Abstract
SIRT1 is a member of the Sir2 family of NAD+ dependent protein deacetylases. The central role of SIRT1 in multiple metabolic and aging-related pathways has pushed SIRT1 to the forefront for the discovery of small molecule activators. Promising compounds including resveratrol and SRT1720 have been reported, however whether these compounds are direct activators and the mechanism by which they activate remains poorly defined. This review will examine the current debate surrounding purported activators, focusing on the assays used in screening compounds, Sirtuin catalysis and the mechanistic basis for their actions. We discuss the potential pathways of SIRT1 activation that could be exploited for the development of novel therapeutics for treating type II diabetes, neurodegeneration and diseases associated with aging.
Keywords: deacetylation, resveratrol, Sir2, SIRT1 activators, mechanism, catalysis, small-molecules
1. Introduction
1.1 Sir2 mechanism and structure
Silent information regulator (Sir2 or Sirtuin) protein deacetylases are a class of evolutionarily conserved enzymes that function in critical cellular processes such as gene silencing, insulin secretion and apoptosis.[1] Sirtuins have been linked with many age-related and metabolic diseases, including type II diabetes, cardiovascular disease, neurodegeneration, inflammation and cancer.[2] The integral regulatory role of Sirtuins in metabolic processes makes them desirable targets for the development of small molecule therapeutics.
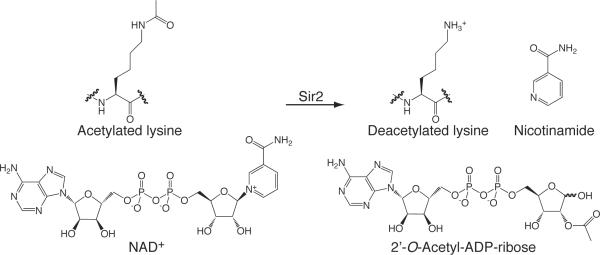
Yeast Sir2 is the founding member of the Sirtuin family of enzymes that function as nicotinamide adenine dinucleotide (NAD+) dependent lysine deacetylases. Sir2 is involved in transcriptional silencing through heterochromatin maintenance[3] and also positively affects lifespan in yeast, worms and flies.[4] Mammals harbor seven distinct Sir2 homologs, referred to as Sirtuins (SIRT1–7), each with diverse cellular locations and targets.[5] The majority of Sirtuin proteins function to catalyze the NAD+ dependent deacetylation of acetylated lysine residues,[6] however this function has not been demonstrated for all Sirtuins. SIRT4 was reported to function as a protein ADP-ribosyl transferase[7] and the molecular function of nuclear SIRT7 is not well established. The products of the Sirtuin catalyzed reaction are 2'-O-acetyl-ADP-ribose (OAADPr), nicotinamide and deacetylated lysine (Scheme 1). Structural studies of various members of the Sir2 family have revealed that Sirtuins contain a structurally conserved elongated core composed of two domains. A Rossmann-fold domain is found at one end and a smaller, more variable, zinc-binding domain is located at the opposite end. Several loops connect the two domains and form a cleft where the nicotinamide and ribose moieties of NAD+ and the acetyl lysine substrate bind. The most conserved amino acids in the Sir2 family are located in the cleft, forming a protein-tunnel, and are responsible for substrate binding and catalysis (reviewed in[8]). Understanding the mechanism of the Sirtuin-catalyzed reaction and the structural basis of Sirtuin function is of major importance to the field and will be discussed in this review.
Scheme 1.
Substrates and products of the Sir2 (Sirtuins) catalyzed deacetylation reaction.
1.2 Small molecule activation of SIRT1
SIRT1, the purported mammalian ortholog of Sir2, will be the focus of this review as its essential role in the maintenance of chromatin structure, cell cycle control, and metabolism is of great interest (other human Sirtuins reviewed in[9]). Much effort has been directed at identifying SIRT1 deacetylation substrates and to date there are at least 34 proposed substrates, each with various roles in cellular function.[10] Among these substrates, SIRT1 specifically deacetylates histone H4 at lysine 16 and histone H3 at lysine K9.[11] Histone tail deacetylation alters electrostatic properties of DNA-histone interactions resulting in transcriptional regulation. Additionally, numerous other non-histone SIRT1 protein targets have been reported and include p53,[12] Forkhead box (FOXO) transcription factors,[13] nuclear factor kappa B (NFκB)[14] and peroxisome proliferator activated gamma coactivater 1α (PGC-1α).[15] These proteins regulate stress resistance, inflammatory responses, fatty acid oxidation and mitochondrial biogenesis. The central role of SIRT1 in these pathways pushed SIRT1 to the forefront for the discovery of small molecule activators and inhibitors. To date, a number of small molecules, with unique scaffolds, have been reported to activate SIRT1.[16]
The observation that whole body overexpression of SIRT1 in mice has characteristics similar to calorie-restricted mice has fueled the search for activators of SIRT1.[17] Caloric restriction was shown to increase lifespan in yeast, worms, flies and mammals and can decrease the incidence of age-related diseases including diabetes, cancer and cardiovascular disorders.[18] Calorie-restricted mice are more metabolically active, are leaner and have lower cholesterol, insulin and glucose levels.[17] Because the overexpression of SIRT1 appeared to mimic these phenotypes, therapeutic activation using small molecules are thought to provide a new approach to treat and prevent age-related diseases.
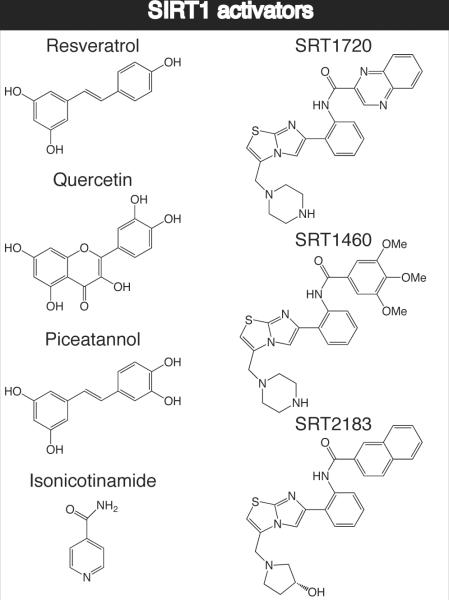
Resveratrol and a series of compounds identified by Sirtris Pharmaceuticals (SRT1720, SRT2183, SRT1460) were originally reported to induce physiological effects consistent with the activation of SIRT1,[2a, 4b] however direct activation has not been established for native substrates. Recently, the controversial issues on the effect and mechanism of small molecule activators of Sirtuins were published.[19] Here we review the current state of knowledge of Sirtuin activators, focusing on the assays used to discover the compounds along with the structural and mechanistic implications of their function. We will also highlight the recent conflicting information over the in vitro and in vivo mechanisms of action of existing small molecule activators of SIRT1 and how this information could be utilized for future discovery of novel small molecule modulators.
