub-GSC interface in only ∼40% of GSCs (Figure 4c), with the remaining GSCs exhibiting either a diffuse or punctate pattern of cytoplasmic Apc2 (Figure 4, b and c). The punctate structure is unlikely to be an artifact of immunofluorescence staining because known partners of Apc2, such as Shaggy (Supplemental Figure S3b) and axin (not shown), colocalized to this structure. To further quantitatively analyze Apc2 localization at the GSC cortex, we measured the pixel intensity of Apc2 staining around the entire GSC cortex in rich versus poor media (Figure 4d; see Materials and Methods for details). This analysis clearly showed a higher Apc2 level at the hub-GSC cortex in rich, but not poor, media. The location of other proteins that are known to localize to the hub-GSC interface, such as DE-cadherin and armadillo (β-catenin), did not change in response to poor media (Supplemental Figure S4), demonstrating the specificity of the change in localization of Apc2. The Apc2 protein level appears to be slightly decreased in poor media, as suggested by Western blotting (Supplemental Figure S3a), which might partly contribute to the decreased Apc2 localization at the hub-GSC interface. We further tested whether InR and Akt regulate centrosome orientation via regulation of Apc2 localization and found that modulation of InR and Akt activity affected Apc2 localization (Figure 5). Expression of a dominant-negative form of InR decreased Apc2 localization at the hub-GSC interface, whereas Apc2 was consistently observed at the hub-GSC cortex when a constitutively active form of InR was expressed, irrespective of media conditions. The same trend was observed upon modulation of Akt activity. However, modulation of these genes did not significantly affect Apc2 protein level (Supplemental Figure S3c), suggesting that these genes influence GSC centrosome orientation primarily through regulating the localization of Apc2.
FIGURE 4:
Apc2 mediates centrosome orientation in response to nutrients. (a, b) Representative Apc2 staining in apical testes tips from flies raised in rich (a) or poor (b) media. Cortical Apc2 localization is indicated with yellow lines. Cytoplasmic punctae of Apc2 are indicated with yellow arrowheads. Green, Apc2; blue, Vasa. (c) Quantification of Apc2 localization in rich or poor media. “Cortical” indicates Apc2 protein at the hub-GSC junction; “diffuse/puncta” indicates Apc2 in the cytoplasm or occasionally at the GSC cortex outside the hub-GSC interface. n = GSCs scored. (d) Pixel intensity analysis of Apc2 protein around the GSC cortex. Circumference of GSC was traced and the pixel intensity was analyzed (see Materials and Methods for detail). Fourteen GSCs from the rich media and 19 GSCs from the poor media were analyzed. Average pixel intensity for rich vs. poor media is shown at the far right. (e) Localization of GFP-Apc2 to the hub-GSC interface following mild expression (nos-gal4>UAS-GFP-Apc2; at 18°C) in poor media. Green, GFP-Apc2; blue, Vasa. (f) The frequency of GSC centrosome misorientation upon mild expression of GFP-Apc2. n > 300/data point. (g) The frequency of GSC centrosome misorientation in the InR mutant that expresses Apc2. n > 300/data point. (h) The frequency of GSC centrosome misorientation in the apc2 mutant. n > 300/data point. (i) The S-phase index (BrdU incorporation) of apc2 mutant GSCs. n > 300/data point.
FIGURE 5:
Apc2 localization is regulated by the insulin signaling pathway. (a–d) Examples of Apc2 staining in apical testes tips from flies that express a dominant-negative (K1409A) or constitutively active (A1325D) form of InR in rich vs. poor media. Cortical localization of Apc2 is indicated with yellow lines. Cytoplasmic punctae of Apc2 are indicated with yellow arrowheads. Green, Apc2; blue, Vasa (germ cells). (e) Quantification of Apc2 localization upon overexpression of a dominant-negative (K1409A) or constitutively active (A1325D) form of InR, Akt-RNAi, or Akt-HA in rich or poor media (mean ± SD). n > 100 testes/data point.
