Abstract
RNAa (RNA activation) is a mechanism by which small dsRNA (double-stranded RNA), termed saRNA (small activating RNA), target promoter sequences to induce gene expression. This technique represents a novel approach to gene overexpression without the use of exogenous DNA. In the present study, we investigated whether RNAa can modulate expression of the development-related gene NANOG and manipulate cell fate. Using a lentivirus-based reporter system as a screening tool, we identified synthetic saRNAs that stimulate NANOG expression in human NCCIT embryonic carcinoma cells. Mismatch mutations to saRNA duplexes define sequence requirement for gene activation. Functional analysis of NANOG induction reveals saRNA treatment predictably modulates the expression of several known downstream target genes, including FOXH1 (forkhead box H1), REST (RE1-silencing transcription factor), OCT4 (octamer-binding protein 4) and REX1 (reduced expression protein 1). Treatment with RA (retinoic acid) triggers NCCIT cell differentiation, reducing NANOG and OCT4 expression and up-regulating several neural markers [i.e. ASCL1 (achaete-scute complex homologue 1), NEUROD1 (neuronal differentiation 1) and PAX6 (paired box 6)]. However, co-treatment with saRNA antagonizes NANOG down-regulation and RA-induced differentiation. Ectopic overexpression of NANOG via lentiviral transduction further recapitulates saRNA results, providing proof-of-concept that RNAa may be utilized to activate development-related genes and manipulate cell fate.
Keywords: cell fate, differentiation, induced pluripotent stem cell (iPS cell), NANOG, RNA activation (RNAa)
Abbreviations: ASCL1, achaete-scute complex homologue 1; dsRNA, double-stranded RNA; ES, embryonic stem; FOXH1, forkhead box H1; GAPDH, glyceraldehyde-3-phosphate dehydrogenase; GFP, green fluorescent protein; HEK, human embryonic kidney; Hsp70, heat-shock protein 70; iPS, induced pluripotent stem; MTS, 3-(4,5-dimethylthiazol-2-yl)-5-(3-carboxymethoxyphenyl)-2-(4-sulfophenyl)-2H-tetrazolium; NEUROD1, neuronal differentiation 1; OCT4, octamer-binding protein 4; PAX6, paired box 6; RA, retinoic acid; REST, RE1-silencing transcription factor; REX1, reduced expression protein 1; RNAa, RNA activation; RT, reverse transcription; saRNA, small activating RNA; SSEA, stage-specific embryonic antigen; TSS, transcription start site
INTRODUCTION
RNAa (RNA activation) is a newly identified phenomenon of gene induction triggered by small dsRNAs (double-stranded RNAs) [1–3]. This class of dsRNA, termed saRNA (small activating RNA), targets non-coding sequences in gene promoters leading to changes in chromatin structure and transcriptional activation [1,3–5]. RNAa is conserved in at least mammalian cells having been shown to activate a number of genes, including CDKN1A (cyclin-dependent kinase inhibitor 1A; p21), CDH1 (E-cadherin), PGR (progesterone receptor), KLF4 (Krüppel-like factor 4), VEGF (vascular endothelial growth factor), NKX3.1 (NK3 homeobox 1), TP53 (p53) etc. [2,6–8]. As such, saRNAs represent a novel tool for stimulating endogenous gene expression.
Viral-mediated overexpression systems are widely used to manipulate cell fate. For instance, ectopic expression of various defined factors [e.g. OCT4 (octamer-binding protein 4), GATA4 (GATA-binding protein 4)] has been used to generate iPS (induced pluripotent stem) cells and direct conversion of fibroblasts into blood progenitor cells or cardiomyocytes [9–11]. However, viral-based systems have inherent drawbacks that interfere with clinical application, including adverse effects on the host genome integrity and immunological consequences. Alternative approaches to gene overexpression that circumvent such problems may improve therapeutic development of cell-based technologies.
NANOG is a transcription factor that plays a critical role in regulating cell fate. In embryonic development, it is involved in maintaining the pluripotent ICM (inner cell mass), sustaining the epiblast and preventing cell differentiation [12–15]. NANOG is also highly expressed in pluripotent cell lines, including ES (embryonic stem), EG (embryonic germ) and EC (embryonic carcinoma) cells. However, NANOG levels sharply decline at the post-implantation stages of embryogenesis and during differentiation in cell culture [12]. In fact, suppression of NANOG alone can cause ES cells to differentiate into extra-embryonic lineages [16,17]. Ectopic expression of NANOG has also been utilized to maintain pluripotent phenotypes in cultured ES cells. For instance, NANOG overexpression has been reported to maintain mouse ES cell pluripotency in the absence of LIF (leukaemia inhibitory factor) [12], as well as to enable human ES cells to grow in a feeder-free environment [18]. NANOG is among the four defined stem cell factors used to reprogram human fibroblasts into iPS cells [19]. Although NANOG is dispensable in some instances of iPS reprogramming, it is a stringent selection marker for germline-competent iPS cells with increased ES cell-like properties [20]. Currently, almost all methods of NANOG overexpression require the use of exogenous genetic material (e.g. viral-based vectors). In the present study, we identify saRNAs that activate NANOG expression and antagonize RA (retinoic acid)-induced differentiation of NCCIT cells. Our results provide proof-of-concept that RNAa may provide an alternative approach to DNA-based overexpression systems for manipulating cell fate.