2. Mechanism
2.1 Mechanism of Sir2 lysine deacetylation
To properly discuss the role of small-molecule probes in Sirtuin biology, a thorough understanding of the Sirtuin catalyzed mechanism is necessary. Sirtuins catalyze the NAD+ dependent deacetylation of acetyl lysine residues resulting in the production of deacetylated lysine, nicotinamide and 2'-O-acetyl-ADP-ribose (Scheme 1).[20] Kinetic studies revealed that the acetyl lysine binds enzyme prior to NAD+ and that nicotinamide is the first product released followed by 2'-OAADPr and deacetylated lysine.[21] Subsequent non-enzymatic intramolecular trans-esterification in aqueous solution yields a mixture of 2'-OAADPr and 3'-OAADPr. The unique use of NAD+ as a substrate distinguishes Sirtuins from class I, II, and IV histone deacetylases, which use a zinc-containing catalytic site to hydrolyze the amide bond, generating free acetate.[22] The coupling of NAD+ consumption to the production of OAADPr is unique and the physiological function of this reaction is not completely understood. The use of NAD+ may provide a link between the metabolic state of the cell and deacetylation status with Sirtuins serving as an NAD+ sensor.[23] Alternatively, the role of NAD+ could be linked to production of OAADPr, which is hypothesized to behave as a secondary messenger in the cell (reviewed in[24]).
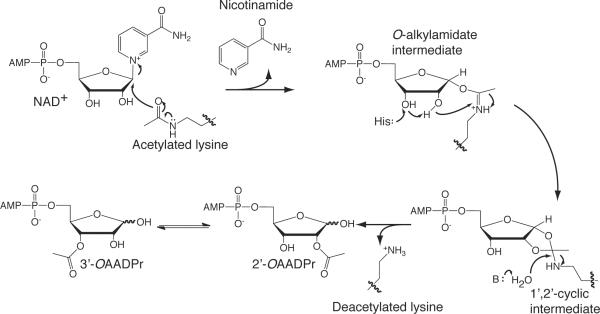
The first chemical step of the reaction involves nucleophilic addition of the acetyl oxygen on the 1'-carbon of the nicotinamide ribose, forming a C1'-O-alkylamidate intermediate (Scheme 2). Several biochemical and structural studies support the formation of the C1'-O-alkylamidate intermediate. These include 18O labeling studies providing evidence for the direct transfer of the acetyl oxygen to the 1'-hydroxyl of OAADPr,[25] nicotinamide exchange reactions,[26] and the use of thioacetyl-lysine containing peptides.[27] Thioacetyl-lysine peptides form stalled C1'-S-alkylamidate intermediates, demonstrating the existence of the alkylamidate intermediate.[28] The mechanism for the nucleophilic attack of the lysine residue is not completely understood. An SN1, concerted SN2 and a dissociative SN2-like reaction have been proposed (reviewed in[29]). After formation of the C1'-O-alkylamidate intermediate, the 2'-hydroxyl group of the NAD+ ribose is activated by a conserved histidine residue in the active site (Scheme 2). The activated hydroxyl attacks the O-alkylamidate carbon to afford the 1'2'-cyclic intermediate.[25b] A base activated water molecule then attacks the cyclic intermediate resulting in the formation of deacetylated lysine and OAADPr[20–21, 25b].
Scheme 2.
Proposed mechanism of Sir2 (Sirtuins) protein deacetylases.
2.2 Structural implications for Sir2 mechanism
Structural studies have been used to provide insight into the Sirtuin catalytic mechanism. Sirtuin structures have been solved containing non-hydrolyzable NAD+ analogs including carba-NAD+ [30] and DADMe-NAD+,[28] which are thought to mimic the transition state of the deacetylation mechanism. The structures suggested conformations consistent with distances that preclude direct nucleophilic attack of the carbonyl oxygen of the acetyl lysine and favor ribocation formation consistent with an SN1 mechanism.[28, 30] However, if carba-NAD+ and DADMe-NAD+ were true mimics of the transition state, they would be expected to bind tighter than substrates or products. Carba-NAD+ and DADMe-NAD+ have Ki values of 200 μM[31] and 360 μM[28] respectively, both of which are substantially higher than the K values of NAD+ for Hst2 and Sir2Tm.[28, 32] Thus, it seems unlikely that these NAD+ analogs are good transition-state mimics.
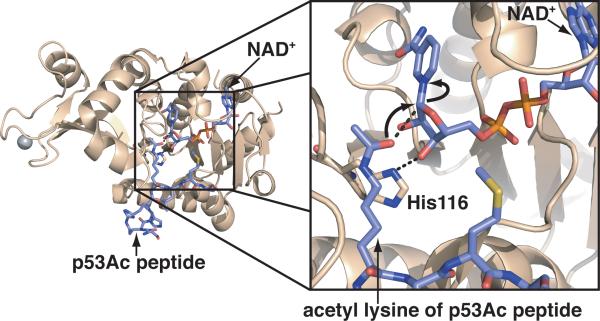
Other solved structures of Sir2 proteins have suggested an SN2-like mechanism for deacetylation. Yeast Hst2, a Sir2 homolog, co-crystallized with ternary complexes containing ADP-ribose and O-acetyl-ADP-ribose revealed an SN2 favored conformation, in which the acetyl lysine and the proposed general base histidine residue are in positions that would enable nucleophilic attack at the nicotinamide-ribosyl bond.[33] To provide further support for an SN2 mechanism Hoff et al. determined the structure of Sir2Tm bound to NAD+ and an acetylated peptide substrate.[34] Unlike the previous structures with carba-NAD+ and DADMe-NAD+, the substrate was positioned to allow nucleophilic attack at the 1'-carbon of the ribose, supporting an SN2-like mechanism (Figure 1).[34] However, because the substrates did not turnover during the course of crystallization (2 hours), it draws into question whether the structure represents the true Michaelis complex.[8, 34]
Figure 1.
Crystal structure of Sir2Tm (PDB: 2H4F)[34] displaying the acetyl lysine poised for attack on C1' of the ribose ring of NAD+. Also highlighted is the conserved histidine residue proposed to act as the catalytic base in the second step of the mechanism.[25b]
Although crystal structures can provide insight into the overall Sirtuin mechanism, they are unlikely to discriminate between an SN1 and SN2 mechanism. This is partially due to the difficulties in crystallizing true Michaelis complexes as well as in generating transition state-like intermediates along the catalytic pathway. There may be significant differences in transition states among Sirtuin enzymes, which could manifest as different degrees of nucleophilic participation in the transitions state for NAD+ glycosidic bond cleavage. Detailed transition state analysis with kinetic isotope effects and computational chemistry approaches, such as those recently performed by Cen et al.,[35] will be critical to discern between the two different mechanisms.