We postulate that delocalization of Apc2 from the hub-GSC interface, due to either a change in Apc2 protein localization or amount, plays a role in regulating GSC centrosome orientation. It is striking that mild overexpression of Apc2 in GSCs (nos-gal4>UAS-GFP-Apc2 at 18°C; Figure 4e; Inaba et al., 2010) was sufficient to suppress centrosome misorientation (Figure 4f). It should be noted that mild overexpression of Apc2 was achieved by raising flies at 18°C, since strong overexpression of Apc2 at 25°C causes cortical localization of Apc2, leading to centrosome misorientation (Inaba et al., 2010). At 18°C, wild-type/control flies showed somewhat lower centrosome misorientation (∼17% compared with ∼25%) in poor media, although it was still significantly higher than in rich media (Figure 4f). Mild overexpression of Apc2 clearly reduced centrosome misorientation in the poor media (Figure 4f). It is interesting to note that mildly overexpressed Apc2 protein can still localize to the hub-GSC interface, supporting correct centrosome orientation. This suggests that delocalization of endogenous Apc2 from the hub-GSC interface in poor media is largely achieved by down-regulation of the protein amount, although additional modes of regulation, such as posttranslational modification of Apc2 protein, may also contribute to changes in Apc2 localization in response to poor nutrient conditions. In this case, an increased amount of Apc2 protein might be recruited to the hub-GSC interface, albeit at a lower affinity. Collectively, these data suggest that Apc2 is a major mediator of nutrient-regulated centrosome orientation downstream of InR signaling. Indeed, Apc2 expression in an InR mutant background considerably suppressed centrosome misorientation (Figure 4g), although not to the extent of the wild type. The incomplete suppression of centrosome misorientation in Inr mutant background may suggest that Apc2 is not the sole target of the InR pathway in mediating GSC centrosome orientation, which is also supported by the observation to be described.
We reasoned that if Apc2 localization to the cortex is required for correct centrosome orientation and thus active GSC divisions, an Apc2 mutant that does not have cortical Apc2 (Yamashita et al., 2003) would have a lower cell cycle index, even in rich media, and would respond less efficiently to poor media. In control GSCs, centrosome misorientation increased 2.7-fold in poor media compared with rich media (Figure 4h), which corresponded to a 2.5-fold decrease in the S-phase index (Figure 4i). In contrast, in the Apc2 mutant, GSCs have high centrosome misorientation, even in rich media (consistent with the previous report using the standard media; Yamashita et al., 2003), which was increased only 1.3-fold in poor media (Figure 4h), leading to a 1.6-fold decrease in the S-phase index (Figure 4i). These results show that Apc2 mutant GSCs are less active in proliferation, even in rich media, and thus are less effective in responding to poor media. However, because Apc2 mutant GSCs can still slow their proliferation to some extent, additional mechanism(s) likely exist that contribute to centrosome misorientation and cell cycle slowdown in response to poor nutrient conditions. The results, taken together, lead us to conclude that Apc2 is a key (but not the sole) component in modulating GSC centrosome orientation and proliferation in response to nutrient conditions.
Centrosome orientation checkpoint is required for GSC cell cycle slowdown in nutrient-poor media
We hypothesized that the cell cycle in GSCs is slowed under limited nutrient conditions through misorientation of GSC centrosomes, which activates the centrosome orientation checkpoint. This hypothesis predicts that mutants defective in this checkpoint would be defective in slowing the cell cycle in poor media. To test this, we examined two mutant conditions that are deficient in the centrosome orientation checkpoint: the centrosomin (cnn) mutant and dominant-negative E-cadherin (dCR4h; Inaba et al., 2010). These two mutant conditions caused high centrosome misorientation, even in rich media (Figure 6a), but maintained a high S-phase index (bromodeoxyuridine [BrdU] incorporation; Figure 6b), confirming that these mutants are defective in the centrosome orientation checkpoint. It is striking that GSCs in these mutants neither increased centrosome misorientation nor slowed the cell cycle in poor media (Figure 6, a and b). These results clearly demonstrate that cell cycle slowdown of GSCs in response to poor nutrients requires an intact GSC centrosome orientation checkpoint and places the centrosome orientation checkpoint upstream of cell cycle slowdown in response to poor nutrient conditions.
