Abstract
Although originally considered toxic, hydrogen sulfide (H2S) has been implicated in mediating various biological processes. Nevertheless, its cellular targets and mode of action are not well understood. Protein Tyrosine Phosphatases (PTPs), which regulate numerous signal transduction pathways, utilize an essential Cys residue at the active site, which is characterized by a low pKa and is susceptible to reversible oxidation. Here, we report that PTP1B, the founding member of this enzyme family, was reversibly inactivated by H2S, in vitro and in vivo, via sulfhydration of the active site Cys residue. Unlike oxidized PTP1B, the sulfhydrated enzyme was preferentially reduced by thioredoxin in vitro, compared to glutathione or dithiothreitol. Sulfhydration of the active site Cys in PTP1B in cells required the presence of cystathionine-γ-lyase (CSE), a critical enzyme in H2S production, and resulted in inhibition of phosphatase activity. Suppression of CSE decreased H2S production and decreased the phosphorylation on Tyr619, and activation, of PERK [protein kinase-like endoplasmic reticulum (ER) kinase] in response to ER stress. PERK, which phosphorylates the eukaryotic translational initiation factor 2 (eIF2α) leading to attenuation of protein translation, was a direct substrate of PTP1B. In addition, CSE knockdown also led to activation of SRC, previously shown to be mediated by PTP1B. These effects of suppressing H2S production on the response to ER stress were abrogated by a small molecule inhibitor of PTP1B. Together, these data define a signaling function for H2S in inhibiting PTP1B activity and thereby promoting PERK activity during the response to ER stress.
Introduction
Gasotransmitters are a class of gaseous signaling molecules that freely permeate membranes and, therefore, unlike classical regulators of signal transduction, act independently of transmembrane receptors [1]. Gasotransmitters are generated enzymatically, in a regulated manner, and act through specific molecular targets. The classic example is nitric oxide (NO), a well-established signaling molecule generated by NO synthases. NO was identified as an endothelium-derived relaxing factor (EDRF) that functions by activating guanylyl cyclase, thereby stimulating the production of guanosine 3′,5′-monophosphate (cGMP). Subsequently NO was shown to influence many cellular processes in various tissues [2]. The study of NO paved the way for identification of other gaseous signaling molecules, including carbon monoxide (CO) and, more recently, hydrogen sulfide (H2S) [1, 3-5]. The latter is known for its toxic properties; nevertheless, at sub-toxic concentrations, H2S regulates a broad spectrum of physiological events [6-8].
The ability to synthesize H2S is found in representatives of all evolutionary kingdoms [9-11] suggesting an ancient metabolic capability. In mammals, H2S biosynthesis is catalyzed predominantly by two pyridoxal phosphate-dependent enzymes, cystathionine-β-synthase (CBS) and cystathionine-γ-lyase (CSE), which together constitute a transulfuration pathway that provides a route for converting dietary methionine to cysteine. CBS predominates in the central nervous system, whereas both enzymes are found in peripheral tissues. Their activity is accompanied by production of H2S at micromolar concentrations (up to ~300 μM) in various tissues [12]. As in the case of NO, the original function characterized for H2S was vasorelaxation; in fact, it has been proposed as a new EDRF, an effect mediated through activation of an ATP-sensitive potassium channel (KATP), which lowers blood pressure [13]. H2S has been implicated in the control of cell proliferation and survival in cardiomyocytes [12]. It is also produced at sites of inflammation; indeed, drugs with the potential to trigger release of H2S have been suggested as potential anti-inflammatories [11, 14, 15]. Intriguingly, treatment with H2S induces a suspended animation-like state in mice by decreasing metabolic rate and core body temperature, an effect that depends on its reversible inhibition of cytochrome c oxidase [16]. The effects of H2S are mediated through its sulfhydration of specific Cys residues in target proteins [17]. For example, in the liver, which produces abundant H2S, 10 to 25% of actin, tubulin, and glyceraldehyde-3-phosphate dehydrogenase (GAPDH) are sulfhydrated in the basal state, a modification that enhances actin polymerization and GAPDH activity [17]. Nevertheless, the identification of additional specific targets that mediate the various physiological functions of H2S and clarification of its roles in signaling remain important quests. Here, we tested the hypothesis that the protein tyrosine phosphatase (PTP) family of enzymes may provide a group of targets for H2S.
The PTPs comprise a large, structurally-diverse family of receptor-like and non-transmembrane enzymes that are specific regulators of signal transduction in their own right and, in conjunction with the protein tyrosine kinases (PTKs), exert exquisite control over various biological functions [18]. Members of the PTP family are characterized by a highly conserved catalytic domain, containing a signature motif, [His-Cys-(X)5-Arg-(Ser/Thr), where X is any amino acid], in which the invariant cysteine has an unusually low pKa. This favors its function as a nucleophile to attack the phosphotyrosine substrate [18]. The presence of this cysteine at the active site has been shown to underlie an important aspect of PTP regulation. Covalent modification of the active site cysteine abrogates its nucleophilic function and thereby inactivates the enzyme. A considerable body of data has now revealed that production of H2O2, in response to a variety of growth factors, hormones and cytokines that act through tyrosine phosphorylation, leads to the transient oxidation and inactivation of those PTPs that normally exert an inhibitory constraint on the signaling pathways. This transient PTP inactivation enhances the kinase-dependent tyrosine phosphorylation of components of the pathway and thereby enables fine-tuning of the response [19]. Other agents that modify the active site cysteine would also be expected to inhibit PTP activity. Indeed, NO inactivates PTPs by S-nitrosylation of the active site cysteine, a modification that is proposed to provide a mechanism to prevent permanent inactivation of PTPs by irreversible oxidation [20]. In this study, we examined whether H2S-induced sulfhydration of PTP1B may also provide a mechanism for regulating tyrosine phosphorylation-dependent signaling.
We have focused on PTP1B, which is the prototypic member of PTP family [21]. It is a well-established inhibitor of insulin and leptin signaling, and is a validated target for therapeutic intervention in diabetes and obesity [21, 22]. PTP1B also promotes HER2-mediated breast tumorigenesis [23, 24], and regulates several other tyrosine phosphorylation-dependent signaling pathways [25]. PTP1B is located on the cytoplasmic face of the endoplasmic reticulum (ER) and has been implicated in ER stress signaling [26]. In this study, we demonstrate that PTP1B is reversibly sulfhydrated following production of H2S in the ER stress response. We show that sulfhydration inhibited the ability of PTP1B to dephosphorylate a critical regulator of the ER stress response, the double-stranded RNA-activated protein kinase-like ER kinase (PERK). Our study identifies sulfhydration as a redox modification for regulation of PTP1B and of the ER stress response and illustrates a further aspect of the control of signaling in response to ER stress.
Results
Inactivation of PTP1B by H2S
As a first step in our analysis, we measured PTP1B phosphatase activity as a function of time and of H2S concentration. We observed an apparent second order rate constant of 22.4 ± 1.8 M−1s−1 for inactivation of PTP1B by H2S (Fig. 1A). The active site cysteine of PTP1B, Cys215, undergoes reversible oxidation by H2O2 and reversible S-nitrosylation by NO [20]; therefore, we compared the kinetics of PTP1B inactivation by H2O2 and NO with the rate of its inactivation by H2S (Fig. S1 and S2). The rate of PTP1B inactivation by H2O2 was 10 ± 1.4 M−1s−1 and that by NO was 2.1 ± 0.5 M−1s−1; the overall extent of reversible inactivation was similar in each case (~90%). Given the physiological roles of H2O2 and NO in regulating PTP1B [14, 15], this comparison suggested that inactivation by H2S may also be physiologically relevant. Furthermore, the data suggest that the active site cysteine of PTP1B may display a degree of specificity in its reaction with oxidants.
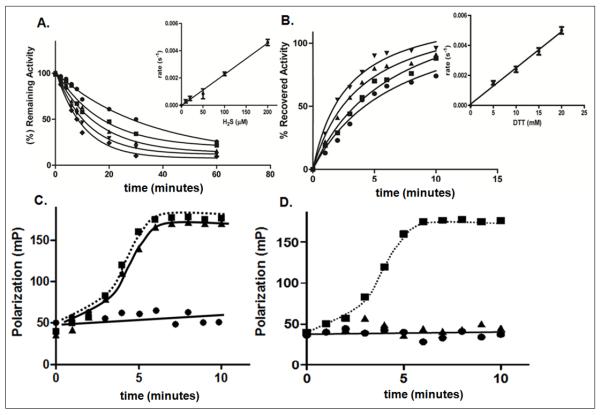
Figure 1. Redox-dependent inactivation and reactivation of PTP1B.
A. Time dependent inactivation of PTP1B by H2S. The phosphatase activity of PTP1B was monitored in the presence of the following concentrations of H2S, 10 μM (●), 20 μM (■), 50 μM (▲), 100 μM (▼), 200 μM (◆). Inset, the concentration dependence of the rate of inactivation was used to derive the second order rate constant, 22.4 ± 1.8 M−1s−1.
B. Time dependent reactivation of PTP1B by DTT. The following concentrations of DTT were used 5 mM (●), 10 mM (■), 15 mM (▲), 20 mM (▼). Inset, the concentration dependence of the rate of reactivation was used to derive the second order rate constant, 0.24 ± 0.1 M−1s−1. Data are representative of three independent determinations.
C and D. Binding of fluorescently-labeled phosphopeptide substrate (5′-FAM-ENDpYINASL) to WT (●), C215S (■), or D181A (▲) mutant forms of PTP1B. The change in fluorescence polarization is plotted as a function of time. The assays were performed in the absence of H2S (C) or following incubation with 100 μM H2S for 10 min (D).
In an alternative approach, we employed substrate-trapping mutant forms of PTP1B to examine the effects of H2S on substrate binding in an in vitro fluorescence polarization assay. In one of these mutants (PTP1B-D181A), the invariant catalytic acid residue Asp181, which protonates the tyrosyl leaving group in the substrate, is mutated to Ala. This mutation impairs catalysis without affecting the affinity for substrate [27]; therefore, the mutant forms stable complexes with tyrosine-phosphorylated substrates. A mutant in which the active site Cys residue is changed to Ser (PTP1B-C215S) also displays substrate trapping properties [28]. The fluorescence polarization assay assesses substrate binding, based on the difference in the rate of rotation of small fluorescently labeled species, such as the peptide substrate, and the larger complex between the PTP1B trapping mutant and the peptide substrate. When a fluorescent molecule is excited with plane-polarized light, it emits light in the same polarized plane only if it is stationary during the excitation of the fluorophore. As the molecule tumbles in solution, the polarization of the emitted light is lost. The large complex of the trapping mutant and peptide tumbles more slowly than the free peptide and so yields a higher fluorescence polarization signal than does the free peptide. Both PTP1B-C215S and PTP1B-D181A, but not the wild-type enzyme, formed a complex with a fluorescently labeled peptide substrate (Fig. 1C); however, only the peptide complex with the D181A mutant was disrupted by treatment with H2S, highlighting the importance of the active site Cys for the inhibitory effects of H2S.