3. Small molecule discovery
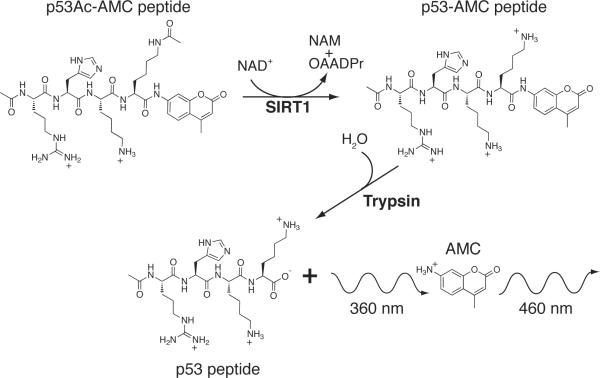
Due to proposed roles in caloric restriction, cell survival, fatty acid metabolism, and glucose homeostasis, SIRT1 has been a popular pharmacological target. SIRT1 activators could play a major role in promoting metabolic homeostasis and positively influencing lifespan. A high-throughput deacetylation assay, known as the Fluor de Lys assay (BIOMOL Research Laboratories,Inc., Plymouth Meeting, Pennsylvania), was developed to screen small molecules for SIRT1 activation (Scheme 3).[4b, 36] The acetyl lysine substrate is a p53-derived peptide containing three amino acids N-terminal of an acetyl lysine residue corresponding to Lys382, and the short peptide is covalently conjugated to a fluorophore. Deacetylation sensitizes the lysine residue to a developer solution composed of trypsin, which cleaves at the C-terminal end of deacetylated lysine and exposes the fluorescent tag, increasing the fluorescent signal.[36] The amount of deacetylation is directly proportional to the fluorescence intensity read on a fluorometer.
Scheme 3.
Diagram of a fluorescence-based assay, like the Fluor de Lys (BIOMOL), displaying reactants and products of the reaction used to monitor Sirtuin activity.[2a, 36a] The p53-derived acetyl lysine peptide is covalently conjugated to a 7-amino-4-methylcoumarin (AMC) fluorophore. Deacetylation sensitizes the lysine residue to a trypsin developer that cleaves at the C-terminal end of deacetylated lysine and exposes the fluorescent tag resulting in an increase in fluorescent signal.
Utilizing the Fluor de Lys assay, Howitz et al. screened a number of small molecule libraries and discovered that two structurally similar polyphenols, quercetin and piceatannol, activated SIRT1 deacetylase activity five- and eight-fold, respectively.[4b] Quercetin and piceatannol are members of a large family of secondary metabolites found in plants, and a secondary screen of this family of compounds identified fifteen additional SIRT1 activators (Scheme 4). The most potent activator was resveratrol, a polyphenol found in red wine that has been linked to a number of health benefits and prevention of age-related diseases.[4b] Dose response experiments, utilizing the Fluor de Lys assay, suggested that at approximately 11 μM, resveratrol doubled the rate of deacetylation by SIRT1.[4b] While these results sparked great interest in the potential mechanism of action of resveratrol, conflicting studies were published subsequently, cautioning the assumption that resveratrol directly activates SIRT1.[19d, 37]
Scheme 4.
In search of more potent activators of SIRT1, Sirtris Pharmaceuticals Inc., performed a different type of high-throughput fluorescence screen, monitoring SIRT1 activity using an engineered 20 amino acid peptide that was N-terminally linked to biotin and C-terminally linked to a fluorescent TAMRA or MR121 tag.[2a] Despite the claim that this peptide is p53 derived,[2a] it shares little sequence similarity to the true p53 Lys382 sequence. As with the high-throughput screen used to discover resveratrol, the newly exposed lysine residue was cleaved with trypsin and the reaction was monitored by a change in fluorescence polarization.[2a] Utilizing the fluorescently-labeled peptide, a high-throughput mass spectrometry assay supported the results from the initial study.[2a]
The effectiveness of the activator compounds was studied by determining the concentration of compound required to increase enzyme activity by 50% (EC1.5) and the maximum percent activation achieved at the highest doses of the compound tested. Three small molecule activators structurally unrelated to resveratrol (SRT1460: EC1.5 2.9 μM, 447% activation, SRT1720: EC1.5 0.16 μM, 781% activation, SRT2183: EC1.5 0.36 μM, 296% activation) were found to be 1,000-fold more potent than resveratrol (EC1.5 46.2 μM, 201% activation using this assay) and were shown to be selective for SIRT1 versus the homologs SIRT2 and SIRT3.[2a] SRT1720 appeared to be the most promising SIRT1 activator and was reported to improve glucose homeostasis, increase insulin sensitivity and increase mitochondrial function in type 2 diabetic mouse models.[2a]
Isothermal titration calorimetry suggested that SRT1460 exhibited binding-site saturation in the presence of SIRT1 and the fluorescently-labeled acetylated peptide. SRT1460 did not bind in the absence of peptide, leading the authors to conclude that SRT1460 and the other activators bind to the SIRT1-peptide substrate complex, promoting a more productive conformation and subsequently enhancing catalytic activity.[2a] N-terminal truncation studies demonstrated that deletion of the N-terminus of SIRT1 decreases the SRT1720-mediated activation of SIRT1 using a fluorescently-labeled peptide.[2a] However, the lack of a SIRT1 structure makes it difficult to determine exactly where the N-terminus will reside in relation to the catalytic domain.[8]
4. Kinetic effects of small molecule activators
Resveratrol and the Sirtris compounds were hypothesized to act by increasing the binding affinity for the acetylated peptide substrate, and therefore the `activation' is revealed through a steady-state kinetic Km effect. Using the Fluor de Lys assay, Howitz et al. determined that resveratrol lowered the Km of the Fluor-de-Lys™-Sirt1 peptide and NAD+ 35- and 5-fold respectively, with no effect on the Vmax.[4b] Utilizing a mass spectrometry assay with the fluorescently-labeled peptide described above, it was proposed that the Sirtris activators bound to the enzyme-substrate complex and lowered the Km for the substrate with no effect on the Km for NAD+ or Vmax.[2a]
The apparent activation described above was reproduced using peptide substrates containing fluorophores, but was not observed with peptides lacking fluorophores or native full length PGC-1α, p53 or acetyl-CoA synthetase 1 (AceS1) as substrates.[19a, 19b, 19d, 37] However, these substrates only represent a small fraction of known SIRT1 substrates. Collectively, these observations suggest the possibility that activator binding to SIRT1 induces a conformational change that promotes tighter binding of the fluorophore on the substrate resulting in the reduced Km of the acetylated peptide.[19d]
5. Complications of the Fluor de Lys assay
5.1 In vitro results
Prior to the discovery of the Sirtris compounds that reportedly led to direct activation of SIRT1,[2a] the necessity of employing a fluorescently-labeled peptide substrate to observe activation by resveratrol was reported.[19d, 37] Utilizing several different assays including the Fluor de Lys assay, coumarin and rhodamine-based fluorescence assays, charcoal binding assay, and a high performance liquid chromatography (HPLC)-based assay, Borra et al. investigated the effect of resveratrol on three p53 peptide substrates. SIRT1 activation was independent of the peptide sequence investigated, but was dependent on the presence of a fluorescent label, showing that resveratrol failed to activate the deacetylase activity of SIRT1 using an unlabeled substrate.[19d] Substrate competition studies demonstrated that the attachment of a fluorophore decreased the binding affinity of the corresponding peptide, but in the presence of resveratrol, the tagged-substrate bound more tightly.[19d] Utilizing [3H]acetate and [14C]nicotinamide release assays, Kaeberlein et al. also demonstrated fluorescent tag-dependent activation of SIRT1 by resveratrol and determined that the Km for the Fluor-de-Lys-p53 substrate was approximately 8.5-fold higher than a p53 peptide lacking the fluorophore.[37]