FIGURE 6:
An intact centrosome orientation checkpoint is required to slow the GSC cell cycle in poor media. (a) Centrosome misorientation frequency in GSCs mutant for cnn or expressing dominant-negative E-cadherin (dCR4h). n > 150/data point. p value (Student's t test, two-tailed) comparing centrosome misorientation in rich vs. poor media is shown. n.s., not statistically significant. (b) The S-phase index (BrdU incorporation frequency) in GSCs from checkpoint-defective mutants and those expressing InRA1325D. n > 450/data point. p value (Student's t test, two-tailed) comparing BrdU incorporation in rich vs. poor media is shown. n.s., not statistically significant. (c) Changes in GSC numbers in testes from control genotypes, cnn mutants, or those expressing dCR4h or InRA1325D after 10 d in rich vs. poor media. p value (Student's t test, two-tailed) comparing the GSC number in rich vs. poor media after 10 d is shown. n.s., not statistically significant. Asterisk indicates that a mild but statistically significant decrease in the GSC number was observed in this control group for unknown reasons. However, the degree of GSC loss was significantly (p < 0.01) more severe in InRA1325D-expressing testis compared with the control. n = 70–100 testes/data point. (d) Model of the regulation of the GSC division rate by nutrient availability and insulin signaling (see the text for details).
We next examined what happens to GSCs under conditions in which the cell cycle does not slow, even in poor media. In addition to GSCs from cnn mutants or those expressing dominant-negative E-cadherin (dCR4h), expression of a constitutively active form of InR (InRA1325D) in the germline (nos-gal4>InRA1325D) resulted in failure of cell cycle slowdown in poor media (Figure 6b), as expected from their low centrosome misorientation in these conditions (Figure 3a). Control testes maintained the GSC number well for 10 d after eclosion in rich and poor nutrient conditions (Figure 6c). In contrast, cnn mutant flies or those expressing InRA1325D showed a significant decrease in GSC number when cultured in poor media for 10 d (Figure 6c). This is not due to a general problem in maintaining the GSC number, because the GSC number did not decrease when these mutants were cultured in rich media. Instead, these results indicate that the slowdown of GSC proliferation is critical to maintain the GSC number in response to poor nutrient conditions. Expression of a dominant-negative form of E-cadherin (dCR4h) significantly decreased the GSC number after 10 d irrespective of media conditions, presumably due to defective cell adhesion to hub cells (Inaba et al., 2010). Together, we concluded that the cell cycle slowdown mediated by the centrosome orientation checkpoint is required to maintain GSC number under poor nutrient conditions.
DISCUSSION
Here we show that male GSC centrosome misorientation is induced by poor nutrient conditions. Such centrosome misorientation is associated with a lower cell proliferation rate, presumably due to activation of the centrosome orientation checkpoint (Figure 6d). Similar to Drosophila male GSCs, Drosophila female GSCs (Drummond-Barbosa and Spradling, 2001; LaFever and Drummond-Barbosa, 2005; Hsu and Drummond-Barbosa, 2009) and intestinal stem cells (McLeod et al., 2010; Choi et al., 2011; O'Brien et al., 2011), as well as C. elegans GSCs (Michaelson et al., 2010), respond to nutrient availability and insulin signaling. However, it is not well understood how nutrient information is transmitted to the cell cycle control of stem cells. Although a considerable amount of knowledge has accumulated over past decades on how nutrient information regulates cell proliferation, and some components of the pathway have been tested in in vivo stem cell settings, it remains unclear how nutrient information might be transduced to the cell cycle control of stem cell divisions in what is a possibly a stem cell–specific manner. The present study provides the first link between nutrient signal, stem cell polarity (i.e., centrosome orientation), and cell cycle regulation. Use of a stem cell–specific mechanism such as the centrosome orientation checkpoint could provide distinct regulation of proliferation in stem cells: it is tempting to speculate that stem cells might respond to a distinct threshold of nutrient levels that is different from other cell types, and thus the centrosome orientation checkpoint functions as an additional layer of regulation of proliferation posed specifically on stem cells. For example, proliferation of stem cells could be more (or less) sensitive to nutrient depletion than proliferation of other cell types. In this regard, it will be interesting to investigate in future studies not only the involvement of other components of the nutrient/insulin signaling pathway, but also the differential regulation of those components between stem cells and other cell types.