EXPERIMENTAL
saRNA design
A 1 kb section of the human NANOG promoter sequence was scanned for saRNA target sites based on rational design rules as reported previously [1,6]. All of the saRNA sequences found are listed in Supplementary Table S1 (available at http://www.BiochemJ.org/bj/443/bj4430821add.htm).
Cell culture and saRNA transfection
NCCIT cells were maintained in RPMI 1640 medium supplemented with 10% FBS (fetal bovine serum), 10 mM Hepes, 1 mM sodium pyruvate, 4.5 g/l glucose, 100 units/ml penicillin and 100 μg/ml streptomycin in a humidified atmosphere of 5% CO2 maintained at 37 °C. Cells were plated in growth medium without antibiotics at a density of ~15%. Transfection of saRNAs was carried out using Lipofectamine™ RNAiMax (Invitrogen) according to the manufacturer's instructions.
RA-induced differentiation
NCCIT cells were dissociated as single cells and maintained at the prescribed cell density in standard growth medium supplemented with 10 μM all-trans-RA (Sigma-Aldrich) for 2 weeks. RA was stored as a solution (10 mM in DMSO) and diluted in growth medium as needed. The medium was replenished every 2 days.
mRNA expression analysis
Total RNA was isolated by using the RNeasy Mini Kit (Qiagen) according to the manufacturer's instructions. RNA (1 μg) was reverse transcribed with oligo(dT) primers. The resulting cDNA samples were amplified by RT (reverse transcription)–PCR using gene-specific primer sets in conjunction with the Power SyBr Green PCR Master Mix (Applied Biosystems). All of the primer sequences are listed in Supplementary Table S1.
NANOG promoter reporter assay
The pGreen-Zeo-NANOG reporter plasmid was obtained from System Biosciences. It contained a 2587 bp fragment of the NANOG promoter (−2452/+135) cloned upstream of the destabilized GFP (green fluorescent protein) copGFP gene. Lentiviral particles were generated by transfecting HEK (human embryonic kidney)-293T cells (Invitrogen) with pGreen-Zeo-NANOG in conjunction with the packaging plasmids pLP1, pLP2 and pLP/VSVG (Invitrogen) for 48 h. The media were harvested and used to infect NCCIT cells. Transduced cells carrying the GFP reporter construct were sorted by FACS (UCSF Laboratory of Cell Analysis, San Francisco, CA, U.S.A.) 3 days after infection. Cells expressing low-to-moderate basal levels of GFP were enriched in the sorted cell population in order to improve the detection of GFP induction. The resulting cells (NCCIT NANOG–GFP) were subcultured and routinely inspected for GFP fluorescence by microscopy. GFP expression remained stable for several months. NCCIT NANOG–GFP cells were transfected with saRNA for 4 days and subsequently analysed on a FACSCalibur flow cytometer (Becton Dickinson) for changes in GFP levels. Forward- and side-scatter plots were used to exclude dead cells, debris and doublets from the histogram analysis. The WinMDI program (http://facs.scripps.edu/software.html) was used to analyse the percentage of GFP positive cells.
Lentivirus-based overexpression of NANOG
A lentiviral human NANOG cDNA overexpression vector (pSin-EF2-NANOG-Pur) was obtained from Addgene (Addgene plasmid 16578). Lentiviral particles were generated by transfecting HEK-293T cells (Invitrogen) with pSin-EF2-NANOG-Pur in conjunction with the packaging plasmids pLP1, pLP2 and pLP/VSVG (Invitrogen) for 48 h. The media were harvested and used to infect NCCIT cells. Puromycin (2 μg/ml) was used to select transduced cells 3 days after infection.
Immunoblot analysis
Immunoblot analysis was performed according to standard laboratory protocol as published previously [21,22]. Primary antibodies included anti-NANOG (1:1000 dilution; 3580S, Cell Signaling Technology), anti-β-3-tubulin (1:1000 dilution; ab18207, Abcam) and anti-OCT-3/4 (C-10) (1:1000 dilution, mouse monoclonal; sc-5279, Santa Cruz Biotechnology) antibodies. Anti-GAPDH (glyceraldehyde-3-phosphate dehydrogenase) (1:5000 dilution, rabbit monoclonal; #2118, Cell Signaling Technology), anti-β-actin (1:3000 dilution; AC-15, Sigma–Aldrich), anti-Hsp70 (heat-shock protein 70) (1:2000 dilution; Thermo Fisher Scientific) and anti-α-tubulin (1:5000 dilution; Sigma–Aldrich) antibodies were used as loading controls.