More recently, Pacholec et al. published an independent study of the Sirtris compounds. Utilizing an HPLC method to separate acetylated and deacetylated products, Pacholec et al. reported that like resveratrol, several Sirtris compounds led to SIRT1 activation in the presence of a peptide substrate covalently linked to a fluorophore, but not with an unlabeled peptide.[19b] Again, raising the question of whether the Sirtris compounds and resveratrol directly interact with the fluorophore. NMR chemical shift studies (CH3) investigated the perturbation of the lysine acetyl group in peptide substrates with and without a covalently linked fluorophore. A resonance shift was detected when SRT1460 was incubated with a TAMRA-p53 peptide, but not an unlabeled p53 peptide, providing evidence that SRT1460 interacted with the fluorophore.[19b] Surface plasmon resonance was used to demonstrate an interaction with the fluorophore, as concentration-dependent binding was observed with a TAMRA-containing peptide and not a native peptide. Isothermal titration calorimetry studies demonstrated that SRT1460 bound to SIRT1 in the presence of the fluorescently-labeled peptide. However, SRT1460 did not bind to a SIRT1 unlabeled native p53 peptide complex, indicating that SRT1460 bound only in the presence of the fluorophore.[19b] Dai et al. confirmed the binding of SRT1460 and SRT1720 to TAMRA-labeled peptides. However, the authors provide evidence that some compounds identified by high-throughput screens similar to Milne et al.,[2a] bind SIRT1 independent of the TAMRA tag, while others appear to increase SIRT1 activity in a substrate-dependent manner.[38]
Whether resveratrol or the Sirtris compounds are direct activators of SIRT1 remains to be established. Use of the most physiologically relevant substrates is a critical part of resolving this controversy. Immunoprecipitated PGC-1α, a known SIRT1 target, was used to show that resveratrol had no effect on the SIRT1 deacetylase activity.[19a] Additionally, small molecule-mediated SIRT1 activation was not observed when full length acetylated p53 and full length acetylated acetyl-CoA synthetase 1 were used as substrates.[19b]
If indeed some activators directly bind SIRT1, the in vivo effects of resveratrol and the Sirtris compounds might reflect a substrate-dependent effect, as an exhaustive analysis of all possible SIRT1 substrates has not been evaluated. Borra et al. proposed a model for the effect of resveratrol on a coumarin-labeled peptide.[19d] Without resveratrol, the coumarin attached to a p53 peptide would be solvent exposed, existing in an energetically unfavorable conformation. It was hypothesized that resveratrol would induce a conformational change that creates a binding pocket that better accommodates coumarin, resulting in enhanced substrate binding.[19d] Therefore, it is possible that a SIRT1 substrate containing the appropriate hydrophobic or aromatic amino acids might in fact be a target for resveratrol- and other small-molecule-based activation. Dai et al. recently reported the substrate-dependent activation of SIRT1 with a few small-molecule compounds. These results suggest that SIRT1 activation may be dependent upon the hydrophobicity of residues located C-terminal of the acetyl lysine.[38]
5.2 In vivo inconsistencies
Resveratrol was been reported to increase the lifespan of Saccharomyces cerevisiae, Drosophila melanogaster and Caenorhabditis elegans in a Sir2-dependent manner.[4b, 4c] A number of additional studies have been performed to investigate the therapeutic potential of a Sir2 dependent function of resveratrol (reviewed in [10, 39]). The in vivo link between resveratrol and Sir2 has also been subject to controversy as increased lifespan has been difficult to replicate in three different yeast strains,[37] D. melanogaster and C. elegans.[40] SIRT1 knockdown studies of resveratrol mediated neuronal protection produced ambiguous results regarding the requirement for SIRT1 in physiologically relevant effects of resveratrol.[41]
The Sirtris activator series were shown to induce numerous healthy phenotypes in obese mice, including improved insulin sensitivity, lower plasma glucose and increased mitochondrial capacity.[2a] These encouraging observations could not be reproduced by Pacholec et al., who reported that treatment at 100 mg/kg of SRT1720 resulted in the death of three of the eight mice used in the study.[2a, 19b] The health benefits of SRT1720 were not reproducible in mice treated at 30 mg/kg.[19b] The basis for the discrepancy in phenotypes is unclear.
6. Alternative mechanisms of SIRT1 activation
6.1 Mechanisms of resveratrol activation
Although the direct link between resveratrol and SIRT1 has been subject to controversy, there are clearly positive metabolic effects and potential therapeutic benefits of resveratrol administration. Recent studies on resveratrol have revealed clinical potential in the treatment of age-related diseases, cancer, cardiovascular disease, and increased insulin sensitivity.[39, 42] Resveratrol treatment is proposed to mimic many positive effects of caloric restriction, leading researchers to hypothesize that the link between caloric restriction and resveratrol is the Sirtuin family of proteins.[43] The issues surrounding the controversial activation of SIRT1 by resveratrol might be rectified through the discovery of an indirect activation pathway or a direct pathway that involves specific substrate activation.
Numerous studies have revealed that resveratrol is a pleiotropic molecule, exhibiting general antioxidant effects, as well as directly inhibiting kinases and cyclooxygenases.[44] Multiple other in vivo targets of resveratrol are known, many of which exhibit higher potency than that reported for SIRT1.[19b, 44] Thus, the in vivo effects of resveratrol cannot be solely attributed to increased SIRT1 activity. Interestingly, while resveratrol exhibits an inhibitory effect on many kinases, AMP-activated kinase (AMPK) is a unique kinase that is activated by resveratrol treatment,[41a, 45] serving as a potential mechanism for the indirect activation of SIRT1 by resveratrol. AMPK is a fuel-sensing enzyme that is responsive to decreases in cellular energy status. AMPK is activated by an increased AMP/ATP ratio and initiates pathways that restore ATP levels, including up-regulation of fatty acid oxidation and inhibition of protein and fatty acid synthesis. Nutrient and oxygen deprivation, as well as increased energy expenditure can activate AMPK.[46] One outcome of these AMPK dependent processes is an increase in NAD+ concentration that might stimulate SIRT1 activity.[47] AMPK deficient mice treated with resveratrol show no changes in mitochondrial biogenesis, exercise performance or insulin sensitivity,[48] indicating the purported resveratrol mediated effects may hinge on AMPK activity.