It is interesting to note that female GSCs do not have stereotypical centrosome orientation (Stevens et al., 2007). In addition, C. elegans GSCs do not undergo stereotypical asymmetric division (Crittenden et al., 2006). Thus the cellular mechanisms that slow cell cycle progression in response to nutrient/insulin signaling are likely to be different in these systems. Other stem cell populations, such as Drosophila neuroblasts (Januschke and Gonzalez, 2008) and mouse radial glia progenitors (Wang et al., 2009), that display stereotypical centrosome orientation might use a mechanism similar to that of male GSCs to modulate cell cycle progression. A recent report described an InR pathway–mediated mechanism that regulates the GSC number in response to protein starvation (McLeod et al., 2010). McLeod et al. (2010) showed that centrosome misorientation does not increase upon protein starvation, in contrast to our observation in poor media. To reconcile these potentially conflicting results, we examined centrosome orientation upon protein starvation and found that centrosome misorientation transiently increases prior to observed GSC loss and then decreases (unpublished data). This demonstrates that the GSC responses to poor media that contain proteins and to protein starvation are different, at least regarding centrosome orientation. The transient increase in centrosome misorientation presumably reflects the systemic environment before protein is completely removed, which is similar to poor media.
Recently it was shown that nutrient status affects the mode of stem cell division in Drosophila intestinal stem cells (ISCs; O'Brien et al., 2011): when the intestine is stimulated to grow by refeeding of a starved animal, the ISCs predominantly divide symmetrically, in contrast to the homeostatic state, when ISCs predominantly divide asymmetrically. Although it is not clear how ISCs divide asymmetrically under homeostatic conditions and how such a mechanism is modulated to divide symmetrically upon stimulation, it is tempting to speculate that ISCs possess a mechanism that links nutrient sensing and asymmetric stem cell division similar to that in male GSCs reported here. However, we have not yet found any physiological conditions that inactivate the centrosome orientation checkpoint and result in symmetric divisions in Drosophila male GSCs. Thus the similarity between ISCs and male GSCs awaits further assessment.
How the InR pathway regulates the GSC division rate via sensing of amino acid levels remains to be elucidated. In Drosophila, many stem cell types respond to amino acid level (but often not to sugar level), and this response is mediated by insulin signaling (LaFever and Drummond-Barbosa, 2005; Hsu et al., 2008; McLeod et al., 2010). Our unpublished results show that TOR is also involved in the regulation of GSC centrosome orientation in response to poor nutrients. However, how the insulin pathway and TOR pathway interact to mediate nutrient response awaits future investigation.
In summary, the present findings illuminate a mechanism for how environmental information (such as nutrient availability) is translated into a cellular response (i.e., slowed cell cycle progression) using a cell-intrinsic mechanism (cell cycle delay/arrest associated with centrosome misorientation). We propose that the centrosome orientation checkpoint, whose primary function is to ensure asymmetric stem cell division, is used to modulate the stem cell proliferation rate in response to nutrient conditions in Drosophila male GSCs.
MATERIALS AND METHODS
Fly husbandry and strains
All fly stocks were raised on rich or poor media (see Supplemental Table S1 for recipe) at 25°C unless otherwise noted. Young adult flies (day 0) were used unless indicated otherwise. The following fly stocks were used: c587-Gal4 from Steven Hou (Kai and Spradling, 2003), Ubi-Pavarotti-GFP from David Glover (Minestrini et al., 2002), UAS-GFP-Apc2 from Mariann Bienz (Hamada and Bienz, 2002), Bam-gal4 from Dennis McKearin (Chen and McKearin, 2003), UAS-Akt-HA from Morris Birnbaum (Verdu et al., 1999), UAS-dilp1-7 from Ernst Hafen and Daniella Drummond-Barbosa (Ikeya et al., 2002), nanos (nos)-Gal4 (Van Doren et al., 1998), UAS-FLP (Duffy et al., 1998), Actin>stop>LacZ (Act5C-FRT-stop-FRT-LacZ) (Struhl and Basler, 1993), UAS-InR (wild type and mutants; Fly Base, http://flybase.org/reports/FBrf0178856.html) from the Bloomington Drosophila Stock Center (Indiana University, Bloomington IN), and UAS-Akt-RNAi (v2902) from the Vienna Drosophila RNAi Center (Vienna, Austria).