Cell proliferation assay
Cells were transfected with saRNA and re-seeded into 96-well plates the next day at a density of 5000 cells/well. Cell proliferation was measured 4 days later by using the CellTiter96 Aqueous one solution cell proliferation assay kit (Promega) containing MTS [3-(4,5-dimethylthiazol-2-yl)-5-(3-carboxymethoxyphenyl)-2-(4-sulfophenyl)-2H-tetrazolium] reagent according to the manufacturer's protocol.
RESULTS AND DISCUSSION
Identification of saRNAs that target the NANOG promoter by using a GFP reporter system
NANOG expression is silenced in most normal primary and cancer cells [12]. However, NCCIT (human embryonic carcinoma) cells are a developmentally pluripotent cell line that express high levels of stem cell markers, including OCT4, NANOG, SSEA (stage-specific embryonic antigen)-1, SSEA-3, SSEA-4, TRA-1-81 etc. [23]. NCCIT cells can also be influenced to differentiate into derivatives of all three embryonic germ layers (i.e. ectoderm, mesoderm and endoderm) and extra-embryonic cell lineages [23,24]. As such, NCCIT cells were selected to determine whether RNAa can be used to activate NANOG expression in vitro and influence cell fate.
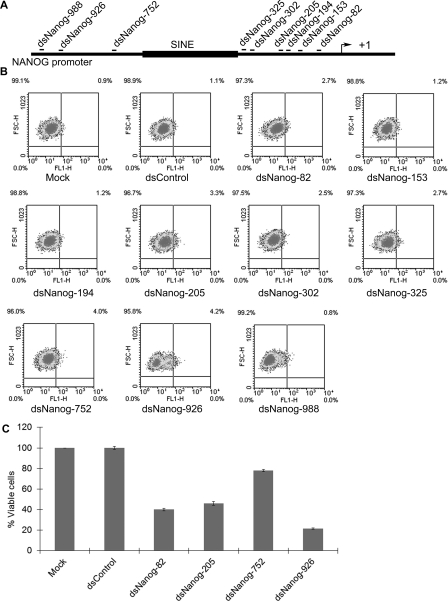
Selecting saRNA target sites within gene promoters is largely a hit-or-miss process. Promoter sequence, chromatin structure, accessibility, overlapping non-coding transcripts etc. may all play a role in influencing transcription and susceptibility to RNAa [5,8]. In order to improve screening efficiency, we generated stable NCCIT (NCCIT NANOG–GFP) cells that contained GFP under control of the human NANOG promoter by lentiviral transduction. Measuring GFP intensity would serve as a readout for the transcriptional activity of the NANOG promoter. We designed a series of saRNAs that targeted the NANOG promoter at sites ranging from −82 to −988 relative to the TSS (transcription start site) by following previously described design rules [6] (Figure 1A and Supplementary Table S1). Each duplex was named according to its target within the NANOG promoter (i.e. dsNanog-82, dsNanog-153, dsNanog-194 etc.). A control saRNA (dsControl) was also synthesized that lacked significant homology with all known human sequences. Each saRNA was transfected into NCCIT NANOG–GFP cells and the GFP intensity was assessed by flow cytometry. Compared with the control treatments, dsNanog-82, -205, -752 and -926 caused at least a 2.7-fold increase in the percentage of the GFP-positive populations, whereas the other saRNAs did not significantly alter GFP expression (Figure 1B). To exclude the possibility that cytotoxic effects may be contributing to the increase in GFP signal, a MTS assay was conducted on the transfected cells (Figure 1C). We found that dsNanog-752 had minimal effects on cell proliferation, whereas dsNanog-82, -205 and -926 similarly inhibited cell growth (Figure 1C). As such, cytotoxic effects may account for the subtle increase in GFP signal in dsNanog-82 and dsNanog-205 transfected cells. However, GFP induction by dsNanog-752 and dsNanog-926, which caused the highest shift among all NANOG dsRNAs (Figure 1B), may not result entirely from cytotoxicity, but rather increases in GFP expression.
Figure 1. Identification of NANOG saRNAs using a GFP reporter system.
(A) Schematic representation of the NANOG promoter. Indicated are the names of each saRNA and its target site relative to the TSS (+1). A SINE (short interspersed element) repetitive element is also indicated in the NANOG promoter. (B) NCCIT cells were infected with lentiviral particles containing GFP under the control of the human NANOG promoter. At 3 days after transduction GFP-positive cells (NCCIT NANOG–GFP) were collected by cell sorting and subcultured for saRNA transfection. NCCIT NANOG–GFP cells were transfected with 50 nM concentrations of the indicated saRNAs for three days and analysed by FACS to assess GFP intensity in each population. Mock samples were transfected in the absence of saRNA. Representative data of n=2. (C) NCCIT cells were transfected with the indicated dsRNA at 100 nM. MTT assay was conducted 96 hours following transfection. Results are plotted as the percentage of viable cells relative to mock transfections (means±S.D. for two independent experiments).