Reactivation of PTP1B by DTT, GSH, and TR/TRR
To examine the reversibility of PTP1B inactivation by H2S, H2O2 and NO, we compared the effects of varying concentrations of the reducing agents dithiothreitol (DTT), reduced glutathione (GSH), or the combination of thioredoxin and thioredoxin reductase (TR/TRR) on the rate of recovery of activity of inactivated PTP1B. In each case, the rate of PTP1B reactivation varied linearly as a function of the concentration of the reducing agent (Figs 1B, S3 & S4) and showed a second-order rate constant (Table 1). The reactivation of H2S-inactivated PTP1B was ~190-fold faster with TR/TRR than with DTT; however, such a difference was not observed for reactivation of oxidized or nitrosylated PTP1B (Table 1). The rates of reactivation of H2O2- and NO-treated PTP1B were similar, although with both H2O2- and NO-treated PTP1B the rate of reactivation with glutathione was at least 10-fold slower than with DTT or TR/TRR. In all cases, >90% of the phosphatase activity was recovered with all three reducing agents. Therefore, the reversibility of H2S-treated PTP1B induced by TR/TRR is kinetically the most favorable, consistent with a potential physiological preference for this reducing agent.
Table 1. Rate of PTP1B reactivation by different reducing agents following inactivation with H2O2, H2S, and NO.
| H2O2 (M−1s−1) | H2s (M−1s−1) | NO (M−1s−1) | ||||||
|---|---|---|---|---|---|---|---|---|
| DTT | GSH | TR/TRR | DTT | GSH | TR/TRR | DTT | GSH | TR/TRR |
| 055 ± 0.1 |
0.042 ± 0.01 |
0.75 ± 0.2 |
0.24 ± 0.1 |
0.013 ± 0.012 |
45.5 ± 2.8 |
0.83 ± 0.12 |
0.012 ± 0.0089 |
0.79 ± 0.1 |
Change in mass of PTP1B following H2S treatment
To define more precisely the nature of the modification to PTP1B induced by incubation with H2S, we used a high-resolution quadrupole time of flight (QTOF) mass spectrometer to analyze a truncated form of PTP1B (residues 1-321) that comprised the catalytic domain of the protein. The intact masses of H2S-treated and untreated PTP1B determined from electrospray ionization mass spectrometry (ESI-MS) spectra were 38407.01 Da and 38374.36 Da, respectively, indicating a modification with a mass addition of 32.65 Da (Fig 2A, upper panel). H2S treatment did not induce such a mass shift with the C215S mutant form of PTP1B (38358.85 Da) (Fig 2A, lower panel). Furthermore, the increase in mass of 32.65 observed for wild-type PTP1B was abolished by treatment with DTT, which suggested that it involved a reversible cysteine modification.
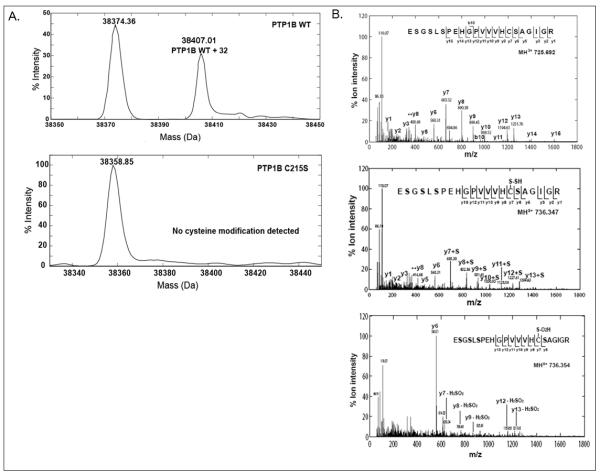
Figure 2. Analysis of H2S-modified PTP1B by mass spectrometry.
A. Direct ESI-MS analysis of wild-type and Cys215-to-Ser mutant forms of PTP1B following H2S treatment. WT or C215S mutant PTP1B (20 μg) was incubated with H2S (100 μM) for 10 min at 25°C and the mass of the protein was determined by ESI-MS analysis. H2S-treated WT PTP1B (upper panel) was resolved into two peaks corresponding to unmodified (38374.36 Da) and H2S-modified (38407.01 Da) protein. The H2S-induced modification was associated with an increase in mass of 32 Da. The deconvoluted spectrum for the active site mutant, PTP1B C215S, showed a single peak of 38358.85 Da and no H2S-induced modification was apparent.
B. LC-MS/MS analysis of the tryptic peptide containing Cys215 of PTP1B. Purified PTP1B (20 μg) was treated with H2S (100 μM) or H2O2 (1mM) for 10 min at 25°C, trypsinized overnight, and analyzed by LC-MS/MS. Untreated peptide, ([M+3H]3+ at m/z 725.692), corresponds to a free Cys215 sulfhydryl group (upper panel); the H2S-treated peptide, ([M+3H]3+ at m/z 736.347, mass error −2.7 ppm), corresponds to sulfhydrated Cys215 (middle panel) and the H2O2-treated peptide, ([M+3H]3+ at m/z 736.354, mass error −1.4ppm), corresponds to the sulfinic acid modification of Cys215 (lower panel). Distinct fragmentation patterns were observed for the tryptic peptide (ESGSLSPEHGPVVVHC(215)SAGIGR) derived from native, H2S-, and H2O2-treated PTP1B.
The active site Cys215 of PTP1B can be oxidized to cysteine-sulfinic acid (SO2H, 64.9697), which has a monoisotopic mass very close to that of sulfhydrated cysteine (S-SH, 64.9520). To distinguish between these subtle mass differences, we trypsinized native, H2O2-, and H2S-treated PTP1B separately, and analyzed the triply charged tryptic peptide containing the catalytic cysteine by mass spectrometry. The three forms were readily distinguishable at the MS1 level by accurate mass measurements as the native (725.692, upper panel), sulfydrated (736.347, mass error −2.7 ppm, middle panel), and sulfinic acid (736.354, mass error −1.4 ppm, lower panel) forms (Fig. 2B). These three precursors were then selected for collision-induced dissociation (CID) to generate signature fragment ions. All three peptides generated confirmatory b-ions (fragment ions from the N-terminus) and y-ions (fragment ions from the C-terminus) for sequence identification. Notably, the H2S-treated peptide displayed a unique increase of 32 Da compared to the native peptide for the y-ion series for residues following Cys215 (y7), suggesting a stable modification of Cys215 with mass addition 32. The MS/MS spectrum for the same tryptic peptide following H2O2 treatment was different, with a hallmark neutral loss of 66 Da for H2SO2 observed for y-ions beyond y7, confirming the sulfinic acid modification of the cysteine. Together, the evidence from the intact protein mass measurements, accurate peptide masses at the MS1 level, and signature MS2 fragmentations suggest that the active site cysteine (Cys215) of PTP1B undergoes sulfhydration upon treatment with H2S.
Measurement of sulfhydration of PTP1B using a thiolate anion-directed probe in HEK293T cells
To demonstrate sulfhydration of PTP1B in a cell model, we treated human embryonic kidney (HEK) 293T cells with varying amounts of a saturated solution of H2S, then lysed the cells in an anaerobic work station to avoid post-lysis oxidation of cysteines. Sulfhydration was followed by applying a cysteinyl labeling assay that utilizes a biotinylated iodoacetic acid probe, which reacts through a nucleophilic substitution of the halide group by the PTP-reactive thiol group, resulting in a stable thioether bond (Fig S5) [29]. After treating the cells with different concentrations of H2S for 30 minutes, we observed minimal labeling in untreated cells, representing the basal amount of reversibly oxidized PTP1B present prior to H2S addition, and saturation of the response at ~25 μM H2S (Fig. 3A). PTP1B sulfhydration could be detected within 2 minutes of exposure of the cells to 100 μM H2S and peaked at 10 min (Fig. 3B). Finally, we used selected ion monitoring (SIM) mass spectrometry to examine the tryptic peptide containing Cys215 from PTP1B. We observed sulfhydration of Cys215in PTP1B from cells treated with H2S(Fig. 3C) but not in untreated cells. Thus, PTP1B was susceptible to H2S-induced sulfhydration in a cellular context.
Figure 3. Detection of H2S-modified PTP1B using a thiolate anion-reactive probe.
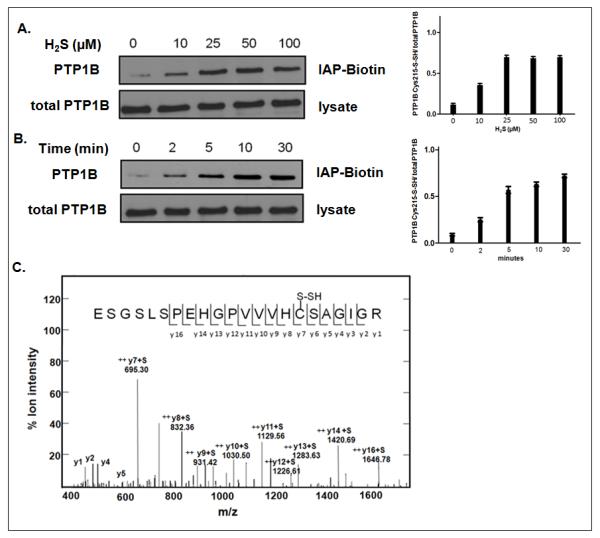
A. HEK293T cells were serum starved, then treated with varying concentrations of H2S (0, 10, 25, 50, or 100 μM) for 30 min at 37°C. Lower panel illustrates total PTP1B in 0.1mg of cell lysate.
B. HEK293T cells were treated with 100 μM H2S for 0, 2, 5, 10, or 30 min at 37°C. In both A and B, cells were lysed and the modification of PTP1B was monitored in 1.0 mg of cell lysate using a sulfhydryl-reactive IAP-Biotin probe (upper panel). Lower illustrates total PTP1B in 0.1mg of cell lysate.
C. PTP1B was immunopurified from HEK293T cells that had been treated with 50 μM H2S for 30 min at 37°C, trypsinized, and analyzed by LC-MS/MS. This is an MS-MS spectrum of the peptide containing the active site cysteine residue, Cys215, illustrating that this residue was in the sulfhydrated form.
