Abstract
Using a newly developed competitive binding assay dependent upon the reassembly of a split reporter protein, we have tested the promiscuity of a panel of reported kinase inhibitors against the AGC group. Many non-AGC targeted kinase inhibitors target multiple members of the AGC group. In general, structurally similar inhibitors consistently exhibited activity toward the same target as well as toward closely related kinases. The inhibition data was analyzed to test the predictive value of either using identity scores derived from residues within 6 Å of the active site or identity scores derived from the entire kinase domain. The results suggest that the active site identity in certain cases may be a stronger predictor of inhibitor promiscuity. The overall results provide general guidelines for establishing inhibitor selectivity, as well as for the future design of inhibitors that either target or avoid AGC kinases.
Introduction
Protein kinases are characterized by their ability to specifically phosphorylate the hydroxyl group of serine, threonine, or tyrosine residues on client proteins, thereby affecting almost all intracellular signal transduction pathways. More than 500 protein kinases comprise the human kinome1 and many kinases have been extensively targeted with small molecule inhibitors as therapeutics for the treatment of disease and also for the development of reagents for elucidating the function of a particular kinase in a signaling pathway.2 The high degree of similarity among kinases often results in off-target inhibition, which can be a significant impediment for correctly interpreting a small molecule’s effect on signal transduction3 as well as resulting in undesirable side-effects in therapeutic applications. Thus there is continued interest in the assessment of the selectivity of small molecule inhibitors to afford appropriately selective biological probes and therapeutics.
The human kinome is commonly divided into seven major groups, based primarily upon function and sequence identity, one of which is the serine/threonine group of AGC kinases.1 The AGC group of protein kinases consists of 60 related proteins and is so named for three key members: cAMP-dependent protein kinase catalytic subunit alpha (PRKACA; also known as PKA), cGMP-dependent protein kinase 1 (PKG1), and protein kinase C (PKC).4,5 As is common among kinases, members of this group are involved in the regulation of cell proliferation, differentiation, and survival. Many of the AGCs are believed to phosphorylate a large number of substrates in vivo, and they play diverse roles in signaling, from the phosphorylation of BCL2-antagonist of cell death to prevent the activation of the apoptotic pathway,6 to the direct control of gene regulation through phosphorylation of transcription factor forkhead box O.7 The consensus substrate motifs recognized by each of the AGC kinases tend to be quite similar within the group, and this redundancy perhaps exists to allow various extra-cellular stimuli to modulate the same downstream effect through different mechanisms.5
A number of AGC kinases have emerged as potential therapeutic drug targets for the treatment of cancer and diabetes.5 Oncogenic mutations resulting in the increased activity of both AKT1 and PDPK1 have been shown to play a role in the survival of certain cancers.8–10 Recent years have seen a push toward multi-kinase targeted inhibitors,11 but the off-target inhibition of kinases critical to normal cellular function can have significant negative consequences.12 For example, the inhibition of AMP-activated protein kinase by sunitinib, a multi-target tyrosine kinase inhibitor used in the treatment of a number of solid tumors, has recently been implicated in cardiotoxic side effects associated with its use.13 Adverse side effects caused by off-target interactions are perhaps acceptable for the short-term treatment of cancer,14 however, long-term therapies will likely require improved selectivity in order to minimize undesirable side effects.
A number of recent publications have detailed significant strides toward screening kinase inhibitors against increasingly larger portions of the kinome. More thorough preclinical screens can be expected to improve clinical outcomes,12 enhance the ability of medicinal chemists to design optimally selective therapeutics,11 and aid in the identification of truly selective small molecule probes for in vivo signal transduction studies. Seminal papers by Cohen and coworkers represent some of the earliest efforts toward developing more complete selectivity profiles of commonly used signal transduction reagents.3,15,16 More recently, several datasets of small molecules profiled against kinase panels have been published by Ambit Biosciences,17,18 GlaxoSmithKline,19,20 and Abbott Laboratories.21 While the Ambit results focused primarily on generating comprehensive selectivity profiles for already characterized kinase inhibitors and therapeutics,17,18 the studies from GlaxoSmithKline and Abbott laboratories sought to identify characteristics common to kinase inhibitors and what types of chemical scaffolds afford the ability to target different, distally related kinases, with particular focus upon the tyrosine kinases.19–21 Taken together, these efforts represent a major step in painting a clearer picture of kinase pharmacology.