saRNA-mediated induction of endogenous NANOG expression in NCCIT cells
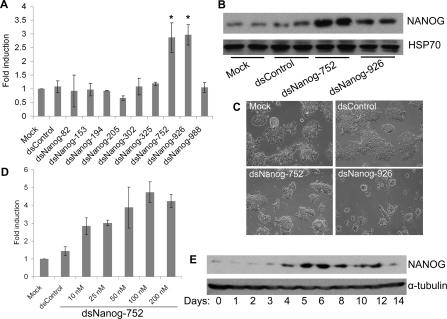
To further validate the screening results, we individually transfected each saRNA into NCCIT cells and evaluated endogenous NANOG expression by quantitative RT–PCR. Compared with the control transfections, dsNanog-752 and dsNanog-926 caused a 2.87- and 2.97-fold increase in NANOG mRNA levels respectively (Figure 2A). The remaining duplexes did not significantly induce NANOG expression. Collectively, analysis of NANOG expression following saRNA treatment correlated with the results from the GFP reporter assay.
Figure 2. saRNA-mediated NANOG overexpression in human embryonic carcinoma NCCIT cells.
(A) NCCIT cells were transfected with 50 nM concentrations of the indicated saRNAs for 96 h. Mock samples were transfected in the absence of saRNA. NANOG expression was assessed by quantitative RT–PCR. Results are the fold changes relative to mock transfections (means±S.D. for three independent experiments). *P< 0.05 compared with the mock. (B) NCCIT cells were transfected with mock, dsControl, dsNanog-752 or dsNanog-926 for 72 h. NANOG protein level was detected by immunoblot analysis. Hsp70 served as a loading control. (C) Representative phase contrast cell images were taken at 40× magnification 72 h after transfection of the indicated saRNAs. (D) NCCIT cells were transfected with dsNanog-752 for 3 days at concentrations ranging from 10 to 200 nM. Expression of NANOG was assessed by quantitative RT–PCR and normalized to GAPDH. Results are fold changes relative to mock transfections (means±S.D. for three independent experiments). (E) NCCIT cells were transfected with 50 nM dsNanog-752 for the indicated lengths of time. The expression of NANOG protein was detected by immunoblot analysis at each time point. α-Tubulin served as a loading control. Representative blot of n=2.
Immunoblot analysis indicated dsNanog-752 and dsNanog-926 also increased NANOG protein levels in NCCIT cells (Figure 2B). Morphologically mock- and dsControl-treated cells remained healthy, whereas dsNanog-752-transfected cells formed compact ES-like colonies (Figure 2C). As found when using the MTS assay, dsNanog-926 caused visible cytotoxic effects as cells appeared considerably less dense compared with other treatments (Figures 2C and 1C). Potential off-target effects and/or stimulation of the innate immune response may contribute to the cytotoxicity of dsNanog-926 in NCCIT cells. As such, all subsequent experiments focused on dsNanog-752.
Analysis of mRNA expression also revealed NANOG was induced in a dose-dependent manner by dsNanog-752 at concentrations ranging from 10 to ~50–100 nM, whereas concentrations higher than ~50–100 nM did not further increase NANOG expression (Figure 2D). Time-course experiments revealed that NANOG protein levels were not induced until day 3–4, with peak activity around day 5 (Figure 2E). NANOG levels remained elevated up to day 12. Previous studies have shown RNAa possesses kinetics distinct from RNAi (RNA interference) as characterized by an initial ~48 h delay with ~10 day continuation in gene activation [1,2,25]. These unique features of RNAa have been attributed to its nuclear nature and consequent epigenetic changes at targeted promoters [1,8,25]. Our observations in the present study reveal that activation NANOG by dsNanog-752 follows the typical kinetics of RNAa.
Sequence requirement of dsNanog-752 for NANOG induction
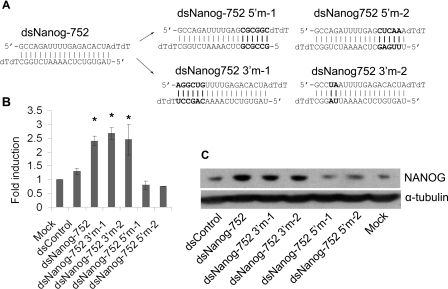
To characterize the sequence requirement for dsNanog-752, we created several mutant saRNAs in which nucleotides at either the 5′-end (dsNanog-752-5′m-1 and dsNanog-752-5′m-2) or 3′-end (dsNanog-752-3′m-1 and dsNanog-52-3′m-2) relative with the antisense strand of dsNanog-752 possessed mismatches with the targeted promoter sequence (Figure 3A). Analysis of mRNA expression revealed that dsNanog-752-3′m-1 and dsNanog-752-3′m-2 possessed activity similar to wild-type dsNanog-752, whereas dsNanog-752-5′m-1 and dsNanog-752-5′m-2 were not capable of inducing NANOG expression (Figure 3B). Immunoblot analysis also confirmed mutant saRNA RT–PCR results (Figure 3C). These findings indicate that mutation to the 3′-end of dsNanog-752 does not interfere with RNAa activity, whereas mutation of the 5′-end abolishes its ability to activate NANOG expression. Similar to our previous results evaluating the sequence requirement for RNAa at the E-cadherin and p21 promoters [1,3], the 5′-end of the antisense strand in dsNaong-752 is important for RNAa specificity. As such, the antisense strand likely serves as the ‘guide’ strand responsible for targeting sequence at the NANOG promoter.