PTP1B sulfhydration under ER stress in control but not in CSE-depleted HeLa cells
We hypothesized that production of H2S may provide a mechanism for enhancing tyrosine phosphorylation, and thereby modulating cell signaling, through the sulfydration and inactivation of PTP1B. PTP1B is localized to the ER and has been implicated in the control of signaling during ER stress [26]. Furthermore, H2S production, mediated by cystathionine-γ-lyase (CSE), has been linked to the ER stress-induced Unfolded Protein Response (UPR) [30]. Therefore, we investigated whether H2S modified PTP1B function and signaling under conditions of ER stress. We exposed HeLa cells to tunicamycin to trigger ER stress and observed a significant increase in H2S production (Fig. 4A). We immunoblotted cell lysates for PTP1B, CSE, and the ER chaperone BiP (binding immunoglobulin protein), which is an ER chaperone that binds to unfolded proteins, regulates the activation of the response pathways and is a marker of ER stress. We detected no significant change in the abundance of PTP1B, but a small increase in the abundance of CSE was observed within 2h of induction of ER stress. There was a marked increase in the expression of BiP, which confirmed the induction of the ER stress response (Fig S6).
Figure 4. H2S produced in response to tunicamycin induced ER stress.
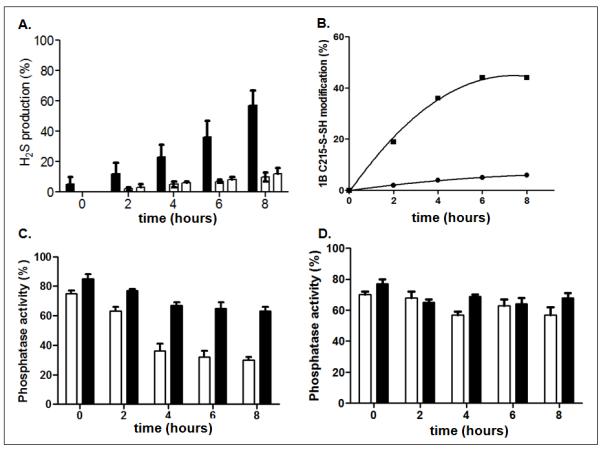
A. ER stress was induced by tunicamycin (10μg/ml) and the production of H2S was measured in control (black bars) or CSE-deficient cells generated using two distinct shRNAs (open bars). ER stress increased H2S production relative to that in untreated cells in control cells but not those in which CSE was knocked down. Data are the mean of three independent experiments and presented as mean ± S.E.M
B. The effect of tunicamycin-induced ER stress on the redox status of PTP1B Cys215 was followed by LC-MS/MS mass spectrometry to quantify the unmodified Cys215 tryptic peptide ([M+3H]3+ 725.692) and the sulfhydrated counterpart ([M+3H]3+ 736.347). A time-dependent increase in sulfhydration (-SSH) was observed for PTP1B immunopurified from control (■) but not CSE-deficient (●) cells.
C & D. PTP1B was immunopurified from control (C) and CSE-deficient (D) cells, using anti-PTP1B antibody DH8, and the enzymatic activity was monitored using pNPP as substrate, in the absence (open bars) or presence (black bars) of 5 mM DTT, following tunicamycin-induced ER stress.
We generated stable HeLa cell lines expressing short hairpin RNAs that targeted CSE mRNA, to suppress H2S production, and examined the effects on sulfhydration of PTP1B under ER stress. Immunoblot analysis indicated that both of the shRNAs we used decreased CSE abundance by >90% (Fig S7). We subjected both control and CSE-depleted cells to ER stress induced by tunicamycin, an inhibitor of protein N-glycosylation, for varying lengths of time and measured H2S production. We detected accumulation of H2S following exposure to tunicamycin in control cells but not in either of the stable cell lines expressing CSE shRNAs (Fig 4A).
To characterize the modifications of the active site cysteine residue that accompanied these changes in H2S production, we used selected ion monitoring (SIM) to quantify the unmodified peptide containing Cys215 ([M+3H]3+ 725.692) and the sulfydrated counterpart ([M+3H]3+ 736.347), which we resolved chromatographically. In the absence of ER stress, 95% of PTP1B was detected in the reduced, active form (Table S1). PTP1B from the control cells that expressed CSE showed an increase in sulfhydration following exposure to tunicamycin, which plateaued within 6 to 8 hours, with a large proportion (~43%) of PTP1B in the sulfhydrated form (Table S1). In contrast, cells in which CSE was depleted showed a much smaller increase in PTP1B sulfhydration (from ~5% to ~10%) together with small increases in oxidized forms (SO2H and SO3H) of the enzyme.
In parallel with the mass spectrometry analysis, we immunopurified PTP1B at various times following exposure to tunicamycin, and measured phosphatase activity in the immunoprecipitates. Treatment with tunicamycin induced a decline in the phosphatase activity of PTP1B immunopurified from control cells. In addition, there was a correlation between the extent of inhibition and the extent of sulfhydration (Fig 4B and C). Furthermore, upon addition of the reducing agent DTT, we were able to restore PTP1B activity in the immunoprecipitate, indicating a reversible, inhibitory redox modification (Fig 4C). In contrast, we did not detect any change in PTP1B activity, with or without reducing agent, in CSE-depleted cells (Fig 4D). These data are consistent with modification of the essential active site cysteine of PTP1B by sulfhydration and suggest that sulfhydration provides a mechanism for inactivation of the enzyme under conditions of ER stress.
H2S-mediated UPR signaling
Our data illustrate that H2S production in cells can lead to inhibition of PTP1B. Consequently, one might anticipate that changes in the levels of H2S may lead to changes in the phosphorylation of tyrosyl residues in proteins. Therefore, we used anti-phosphotyrosine antibody immunoblotting to assess tyrosine phosphorylation in control and CSE-depleted HeLa cells following tunicamycin-induced ER stress (Fig. 5). We anticipated that in the control cells with CSE, H2S would be produced, thereby inhibiting PTP activity and promoting tyrosine phosphorylation, whereas in CSE-depleted cells, in which the production of H2S was decreased, the result would be increased PTP activity, and thus lower cellular phosphotyrosine, compared to the control cells. Instead, we detected an unexpected increase in phosphorylation of proteins of 30-60 kDa in CSE-depleted cells upon tunicamycin-induced ER stress (Fig 5A). PTP1B activates the PTK SRC, a 60kDa protein, by dephosphorylating the C-terminal inhibitory phosphorylation site, Tyr527, thereby promoting autophosphorylation of SRC on Tyr416, a process that is required for robust kinase activity [31, 32]. Therefore, we used phospho-specific antibodies to assess the phosphorylation status of Tyr527and Tyr416in SRC (Fig 5B). We noted that there was a marked decrease in phosphorylation of Tyr527, with concomitant increase in phosphorylation of Tyr416, in CSE-depleted cells upon tunicamycin-induced ER stress. These effects were abrogated by inclusion of MSI-1436, a small molecule, non-competitive inhibitor of PTP1B that has been shown to inhibit the phosphatase in mouse models of obesity [33], consistent with the identification of SRC as a substrate of PTP1B in this context.
Figure 5. H2S-mediated signaling in HeLa cells in response to ER stress.
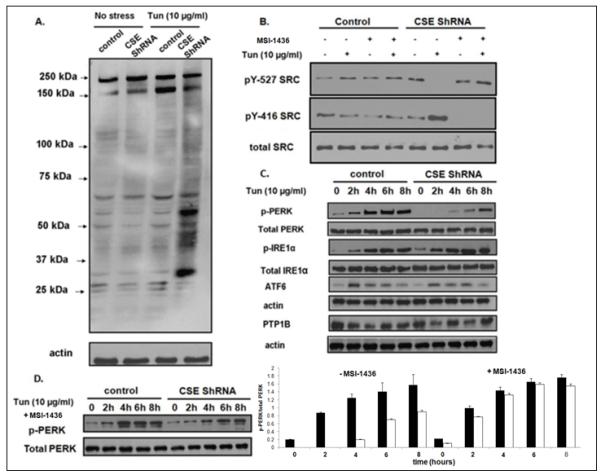
A. Immunoblot showing the overall change in tyrosine phosphorylation induced by ER stress, visualized using 4G10 anti-phosphotyrosine antibody. The left two lanes show tyrosine phosphorylation in the absence of ER stress, the right two lanes show tyrosine phosphorylation following four hours of treatment with tunicamycin. Control and CSE-depleted cells are as indicated.
B. SRC activation was monitored in control and CSE-depleted cells using phospho-specific antibodies to the inhibitory C-terminal site (Tyr527, upper panel) and the autophosphorylation site (Tyr527, middle panel). The lower panel illustrates total SRC. ER stress was induced for 4h with tunicamycin. Cells were treated with PTP1B inhibitor MSI-1436 (5 μM) as indicated.
C. Immunoblot analyses to examine the activation of components of the unfolded protein response. The data illustrate a time course, over 0-8 h, to examine activation of IRE1 [anti-phospho-IRE1α (Ser724) compared to total IRE1 (anti-IRE1α)] and the activation of PERK [anti-phospho-PERK (Thr980) compared to total PERK (anti-PERK)] in control and CSE-deficient cells. There were no obvious changes in the abundance of ATF6 or PTP1B.
D. Immunoblot illustrating that the PTP1B inhibitor MSI-1436 (5 μM) restored PERK activation in response to tunicamycin-induced ER stress in CSE-depleted cells.
The anti-phosphotyrosine blot also revealed that an increase in tyrosine phosphorylation of a protein of ~150kDa was induced by ER stress in response to tunicamycin in control cells, which was not detected in CSE-depleted cells (Fig 5A). This is consistent with attenuation of phosphatase activity by H2S in the presence of CSE under ER stress. To identify this protein, we examined the three main branches of ER stress-induced signaling in control and CSE-depleted HeLa cells. We observed that ER stress increased the phosphorylation of PERK on Thr980, a marker of PERK activation [34-36], and that this response was delayed and attenuated in CSE-depleted cells (Fig. 5C, Fig. S8. In contrast, changes in the abundance of ATF6 or phosphorylation of IRE1 were considerably less pronounced. To explore further the importance of PTP1B to this impaired activation of PERK, we tested the effects of the small molecule inhibitor MSI-1436 [33]. We observed that whereas MSI-1436 did not affect PERK activation in response to ER stress in control cells, it restored PERK activation in CSE-depleted cells to a similar level to that observed in control cells (Fig. 5D). These data suggest that PERK activation was regulated in a manner that depends on sulfhydration of PTP1B.
PTP1B dephosphorylation of PERK
The cytoplasmic portion of PERK contains a protein kinase domain that is activated by oligomerization, leading to enhanced autophosphorylation [37]. The phosphorylation of Thr 980, which was measured in Figure 5C and D, is a marker of its activation status; however, considering the specificity of PTP1B for tyrosyl residues in proteins, this particular site will not be the direct substrate of the phosphatase. Autophosphorylation of Tyr619, which is located in a highly conserved segment of the kinase domain, promotes phosphorylation of Thr980 and activation of PERK [30]. Therefore, we investigated the possibility that phospho-Tyr619-PERK was a physiological substrate of PTP1B.