Immunofluorescence staining
Immunofluorescence staining was performed as described previously (Cheng et al., 2008). The following antibodies were used: mouse anti–γ-tubulin (1:100; GTU-88, Sigma-Aldrich, St. Louis, MO), mouse anti–fasciclin III (1:20; developed by C. Goodman and obtained from the Developmental Studies Hybridoma Bank, University of Iowa, Iowa City, IA), mouse anti–adducin-like (1:20; developed by H. D. Lipshitz (Ding et al., 1993) and obtained from the Developmental Studies Hybridoma Bank), rabbit anti–Thr 3-phosphorylated histone H3 (1:200; Upstate, Millipore, Billerica, CA), mouse anti–β-galactosidase (1:200; G4644; Sigma-Aldrich), goat anti-Vasa (1:100; dC-13; Santa Cruz Biotechnology, Santa Cruz, CA), rabbit anti-Vasa (1:100; d-26; Santa Cruz Biotechnology), mouse anti-BrdU (1:200; BU-33; Sigma-Aldrich), mouse anti–acetylated tubulin (1:100; Sigma-Aldrich), and rabbit anti-Apc2 (1:5000; a generous gift from Marian Bienz). GSCs were scored as misoriented when neither of the two centrosomes was observed away from the hub-GSC interface, as described previously (Cheng et al., 2008). MetaMorph software (Molecular Devices, Sunnyvale, CA) was used to compare the intensity of Apc2 staining near the hub with staining around the rest of the cell circumference. Briefly, we traced the cells of interest with the multiline tool set at a thickness of five pixels and made a data log of the pixel intensity values. This was done for GSCs from flies grown in rich media and in poor media. We then used Excel (Microsoft, Redmond, WA) to make line graphs using the pixel intensity data from MetaMorph. The average intensity for each pixel point was used to make a line graph to compare the nutrient-rich and -poor sets. GSC centrosome misorientation was defined as GSCs in which neither of the two centrosomes is closely associated with the hub-GSC junction (Cheng et al., 2008).
BrdU incorporation
Day 0 adult flies were dissected into 1× phosphate-buffered saline (PBS) within 15 min and incubated with 0.1 μg/ml BrdU (final concentration) for exactly 45 min. Testes were then fixed in 4% formaldehyde in 1× PBS for 30–60 min, treated with DNaseI in 1× DNaseI buffer (Invitrogen, Carlsbad, CA) for 2 h, and incubated with primary antibody (anti-BrdU and anti-Vasa) overnight at 4°C. Finally, the samples were processed using the standard immunofluorescence staining protocol described earlier.
Western blotting
Testes (60 pairs/sample) were dissected in PBS at room temperature within 20–30 min. Testes were then dissolved in lithium dodecyl sulfate sample buffer (NuPAGE; Invitrogen) supplemented with 2% SDS and protease inhibitor cocktail (EDTA-free; Roche, Indianapolis, IN). The samples were separated on NuPAGE Bis-Tris gels (4–12%; Invitrogen) and transferred onto polyvinylidene fluoride membranes (Immobilon-P; Millipore). Blots were blocked in PBS containing 5% nonfat milk powder and 0.1% Triton X-100, followed by incubation with primary antibodies diluted in PBS containing 5% nonfat milk powder and 0.1% Triton X-100. Next the blots were washed with PBS containing 0.1% Triton X-100, followed by incubation with the secondary antibody. After washing, detection was performed using the enhanced chemiluminescence system (Amersham-Pharmacia Biotech, GE Healthcare Bio-Sciences, Piscataway, NJ). Primary antibodies were anti-Apc2 (1:100; rabbit, a kind gift from Mariann Bienz) and anti–α-tubulin (1:200; mouse, monoclonal, clone DM1a; Sigma-Aldrich). Secondary antibodies were horseradish peroxidase (HRP)-conjugated anti-mouse and anti-rabbit (1:4000; Jackson ImmunoResearch Laboratories, West Grove, PA).