Figure 3. Sequence requirement for dsNanog-752.
(A) Mutations to the 5′-end (relative of the antisense strand) of dsNanog-752 resulted in duplexes dsNanog-752-5′m-1 and dsNanog-752-5′m-2. Mutations to the 3′-end of dsNanog-752 created mutant derivatives dsNanog-752-3′m-1 and dsNanog-752-3′m-2. The mutated bases are shown in bold. (B) NCCIT cells were transfected with 50 nM of the indicated saRNA molecules for 96 h. Expression of NANOG was accessed by quantitative RT–PCR and normalized to GAPDH. Results are fold changes relative to mock transfections (means±S.D. for two independent experiments). *P<0.05 compared with the mock. (C) NCCIT cells were transfected as in (B) and NANOG protein levels were detected by immunoblot analysis. α-Tubulin served as a loading control.
NANOG induction by saRNA modulates the expression of its downstream genes
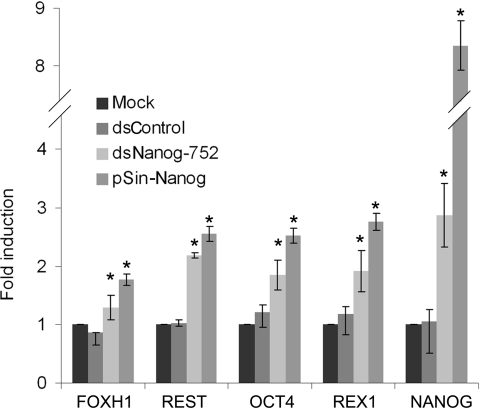
NANOG is a key transcription factor involved in regulating embryonic development and self-renewal of pluripotent stem cells. Using genome-wide location analyses, hundreds of genes have been identified as downstream targets of NANOG [26–28]. To determine if RNAa-based overexpression of NANOG influences downstream gene expression, we evaluated mRNA levels of several known downstream genes [i.e. OCT4, REX1 (reduced expression protein 1), REST (RE1-silencing transcription factor) and FOXH1 (forkhead box H1)] [29,30] following dsNanog-752 transfection in NCCIT cells. As a positive control, NANOG cDNA was also overexpressed in NCCIT cells using a lentiviral vector (pSin-Nanog). As expected, ectopic overexpression via pSin-Nanog elevated NANOG mRNA levels by 8.3-fold and predictably increased expression of all four downstream genes (Figure 4). Similarly, albeit to a lesser extent, dsNanog-752 also up-regulated the expression of each downstream gene (Figure 4). This result suggests restoration of endogenous NANOG expression by RNAa results in a functional protein capable of predictably modulating the expression of its downstream genes.
Figure 4. dsNanog-752 induces expression of downstream NANOG-regulated genes.
NCCIT cells were transfected with 50 nM concentrations of the indicated saRNAs for 96 h. Mock samples were transfected in the absence of saRNA. Expression of NANOG and its downstream genes (OCT4, REX1, REST and FOXH1) was assessed by quantitative RT–PCR and normalized to GAPDH. Results are fold changes relative to mock transfections (means±S.D. for three independent experiments). *P< 0.05 compared with the mock. cDNA generated from NCCIT cells transduced with lentivirus (pSin-Nanog) served as a positive control for NANOG overexpression.
NANOG activation by saRNA antagonizes RA-induced differentiation in NCCIT cells
Since exogenous NANOG expression can sustain pluripotency in ES cells [12,18], it would be interesting to determine if RNAa-mediated overexpression of NANOG can also prevent cell differentiation. It has been reported that certain chemical compounds (e.g. RA) induce neuronal-like features in human embryonic carcinoma cells [23]. To analyse the effects of dsNanog-752 on RA-induced differentiation, we treated NCCIT cells with 10 μM RA for 2 weeks. During this period, the cells were transfected with dsNanog-752 every 4 days for a total of 12 days. NCCIT cells transduced with pSin-Nanog lentivirus particles were also treated with RA and served as a positive control for NANOG overexpression. Phenotypically, RA forced mock- and dsControl-treated cells to differentiate as evident by the loss of compact, ES-like colony structure, whereas dsNanog-752 and lentiviral Nanog overexpression allowed cells to retain ES-like colony morphology (Figure 5). Expression analysis by quantitative RT–PCR revealed RA also caused a 98.7% decrease in NANOG mRNA levels compared with the untreated cells; however, dsNanog-752 sustained NANOG expression in the presence of RA treatment resulting in ~15.7-fold higher NANOG mRNA levels compared with dsControl transfected cells (Figure 6A). Lentiviral transduced cells (pSin-Nanog) produced ~1500-fold higher NANOG mRNA levels compared with dsControl treatments (Figure 6A). Since OCT4 is a key downstream gene of NANOG, we also evaluated its expression following RA-induced differentiation in NCCIT cells. As shown in Figures 6(B) and 6(C), expression of OCT4 exhibited a pattern in concordance with NANOG (Figure 6A), suggesting tight regulation of OCT4 by NANOG in NCCIT cells. Interestingly, despite differences in magnitude between RNAa- and lentiviral-mediated overexpression of NANOG in RA-treated cells (Figure 6A), both techniques maintained similar levels of OCT4 expression (Figure 6B). It has been proposed that RNAa may represent a more natural approach to gene overexpression capable of facilitating similar measurable effects on downstream phenotypes as ectopic overexpression systems without the need for massive product output [6].