To address this possibility, we used the D181A substrate-trapping mutant form of PTP1B for in vitro and transfection studies. We generated lysates from untreated cells or cells treated with tunicamycin, and incubated them with wild-type PTP1B or PTP1B-D181A. PTP1B was then immunoprecipitated and its interaction with PERK was monitored with anti-PERK antibody. We observed that the PTP1B-D181A substrate-trapping mutant, but not the wild-type enzyme, co-precipitated PERK (Fig 6A). Furthermore, this association was observed only from lysates of tunicamycin-treated cells. Treatment of the PTP protein with pervanadate, which promotes oxidation of the active site cysteine of PTP family members, also disrupted the interaction between PTP1B-D181A and PERK (Fig. 6A); this highlights the importance of the active site cysteine, consistent with an enzyme-substrate interaction. In addition, to examine the importance of phosphorylation of PERK for its interaction with the substrate-trapping mutant form of PTP1B, we treated the lysate with calf intestinal alkaline phosphatase (CIAP), to eliminate PERK phosphorylation. We observed that this treatment abrogated the association (Fig. 6B). We examined the complexes that co-immunoprecipitated with wild-type or substrate-trapping mutant PTP1B by LC-MS/MS mass spectrometry and identified the pTyr619 PERK peptide in complexes associated with PTP1B-D181A but not wild-type PTP1B (Fig. 6C). In addition, to examine the association between PTP1B and PERK in a cellular context, we overexpressed wild-type and substrate-trapping mutant forms of PTP1B in HEK293T cells and subjected the cells to ER stress. We immunoprecipitated PTP1B and assessed its association with PERK by immunoblotting using anti-PERK antibody. Wild-type and substrate-trapping mutant PTP1B were expressed and immunoprecipitated at similar levels (Fig. 6D, lower panel); however, as seen in vitro, only the PTP1B-D181A substrate-trapping mutant co-precipitated PERK (Fig. 6D, upper panel). The fact that we can detect such complex formation within the cell is most likely explained by the catalytically impaired, substrate-trapping mutant being overexpressed to levels that exceed the ability of H2S to modify it and thus it is able to form a complex with pTyr-PERK produced in response to ER stress. Considering the specificity of PTP1B for tyrosyl residues in proteins, we tested the importance of Tyr619 in PERK for the association with the substrate-trapping mutant. Myc-tagged murine PERK (wild-type or Tyr615, which is equivalent to Tyr619 in the human protein, substituted with Phe) was expressed in HEK293T cells, which were then treated with tunicamycin, lysed and incubated with either wild-type or D181A mutant PTP1B. As shown in Figure 6E, mutation of the pTyr619 site of phosphorylation in PERK abrogated its interaction with the PTP1B-D181A substrate-trapping mutant. Finally, we demonstrated that wild-type PTP1B directly dephosphorylated myc-tagged PERK that had been isolated from tunicamycin-treated HEK293T cells by immunoprecipitation (Fig. 6F). Collectively, these data establish a direct enzyme-substrate interaction between PTP1B and pTyr619-PERK.
Figure 6. Interaction of the D181A substrate-trapping mutant form of PTP1B and tyrosine-phosphorylated PERK.
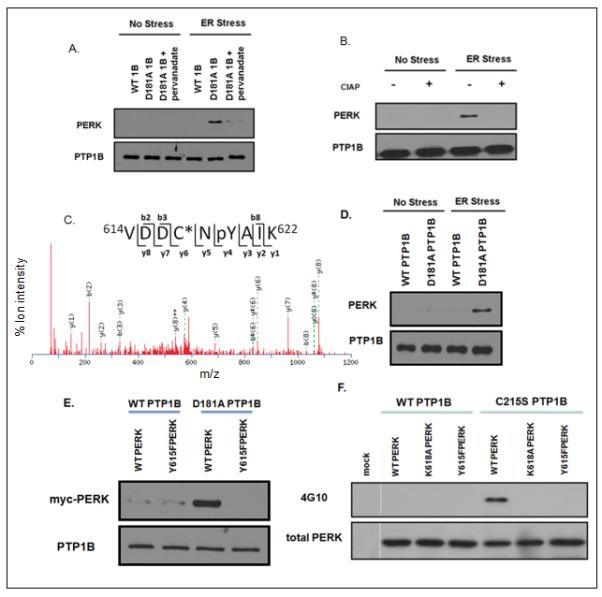
A. Lysates were prepared from untreated control cells (No Stress), or cells treated with tunicamycin for 4h (ER Stress), and incubated with PTP1B or PTP1B-D181Ain the presence or absence of 1mM pervanadate. PTP1B immunoprecipitates were subjected to SDS-PAGE and immunoblotted with anti-PERK antibody (upper panel). The blot was then stripped and re-blotted with anti-PTP1B antibody FG6 (lower panel).
B. Lysates were prepared from untreated cells (No Stress), or cells treated with tunicamycin for 4h (ER Stress), and then incubated in the absence (−) or presence (+) of calf intestine alkaline phosphatase (CIAP). The treated lysates were then incubated with wild-type PTP1B or PTP1B-D181A, PTP1B immunoprecipitates were resolved by SDS-PAGE and immunoblotted with anti-PERK antibody (upper panel). The blot was then stripped and re-blotted with anti-PTP1B antibody FG6 (lower panel).
C. PERK was identified as a PTP1B substrate by mass spectrometry. Immunopurified protein complex from the substrate trapping experiment was treated with vanadate to release the bound substrate, which was then subjected to trypsinization and the peptides obtained were analyzed by MUDPIT. This is an MS-MS spectrum of the peptide containing phosphorylated Tyr619 in PERK, which bound to the substrate-trapping mutant D181A form of PTP1B.
D. Wild-type PTP1B or PTP1B-D181Awere overexpressed in HEK293T cells and immunoprecipitated following tunicamycin treatment for 4 hours, resolved by SDS-PAGE, and immunoblotted with anti-PERK rabbit polyclonal antibody (upper panel) and also stripped and re-blotted using anti-PTP1B antibody FG6 (lower panel).
E. Wild-type and mutant (Y615F) myc-tagged forms of mouse PERK were overexpressed in HEK293T cells and immunoprecipitated using 9E10 anti-myc antibody following tunicamycin treatment for 4 hours. PERK was eluted with myc peptide, then incubated with Ni-NTA bead-bound wild-type or D181A substrate-trapping mutant forms of PTP1B. Complexes were resolved by SDS-PAGE, immunoblotted with antimyc antibody (9E10), then stripped and re-blotted using anti-PTP1B antibody FG6 (lower panel).
F. Wild-type and mutant, K618A (catalytically inactive), and Y615F forms of myc-tagged mouse PERK were overexpressed in HEK293T cells and immunoprecipitated following four hours of tunicamycin treatment. Mock represents the vector control. The immunoprecipitated proteins were incubated with wild-type or C215S catalytically inactive mutant forms of PTP1B, resolved by SDS-PAGE, and immunoblotted with antiphosphotyrosine antibody (4G10). The lower panel is a loading control obtained by stripping and re-blotting with anti-PERK antibody.
Discussion
The essential role of the active site cysteine residue in PTP-mediated catalysis provides a mechanism for redox-based regulation of PTP activity. The reversible oxidation and inactivation of PTPs in response to the regulated production of H2O2 provides a well-established mechanism for control of tyrosine phosphorylation-dependent signaling [18, 19]. NO-induced nitrosylation of the active site cysteine in PTPs has also been reported [20]. In this report, we demonstrate that PTP1B function is regulated by reversible sulfhydration in response to endogenously generated H2S and that this is an important component of the control of the ER stress response.
Consistent with such previously identified targets as actin, tubulin and glyceraldehyde-3-phosphate dehydrogenase [17, 38], we observed that H2S sulfhydrated the active site cysteine of PTP1B. However, whereas sulfhydration is generally thought to activate target enzymes [6], we found that sulfhydration of Cys215 inhibited PTP1B activity. We observed sulfhydration of PTP1B in vitro at concentrations of H2S encountered in vivo [12] and found that it occurred exclusively on the catalytic cysteine, Cys215, leaving the other cysteine residues in the protein unmodified. This is consistent with the unique properties of this residue, which distinguish it from other cysteine residues in the protein. Cys215 is present as a thiolate, rather than as a free thiol, a feature conferred by the architecture of the PTP active site [39] that renders these enzymes exquisitely sensitive to inactivation by modification of the catalytic cysteine. The observation that PTP1B was sulfhydrated and inactivated simply following addition of H2S in vitro was nevertheless surprising as direct reaction of the cysteine with H2S is unlikely. This raises questions as to the underlying mechanism. High throughput screens of chemical libraries have shown that certain inhibitors function as “peroxide generators” when included with the other constituents of a standard phosphatase activity assay and that any peroxide thus generated inhibits PTP activity [40]. It is possible that a similar situation may apply here and any peroxide could affect the form in which H2S is encountered by PTP1B. For example, any H2O2 in the assay could interact with H2S/HS− to generate HSSH [41], which could react directly with the PTP active site. Alternatively, any oxidized PTP1B produced in the reaction, which would adopt the cyclic sulphenamide conformation [39], could also react directly with H2S. We observed a striking selectivity in sensitivity of sulfhydrated PTP1B to different reducing agents. Although oxidized and nitrosylated PTP1B displayed similar sensitivities to DTT, GSH, and thioredoxin, the sulfhydrated enzyme, which features a –SSH moiety on the active site cysteine, displayed ~190-fold faster reduction and reactivation by thioredoxin, the major intracellular disulfide bond reductase, compared to DTT and GSH. Therefore sulfhydration of PTP1B may play a role in controlling the mechanism for reduction and reactivation of the enzyme (Fig. S9). Nevertheless, these mechanistic considerations do not change the essential conclusions of our study. The sulfhydrated form of PTP1B, which was produced to high stoichiometry in cells under conditions of ER stress, is inactive and, therefore, provides a potential physiological mechanism for regulating pTyr-dependent signaling.
The ER plays a major role in controlling the folding and modification of newly synthesized transmembrane and secreted proteins. The folding capacity of the ER is modulated in response to the environmental and physiological status of the cell; thus, a cell can match its capacity for protein folding to its physiological requirements. However, if that capacity is exceeded, resulting in a condition known as ER stress, then an integrated set of three signaling pathways, the Unfolded Protein Response (UPR), is activated [37]. These pathways are triggered by transmembrane sensor proteins that detect unfolded proteins in the ER and transduce a signal that initiates a response in the cytosol or nucleus. This allows the cell to fine-tune the rates of protein synthesis and the folding capacity to maintain homeostasis and, if that is not successful, to trigger cell death. PTP1B is resident in the ER and has been implicated in the control of ER stress [26]. Furthermore, the production of H2S by cystathionine-γ-lyase has been implicated as a facet of the regulation of the ER stress response [42]. Therefore, we explored further the potential for integration of the effects of PTP1B and H2S in the UPR.