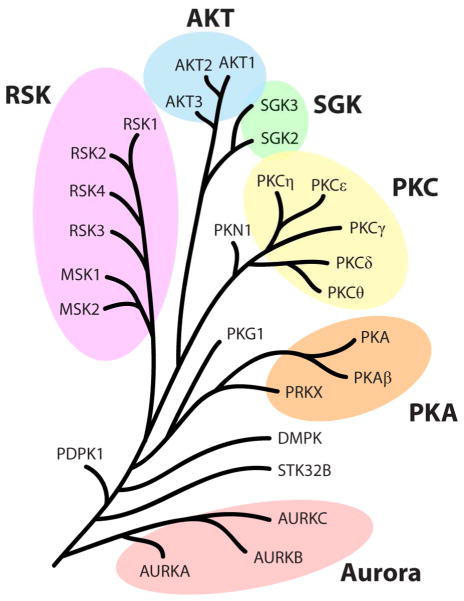
Many commercially available small molecule sets are used to dissect signal transduction pathways, though their potential off-target effects have not been systematically investigated. Herein we seek to improve the knowledge base regarding kinase inhibitor selectivity, particularly with regard to understanding potential off target effects against the AGC family. To this end we have screened a library of 80 previously characterized kinase inhibitors against a panel of 27 protein kinases. This panel was comprised of 23 AGC kinases as well as the three Aurora kinase isoforms and STK32B because of their relatively high identity to this group (Figure 1). Of the 80 compounds tested, only 10 of them have been reported to selectively target members of the AGC group. We employed a recently reported cell-free kinase inhibition assay which relies upon competitive active-site interactions to effect luminescence generation.22 This method allows for the rapid interrogation of many kinases without first having to optimize recombinant protein expression or identify substrates for poorly studied kinases. The selectivities of each compound were evaluated by examining how similarly structured small molecules affected highly similar kinases. In order to appraise the relationship between kinase identity and inhibitor promiscuity, kinase identity groups of either the kinase domain or only active-site residues were scored for inhibition frequency and compared between identity groups.
Figure 1.
A dendrogram of the 27 protein kinases screened in this study. Six families are highlighted.
Results and Discussion
Kinase Library Construction and Screening Assay
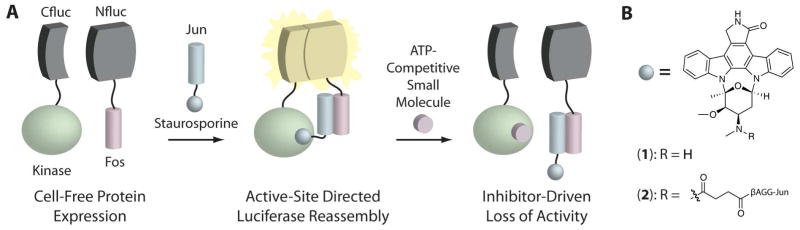
In order to utilize the aforementioned competitive binding assay, each kinase was prepared by first fusing the protein kinase domain of 27 kinases to the C-terminal half of firefly luciferase (Cfluc) through a 13-residue linker (Supporting Information, Table S1). Only the kinase domain and the AGC C-terminal domain,23 where relevant, were included for these constructs. Because we were interested in interactions at the active site of the kinases, and in particular the ATP-binding site, peripheral domains were excluded to prevent potential interference. Several of the kinases used in this study contain two kinase domains, namely the ribosomal protein S6 kinases (RSKs), and in these instances only the N-terminal kinase domain was attached to the appropriate luciferase half. A second construct consisting of the complementary N-terminal half of luciferase (Nfluc) was attached to the coiled-coil Fos and translated in reticulocyte lysate alongside each Cfluc-kinase chimera. The Jun peptide, which binds Fos, was conjugated to an ATP-competitive kinase inhibitor, a staurosporine analog, and added to a mixture of these two proteins, resulting in increased luminescence due to a functional ternary complex (Figure 2A). Because of its promiscuity, staurosporine (1) provides an ideal active-site anchor, allowing us to interrogate any kinase that binds our modified staurosporine conjugated to Jun (2).24,25 Following the formation of the light-generating ternary complex, the addition of free kinase inhibitors targeting the ATP-binding site could be used to outcompete staurosporine binding, resulting in a loss of luminescence. A library of 80 common kinase inhibitors, designed to target a diverse array of kinases, was screened against each of the 27 kinases in a 96-well format to identify potential interactions. All inhibitors were tested at a final concentration of 10 μM in order to qualitatively generate selectivity profiles for each small molecule against the AGC group of kinases. The extent to which luminescent signal was abrogated by the addition of a compound was tabulated as percent inhibition values; a higher percent inhibition means a greater relative loss of luminescence. A full table of all of the results can be found in the Supplementary Information (Table S2).