Supplementary Material
Acknowledgments
We thank Mariann Bienz, Morris Birnbaum, David Glover, Dennis McKearin, Eric Rulifson, Daniella Drummond-Barbosa, Ernst Hafen, the Bloomington Stock Center, the Vienna Drosophila RNAi Center, and the Developmental Studies Hybridoma Bank for reagents. We also thank Yamashita lab members, Alan Saltiel, and Ken Inoki for comments on the manuscript and Samuel Straight (University of Michigan Center for Live Cell Imaging, University of Michigan Ann Arbor, MI) for help with image analysis.
Abbreviations used:
- BrdU
5-bromo-2′-deoxyuridine
- GFP
green fluorescent protein
- GSC
germline stem cell
Footnotes
This article was published online ahead of print in MBoC in Press (http://www.molbiolcell.org/cgi/doi/10.1091/mbc.E11-12-0999) on February 22, 2012.
REFERENCES
- Brawley C, Matunis E. Regeneration of male germline stem cells by spermatogonial dedifferentiation in vivo. Science. 2004;304:1331–1334. doi: 10.1126/science.1097676. [DOI] [PubMed] [Google Scholar]
- Brogiolo W, Stocker H, Ikeya T, Rintelen F, Fernandez R, Hafen E. An evolutionarily conserved function of the Drosophila insulin receptor and insulin-like peptides in growth control. Curr Biol. 2001;11:213–221. doi: 10.1016/s0960-9822(01)00068-9. [DOI] [PubMed] [Google Scholar]
- Buttrick GJ, Beaumont LM, Leitch J, Yau C, Hughes JR, Wakefield JG. Akt regulates centrosome migration and spindle orientation in the early Drosophila melanogaster embryo. J Cell Biol. 2008;180:537–548. doi: 10.1083/jcb.200705085. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chen D, McKearin DM. A discrete transcriptional silencer in the bam gene determines asymmetric division of the Drosophila germline stem cell. Development. 2003;130:1159–1170. doi: 10.1242/dev.00325. [DOI] [PubMed] [Google Scholar]
- Cheng J, Turkel N, Hemati N, Fuller MT, Hunt AJ, Yamashita YM. Centrosome misorientation reduces stem cell division during ageing. Nature. 2008;456:599–604. doi: 10.1038/nature07386. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Choi NH, Lucchetta E, Ohlstein B. Nonautonomous regulation of Drosophila midgut stem cell proliferation by the insulin-signaling pathway. Proc Natl Acad Sci USA. 2011;108:18702–18707. doi: 10.1073/pnas.1109348108. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Crittenden SL, Leonhard KA, Byrd DT, Kimble J. Cellular analyses of the mitotic region in the Caenorhabditis elegans adult germ line. Mol Biol Cell. 2006;17:3051–3061. doi: 10.1091/mbc.E06-03-0170. [DOI] [PMC free article] [PubMed] [Google Scholar]
- David J, Van Herrewege J, Fouillet P. Quantitative under-feeding of Drosophila: effects on adult longevity and fecundity. Exp Gerontol. 1971;6:249–257. doi: 10.1016/0531-5565(71)90037-4. [DOI] [PubMed] [Google Scholar]
- Ding D, Parkhurst SM, Lipshitz HD. Different genetic requirements for anterior RNA localization revealed by the distribution of adducin-like transcripts during Drosophila oogenesis. Proc Natl Acad Sci USA. 1993;90:2512–2516. doi: 10.1073/pnas.90.6.2512. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Drummond-Barbosa D. Stem cells, their niches and the systemic environment: an aging network. Genetics. 2008;180:1787–1797. doi: 10.1534/genetics.108.098244. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Drummond-Barbosa D, Spradling AC. Stem cells and their progeny respond to nutritional changes during Drosophila oogenesis. Dev Biol. 2001;231:265–278. doi: 10.1006/dbio.2000.0135. [DOI] [PubMed] [Google Scholar]
- Duffy JB, Harrison DA, Perrimon N. Identifying loci required for follicular patterning using directed mosaics. Development. 1998;125:2263–2271. doi: 10.1242/dev.125.12.2263. [DOI] [PubMed] [Google Scholar]