The three pathways that constitute the UPR are initiated and controlled by the transmembrane ER proteins inositol requiring-1 (IRE1), activating transcription factor-6 (ATF6) and double-stranded RNA-activated protein kinase-like ER kinase (PERK) [34, 37, 43, 44]. IRE1 is a bifunctional protein kinase and site-specific endoribonuclease. When its luminal segment senses the accumulation of unfolded protein it undergoes oligomerization in the ER membrane, which induces autophosphorylation of the cytoplasmic kinase domain and activation of the endoribonuclease activity. The latter promotes splicing of X-box binding protein mRNA to generate XBP1s, which is a potent transcriptional activator that induces expression of genes encoding both chaperones and proteins associated with degradation. Furthermore, binding of TRAF2 by phosphorylated IRE1 promotes activation of the JNK MAP kinase and control of apoptosis. The stress sensor ATF6 also presents a segment to the ER lumen that detects unfolded proteins, resulting in activation and transport to the Golgi where it is cleaved by proteases. This cleaved ATF6 is released from the membrane and imported into the nucleus where it directs the transcription of a subset of genes that regulate the UPR. Finally, PERK, like IRE1, is an ER transmembrane kinase that oligomerizes under conditions of ER stress leading to autophosphorylation and activation. The effects of PERK are exerted through phosphorylation of Ser51 in eIF2α, which catalyzes the rate-limiting step of initiation of protein translation [35]. This phosphorylation traps eIF2α in an inactive, GDP-bound state, which decreases protein synthesis and limits the load of unfolded protein entering the ER. Under conditions of eIF2α phosphorylation, translation of the transcription factor ATF4 is also induced, which regulates UPR gene expression. The phosphorylation status of eIF2α is a major point of signal integration, such that downstream events are referred to as the Integrated Stress Response [37]. This places considerable importance on maintaining tight control over the phosphorylation status of eIF2α. The reversibility of PERK phosphorylation is an important component of this regulatory mechanism. Our data demonstrate that PTP1B exerts a major influence on the activity of PERK, but not IRE1 or ATF6, under conditions of ER stress. There is evidence to suggest crosstalk between these three pathways [45-47]; however differential regulation of the pathways is also thought to circumvent functional redundancy [37]. Our study reveals that PTP1B underlies a new aspect of differential regulation of UPR signaling and illustrates another example of specificity in the function of a member of the PTP family.
PERK belongs to the RNA-dependent protein kinase (PKR) family of proteins. Phosphorylation of a conserved tyrosine residue within the catalytic domain has been implicated in activation of PKR kinases [48]. In human PERK, this residue is Tyr619, which corresponds to mouse PERK Tyr615. PERK is a dual-specificity kinase that can autophosphorylate on this tyrosyl residue, coincident with autophosphorylation of Thr980 in its activation loop. Mutation of human Tyr619 compromises PERK activation and its phosphorylation of eIF2α [36]. Our data reveal that Tyr619 is a substrate of PTP1B through which PTP1B can attenuate PERK activity and thereby the UPR. PTP1B has a preference for acidic residues N-terminal to the phosphorylated tyrosine [28, 49]; the sequence flanking PERK Tyr619 resembles that of other PTP1B substrates, with two aspartic acid residues N-terminal to the phosphorylated tyrosine [49, 50]. Our observations are consistent with a study showing that PTP1B overexpression suppressed tunicamycin-induced PERK signaling and eIF2α phosphorylation in MIN6 insulinoma β cells, whereas RNAi-mediated PTP1B deficiency enhanced this ER stress response [51]. A previous study comparing ER stress signaling in fibroblasts from wild-type and PTP1B-deficient mice, however, suggested that PTP1B was required for activation of IRE1 signaling, as measured by XBP1 splicing and JNK activation, but did not affect PERK phosphorylation of Ser51 in eIF2α [26]. The reason for this discrepancy is unclear; however, there have been no further reports defining the mechanism underlying these effects of PTP1B on IRE1 signaling. The difference may be related to the chronic loss of PTP1B in these animals, which, in itself, could be interpreted by the cells as a stress that may affect the UPR.
In summary, we have identified PTP1B as a phosphatase that specifically inhibited the PERK arm of the UPR. The ability of PTP1B to dephosphorylate Tyr619 and inactivate PERK is fine-tuned by the production of H2S by CSE in response to ER stress. The resulting sulfhydration and inactivation of PTP1B facilitated PERK phosphorylation and activation, promoting restoration of ER homeostasis. The activity of PERK was in turn kept in check by the reduction and reactivation of PTP1B, likely mediated by thioredoxin, which ensured reversibility of PERK phosphorylation and flexibility in the response to ER stress (Fig. 7). In addition to acute regulation of PTP1B function, sulfhydration may also act as a protective mechanism, adapted by the enzyme to prevent irreversible oxidation and inactivation. Even mild oxidative stress can rapidly convert the PTP active site cysteine to irreversibly oxidized sulfinic and sulfonic acid forms [39]. NO-induced S-nitrosylation of the active site cysteine protects PTP1B from permanent inactivation by reactive oxygen species (ROS) [20]. Given the interplay among the different gasotransmitters and their regulation of similar functions, it is conceivable that H2S may also be involved in preventing irreversible PTP oxidation. This could be particularly important in the context of the ER, which is a highly oxidizing environment due to the redox dependence of protein folding, and ER stress, which also leads to an increase in ROS production [52, 53].
Figure 7. Proposed model for the role of sulfhydration of PTP1B in regulating the response to ER stress.
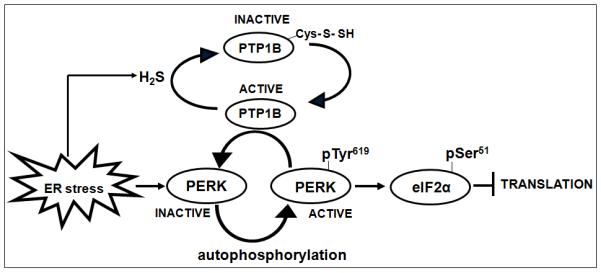
Induction of ER stress increases the production of H2S by cystathionine-γ-lyase, which leads to sulfhydration of the essential active site cysteine residue in PTP1B and concomitant inactivation of the phosphatase. This transiently protects pTyr619 of PERK from dephosphorylation, promoting PERK activation and its ability to inhibit global translation by phosphorylating eIF2α. Reduction and reactivation of sulfhydrated PTP1B restores phosphatase activity and its ability to dephosphorylate and inactivate PERK.
MATERIALS & METHODS
Materials
Antibodies were purchased from Cell Signaling (IRE1, PERK, phospho-PERK-T980, eIF2α, phospho-eIF2α-S51, pY419-SRC and SRC), Sigma (β-actin), Thermo Scientific (phospho-IRE1-S724), Millipore (phosphotyrosine 4G10 and ATF6), and Abcam (CSE). Chemicals were obtained from Sigma unless otherwise noted. The Y615F-PERK (murine, equivalent to Y619F in human) was a gift from Anotonios Koromilas (McGill University).
Cell Culture
Short hairpin RNAs targeting luciferase and CSE were obtained from Open Biosystems and cloned into the pSVG vector between the restriction sites EcoRI and XhoI. The sequences of the CSE shRNAs were AGGAGCTGATATTTCTATGTATTAGTGAAGCCACAGATGTAATACATAGAAATATCAGCTCCC and CCCTCAAGAACCTAAAGCTATTTAGTGAAGCCACAGATGTAAATAGCTTTAGGTTCTTGAGGA. Stable plasmid expression was achieved via sequential retroviral transduction, followed by selection with 2 μg/ml puromycin. ER stress was induced with tunicamycin at a final concentration of 10 μg/ml or thapsigargin (1 μM) for 0 to 8h. MSI-1436 was used at a concentration of 5 μM to inhibit PTP1B. For transient transfection, 293T cells were plated in DMEM medium supplemented with 10% FBS for 16 h. The culture medium was replaced by OptiMEM (Life Technology) without serum, then plasmid (5 μg/dish) was introduced using Fugene (Roche) according to the manufacturer’s recommendations. The transfection efficiency was routinely 80%.
Examination of PERK recognition as a substrate by PTP1B
To demonstrate direct dephosphorylation of PERK by PTP1B, WT and mutant (K618A and Y615F) forms of myc-tagged mouse PERK were overexpressed in HEK293T cells for 36h, after which cells (either untreated or treated with tunicamycin for 4h) were lysed on ice for 30 min. Overexpressed PERK was immunopurified with anti-myc antibody (9E10). After extensive washing, the immunoprecipitate was incubated with wild-type or C215S catalytically-inactive mutant forms of PTP1B (10 ng) for 30 minutes at room temperature, following which the beads were centrifuged at 3000 × g for 10 min, resolved by SDS-PAGE and immunoblotted with anti-phosphotyrosine antibody (4G10). For substrate trapping in vitro, lysates prepared from untreated control cells, or cells treated with tunicamycin (10 μg/ml) for 4h, were incubated overnight at 4°C with 20 μl His-tagged WT PTP1B and D181A PTP1B fusion protein coupled to beads (10 μg/μl) in the presence and absence of 1 mM pervanadate. After several washes, complexes were analyzed by immunoblotting. In order to examine the importance of phosphorylation of PERK for the interaction, immunopurified WT and Y615F PERK was also subjected to in vitro substrate trapping with WT PTP1B and D181A PTP1B as described above. For trapping in cells, WT and D181A mutant PTP1B were overexpressed in HEK293T cells for 36h, after which cells (either untreated or treated with tunicamycin for 4h) were lysed on ice for 30 min. The PTP was then immunopurified using anti-PTP1B antibody FG6. After extensive washing in lysis buffer, the precipitated proteins were examined by immunoblotting.
Rate of inactivation and reactivation of PTP1B
The rate of inactivation of PTP1B was measured by a standard spectrophotometric assay using p-nitrophenyl phosphate (p-NPP) as substrate. PTP1B (1 μM) was incubated at 25 °C with varying concentrations of H2S (1-200 μM), H2O2 (1–5 mM), or NO donor SNAP (1-20 mM) in 50 mM HEPES pH 7.0, 100 mM NaCl and 0.1 % BSA. Phosphatase activities were measured immediately in a continuous assay that monitors the production of nitrophenol at 405 nm (18000 M−1 cm−1). To calculate the rates of reactivation by DTT, GSH, or TR/TRR, PTP1B (10 μM) was initially inactivated by incubation with H2S (1 - 100 μM), H2O2 (0.5 – 2 mM), SNAP or GSNO (1 - 20 mM); aliquots were then diluted 2-fold into 50 mM HEPES pH 7.5, 100 mM NaCl, 0.1 % BSA, and 1 mM EDTA containing varying amounts of DTT (10 – 200 mM), GSH (50–200 mM), or TR/TRR (0.2–5 equivalents of TR to PTP1B). Following incubation under reducing conditions for varying amounts of time (1–60 min), aliquots were assayed for phosphatase activity using pNPP (2 mM). For assays using TR/TRR as the reductant, NADPH (25 - 300 μM) was also included with a ratio of TR to TRR of 200:1.