6.2 Endogenous SIRT1 regulators
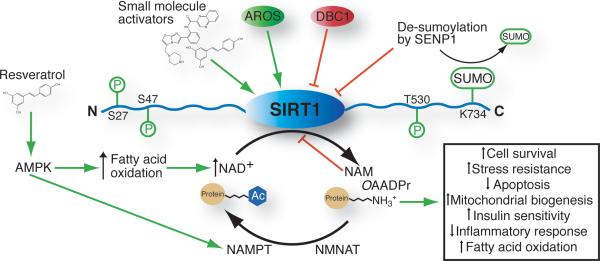
Evidence for endogenous regulators of SIRT1 activity exists and may provide alternative mechanisms for the activation of SIRT1 (Figure 2). The levels of intracellular substrate NAD+ and product nicotinamide influence SIRT1 activity. Nicotinamide is a potent product inhibitor of the deacetylation reaction, and is widely used in laboratory research as a general Sirtuin inhibitor. Nicotinamide is able to enter the active site and react with the alkylamidate intermediate (Scheme 2), reforming NAD+ and preventing the forward reaction.[26, 49] High concentrations of isonicotinamide can partially relieve the product inhibition by competing with nicotinamide for active site binding, without substantially affecting the forward deacetylation reaction.[26, 49] The high concentrations of isonicotinamide needed to successfully compete with nicotinamide provide challenges for its use,[10] but present an interesting scaffold for the design of new synthetic activators. Relief of nicotinamide inhibition is one example of how understanding the catalytic mechanism of Sirtuins allows for the design of an activator that may be able to increase the rate of chemical catalysis.
Figure 2.
Potential regulatory pathways of SIRT1 that could be exploited to increase SIRT1-mediated deacetylation. Phosphorylation, sumoylation, AMPK, small molecules, increased NAD+ levels and AROS binding are purposed activators (depicted in green). Nicotinamide, DBC1 binding, and de-sumoylation are purposed inhibitory pathways that could be regulated to increase SIRT1 activity (depicted in red). AMPK: AMP-activated kinase, AROS: Active regulator of SIRT1, DBC1: Deleted in breast cancer-1, NAM: Nicotinamide, NAMPT: Nicotinamide phosphoribosyltransferase, NMANT: Nicotinamide mononucleotide adenylyltransferase, OAADPr: O-Acetyl-ADP-ribose, SENP1: Sentrin specific protease 1, SUMO: Small ubiquitin-like modifier.
The NAD+ salvage pathway might also serve to increase SIRT1 activity by increasing NAD+ synthesis.[50] In mammals, the NAD+ salvage pathway relies on two enzymes to convert nicotinamide to NAD+. Nicotinamide phosphoribosyltransferase (NAMPT) catalyzes the addition of 5-phosphoribosylpyrophosphate to nicotinamide to form nicotinamide mononucleotide (NMN), the rate-limiting step of the biosynthetic pathway.[50a] Nicotinamide mononucleotide adenylyltransferase (NMNAT) subsequently converts NMN to NAD+.[50a]
Enhanced NAMPT activity resulted in increased cellular lifespan in a SIRT1 dependent manner in human vascular smooth muscle cells.[50c] The ability to increase NAD+ biosynthesis through the activation of NAMPT or NMNAT provides a unique mechanism of activating SIRT1.[51] In addition to AMPK's ability to increase intracellular NAD+ concentration though the control of cellular energy status, AMPK also increases the production of NAD+ through stimulated transcription of NAMPT.[52] A recent article highlights the relationship between NAD+ metabolism and SIRT1 function.[23]
Other proposed endogenous regulators of SIRT1 include the binding of AROS protein (active regulator of SIRT1) to the N-terminal domain of SIRT1 (amino acids 114–217)[53] and the binding of an inhibitory protein, DBC1 (Deleted in Breast Cancer 1), to the catalytic domain of SIRT1.[54] Using a yeast two hybrid assay Kim et al. identified AROS and determined that it could stimulate p53 deacetylation in vivo and enhance deacetylase activity two fold in vitro.[53] In response to DNA damage, cell lines over expressing AROS show lower p53 acetylation, attenuating the ability of p53 to initiate apoptosis.[53] Understanding the mechanistic basis for the SIRT1 activation upon AROS binding could allow for the design of small molecule activators capable of binding to the same region, mimicking the effects of AROS.
Until recently, the negative regulatory mechanisms of SIRT1 were poorly understood. Through large-scale affinity purification, it was discovered that DBC1 forms a complex with SIRT1.[54] Zhao et al. and Kim et al. determined that DBC1 represses SIRT1 deacetylation of p53 and FOXO3 in vitro and in vivo.[54] It was found that the leucine zipper motif of DBC1[54a] binds to the catalytic core domain of SIRT1, but not to other Sirtuins.[54] Co-expression of DBC1 reduced the binding of SIRT1 to p53 and FOXO, suggesting that blocking the substrate-binding site is one mechanism for DBC1-mediated SIRT1 inhibition.[54] Co-immunoprecipitation of DBC1-SIRT1 from mouse liver extracts demonstrated that the interaction is regulated by the metabolic state of the cell, with increased interaction during a high-fat diet, correlating with decreased SIRT1 activity.[55] Interestingly, this interaction was disrupted following 24 hours of fasting, however the details of the mechanism are not known.[55] In addition, DBC1 can disrupt the interaction between the methyltransferase SUV39H1 and SIRT1 and therefore, plays an important role in regulating heterochromatin formation.[56] For these reasons, DBC1 is a potential therapeutic target for regulating SIRT1 activity. Molecules that disrupt the interaction between DBC1 and SIRT1 would function essentially as SIRT1 activators.
Another mechanism for controlling SIRT1 function is post-translational modification, which to date includes C-terminal sumoylation at K734[57] and phosphorylation.[58] Yang et al. reported SIRT1 sumoylation leads to a two fold increase in deacetylase activity in vitro compared to a SIRT1 mutant that is unable to be sumoylated.[57] Sumoylation of SIRT1 is further regulated by SENP1, a desumoylase that associates with SIRT1 in response to cellular stress and lowers its activity towards pro-apoptotic proteins.[57] Three independent studies reported that SIRT1 phosphorylation on different sites located in the C and N-terminal regions results in SIRT1 activation.[58] These reports suggest that different kinases including cyclinB/Cdk1,[58a] DYRK (dual specificity tyrosine phosphorylation regulated kinase),[58b] and JNK1 (cJUN N-terminal kinase)[58c] are capable of phosphorylating serine and threonine residues. These phosphorylation events are proposed to induce conformational changes of SIRT1, placing it in a more active state. Further molecular studies are necessary to understand how different phosphorylation events can stimulate SIRT1 activity, and to uncover the upstream activation of the protein kinases involved. These biochemical details will be essential to harness their potential for both direct and upstream modulation of SIRT1 activity.
7. Concluding remarks
The Fluor de Lys and related fluorescence based assays have been shown to be a very efficient approach for screening a large number of compounds. However, as with all high-throughput screening techniques, the results are best interpreted within the limiting context of the physical assay used. Additional independent assays, using native substrates, will validate the results of positive hits. Numerous Sirtuin assays have been described, with each measuring different components of the reaction. To directly monitor deacetylation, MS, HPLC[19b, 59] and microfluidic mobility shift assays[60] have been described. A continuous microplate assay,[61] [14C]nicotinamide release assay,[37, 62] nicotinamide exchange assay[31, 63] and TLC methods[59, 63–64] measure nicotinamide formation. To monitor the production of OAADPr, charcoal binding,[59] TLC[63–64] and HPLC assays[59] can be used. These published assays afford a range of techniques capable of validating small molecule effects.