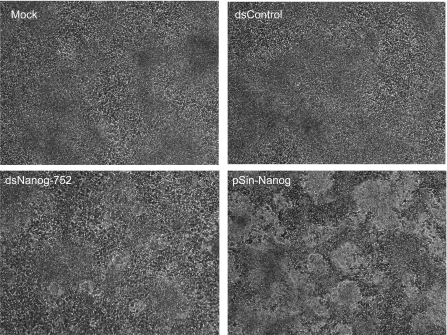
Figure 5. dsNanog-752 maintains an ES-like morphology in NCCIT cells treated with RA.
NCCIT cells treated with 10 μM RA were transfected with 50 nM of the indicated dsRNAs every 4 days. Representative phase-contrast cell images were taken at 40× magnification on day 14 after transfection.
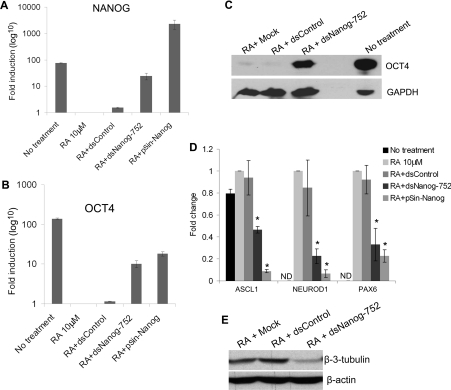
Figure 6. dsNanog-752 antagonizes RA-induced differentiation in NCCIT cells.
NCCIT cells were cultured with 10 μM RA and co-treated with 50 nM concentrations of the indicated saRNAs every 96 h for 14 days. NCCIT cells transduced with lentivirus (pSin-Nanog) served as a positive control for NANOG overexpression. mRNA expression levels of NANOG (A) and OCT4 (B) were assessed by quantitative RT–PCR and normalized to that of GAPDH. Results are fold changes relative to the 10 μM RA treatment (means±S.D. for three independent experiments). (C) Protein levels of OCT4 were detected by immunoblot analysis. GAPDH served as a loading control. Representative blot of n=2. (D) Neural markers ASCL1, NEUROD1 and PAX6 were assessed by quantitative RT–PCR and normalized to that of GAPDH. Results are fold changes relative to the 10 μM RA treatment (means±S.D. for three independent experiments). * P< 0.05 compared with RA. ND, not detectable. (E) Protein levels of β-3-tubulin were detected by immunoblot analysis. β-actin served as a loading control. Representative blot of n=2.
Consistent with the findings from previous studies [23,31], RA treatment also up-regulated several markers for neural differentiation [i.e. ASCL1 (achaete-scute complex homologue 1), NEUROD1 (neuronal differentiation 1) and PAX6 (paired box 6)], suggesting RA promotes differentiation of NCCIT cells towards neurons (Figure 6D). However, co-treatment with dsNanog-752 significantly curbed neural marker gene up-regulation by RA in a manner similar to pSin-Nanog-transduced cells (Figure 6D). Although not sufficient to completely halt RA activity, these results demonstrate that dsNanog-752 antagonized the differentiation phenotype induced by RA treatment in NCCIT cells. In support of this, immunoblot analysis revealed dsNanog-752 also decreased expression of neural differentiation marker β-3-tubulin (Figure 6E).
Taken together, this study utilizes RNAa as a molecular tool to enhance expression of NANOG in human NCCIT cells. NANOG overexpression by dsNanog-752 led to the predictable modulation of several downstream genes and antagonized RA-induced differentiation. Furthermore, these results were recapitulated by vector-mediated overexpression of NANOG. The findings of the present study provide proof-of-concept that RNAa may provide an alternative approach to DNA-based overexpression systems for manipulating cell fate and activating development-related genes.
Online data
AUTHOR CONTRIBUTION
Long-Cheng Li and Xiaoling Wang conceived the study, designed the experiments, analysed the data and wrote the paper. Xiaoling Wang and Ji Wang performed the experiments. Vera Huang and Robert Place contributed to the experimental design and wrote the paper.