Fluorescence Polarization assay
Wild-type and mutant forms of PTP1B were incubated with or without 100 μM H2S for 10 minutes at room temperature and binding was measured with 25 nM substrate 5′-FAM-ENDpYINASL in 50 mM HEPES pH 7.0 and 100 mM NaCl, using the Spectramax Gemini xps fluorescence plate reader (Molecular devices). Excitation was set at 494 nm and emission was collected at 522 nm. Fluorescence polarization is plotted as fluorescence intensity (vertical)/fluorescence intensity (horizontal). As it is a ratio of intensities, the polarization value is dimensionless; it is expressed as arbitrary millipolarization units.
Cysteinyl labeling assay
H2S stock solution was prepared freshly by directly bubbling pure H2S gas into distilled water to generate a saturated solution (0.09M H2S at 30 °C) [54]. H2S stock solution was diluted to different concentrations in cell culture medium or appropriate buffer. Cells were grown to confluence and serum-starved for 16 h before stimulation with H2S. Following stimulation, cells were placed in an anaerobic work-station where the pO2 level was maintained at 1-2 ppm. After removing DMEM, cells were rapidly lysed with ice-cold degassed lysis buffer (25 mM sodium acetate pH 5.5, 1% Nonidet P-40, 150 mM NaCl, 10% (v/v) glycerol, 25 μg/ml aprotinin, 25 μg/ml leupeptin) and transferred to brown-colored tubes. Lysates were shaken for 1 h at room temperature to allow complete alkylation of free thiols. Cell debris was then cleared by centrifugation at 12,000 × g for 3 min at room temperature. Protein concentrations were determined by the method of Bradford, and 1 mg of cell lysate was slowly applied to desalting columns that had been equilibrated with IAA-free lysis buffer. Buffer exchange was performed by centrifuging at 2,000 × g for 2 min at 4°C. IAA-cleared lysates were then supplemented with 1 mM DTT and allowed to incubate for 30 min on a shaker at room temperature. During this phase, the persulfide and other reversibly oxidized forms of the active site Cys residue, which were protected from alkylation in the previous step, were reduced back to their thiolate states. The lysates were then incubated with biotinylated IAP probe (5 mM) for 1 h on a shaker at room temperature. Biotinylated proteins were enriched by using streptavidin-Sepharose beads for 16 h at 4°C on a rotating wheel, with sequential rounds of centrifugation (12,000 × g, 1 min, 4°C) using PBS to wash the beads. The beads were resuspended in 20 μl of 4×Laemmli sample buffer and heated at 90°C for 1 min. PTP1B was then identified by immunoblotting.
Measurement of PTP1B activity by immunophosphatase assay
Cells untreated or treated with tunicamycin (10 μg/ml) were lysed on ice using 20 mM Tris-HCl pH 7.5, 100 mM NaCl, 0.5% Nonidet P-40, supplemented with 5 μg/ml aprotinin, 5 μg/ml leupeptin, and 0.2 mm phenylmethylsulfonyl fluoride. PTP1B was immunoprecipitated from 500 μg of cell lysate with 2 μg of antibody DH8 for 90 min at 4°C, followed by incubation with protein G-Sepharose beads for 1 h. The immunoprecipitates were washed twice, first with lysis buffer, then PTP assay buffer (50 mM HEPES, pH 7.0, 100 mM NaCl, 2 mM EDTA and 0.1 mg/ml bovine serum albumin). Following immunoprecipitation, PTP1B was assayed using 10 mM p-nitrophenyl phosphate as substrate for 30 min at 25°C in the absence and presence of 5 mM DTT. The reaction was stopped by addition of NaOH to a final concentration of 1 M. The increase in absorbance at 405 nm was measured to determine the enzyme activity. Linearity of activity was demonstrated using different amounts of the immunoprecipitate. The values in the absence of enzyme were taken as background and were always subtracted for correction. Data are presented as % phosphatase activity, in which the activity in the immunoprecipitate is expessed relative to that of an equal amount of purified recombinant PTP1B.
H2S measurement
H2S was measured as described previously [54, 55]. Cells were lysed in 50 mM phosphate buffer pH 7.5 with zinc acetate (1% w/v) to trap H2S. The reaction was stopped after 10 min by adding 20 μM N,N-dimethyl-p-phenylenediamine sulfate and 30 mM FeCl3, incubated in the dark for 30 min, and 10% trichloroacetic acid was added to precipitate any protein. Subsequently, the mixture was centrifuged at 10,000 × g for 10 min and absorbance of the supernatant at 670 nm was measured. H2S concentration was calculated from a calibration curve of standard H2S solutions. Data are presented as percent increase in H2S relative to untreated cells.
Mass Spectrometry
Recombinant PTP1B proteins (~20 μg each of WT, H2S-treated, and H2O2-treated) were digested overnight with trypsin (1:50 w/w) at 37°C. Tryptic peptides were acidified by the addition of formic acid (0.1% v/v final), and aliquots of 2 μl peptide samples were injected onto a microfluidic RPLC chip (43mm × 75μm C18 chip column with 40 nL trap column) and acquired on a nano-LC QTOF system (Agilent QTOF 6520). Both data-dependent and targeted acquisition modes were applied to PTP1B peptides. Peaklist files were generated with MassHunter software (version B.02.00) and searched against the IPI human database (87,130 sequences) using MASCOT v2.3 [56]. Manual interpretation was also applied to validate peptide spectra. For the time-course study in tunicamycin-treated cells, selected ion monitoring (SIM) was used to quantify specifically the native and modified Cys215-containing peptides. Chromatography peak detection, integration and quantitation were performed with MassHunter software. For intact protein mass measurement, proteins were first buffer-exchanged into 20 mM triethylammonium bicarbonate buffer with a PD10 column. Protein samples (~1 μg) were injected onto a customized C8 protein chip (43mm × 75μm C8 chip column with 40 nL trap column) and acquired on the QTOF 6520. Acquisition parameters were the same as for peptide MS, except that data were acquired in profile mode and the fragmentor voltage was set at 200V. Intact protein spectra were integrated and deconvoluted with the maximum entropy workflow in the Bioconfirm software. For examination of sulfhydration of Cys215-containing peptides in PTP1B from H2S-treated 293T cells, the samples were analyzed using an Orbitrap Velos mass spectrometer (Thermo) using capillary RPHPLC and acquisition parameters essentially as described in [57].
PTP1B substrate identification
Immunopurified protein complexes from the substrate-trapping experiments were treated with 5 mM vanadate to release the bound substrates and subjected to trypsin digestion overnight at 37°C after reduction with DTT and alkylation with iodoacetamide [58]. The digested peptides were then acidified by 0.1% v/v TFA and desalted by C18 stagetip [59]. Peptides samples were then dried and resuspended in loading buffer (5% ACN and 0.1% FA). Peptides were identified by a four-step multi-dimensional protein identification technology (MUDPIT) [60] at ammonium acetate salt concentrations of 10%, 45%, 60% and 100% respectively, each followed by a 120 minute RPLC gradient on a Proxeon nano LC system. Data were acquired on a LTQ-Orbitrap XL with a survey scan at 30,000 resolution and followed by 6 data-dependent CID MS2 scans at a resolution of 7,500, 10 ms activation time, NCE at 25, and q at 0.25. Peaklist files were generated by Mascot Distiller (v2.3) and searched against the IPI human database (87,130 sequences) using MASCOT v2.3. Search parameters are 15 ppm for MS1 and 0.6 Da for MS2, carbamidomethyl (Cys) as fixed modification, with methionine oxidation and tyrosine phosphorylation as variable modifications. Peptide scores with p-value below 0.05 are considered as confident identifications.
Statistical analysis
Immunoblotting was performed by standard methods. All blots are representative of experiments that were performed at least three times. In figures where blots where quantitated, we used ImageJ software and have represented the data as a bar graph (+/− standard error of mean (SEM)).
Supplementary Material
Figure S1. Time-dependent inactivation of PTP1B by H2O2.
The phosphatase activity of PTP1B was monitored in the presence of H2O2, 0.5 mM (●), 1 mM (■), 1.5 mM (▲), 2 mM (▼). Inset, the concentration dependence of the rate of inactivation was used to derive the second order rate constant, 10 ± 1.4 M−1s−1.
Table S1. Quantitation of the different redox forms of Cys215 in PTP1B observed following induction of ER stress with tunicamycin.
Figure S2. Time-dependent inactivation of PTP1B by NO.
The phosphatase activity of PTP1B was monitored in the presence of GSNO, 1 mM (●), 2 mM (■), 5 mM (▲), 10 mM (▼), 20 mM (◆). Inset, the concentration dependence of the rate of inactivation was used to derive the second order rate constant, 2.1 ± 0.5 M−1s−1.
Figure S3. Time-dependent reactivation of PTP1B by TR/TRR.
NADPH was used at 10 μM (○), 25 μM (◆), 50 μM (▼), 150 μM (▲) or 300 μM (●). Inset: the second order rate constant, 45.5 ± 2.8 M−1s−1, was derived from the concentration dependence of the rate of inactivation. Data are representative of three independent determinations.
Figure S4. Time-dependent reactivation of PTP1B by GSH.
GSH was used at 50 mM (●), 75 mM (■), 100 mM (▲), or 200 (▼) mM. Inset: the second order rate constant, 0.013 ± 0.012 M−1s−1, was derived from the concentration dependence of the rate of inactivation. Data are representative of three independent determinations.
Figure S5. Mechanism of PTP labeling by the IAP probe.
Schematic mechanism of PTP reactivity towards a biotinylated iodoacetyl-polyethylene glycol(IAP) probe Adapted from [29]
Figure S6 Induction of ER stress by tunicamycin.
Representative immunoblots of cystathionine-γ-lyase (CSE), PTP1B, and the ER chaperone BiP from cell lysates following exposure to tunicamycin (10 μg/ml) for 0, 2, 4, 6, or 8 hours. The blots of actin were included as loading controls.
Figure S7 Decrease in cystathionine-γ-lyase (CSE) by RNAi.
Representative immunoblot to show the abundance of cystathionine-γ-lyase (CSE) in cells expressing specific shRNAs (shRNA1 and shRNA2) compared to control luciferase shRNA-expressing cells. The blots of actin were included as loading controls.
Figure S8 Changes in Components of the Unfolded Protein Response (UPR) following exposure to thapsigargin. ER stress was induced by thapsigargin (TG, 1 μM) for 0, 2, 4, 6, or 8 hours. Immunoblot showing activation of components of the UPR.. The data illustrate a time course, over 0-8 h, to examine activation of PERK [anti-phospho-PERK (Thr980) compared to total PERK (anti-PERK)], and the activation of IRE1 [anti-phospho-IRE1α (Ser724) compared to total IRE1 (anti-IRE1α)]in control and CSE-deficient cells. There was no obvious difference in the abundance of ATF6.