To help resolve the debate over resveratrol and the Sirtris activators it is critical to determine their mechanism of action. The stimulatory effect of SIRT1 activators might be selective for certain substrates through a steady-state Km effect (increased productive binding) or through the enhancement of the rate of chemical catalysis. If these molecules enhance chemical catalysis, elucidating the complete mechanism of NAD+ dependent deacetylation, including whether the first step of the reaction is SN1 or SN2-like, becomes increasingly important. Understanding which step of the mechanism is up-regulated allows for the design of molecules that specifically enhance the chemistry steps of catalysis. Alternatively, if the activation is through a Km effect, activators could be developed that are specific for certain Sirtuin substrates.
Furthermore, it is critical to solve sirtuin structures with activators bound. Unfortunately, there are no published examples of such compounds bound to Sirtuin proteins. With a SIRT1 structure, researchers will be able use rational design methods to create structure-based inhibitors and activators. Conformational changes induced by endogenous SIRT1 modulators likely place SIRT1 in an `activated' state through a yet undiscovered pathway.[53, 57–58] AROS, post-translational modifications, and small molecule activators might utilize unique mechanisms or a common allosteric mechanism to modulate SIRT1 between a less-active and a more-active conformation. With new structural and mechanistic information, an activator could be developed that induces these conformational changes and stabilizes the more-active conformation.
By analogy to the possible indirect activation of SIRT1 by resveratrol, it would seem plausible that some of the Sirtris compounds might utilize an indirect pathway for SIRT1 activation. Therefore, it is prudent to elucidate the endogenous regulatory mechanisms of SIRT1 control. Fundamental knowledge, such as understanding the kinases and phosphatases that regulate the post-translational modifications of SIRT1, will provide new targets for the development of compounds that lead to SIRT1 activation.
Sirtuins play an extremely important role in diverse cellular processes. As further structural and mechanistic details are uncovered, new ways to modulate the activity of these vital deacetylases will be revealed. This essential information will afford an excellent opportunity for the design of therapeutics to treat metabolic and age-related diseases.
References
- [1].a) Frye R. Biochem. Biophys. Res. Commun. 2000;273:793. doi: 10.1006/bbrc.2000.3000. [DOI] [PubMed] [Google Scholar]; b) Elliott P, Jirousek M. Curr. Opin. Invest. Drugs (BioMed Cent.) 2008;9:371. [PubMed] [Google Scholar]
- [2].a) Milne JC, Lambert PD, Schenk S, Carney DP, Smith JJ, Gagne DJ, Jin L, Boss O, Perni RB, Vu CB, Bemis JE, Xie R, Disch JS, Ng PY, Nunes JJ, Lynch AV, Yang H, Galonek H, Israelian K, Choy W, Iffland A, Lavu S, Medvedik O, Sinclair DA, Olefsky JM, Jirousek MR, Elliott PJ, Westphal CH. Nature. 2007;450:712. doi: 10.1038/nature06261. [DOI] [PMC free article] [PubMed] [Google Scholar]; b) Milne J, Denu J. Curr. Opin. Chem. Biol. 2008;12:11. doi: 10.1016/j.cbpa.2008.01.019. [DOI] [PubMed] [Google Scholar]
- [3].a) Braunstein M, Sobel RE, Allis CD, Turner BM, Broach JR. Mol. Cell. Biol. 1996;16:4349. doi: 10.1128/mcb.16.8.4349. [DOI] [PMC free article] [PubMed] [Google Scholar]; b) Kaeberlein M, McVey M, Guarente L. Genes Dev. 1999;13:2570. doi: 10.1101/gad.13.19.2570. [DOI] [PMC free article] [PubMed] [Google Scholar]; c) Rine J, Herskowitz I. Genetics. 1987;116:9. doi: 10.1093/genetics/116.1.9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [4].a) Gasser S, Cockell M. Gene. 2001;279:1. doi: 10.1016/s0378-1119(01)00741-7. [DOI] [PubMed] [Google Scholar]; b) Howitz KT, Bitterman KJ, Cohen HY, Lamming DW, Lavu S, Wood JG, Zipkin RE, Chung P, Kisielewski A, Zhang LL, Scherer B, Sinclair DA. Nature. 2003;425:191. doi: 10.1038/nature01960. [DOI] [PubMed] [Google Scholar]; c) Wood JG, Rogina B, Lavu S, Howitz K, Helfand SL, Tatar M, Sinclair D. Nature. 2004;430:686. doi: 10.1038/nature02789. [DOI] [PubMed] [Google Scholar]
- [5].Smith B, Hallows W, Denu J. Chem Biol. 2008;15:1002. doi: 10.1016/j.chembiol.2008.09.009. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [6].Blander G, Guarente L. Annu. Rev. Biochem. 2004;73:417. doi: 10.1146/annurev.biochem.73.011303.073651. [DOI] [PubMed] [Google Scholar]
- [7].Haigis MC, Mostoslavsky R, Haigis KM, Fahie K, Christodoulou DC, Murphy AJ, Valenzuela DM, Yancopoulos GD, Karow M, Blander G, Wolberger C, Prolla TA, Weindruch R, Alt FW, Guarente L. Cell. 2006;126:941. doi: 10.1016/j.cell.2006.06.057. [DOI] [PubMed] [Google Scholar]
- [8].Sanders BD, Jackson B, Marmorstein R. Biochim. Biophys. Acta, Proteins Proteomics. 2009;1 [Google Scholar]
- [9].Haigis MC, Sinclair DA. Annu. Rev. Pathol. Mech. Dis. 2010;5:253. doi: 10.1146/annurev.pathol.4.110807.092250. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [10].Baur JA. Biochim. Biophys. Acta. 2010;1804:1626. doi: 10.1016/j.bbapap.2009.10.025. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [11].Vaquero A, Scher M, Lee D, Erdjument-Bromage H, Tempst P, Reinberg D. Mol. Cell. 2004;16:93. doi: 10.1016/j.molcel.2004.08.031. [DOI] [PubMed] [Google Scholar]