Figure 2.
Scheme of the competitive binding assay using split-luciferase reassembly. (A) A protein kinase and the Fos peptide were fused to complementary halves of firefly luciferase, Cfluc and Nfluc respectively. The addition of a Jun-staurosporine conjugate leads to luciferase activation. Small molecules capable of binding in the active site of the kinase prevent complex formation and result in a loss of luminescence. (B) Structures of staurosporine (1) and its modified analog attached to Jun (2).
Nonselective Kinase Inhibitors
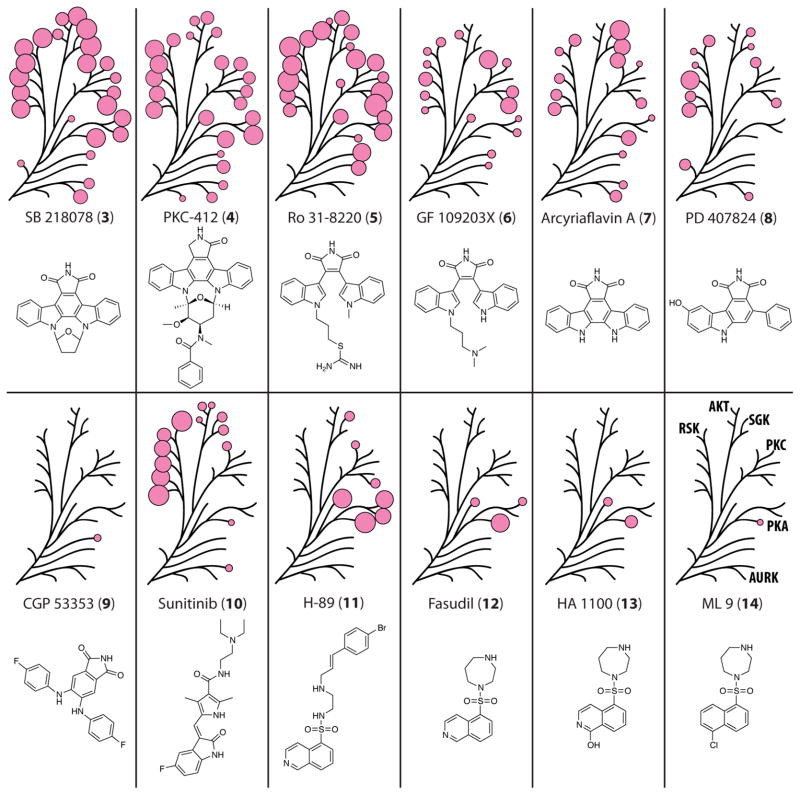
Several of the small molecules screened in this panel were quite promiscuous and were found to have activity against a relatively large fraction of the kinases tested. Many of these “nonselective” inhibitors share very similar structural elements to 1, containing a bisindolylmaleimide or indolocarbazole scaffold. Interest in these structural motifs has not waned as can be seen from recent drug discovery efforts by Novartis26 and ArQule.27 Two inhibitors, SB 218078 (3) and PKC-412 (4), possess the most staurosporine-like structural features and were also the most promiscuous compounds in this set (Figure 3). Interestingly, 3 is marketed as being a selective inhibitor of checkpoint kinase (CHK1),28 and 4, also known as midostaurin, is currently in phase III clinical trials for the treatment several cancer types.14 Every kinase in the panel was inhibited at least 20% by one or both of these compounds. Most of the kinases were inhibited relatively equally by both compounds, but some of them demonstrated a preference for one over the other. For example, 4 was much more active against PKG1 and STK32B, while SGK2 showed >60% more inhibition by 3.
Figure 3.