Figure S9 Proposed mechanism for persulfide modification of PTP1B. The thiolate anion (Cys215) in the active site of PTP1B is highly susceptible to oxidation [39]. Upon oxidation, it forms a stable cyclic sulphenamide intermediate, which has been postulated to promote reversibility of oxidation [39]. Higher oxidation, to sulfinic and sulfonic acid, generates irreversibly oxidized forms of PTP1B. Reaction of H2S with the cyclic sulphenamide would be expected to generate the persulfide modification we detected, which could protect PTP1B from higher irreversible oxidation and may also promote reactivation through thioredoxin. Alternatively H2S could also form a persulfide on reaction with hydrogen peroxide, which could react directly with reduced PTP1B.
ONE SENTENCE SUMMARY.
We demonstrate that the protein tyrosine phosphatase PTP1B is reversibly sulfhydrated and inactivated by the “gasotransmitter” H2S in response to ER stress, which transiently protects pTyr 619 of PERK from dephosphorylation by PTP1B, promoting activation of PERK and its ability to inhibit global translation.
ACKNOWLEDGMENTS
We thank Cristian Ruse and the members of the CSHL Proteomics Shared Resource for their invaluable assistance.
Funding: This work was supported by grants CA53840 and GM55989 from NIH (to NKT) and the CSHL Cancer Centre Support Grant CA45508. The authors are also grateful for support from the Irving Hansen Foundation and the Masthead Cove Yacht Club Carol Marcincuk Fund. PTP1B inhibitor MSI-1436 was provided by Ohr Pharmaceuticals and Genaera Corporation.
Footnotes
AUTHOR CONTRIBUTIONS
N.K. and N.K.T. designed the experiments and analyzed the data. N.K. performed the experiments. C.F. and D.P. were responsible for the mass spectrometry analysis. N.K. and N.K.T. wrote the manuscript, with contributions from C.F. and D.P.
COMPETING INTERESTS
The authors have no competing interests.
REFERENCES
- 1.Wang R. Two’s company, three’s a crowd: can H2S be the third endogenous gaseous transmitter? The FASEB journal: official publication of the Federation of American Societies for Experimental Biology. 2002;16(13):1792–8. doi: 10.1096/fj.02-0211hyp. [DOI] [PubMed] [Google Scholar]
- 2.Foster MW, Hess DT, Stamler JS. Protein S-nitrosylation in health and disease: a current perspective. Trends in molecular medicine. 2009;15(9):391–404. doi: 10.1016/j.molmed.2009.06.007. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Mancardi D, et al. Old and New Gasotransmitters in the Cardiovascular System: Focus on the Role of Nitric Oxide and Hydrogen Sulfide in Endothelial Cells and Cardiomyocytes. Current pharmaceutical biotechnology. 2011 doi: 10.2174/138920111798281090. [DOI] [PubMed] [Google Scholar]
- 4.Wang R. Hydrogen sulfide: the third gasotransmitter in biology and medicine. Antioxidants & redox signaling. 2010;12(9):1061–4. doi: 10.1089/ars.2009.2938. [DOI] [PubMed] [Google Scholar]
- 5.Boehning D, Snyder SH. Novel neural modulators. Annual review of neuroscience. 2003;26:105–31. doi: 10.1146/annurev.neuro.26.041002.131047. [DOI] [PubMed] [Google Scholar]
- 6.Gadalla MM, Snyder SH. Hydrogen sulfide as a gasotransmitter. Journal of neurochemistry. 2010;113(1):14–26. doi: 10.1111/j.1471-4159.2010.06580.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Kabil O, Banerjee R. Redox biochemistry of hydrogen sulfide. The Journal of biological chemistry. 2010;285(29):21903–7. doi: 10.1074/jbc.R110.128363. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Szabo C. Hydrogen sulphide and its therapeutic potential. Nature reviews. Drug discovery. 2007;6(11):917–35. doi: 10.1038/nrd2425. [DOI] [PubMed] [Google Scholar]
- 9.Sakurai H, et al. Inorganic sulfur oxidizing system in green sulfur bacteria. Photosynthesis research. 2010;104(2-3):163–76. doi: 10.1007/s11120-010-9531-2. [DOI] [PubMed] [Google Scholar]
- 10.Lee HS, Hwang BJ. Methionine biosynthesis and its regulation in Corynebacterium glutamicum: parallel pathways of transsulfuration and direct sulfhydrylation. Applied microbiology and biotechnology. 2003;62(5-6):459–67. doi: 10.1007/s00253-003-1306-7. [DOI] [PubMed] [Google Scholar]
- 11.Kimura H. Hydrogen sulfide: from brain to gut. Antioxidants & redox signaling. 2010;12(9):1111–23. doi: 10.1089/ars.2009.2919. [DOI] [PubMed] [Google Scholar]
- 12.Baskar R, Bian J. Hydrogen sulfide gas has cell growth regulatory role. European journal of pharmacology. 2011;656(1-3):5–9. doi: 10.1016/j.ejphar.2011.01.052. [DOI] [PubMed] [Google Scholar]
- 13.Wang R. Hydrogen sulfide: a new EDRF. Kidney international. 2009;76(7):700–4. doi: 10.1038/ki.2009.221. [DOI] [PubMed] [Google Scholar]
- 14.Liu YY, et al. H2S releasing aspirin protects amyloid beta induced cell toxicity in BV-2 microglial cells. Neuroscience. 2011;193:80–8. doi: 10.1016/j.neuroscience.2011.07.023. [DOI] [PubMed] [Google Scholar]
- 15.Caliendo G, et al. Synthesis and biological effects of hydrogen sulfide (H2S): development of H2S-releasing drugs as pharmaceuticals. Journal of medicinal chemistry. 2010;53(17):6275–86. doi: 10.1021/jm901638j. [DOI] [PubMed] [Google Scholar]
- 16.Blackstone E, Morrison M, Roth MB. H2S induces a suspended animation-like state in mice. Science. 2005;308(5721):518. doi: 10.1126/science.1108581. [DOI] [PubMed] [Google Scholar]
- 17.Mustafa AK, et al. H2S signals through protein S-sulfhydration. Science signaling. 2009;2(96):ra72. doi: 10.1126/scisignal.2000464. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Tonks NK. Protein tyrosine phosphatases: from genes, to function, to disease. Nature reviews. Molecular cell biology. 2006;7(11):833–46. doi: 10.1038/nrm2039. [DOI] [PubMed] [Google Scholar]
- 19.Tonks NK. Redox redux: revisiting PTPs and the control of cell signaling. Cell. 2005;121(5):667–70. doi: 10.1016/j.cell.2005.05.016. [DOI] [PubMed] [Google Scholar]
- 20.Chen YY, et al. Cysteine S-nitrosylation protects protein-tyrosine phosphatase 1B against oxidation-induced permanent inactivation. The Journal of biological chemistry. 2008;283(50):35265–72. doi: 10.1074/jbc.M805287200. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Tonks NK. PTP1B: from the sidelines to the front lines. FEBS letters. 2003;546(1):140–8. doi: 10.1016/s0014-5793(03)00603-3. [DOI] [PubMed] [Google Scholar]
- 22.Cook WS, Unger RH. Protein tyrosine phosphatase 1B: a potential leptin resistance factor of obesity. Developmental cell. 2002;2(4):385–7. doi: 10.1016/s1534-5807(02)00158-2. [DOI] [PubMed] [Google Scholar]
- 23.Tonks NK, Muthuswamy SK. A brake becomes an accelerator: PTP1B--a new therapeutic target for breast cancer. Cancer cell. 2007;11(3):214–6. doi: 10.1016/j.ccr.2007.02.022. [DOI] [PubMed] [Google Scholar]
- 24.Lessard L, Stuible M, Tremblay ML. The two faces of PTP1B in cancer. Biochimica et biophysica acta. 2010;1804(3):613–9. doi: 10.1016/j.bbapap.2009.09.018. [DOI] [PubMed] [Google Scholar]
- 25.Dube N, Tremblay ML. Beyond the metabolic function of PTP1B. Cell cycle. 2004;3(5):550–3. [PubMed] [Google Scholar]
- 26.Gu F, et al. Protein-tyrosine phosphatase 1B potentiates IRE1 signaling during endoplasmic reticulum stress. The Journal of biological chemistry. 2004;279(48):49689–93. doi: 10.1074/jbc.C400261200. [DOI] [PubMed] [Google Scholar]
- 27.Flint AJ, et al. Development of “substrate-trapping” mutants to identify physiological substrates of protein tyrosine phosphatases. Proceedings of the National Academy of Sciences of the United States of America. 1997;94(5):1680–5. doi: 10.1073/pnas.94.5.1680. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Barford D, Flint AJ, Tonks NK. Crystal structure of human protein tyrosine phosphatase 1B. Science. 1994;263(5152):1397–404. [PubMed] [Google Scholar]
- 29.Boivin B, et al. A modified cysteinyl-labeling assay reveals reversible oxidation of protein tyrosine phosphatases in angiomyolipoma cells. Proceedings of the National Academy of Sciences of the United States of America. 2008;105(29):9959–64. doi: 10.1073/pnas.0804336105. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Wei H, et al. Hydrogen sulfide attenuates hyperhomocysteinemia-induced cardiomyocytic endoplasmic reticulum stress in rats. Antioxidants & redox signaling. 2010;12(9):1079–91. doi: 10.1089/ars.2009.2898. [DOI] [PubMed] [Google Scholar]
- 31.Bjorge JD, Pang A, Fujita DJ. Identification of protein-tyrosine phosphatase 1B as the major tyrosine phosphatase activity capable of dephosphorylating and activating c-Src in several human breast cancer cell lines. The Journal of biological chemistry. 2000;275(52):41439–46. doi: 10.1074/jbc.M004852200. [DOI] [PubMed] [Google Scholar]
- 32.Zhu S, Bjorge JD, Fujita DJ. PTP1B contributes to the oncogenic properties of colon cancer cells through Src activation. Cancer research. 2007;67(21):10129–37. doi: 10.1158/0008-5472.CAN-06-4338. [DOI] [PubMed] [Google Scholar]
- 33.Lantz KA, et al. Inhibition of PTP1B by trodusquemine (MSI-1436) causes fat-specific weight loss in diet-induced obese mice. Obesity. 2010;18(8):1516–23. doi: 10.1038/oby.2009.444. [DOI] [PubMed] [Google Scholar]
- 34.Mori K. Signalling pathways in the unfolded protein response: development from yeast to mammals. Journal of biochemistry. 2009;146(6):743–50. doi: 10.1093/jb/mvp166. [DOI] [PubMed] [Google Scholar]
- 35.Harding HP, et al. Perk is essential for translational regulation and cell survival during the unfolded protein response. Molecular cell. 2000;5(5):897–904. doi: 10.1016/s1097-2765(00)80330-5. [DOI] [PubMed] [Google Scholar]