- [12].Vaziri H, Dessain S, Eaton E, Imai S, Frye R, Pandita T, Guarente L, Weinberg R. Cell. 2001;107:149. doi: 10.1016/s0092-8674(01)00527-x. [DOI] [PubMed] [Google Scholar]
- [13].Brunet A, Sweeney L, Sturgill J, Chua K, Greer P, Lin Y, Tran H, Ross S, Mostoslavsky R, Cohen H. Science. 2004;303:2011. doi: 10.1126/science.1094637. [DOI] [PubMed] [Google Scholar]
- [14].Yeung F, Hoberg J, Ramsey C, Keller M, Jones D, Frye R, Mayo M. EMBO J. 2004;23:2369. doi: 10.1038/sj.emboj.7600244. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [15].Rodgers J, Lerin C, Haas W, Gygi S, Spiegelman B, Puigserver P. Nature. 2005;434:113. doi: 10.1038/nature03354. [DOI] [PubMed] [Google Scholar]
- [16].a) Mai A, Valente S, Meade S, Carafa V, Tardugno M, Nebbioso A, Galmozzi A, Mitro N, De Fabiani E, Altucci L, Kazantsev A. J. Med. Chem. 2009;52:5496. doi: 10.1021/jm9008289. [DOI] [PubMed] [Google Scholar]; b) Bemis JE, Vu CB, Xie R, Nunes JJ, Ng PY, Disch JS, Milne JC, Carney DP, Lynch AV, Jin L, Smith JJ, Lavu S, Iffland A, Jirousek MR, Perni RB. Bioorg. Med. Chem. Lett. 2009;19:2350. doi: 10.1016/j.bmcl.2008.11.106. [DOI] [PubMed] [Google Scholar]
- [17].Bordone L, Cohen D, Robinson A, Motta MC, van Veen E, Czopik A, Steele AD, Crowe H, Marmor S, Luo J, Gu W, Guarente L. Aging Cell. 2007;6:759. doi: 10.1111/j.1474-9726.2007.00335.x. [DOI] [PubMed] [Google Scholar]
- [18].Bordone L, Guarente L. Nat. Rev. Mol. Cell Biol. 2005;6:298. doi: 10.1038/nrm1616. [DOI] [PubMed] [Google Scholar]
- [19].a) Beher D, Wu J, Cumine S, Kim KW, Lu S-C, Atangan L, Wang M. Chemical Biology & Drug Design. 2009;74:619. doi: 10.1111/j.1747-0285.2009.00901.x. [DOI] [PubMed] [Google Scholar]; b) Pacholec M, Bleasdale JE, Chrunyk B, Cunningham D, Flynn D, Garofalo RS, Griffith D, Griffor M, Loulakis P, Pabst B, Qiu X, Stockman B, Thanabal V, Varghese A, Ward J, Withka J, Ahn K. J. Biol. Chem. 2010;285:8340. doi: 10.1074/jbc.M109.088682. [DOI] [PMC free article] [PubMed] [Google Scholar]; c) Schmidt C. Nature. 2010;28:185. doi: 10.1038/nbt0310-185. [DOI] [PubMed] [Google Scholar]; d) Borra MT, Smith BC, Denu JM. J. Biol. Chem. 2005;280:17187. doi: 10.1074/jbc.M501250200. [DOI] [PubMed] [Google Scholar]
- [20].Jackson M, Denu J. J. Biol. Chem. 2002;277:18535. doi: 10.1074/jbc.M200671200. [DOI] [PubMed] [Google Scholar]
- [21].Borra M, Langer M, Slama J, Denu J. Biochemistry. 2004;43:9877. doi: 10.1021/bi049592e. [DOI] [PubMed] [Google Scholar]
- [22].Hodawadekar S, Marmorstein R. Oncogene. 2007;26:5528. doi: 10.1038/sj.onc.1210619. [DOI] [PubMed] [Google Scholar]
- [23].Zhang T, Kraus WL. Biochim. Biophys. Acta, Proteins Proteomics. 2010;1804:1666. doi: 10.1016/j.bbapap.2009.10.022. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [24].Tong L, Denu JM. Biochim. Biophys. Acta, Proteins Proteomics. 2010:1. [Google Scholar]
- [25].a) Smith B, Denu J. Biochemistry. 2006;45:272. doi: 10.1021/bi052014t. [DOI] [PMC free article] [PubMed] [Google Scholar]; b) Sauve A, Celic I, Avalos J, Deng H, Boeke J, Schramm V. Biochemistry. 2001;40:15456. doi: 10.1021/bi011858j. [DOI] [PubMed] [Google Scholar]
- [26].Jackson M, Schmidt M, Oppenheimer N, Denu J. J. Biol. Chem. 2003;278:50985. doi: 10.1074/jbc.M306552200. [DOI] [PubMed] [Google Scholar]
- [27].a) Smith B, Denu J. Biochemistry. 2007;46:14478. doi: 10.1021/bi7013294. [DOI] [PubMed] [Google Scholar]; b) Fatkins DG, Monnot AD, Zheng W. Bioorg. Med. Chem. Lett. 2006;16:3651. doi: 10.1016/j.bmcl.2006.04.075. [DOI] [PubMed] [Google Scholar]
- [28].Hawse W, Hoff K, Fatkins D, Daines A, Zubkova O, Schramm V, Zheng W, Wolberger C. Structure. 2008;16:1368. doi: 10.1016/j.str.2008.05.015. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [29].Sauve AA. Biochim. Biophys. Acta. 2010;1804:1591. doi: 10.1016/j.bbapap.2010.01.021. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [30].Zhao K, Harshaw R, Chai X, Marmorstein R. Proc. Natl. Acad. Sci. U.S.A. 2004;101:8563. doi: 10.1073/pnas.0401057101. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [31].Landry J, Slama JT, Sternglanz R. Biochem. Biophys. Res. Commun. 2000;278:685. doi: 10.1006/bbrc.2000.3854. [DOI] [PubMed] [Google Scholar]
- [32].Tanner KG, Landry J, Sternglanz R, Denu JM. Proc. Natl. Acad. Sci. U.S.A. 2000;97:14178. doi: 10.1073/pnas.250422697. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [33].Zhao K, Chai X, Clements A, Marmorstein R. Nat. Struct. Biol. 2003;10:864. doi: 10.1038/nsb978. [DOI] [PubMed] [Google Scholar]
- [34].Hoff KG, Avalos JL, Sens K, Wolberger C. Structure. 2006;14:1231. doi: 10.1016/j.str.2006.06.006. [DOI] [PubMed] [Google Scholar]
- [35].Cen Y, Sauve AA. J. Am. Chem. Soc. 2010 doi: 10.1021/ja910342d. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [36].a) Wegener D, Wirsching F, Riester D, Schwienhorst A. Chem. Biol. 2003;10:61. doi: 10.1016/s1074-5521(02)00305-8. [DOI] [PubMed] [Google Scholar]; b) Heltweg B, Trapp J, Jung M. Methods. 2005;36:332. doi: 10.1016/j.ymeth.2005.03.003. [DOI] [PubMed] [Google Scholar]
- [37].Kaeberlein M, McDonagh T, Heltweg B, Hixon J, Westman EA, Caldwell SD, Napper A, Curtis R, DiStefano PS, Fields S, Bedalov A, Kennedy BK. J. Biol. Chem. 2005;280:17038. doi: 10.1074/jbc.M500655200. [DOI] [PubMed] [Google Scholar]