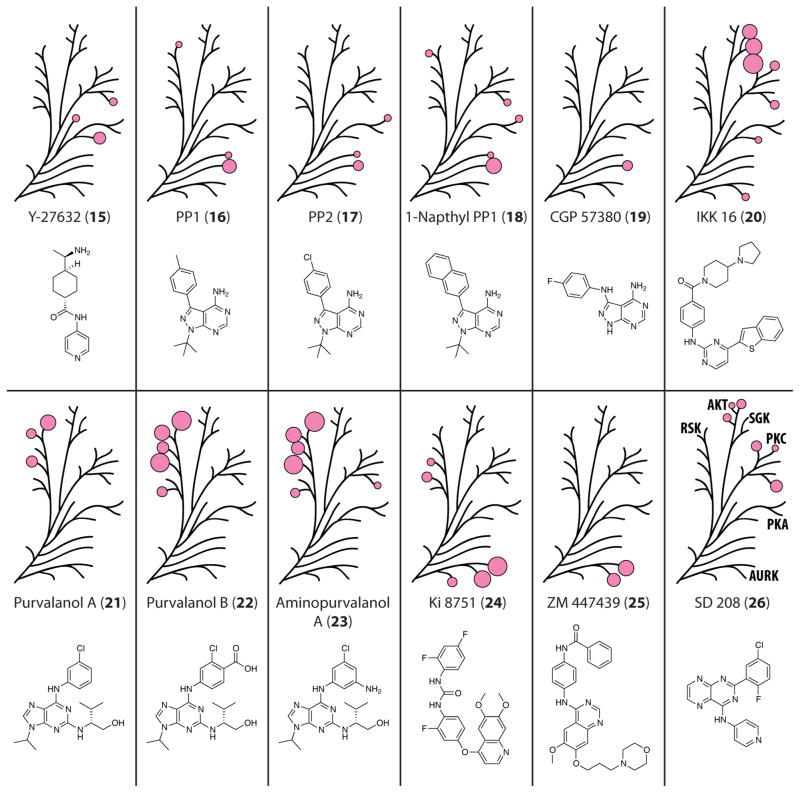
Selectivity profiles for the screened compounds 3–14, with the results mapped to the AGC dendrogram. Each pink circle is proportional in size to the measured % inhibition for each kinase, with only % inhibition >25% being shown.
Two bisindolylmaleimides, Ro 31-8220 (5) and GF 109203X (6), constitute a second pair of staurosporine-like compounds sharing similar structural features, but these two demonstrated more selectivity than 3 and 4. Both 5 and 6 were originally developed as PKC inhibitors29,30 with the former inhibiting all five of the PKC isoforms tested at least 47%. Across the board, 5 was the more potent and less selective inhibitor of the two, with no kinase exhibiting greater inhibition by compound 6. Three of the PKC isoforms, γ, η, and θ, appeared relatively tolerant to differences between the two compounds and showed less loss in inhibitory activity by 6 than did many of the other kinases. It is significant to note that only PDPK1 and the three Aurora kinases were not appreciably inhibited by either of these compounds.
Arcyriaflavin A (7), PD 407824 (8), and CGP 53353 (9), represent minimal analogs of staurosporine, where 7 and 8, containing an indolocarbazole scaffold, have been reported to selectively inhibit cyclin-dependent kinase (CDK) 4/cylin D1 and CHK1 and Wee1 respectively.31,32 Eight of the kinases tested showed no inhibition by either compound, but more than half were inhibited >25% by one or both. 9, a PKC inhibitor selective for the βII isoform,33 was the lone selective compound within this group, possibly because of lack of potency, inhibiting only PKCγ and DMPK at 22% and 30% respectively. In contrast with the other compounds similar to staurosporine, 9 lacks the indole ring and is by far the most conformationally flexible of this class of compounds. Two other maleimide-based compounds, SB 415286 (39) and SB 216763 (40), were also tested, and neither molecule exhibited better than 25% inhibition against any of the kinases tested.
Sunitinib (10), a tyrosine kinase inhibitor currently FDA-approved for the treatment of gastrointestinal stromal tumors, was the most promiscuous inhibitor lacking significant structural similarities with staurosporine, aside from an indolone ring. All six of the members of the RSK family were inhibited >50%, with eight additional kinases inhibited >25%.
Selective Kinase Inhibitors
In contrast with the staurosporine-like group of inhibitors, the overwhelming majority of compounds in our library exhibited more limited selectivity profiles. In fact, a large number of the small molecules showed no measurable activity at 10 μM against any of the kinases tested here. While some of the compounds possess decidedly unique structures relative to other library members, several groups of molecules sharing conserved or similar substructures can be readily identified. Similarly structured inhibitors consistently demonstrated activity toward the same protein kinase and frequently against groups of proteins sharing high identity.