FUNDING
This work was supported by the California Institute for Regenerative Medicine [grant number RL1-00660-1] and the National Institutes of Health [grant number 1R01GM090293-0109].
References
- 1.Li L. C., Okino S. T., Zhao H., Pookot D., Place R. F., Urakami S., Enokida H., Dahiya R. Small dsRNAs induce transcriptional activation in human cells. Proc. Natl. Acad. Sci. U.S.A. 2006;103:17337–17342. doi: 10.1073/pnas.0607015103. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Janowski B. A., Younger S. T., Hardy D. B., Ram R., Huffman K. E., Corey D. R. Activating gene expression in mammalian cells with promoter-targeted duplex RNAs. Nat. Chem. Biol. 2007;3:166–173. doi: 10.1038/nchembio860. [DOI] [PubMed] [Google Scholar]
- 3.Place R. F., Li L. C., Pookot D., Noonan E. J., Dahiya R. MicroRNA-373 induces expression of genes with complementary promoter sequences. Proc. Natl. Acad. Sci. U.S.A. 2008;105:1608–1613. doi: 10.1073/pnas.0707594105. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Morris K. V., Santoso S., Turner A. M., Pastori C., Hawkins P. G. Bidirectional transcription directs both transcriptional gene activation and suppression in human cells. PLoS Genet. 2008;4:e1000258. doi: 10.1371/journal.pgen.1000258. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Schwartz J. C., Younger S. T., Nguyen N. B., Hardy D. B., Monia B. P., Corey D. R., Janowski B. A. Antisense transcripts are targets for activating small RNAs. Nat. Struct. Mol. Biol. 2008;15:842–848. doi: 10.1038/nsmb.1444. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Huang V., Qin Y., Wang J., Wang X., Place R. F., Lin G., Lue T. F., Li L. C. RNAa is conserved in mammalian cells. PLoS ONE. 2010;5:e8848. doi: 10.1371/journal.pone.0008848. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Chu Y., Yue X., Younger S. T., Janowski B. A., Corey D. R. Involvement of argonaute proteins in gene silencing and activation by RNAs complementary to a non-coding transcript at the progesterone receptor promoter. Nucleic Acids Res. 2010;38:7736–7748. doi: 10.1093/nar/gkq648. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Portnoy V., Huang V., Place R. F., Li L.-C. Small RNA and transcriptional upregulation. Wiley Interdiscip. Rev.: RNA. 2011;2:748–60. doi: 10.1002/wrna.90. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Ieda M., Fu J. D., Delgado-Olguin P., Vedantham V., Hayashi Y., Bruneau B. G., Srivastava D. Direct reprogramming of fibroblasts into functional cardiomyocytes by defined factors. Cell. 2010;142:375–386. doi: 10.1016/j.cell.2010.07.002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Szabo E., Rampalli S., Risueno R. M., Schnerch A., Mitchell R., Fiebig-Comyn A., Levadoux-Martin M., Bhatia M. Direct conversion of human fibroblasts to multilineage blood progenitors. Nature. 2010;468:521–526. doi: 10.1038/nature09591. [DOI] [PubMed] [Google Scholar]
- 11.Vierbuchen T., Ostermeier A., Pang Z. P., Kokubu Y., Sudhof T. C., Wernig M. Direct conversion of fibroblasts to functional neurons by defined factors. Nature. 2010;463:1035–1041. doi: 10.1038/nature08797. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Chambers I., Colby D., Robertson M., Nichols J., Lee S., Tweedie S., Smith A. Functional expression cloning of Nanog, a pluripotency sustaining factor in embryonic stem cells. Cell. 2003;113:643–655. doi: 10.1016/s0092-8674(03)00392-1. [DOI] [PubMed] [Google Scholar]
- 13.Mitsui K., Tokuzawa Y., Itoh H., Segawa K., Murakami M., Takahashi K., Maruyama M., Maeda M., Yamanaka S. The homeoprotein Nanog is required for maintenance of pluripotency in mouse epiblast and ES cells. Cell. 2003;113:631–642. doi: 10.1016/s0092-8674(03)00393-3. [DOI] [PubMed] [Google Scholar]
- 14.Chambers I., Silva J., Colby D., Nichols J., Nijmeijer B., Robertson M., Vrana J., Jones K., Grotewold L., Smith A. Nanog safeguards pluripotency and mediates germline development. Nature. 2007;450:1230–1234. doi: 10.1038/nature06403. [DOI] [PubMed] [Google Scholar]