- Fuller MT, Spradling AC. Male and female Drosophila germline stem cells: two versions of immortality. Science. 2007;316:402–404. doi: 10.1126/science.1140861. [DOI] [PubMed] [Google Scholar]
- Grandison RC, Piper MD, Partridge L. Amino-acid imbalance explains extension of lifespan by dietary restriction in Drosophila. Nature. 2009;462:1061–1064. doi: 10.1038/nature08619. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hamada F, Bienz M. A Drosophila APC tumour suppressor homologue functions in cellular adhesion. Nat Cell Biol. 2002;4:208–213. doi: 10.1038/ncb755. [DOI] [PubMed] [Google Scholar]
- Hsu HJ, Drummond-Barbosa D. Insulin levels control female germline stem cell maintenance via the niche in Drosophila. Proc Natl Acad Sci USA. 2009;106:1117–1121. doi: 10.1073/pnas.0809144106. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hsu HJ, LaFever L, Drummond-Barbosa D. Diet controls normal and tumorous germline stem cells via insulin-dependent and -independent mechanisms in Drosophila. Dev Biol. 2008;313:700–712. doi: 10.1016/j.ydbio.2007.11.006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ikeya T, Galic M, Belawat P, Nairz K, Hafen E. Nutrient-dependent expression of insulin-like peptides from neuroendocrine cells in the CNS contributes to growth regulation in Drosophila. Curr Biol. 2002;12:1293–1300. doi: 10.1016/s0960-9822(02)01043-6. [DOI] [PubMed] [Google Scholar]
- Inaba M, Yuan H, Salzmann V, Fuller MT, Yamashita YM. E-cadherin is required for centrosome and spindle orientation in Drosophila male germline stem cells. PLoS ONE. 2010;5:e12473. doi: 10.1371/journal.pone.0012473. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Januschke J, Gonzalez C. Drosophila asymmetric division, polarity and cancer. Oncogene. 2008;27:6994–7002. doi: 10.1038/onc.2008.349. [DOI] [PubMed] [Google Scholar]
- Kai T, Spradling A. An empty Drosophila stem cell niche reactivates the proliferation of ectopic cells. Proc Natl Acad Sci USA. 2003;100:4633–4638. doi: 10.1073/pnas.0830856100. [DOI] [PMC free article] [PubMed] [Google Scholar]
- LaFever L, Drummond-Barbosa D. Direct control of germline stem cell division and cyst growth by neural insulin in Drosophila. Science. 2005;309:1071–1073. doi: 10.1126/science.1111410. [DOI] [PubMed] [Google Scholar]
- Mair W, Sgro CM, Johnson AP, Chapman T, Partridge L. Lifespan extension by dietary restriction in female Drosophila melanogaster is not caused by a reduction in vitellogenesis or ovarian activity. Exp Gerontol. 2004;39:1011–1019. doi: 10.1016/j.exger.2004.03.018. [DOI] [PubMed] [Google Scholar]
- McLeod CJ, Wang L, Wong C, Jones DL. Stem cell dynamics in response to nutrient availability. Curr Biol. 2010;20:2100–2105. doi: 10.1016/j.cub.2010.10.038. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Michaelson D, Korta DZ, Capua Y, Hubbard EJ. Insulin signaling promotes germline proliferation in C. elegans. Development. 2010;137:671–680. doi: 10.1242/dev.042523. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Minestrini G, Mathe E, Glover DM. Domains of the Pavarotti kinesin-like protein that direct its subcellular distribution: effects of mislocalisation on the tubulin and actin cytoskeleton during Drosophila oogenesis. J Cell Sci. 2002;115:725–736. doi: 10.1242/jcs.115.4.725. [DOI] [PubMed] [Google Scholar]
- Morrison SJ, Kimble J. Asymmetric and symmetric stem-cell divisions in development and cancer. Nature. 2006;441:1068–1074. doi: 10.1038/nature04956. [DOI] [PubMed] [Google Scholar]
- Narbonne P, Roy R. Inhibition of germline proliferation during C. elegans dauer development requires PTEN, LKB1 and AMPK signalling. Development. 2006;133:611–619. doi: 10.1242/dev.02232. [DOI] [PubMed] [Google Scholar]