One such group of structurally similar small molecules found in this library is the sulfonylisoquinoline-containing molecules: H-89 (11), fasudil (12), and HA-1100 (13; also known as hydroxyfasudil). Two other compounds can be included in this group because of structural similarity (ML-9, 14) and a common identified target (Y-27632, 15). 11 has been marketed as a relatively selective inhibitor of PKA, but is known to exhibit activity against a number of other kinases,3,15 and AKT1 and eight other AGC kinases were inhibited at least 20%. Among those inhibited were both isoforms of serum/glucocorticoid-regulated kinase (SGK), PKCη, and PKCθ. Additionally, all three members of the PKA family (PKA, PKAβ, and PRKX) and the highly similar PKG1 were inhibited by more than 65%. 12, its active metabolite 13, and 15 have been identified as potent inhibitors of Rho-associated protein kinase 1 (ROCK1),34–36 and all of them exhibited activity toward PKG1 and PRKX, with 12 also inhibiting PKA and PKAβ. All four of these targets are fairly similar, based on kinase domain identity, and some cross-kinase activity for relatives is not unexpected. Interestingly, 14 is structurally similar to 13 but is a considerably less potent inhibitor of PKG1 and PRKX. This is likely due to the replacement of the isoquinoline nitrogen with a carbon and the substitution of a hydroxyl for a chloro group. Based on the crystal structures of 13 bound to ROCK1 and PKA, the nitrogen and hydroxyl group make important hydrogen bonds to a backbone carbonyl and amide nitrogen respectively.37,38 The inability of ML-9 to form this hydrogen bond is perhaps the basis for the low activity of this compound toward this set of kinases.
A second group of compounds sharing a pyrazolopyrimidine core includes PP1 (16), PP2 (17), 1-naphthyl PP1 (18), and CGP 57380 (19). 16 and 17 were first identified as potent inhibitors of Src family kinases,184 but further studies revealed activity toward several non-tyrosine kinases and that this is controlled by the residue size at a putative gatekeeper site.185,186 The kinases most potently inhibited by PP1 possess either a valine or threonine at this position (Figure 5.4), while those which are weakly inhibited usually contain a larger hydrophobic residue, such as isoleucine, leucine, or methionine. Using a chemical genetics approach, 18 was developed to target mutant kinases with a glycine in the gatekeeper position, enabling the active site of such mutants to accommodate the larger naphthyl ring,42 but has also demonstrated activity against a number of wild-type kinases.3 Among the members of the panel tested here, STK32B was the most potently inhibited by 16, 17, and 18 and was the sole kinase to contain a valine at the gatekeeper site. The other 26 kinases tested have either a leucine or methionine at this position. The only other kinases to be inhibited by all three of these compounds were PKA and DMPK, though weakly (between 19% and 32% inhibition). In spite of it having been designed to be more selective, 18 exhibited >20% inhibition against seven kinases. While it lacks the t-butyl functional group and contains a secondary amine linkage to a fluorophenyl modification, 19 can be included in this group as well because it contains the same pyrazolopyrimidine substructure. Reportedly selective for MNK1 over Src and several other kinases inhibited by 16,43 19 was significantly active only against STK32B (42% inhibition). STK32B was the only kinase to be inhibited >40% by any and all of the four pyrazolopyrimidine-based inhibitors.
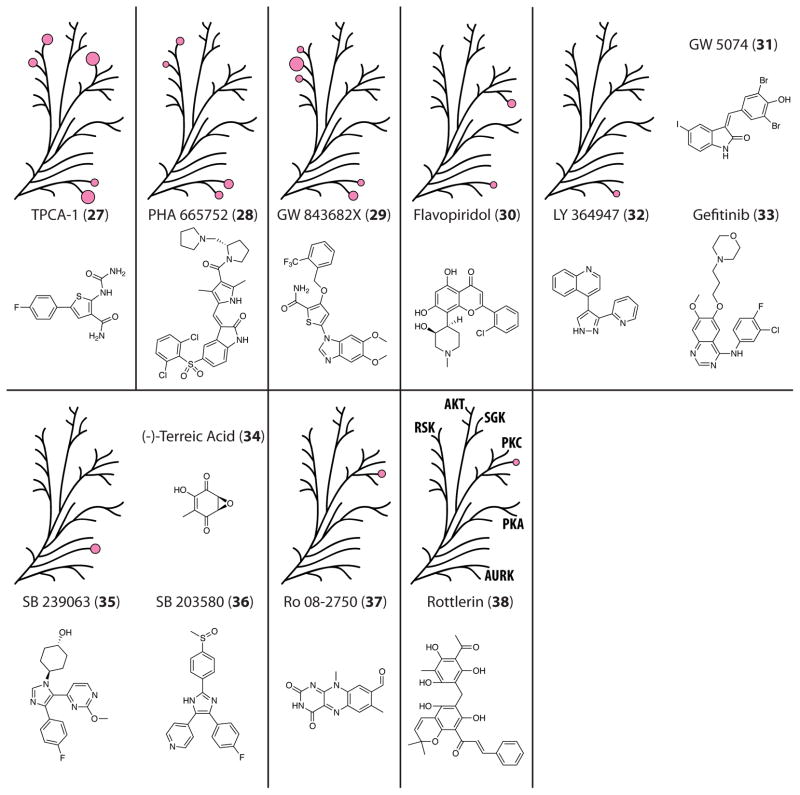
Figure 5.