- 36.Su Q, et al. Modulation of the eukaryotic initiation factor 2 alpha-subunit kinase PERK by tyrosine phosphorylation. The Journal of biological chemistry. 2008;283(1):469–75. doi: 10.1074/jbc.M704612200. [DOI] [PubMed] [Google Scholar]
- 37.Ron D, Walter P. Signal integration in the endoplasmic reticulum unfolded protein response. Nature reviews. Molecular cell biology. 2007;8(7):519–29. doi: 10.1038/nrm2199. [DOI] [PubMed] [Google Scholar]
- 38.Jiang B, et al. Molecular mechanism for H(2)S-induced activation of K(ATP) channels. Antioxidants & redox signaling. 2010;12(10):1167–78. doi: 10.1089/ars.2009.2894. [DOI] [PubMed] [Google Scholar]
- 39.Salmeen A, et al. Redox regulation of protein tyrosine phosphatase 1B involves a sulphenyl-amide intermediate. Nature. 2003;423(6941):769–73. doi: 10.1038/nature01680. [DOI] [PubMed] [Google Scholar]
- 40.Tjernberg A. Hallen, Dan, Schultz, Johan, James, Stephen, Benkestock, Kurt, Bystrom, Styrbjorn, Weigelt, Johan, Mechanisms of action of pyridazine analogues on protein tyrosine phosphatase 1B (PTP1B) Biorganic & Medicinal Chemistry Letters. 2004;14:891–895. doi: 10.1016/j.bmcl.2003.12.014. [DOI] [PubMed] [Google Scholar]
- 41.Carballal S, et al. Reactivity of hydrogen sulfide with peroxynitrite and other oxidants of biological interest. Free radical biology & medicine. 2011;50(1):196–205. doi: 10.1016/j.freeradbiomed.2010.10.705. [DOI] [PubMed] [Google Scholar]
- 42.Yang G, et al. H2S, endoplasmic reticulum stress, and apoptosis of insulin-secreting beta cells. The Journal of biological chemistry. 2007;282(22):16567–76. doi: 10.1074/jbc.M700605200. [DOI] [PubMed] [Google Scholar]
- 43.Yoshida H. ER stress and diseases. The FEBS journal. 2007;274(3):630–58. doi: 10.1111/j.1742-4658.2007.05639.x. [DOI] [PubMed] [Google Scholar]
- 44.Kohno K. Stress-sensing mechanisms in the unfolded protein response: similarities and differences between yeast and mammals. Journal of biochemistry. 2010;147(1):27–33. doi: 10.1093/jb/mvp196. [DOI] [PubMed] [Google Scholar]
- 45.Madeo F, Kroemer G. Intricate links between ER stress and apoptosis. Molecular cell. 2009;33(6):669–70. doi: 10.1016/j.molcel.2009.03.002. [DOI] [PubMed] [Google Scholar]
- 46.Groenendyk J, Michalak M. Endoplasmic reticulum quality control and apoptosis. Acta biochimica Polonica. 2005;52(2):381–95. [PubMed] [Google Scholar]
- 47.Shen X, et al. Genetic interactions due to constitutive and inducible gene regulation mediated by the unfolded protein response in C. elegans. PLoS genetics. 2005;1(3):e37. doi: 10.1371/journal.pgen.0010037. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Su Q, et al. Tyrosine phosphorylation acts as a molecular switch to full-scale activation of the eIF2alpha RNA-dependent protein kinase. Proceedings of the National Academy of Sciences of the United States of America. 2006;103(1):63–8. doi: 10.1073/pnas.0508207103. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Salmeen A, et al. Molecular basis for the dephosphorylation of the activation segment of the insulin receptor by protein tyrosine phosphatase 1B. Molecular cell. 2000;6(6):1401–12. doi: 10.1016/s1097-2765(00)00137-4. [DOI] [PubMed] [Google Scholar]
- 50.Garaud M, Pei D. Substrate profiling of protein tyrosine phosphatase PTP1B by screening a combinatorial peptide library. Journal of the American Chemical Society. 2007;129(17):5366–7. doi: 10.1021/ja071275i. [DOI] [PubMed] [Google Scholar]
- 51.Bettaieb A, et al. Differential regulation of endoplasmic reticulum stress by protein tyrosine phosphatase 1B and T cell protein tyrosine phosphatase. The Journal of biological chemistry. 2011;286(11):9225–35. doi: 10.1074/jbc.M110.186148. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Santos CX, et al. Mechanisms and implications of reactive oxygen species generation during the unfolded protein response: roles of endoplasmic reticulum oxidoreductases, mitochondrial electron transport, and NADPH oxidase. Antioxidants & redox signaling. 2009;11(10):2409–27. doi: 10.1089/ars.2009.2625. [DOI] [PubMed] [Google Scholar]
- 53.Malhotra JD, Kaufman RJ. Endoplasmic reticulum stress and oxidative stress: a vicious cycle or a double-edged sword? Antioxidants & redox signaling. 2007;9(12):2277–93. doi: 10.1089/ars.2007.1782. [DOI] [PubMed] [Google Scholar]
- 54.Zhao W, et al. The vasorelaxant effect of H(2)S as a novel endogenous gaseous K(ATP) channel opener. The EMBO journal. 2001;20(21):6008–16. doi: 10.1093/emboj/20.21.6008. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Siegel LM. A Direct Microdetermination for Sulfide. Analytical biochemistry. 1965;11:126–32. doi: 10.1016/0003-2697(65)90051-5. [DOI] [PubMed] [Google Scholar]
- 56.Perkins DN, et al. Probability-based protein identification by searching sequence databases using mass spectrometry data. Electrophoresis. 1999;20(18):3551–67. doi: 10.1002/(SICI)1522-2683(19991201)20:18<3551::AID-ELPS3551>3.0.CO;2-2. [DOI] [PubMed] [Google Scholar]
- 57.Obad S, et al. Silencing of microRNA families by seed-targeting tiny LNAs. Nature genetics. 2011;43(4):371–8. doi: 10.1038/ng.786. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58.Chang YC, et al. Tyrosine phosphoproteomics and identification of substrates of protein tyrosine phosphatase dPTP61F in Drosophila S2 cells by mass spectrometry-based substrate trapping strategy. Journal of proteome research. 2008;7(3):1055–66. doi: 10.1021/pr700801p. [DOI] [PubMed] [Google Scholar]
- 59.Rappsilber J, Mann M, Ishihama Y. Protocol for micro-purification, enrichment, pre-fractionation and storage of peptides for proteomics using StageTips. Nature protocols. 2007;2(8):1896–906. doi: 10.1038/nprot.2007.261. [DOI] [PubMed] [Google Scholar]
- 60.Washburn MP, Wolters D, Yates JR., 3rd Large-scale analysis of the yeast proteome by multidimensional protein identification technology. Nature biotechnology. 2001;19(3):242–7. doi: 10.1038/85686. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Figure S1. Time-dependent inactivation of PTP1B by H2O2.
The phosphatase activity of PTP1B was monitored in the presence of H2O2, 0.5 mM (●), 1 mM (■), 1.5 mM (▲), 2 mM (▼). Inset, the concentration dependence of the rate of inactivation was used to derive the second order rate constant, 10 ± 1.4 M−1s−1.
Table S1. Quantitation of the different redox forms of Cys215 in PTP1B observed following induction of ER stress with tunicamycin.
Figure S2. Time-dependent inactivation of PTP1B by NO.
The phosphatase activity of PTP1B was monitored in the presence of GSNO, 1 mM (●), 2 mM (■), 5 mM (▲), 10 mM (▼), 20 mM (◆). Inset, the concentration dependence of the rate of inactivation was used to derive the second order rate constant, 2.1 ± 0.5 M−1s−1.
Figure S3. Time-dependent reactivation of PTP1B by TR/TRR.
NADPH was used at 10 μM (○), 25 μM (◆), 50 μM (▼), 150 μM (▲) or 300 μM (●). Inset: the second order rate constant, 45.5 ± 2.8 M−1s−1, was derived from the concentration dependence of the rate of inactivation. Data are representative of three independent determinations.
Figure S4. Time-dependent reactivation of PTP1B by GSH.
GSH was used at 50 mM (●), 75 mM (■), 100 mM (▲), or 200 (▼) mM. Inset: the second order rate constant, 0.013 ± 0.012 M−1s−1, was derived from the concentration dependence of the rate of inactivation. Data are representative of three independent determinations.
Figure S5. Mechanism of PTP labeling by the IAP probe.
Schematic mechanism of PTP reactivity towards a biotinylated iodoacetyl-polyethylene glycol(IAP) probe Adapted from [29]
Figure S6 Induction of ER stress by tunicamycin.
Representative immunoblots of cystathionine-γ-lyase (CSE), PTP1B, and the ER chaperone BiP from cell lysates following exposure to tunicamycin (10 μg/ml) for 0, 2, 4, 6, or 8 hours. The blots of actin were included as loading controls.
Figure S7 Decrease in cystathionine-γ-lyase (CSE) by RNAi.
Representative immunoblot to show the abundance of cystathionine-γ-lyase (CSE) in cells expressing specific shRNAs (shRNA1 and shRNA2) compared to control luciferase shRNA-expressing cells. The blots of actin were included as loading controls.
Figure S8 Changes in Components of the Unfolded Protein Response (UPR) following exposure to thapsigargin. ER stress was induced by thapsigargin (TG, 1 μM) for 0, 2, 4, 6, or 8 hours. Immunoblot showing activation of components of the UPR.. The data illustrate a time course, over 0-8 h, to examine activation of PERK [anti-phospho-PERK (Thr980) compared to total PERK (anti-PERK)], and the activation of IRE1 [anti-phospho-IRE1α (Ser724) compared to total IRE1 (anti-IRE1α)]in control and CSE-deficient cells. There was no obvious difference in the abundance of ATF6.
Figure S9 Proposed mechanism for persulfide modification of PTP1B. The thiolate anion (Cys215) in the active site of PTP1B is highly susceptible to oxidation [39]. Upon oxidation, it forms a stable cyclic sulphenamide intermediate, which has been postulated to promote reversibility of oxidation [39]. Higher oxidation, to sulfinic and sulfonic acid, generates irreversibly oxidized forms of PTP1B. Reaction of H2S with the cyclic sulphenamide would be expected to generate the persulfide modification we detected, which could protect PTP1B from higher irreversible oxidation and may also promote reactivation through thioredoxin. Alternatively H2S could also form a persulfide on reaction with hydrogen peroxide, which could react directly with reduced PTP1B.