- 15.Silva J., Nichols J., Theunissen T. W., Guo G., van Oosten A. L., Barrandon O., Wray J., Yamanaka S., Chambers I., Smith A. Nanog is the gateway to the pluripotent ground state. Cell. 2009;138:722–737. doi: 10.1016/j.cell.2009.07.039. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Hamazaki T., Kehoe S. M., Nakano T., Terada N. The Grb2/Mek pathway represses Nanog in murine embryonic stem cells. Mol. Cell. Biol. 2006;26:7539–7549. doi: 10.1128/MCB.00508-06. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Hyslop L., Stojkovic M., Armstrong L., Walter T., Stojkovic P., Przyborski S., Herbert M., Murdoch A., Strachan T., Lako M. Downregulation of NANOG induces differentiation of human embryonic stem cells to extraembryonic lineages. Stem Cells. 2005;23:1035–1043. doi: 10.1634/stemcells.2005-0080. [DOI] [PubMed] [Google Scholar]
- 18.Darr H., Mayshar Y., Benvenisty N. Overexpression of NANOG in human ES cells enables feeder-free growth while inducing primitive ectoderm features. Development. 2006;133:1193–1201. doi: 10.1242/dev.02286. [DOI] [PubMed] [Google Scholar]
- 19.Yu J., Vodyanik M. A., Smuga-Otto K., Antosiewicz-Bourget J., Frane J. L., Tian S., Nie J., Jonsdottir G. A., Ruotti V., Stewart R., et al. Induced pluripotent stem cell lines derived from human somatic cells. Science. 2007;318:1917–1920. doi: 10.1126/science.1151526. [DOI] [PubMed] [Google Scholar]
- 20.Okita K., Ichisaka T., Yamanaka S. Generation of germline-competent induced pluripotent stem cells. Nature. 2007;448:313–317. doi: 10.1038/nature05934. [DOI] [PubMed] [Google Scholar]
- 21.Noonan E. J., Place R. F., Basak S., Pookot D., Li L. C. miR-449a causes Rb-dependent cell cycle arrest and senescence in prostate cancer cells. Oncotarget. 2010;1:349–358. doi: 10.18632/oncotarget.167. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Huang V., Place R. F., Portnoy V., Wang J., Qi Z., Jia Z., Yu A., Shuman M., Yu J., Li L. C. Upregulation of Cyclin B1 by miRNA and its implications in cancer. Nucleic Acids Res. 2012;40:1695–1707. doi: 10.1093/nar/gkr934. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Damjanov I., Horvat B., Gibas Z. Retinoic acid-induced differentiation of the developmentally pluripotent human germ cell tumor-derived cell line, NCCIT. Lab. Invest. 1993;68:220–232. [PubMed] [Google Scholar]
- 24.Jung K. H., Das N. D., Park J. H., Lee H. T., Choi M. R., Chung M. K., Park K. S., Jung M. H., Lee B. C., Choi I. G., Chai Y. G. Effects of acute ethanol treatment on NCCIT cells and NCCIT cell-derived embryoid bodies (EBs) Toxicol. In Vitro. 2010;24:1696–1704. doi: 10.1016/j.tiv.2010.05.017. [DOI] [PubMed] [Google Scholar]
- 25.Place R. F., Noonan E. J., Foldes-Papp Z., Li L. C. Defining features and exploring chemical modifications to manipulate RNAa activity. Curr. Pharm. Biotechnol. 2010;11:518–526. doi: 10.2174/138920110791591463. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Sharov A. A., Masui S., Sharova L. V., Piao Y., Aiba K., Matoba R., Xin L., Niwa H., Ko M. S. Identification of Pou5f1, Sox2, and Nanog downstream target genes with statistical confidence by applying a novel algorithm to time course microarray and genome-wide chromatin immunoprecipitation data. BMC Genomics. 2008;9:269. doi: 10.1186/1471-2164-9-269. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Loh Y. H., Wu Q., Chew J. L., Vega V. B., Zhang W., Chen X., Bourque G., George J., Leong B., Liu J., et al. The Oct4 and Nanog transcription network regulates pluripotency in mouse embryonic stem cells. Nat. Genet. 2006;38:431–440. doi: 10.1038/ng1760. [DOI] [PubMed] [Google Scholar]
- 28.Boyer L. A., Lee T. I., Cole M. F., Johnstone S. E., Levine S. S., Zucker J. P., Guenther M. G., Kumar R. M., Murray H. L., Jenner R. G., et al. Core transcriptional regulatory circuitry in human embryonic stem cells. Cell. 2005;122:947–956. doi: 10.1016/j.cell.2005.08.020. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Shi W., Wang H., Pan G., Geng Y., Guo Y., Pei D. Regulation of the pluripotency marker Rex-1 by Nanog and Sox2. J. Biol. Chem. 2006;281:23319–23325. doi: 10.1074/jbc.M601811200. [DOI] [PubMed] [Google Scholar]
- 30.Pan G., Li J., Zhou Y., Zheng H., Pei D. A negative feedback loop of transcription factors that controls stem cell pluripotency and self-renewal. FASEB J. 2006;20:1730–1732. doi: 10.1096/fj.05-5543fje. [DOI] [PubMed] [Google Scholar]
- 31.Tonge P. D., Andrews P. W. Retinoic acid directs neuronal differentiation of human pluripotent stem cell lines in a non-cell-autonomous manner. Differentiation. 2010;80:20–30. doi: 10.1016/j.diff.2010.04.001. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.