Selectivity profiles for the screened compounds 27–38, with the results mapped to the AGC dendrogram. Each pink circle is proportional in size to the measured % inhibition for each kinase, with only % inhibition >25% being shown.
Figure 4.
Selectivity profiles for the screened compounds 15–26, with the results mapped to the AGC dendrogram. Each pink circle is proportional in size to the measured % inhibition for each kinase, with only % inhibition >25% being shown.
Due to their involvement in NFκB signaling, a number of protein kinases are potential targets for the treatment of rheumatism and inflammation.44 Recent work by Novartis led to the development of a selective inhibitor for IκB kinases 1 and 2 (IKK1 and IKK2), IKK 16 (20).45 In our assay, this molecule was found to be one of the few non-staurosporine-like compounds to potently inhibit SGK2 and SGK3, both at >60% inhibition. Compound 20 was also observed to inhibit Aurora kinase B (AURKB), PRKX, and three of the five PKC isoforms (ε, η, and θ) >29%. PKCη was the most potently inhibited of these 7 kinases at 83% inhibition, which was the greatest inhibition measured by any of the compounds against this kinase.
Several purvalanol derivatives, purvalanol A (21), purvalanol B (22), and aminopurvalanol A (23), were also included in the inhibitor screen. All three of these compounds were engineered to target cyclin-dependent kinases46,47 but have been reported to have significant, though less potent, activity toward several other kinases, including RSK1.3,16 Each of the three purvalanol compounds was found to inhibit at least three of the six RSK family members at >40% inhibition. 21 was the least potent, with 22 and 23 exhibiting comparable activity against five RSKs. Nuclear mitogen- and stress-activated protein kinase 1 (MSK1) was the only member of this family not to be inhibited >40% by at least two of these compounds. Roscovitine (41) and olomoucine (42), two other CDK-targeted inhibitors with structures similar to the purvalanols,48,49 were seen to have negligible activity against any of the kinases tested.
Two inhibitors, Ki 8751 (24) and ZM 447439 (25), were among several compounds to show activity against one or more of the Aurora kinases. Compound 24, reported to be selective for vascular endothelial growth factor receptor 2 over a number of other receptor tyrosine kinases,50 was quite effective at inhibiting several AGC kinases. In particular, all three Aurora kinases were inhibited the most, between 41–80%, and four of the RSK family kinases were inhibited >20%. 25, an inhibitor found to preferentially target AURKB and AURKC over AURKA and a number of other kinases,51 was found to be very selective for its targets. Within the subset of protein kinases assayed, both AURKB and AURKC were inhibited >50% at 10 μM, with the compound failing to show appreciable activity toward any other kinase.
SD 208 (26) was first derived as an inhibitor of transforming growth factor-beta receptor 1 (TGFβR1).52 Transforming growth factor-beta signaling has been implicated in playing a role in the migration and invasion of malignant glioma,53 and as such, its receptor, TGFβR1, has drawn interest as a target in order to block signaling by this ligand. In our assay, 26 was shown to have >25% inhibition toward all three AKTs and as well as three of the PKC isoforms (ε, η, and θ).
Though quite structurally distinct inhibitors, TPCA-1 (27), PHA 665752 (28), and GW 843682X (29) demonstrated somewhat similar patterns of inhibition. Intended to target IKK2, c-MET, and polo-like kinase 1 respectively,54–56 each of these molecules showed activity against AURKB, AURKC, and at least two of the RSKs at >25% inhibition. Compound 27 was the only one of these to also significantly inhibit PKCη.