- O'Brien LE, Soliman SS, Li X, Bilder D. Altered modes of stem cell division drive adaptive intestinal growth. Cell. 2011;147:603–614. doi: 10.1016/j.cell.2011.08.048. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Parrish JZ, Xu P, Kim CC, Jan LY, Jan YN. The microRNA bantam functions in epithelial cells to regulate scaling growth of dendrite arbors in Drosophila sensory neurons. Neuron. 2009;63:788–802. doi: 10.1016/j.neuron.2009.08.006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Saltiel AR, Kahn CR. Insulin signalling and the regulation of glucose and lipid metabolism. Nature. 2001;414:799–806. doi: 10.1038/414799a. [DOI] [PubMed] [Google Scholar]
- Sheng XR, Brawley CM, Matunis EL. Dedifferentiating spermatogonia outcompete somatic stem cells for niche occupancy in the Drosophila testis. Cell Stem Cell. 2009;5:191–203. doi: 10.1016/j.stem.2009.05.024. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Stevens NR, Raposo AA, Basto R, St Johnston D, Raff JW. From stem cell to embryo without centrioles. Curr Biol. 2007;17:1498–1503. doi: 10.1016/j.cub.2007.07.060. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Struhl G, Basler K. Organizing activity of wingless protein in Drosophila. Cell. 1993;72:527–540. doi: 10.1016/0092-8674(93)90072-x. [DOI] [PubMed] [Google Scholar]
- Taguchi A, White MF. Insulin-like signaling, nutrient homeostasis, and life span. Annu Rev Physiol. 2008;70:191–212. doi: 10.1146/annurev.physiol.70.113006.100533. [DOI] [PubMed] [Google Scholar]
- Ueishi S, Shimizu H, Y HI. Male germline stem cell division and spermatocyte growth require insulin signaling in Drosophila. Cell Struct Funct. 2009;34:61–69. doi: 10.1247/csf.08042. [DOI] [PubMed] [Google Scholar]
- Van Doren M, Williamson AL, Lehmann R. Regulation of zygotic gene expression in Drosophila primordial germ cells. Curr Biol. 1998;8:243–246. doi: 10.1016/s0960-9822(98)70091-0. [DOI] [PubMed] [Google Scholar]
- Verdu J, Buratovich MA, Wilder EL, Birnbaum MJ. Cell-autonomous regulation of cell and organ growth in Drosophila by Akt/PKB. Nat Cell Biol. 1999;1:500–506. doi: 10.1038/70293. [DOI] [PubMed] [Google Scholar]
- Wang X, Tsai JW, Imai JH, Lian WN, Vallee RB, Shi SH. Asymmetric centrosome inheritance maintains neural progenitors in the neocortex. Nature. 2009;461:947–955. doi: 10.1038/nature08435. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Werz C, Kohler K, Hafen E, Stocker H. The Drosophila SH2B family adaptor Lnk acts in parallel to chico in the insulin signaling pathway. PLoS Genetics. 2009;5:e1000596. doi: 10.1371/journal.pgen.1000596. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wu Q, Zhang Y, Xu J, Shen P. Regulation of hunger-driven behaviors by neural ribosomal S6 kinase in Drosophila. Proc Natl Acad Sci USA. 2005;102:13289–13294. doi: 10.1073/pnas.0501914102. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yamashita YM, Jones DL, Fuller MT. Orientation of asymmetric stem cell division by the APC tumor suppressor and centrosome. Science. 2003;301:1547–1550. doi: 10.1126/science.1087795. [DOI] [PubMed] [Google Scholar]
- Yamashita YM, Mahowald AP, Perlin JR, Fuller MT. Asymmetric inheritance of mother versus daughter centrosome in stem cell division. Science. 2007;315:518–521. doi: 10.1126/science.1134910. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yuan H, Chiang CY, Cheng J, Salzmann V, Yamashita YM. Regulation of cyclin A localization downstream of Par-1 function is critical for the centrosome orientation checkpoint in Drosophila male germline stem cells. Dev Biol. 2012;361:57–67. doi: 10.1016/j.ydbio.2011.10.010. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.