- [38].Dai H, Kustigian L, Carney D, Case A, Considine T, Hubbard BP, Perni RB, Riera TV, Szczepankiewicz B, Vlasuk GP, Stein RL. J. Biol. Chem. 2010 doi: 10.1074/jbc.M110.133892. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [39].Baur J, Sinclair D. Nat. Rev. Drug Discovery. 2006;5:493. doi: 10.1038/nrd2060. [DOI] [PubMed] [Google Scholar]
- [40].Bass TM, Weinkove D, Houthoofd K, Gems D, Partridge L. Mech. Ageing Dev. 2007;128:546. doi: 10.1016/j.mad.2007.07.007. [DOI] [PubMed] [Google Scholar]
- [41].a) Dasgupta B, Milbrandt J. Proc. Natl. Acad. Sci. U.S.A. 2007;104:7217. doi: 10.1073/pnas.0610068104. [DOI] [PMC free article] [PubMed] [Google Scholar]; b) Tang B. Brain Res. Bull. 2009 [Google Scholar]
- [42].Marques F, Markus M, Morris B. Int. J. Biochem. Cell Biol. 2009 doi: 10.1016/j.biocel.2009.06.003. [DOI] [PubMed] [Google Scholar]
- [43].Baur JA. Mech. Ageing Dev. 2010;131:261. doi: 10.1016/j.mad.2010.02.007. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [44].Pirola L, Frojdo S. IUBMB Life. 2008;60:323. doi: 10.1002/iub.47. [DOI] [PubMed] [Google Scholar]
- [45].a) Zang M, Xu S, Maitland-Toolan KA, Zuccollo A, Hou X, Jiang B, Wierzbicki M, Verbeuren TJ, Cohen RA. Diabetes. 2006;55:2180. doi: 10.2337/db05-1188. [DOI] [PubMed] [Google Scholar]; b) Baur J, Pearson K, Price N, Jamieson H, Lerin C, Kalra A, Prabhu V, Allard J, Lopez-Lluch G, Lewis K. Nature. 2006;444:337. doi: 10.1038/nature05354. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [46].Ruderman NB, Julia Xu X, Nelson L, Cacicedo JM, Saha AK, Lan F, Ido Y. AJP: Endocrinol. Metab. 2010;298:E751. doi: 10.1152/ajpendo.00745.2009. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [47].Cantó C, Gerhart-Hines Z, Feige JN, Lagouge M, Noriega L, Milne JC, Elliott PJ, Puigserver P, Auwerx J. Nature. 2009;458:1056. doi: 10.1038/nature07813. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [48].Um J, Park S, Kang H, Yang S, Foretz M, McBurney M, Kim M, Viollet B, Chung J. Diabetes. 2010;59:554. doi: 10.2337/db09-0482. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [49].a) Bitterman K, Anderson R, Cohen H, Latorre-Esteves M, Sinclair D. J. Biol. Chem. 2002;277:45099. doi: 10.1074/jbc.M205670200. [DOI] [PubMed] [Google Scholar]; b) Sauve AA, Moir RD, Schramm VL, Willis IM. Mol. Cell. 2005;17:595. doi: 10.1016/j.molcel.2004.12.032. [DOI] [PubMed] [Google Scholar]
- [50].a) Revollo JR, Grimm AA, Imai S-I. J. Biol. Chem. 2004;279:50754. doi: 10.1074/jbc.M408388200. [DOI] [PubMed] [Google Scholar]; b) Grubisha O, Smith B, Denu J. FEBS JOURNAL. 2005;272:4607. doi: 10.1111/j.1742-4658.2005.04862.x. [DOI] [PubMed] [Google Scholar]; c) van der Veer E, Ho C, O'Neil C, Barbosa N, Scott R, Cregan S, Pickering J. J. of Biol. Chem. 2007;282:10841. doi: 10.1074/jbc.C700018200. [DOI] [PubMed] [Google Scholar]
- [51].Imai S, Kiess W. Frontiers in bioscience: a journal and virtual library. 14:2983. doi: 10.2741/3428. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [52].Fulco M, Cen Y, Zhao P, Hoffman EP, McBurney MW, Sauve AA, Sartorelli V. Dev. Cell. 2008;14:661. doi: 10.1016/j.devcel.2008.02.004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [53].Kim E, Kho J, Kang M, Um S. Mol. cell. 2007;28:277. doi: 10.1016/j.molcel.2007.08.030. [DOI] [PubMed] [Google Scholar]
- [54].a) Kim JE, Chen J, Lou Z. Nature. 2008;451:583. doi: 10.1038/nature06500. [DOI] [PubMed] [Google Scholar]; b) Zhao W, Kruse J, Tang Y, Jung S, Qin J, Gu W. Nature. 2008;451:587. doi: 10.1038/nature06515. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [55].Escande C, Chini CC, Nin V, Dykhouse KM, Novak CM, Levine J, van Deursen J, Gores GJ, Chen J, Lou Z, Chini EN. J. Clin. Invest. 2010;120:545. doi: 10.1172/JCI39319. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [56].Li Z, Chen L, Kabra N, Wang C, Fang J, Chen J. J. Biol. Chem. 2009;284:10361. doi: 10.1074/jbc.M900956200. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [57].Yang Y, Fu W, Chen J, Olashaw N, Zhang X, Nicosia S, Bhalla K, Bai W. Nat. Cell Biol. 2007;9:1253. doi: 10.1038/ncb1645. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [58].a) Sasaki T, Maier B, Koclega K, Chruszcz M, Gluba W, Stukenberg P, Minor W, Scrable H. PLoS One. 2008;3 doi: 10.1371/journal.pone.0004020. [DOI] [PMC free article] [PubMed] [Google Scholar]; b) Guo X, Williams JG, Schug TT, Li X. J. of Biol. Chem. 2010;285:13223. doi: 10.1074/jbc.M110.102574. [DOI] [PMC free article] [PubMed] [Google Scholar]; c) Nasrin N, Kaushik V, Fortier E, Wall D, Pearson K, de Cabo R, Bordone L. 2009 doi: 10.1371/journal.pone.0008414. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [59].Borra MT, Denu JM. Methods Enzymol. 2004;376:171. doi: 10.1016/S0076-6879(03)76011-X. [DOI] [PubMed] [Google Scholar]
- [60].Liu Y, Gerber R, Wu J, Tsuruda T, McCarter J. Anal. Biochem. 2008;378:53. doi: 10.1016/j.ab.2008.02.018. [DOI] [PubMed] [Google Scholar]
- [61].Smith BC, Hallows WC, Denu JM. Anal. Biochem. 2009;394:101. doi: 10.1016/j.ab.2009.07.019. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [62].McDonagh T, Hixon J, DiStefano PS, Curtis R, Napper AD. Methods. 2005;36:346. doi: 10.1016/j.ymeth.2005.03.005. [DOI] [PubMed] [Google Scholar]
- [63].Landry J, Sutton A, Tafrov ST, Heller RC, Stebbins J, Pillus L, Sternglanz R. Proc. Natl. Acad. Sci. U.S.A. 2000;97:5807. doi: 10.1073/pnas.110148297. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [64].Tanny JC, Moazed D. Proc. Natl. Acad. Sci. U.S.A. 2001;98:415. doi: 10.1073/pnas.031563798. [DOI] [PMC free article] [PubMed] [Google Scholar]