A number of compounds showed activity against just one or two of the kinases tested. This group of inhibitors included flavopiridol (30),57 which hit PKCθ and AURKC; GW 5074 (31),58 LY 364947 (32),59 and gefitinib/Iressa (33),60 which hit AURKB; terreic acid (34),61 SB 239063 (35),62 and SB 203580 (36),63 which hit STK32B; and Ro 08-2750 (37);64 and rottlerin (38),65 which hit PKCγ. Very few structural similarities exist between these molecules, and their activities were relatively lower than some of the other inhibitors, with no inhibition >40% being measured.
Interestingly, 36 of the 80 compounds tested showed little to no activity (<25% inhibition) at 10 μM against any of the kinases tested (Figure 6). Given the conserved nature of protein kinase active sites, this level of selectivity against the AGC family is encouraging for the future development of highly selective molecular probes. These scaffolds may provide a starting point for designing new inhibitors that avoid the off-target inhibition of the AGC family of kinases tested here. Despite many of these compounds having unusual scaffolds for kinase inhibitors, all of the compounds tested are marketed as potent and selective kinase inhibitors. It is worth noting that several of these compounds, namely 51 and 54–58, can potentially function as Michael acceptors, an activity that could be quenched by any number of components found in the lysate assay milieu.
Figure 6.
Structures of compounds 39–74, which were found to demonstrate <25% inhibition for every kinase tested.
Trends in Inhibition
To analyze the extent to which kinase identity plays a role in the patterns of inhibition observed among the AGC kinases tested, we compared the relationship between kinase domain identity and the likelihood of cross-kinase activity. A cursory examination of the data already discussed suggests that more similar kinases tend to be inhibited consistently by the same inhibitors. In attempting to generate a more quantitative evaluation of this phenomenon, we sought to answer the question “If an inhibitor shows activity against any given kinase, what is the likelihood that it will inhibit other similar kinases?” Toward this goal, we aligned each kinase against every other kinase tested to tabulate all possible pairwise identity scores using only their respective kinase domains (Supporting Information, Table S3). Kinase identity groups were defined based upon what set of kinase domains are linked to each other through a minimum percent identity score. We then analyzed the inhibition data using the following equation that describes the probability of an inhibitor hitting multiple kinases (F) within a given identity group:
| (1) |
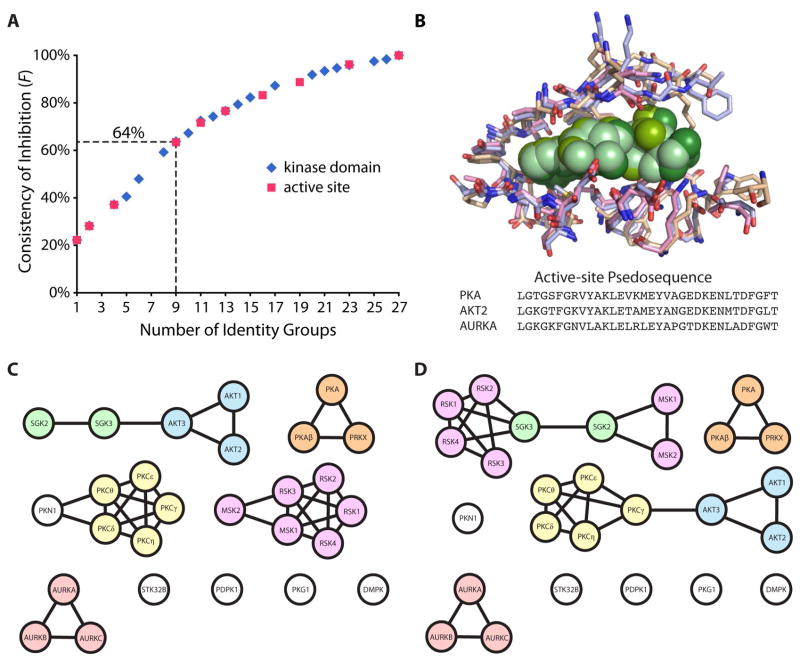
For a group of kinases connected through a given percent identity, x is defined as the number of inhibitors showing >25% inhibition against each kinase in that group, n is the number of kinases in that identity group, and T is the total number of unique inhibitors to demonstrate >25% inhibition against at least one of the kinases within the identity group. This function was applied to each kinase group at several different identity cutoffs, and the aggregate F values at each cutoff were averaged to observe general trends across the identity groups (Figure 7A). The identity cutoffs were selected based upon what minimum percent identity would result in a change in the number of possible identity groups. For example, at 100% identity, each kinase is related only to itself, resulting in 27 groups consisting of one kinase each and an F value of 100%. At the opposite extreme, all kinases would be lumped into one group and necessarily have the same F value, in this case 22%. The data for all intermediate numbers of groups, including the percent identity cutoffs used to acquire that group number, can be found in Supporting Information, Table S4. The spread of pairwise identity scores for the kinase domains ranged from 95% to 29%. In general, these results confirm that as the identity cutoff is lowered and the relationship between kinases becomes more different (fewer groups) the calculated F values also decrease.
Figure 7.
Comparison of inhibition frequencies between kinase domain or active site identity groups. (A) Average F values for incrementally binned identity groups were plotted for either the full kinase domain or active site residues. (B) The crystal structures and pseudosequence of residues within 6 Å of an ATP analog (green) aligned for PKA, AKT2, and AURKA. Homology maps generated using a percent identity cutoff that results in the formation of nine groups for the (C) kinase domain and (D) active site residues.
In order to compare how the consistency of inhibition might trend differently for active site residues relative to the full kinase domain, we also rescored the F values using identity groups based on active site homology (Figure 7A). A pseudosequence of active site residues was assigned to each kinase by identifying any residues within 6 Å of the kinase active site. The crystal structure of PKA was aligned with the structures of two other AGC kinases, AKT2 and AURKA, and any amino acids that were within 6 Å of the ATP analogs bound in the active site of all three structures were included in the 34 residue pseudosequence (Figure 7B). AKT2 and AURKA were chosen to ensure that structural elements important for substrate binding in kinases more distantly related to PKA were not neglected. The corresponding pseudosequence residues in all 27 kinases were used to generated pairwise percent identity values based on the active site only (Supporting Information, Table S5). Newly defined identity groups were then used to regenerate the frequency of inhibition values for the same percent identity cutoffs used with the full kinase domain (Supporting Information, Table S6). Relative to the full kinase domain, the range of percent identity values for the active site pseudosequence alignment was much narrower, ranging from 100% to 47%. By binning the kinases into groups according to what minimum percent identity results in new connectivities, any bias that would otherwise be introduced by trying to directly compare the two sets of identity scores is normalized. As is clearly illustrated by a comparison of this data with that for the full kinase domain (Figure 7A), the aggregate F values follow a nearly identical trend. This is somewhat surprising, given that it may be expected that a different curve would result for the active site residues alone, which more directly dictate active site structure, and therefore the shape of inhibitor binding pockets, than the more subtle structural constraints imposed by distal residues.
Even so, important differences still exist between the identity groups defined by the full kinase domain or the active site alone. This shift in identity connectivities can be more readily illustrated by comparing the homology maps when 9 groups are present using the pairwise kinase-to-kinase identity scores of either the full kinase domain (Figure 7C) or the active site pseudosequence (Figure 7D). The percent identity cutoffs used to generate these groups were 54% and 82% respectively. Some kinases are more closely associated with alternative sets of nearest-neighbor kinases when comparing the two homology maps. For example, the kinase domains for SGK2 and SGK3 share a higher identity with the three AKT kinases (52–55%) than they do with the six RSKs (44–49%), but when looking at only the active site proximal residues, they appear more identical to the latter (73–85%) rather than the former (73–76%). This difference in sequence could potentially explain why both SGKs and the RSKs are inhibited by the staurosporine analogs 7 and 8, while the AKTs are not. Likewise, several of the PKCs (δ, γ, ε) exhibited no inhibition by 7 and 8, similar to the three AKT isoforms. With respect to kinase domain identity, the AKTs are more closely related to the SGKs than the PKCs. In terms of active site residues, all three AKTs are closer in identity to PKCγ and PKCε (76–82%) than to either SGK (73–76%), possibly providing an explanation as to why only the SGKs were inhibited by 7 and 8. Interestingly, PKA, shares >70% identity with the active site residues of 20 other kinases, more than any other kinase used in the present study, and thus may provide a good general model for the routine testing of off-target inhibition of the AGC family. Importantly, a comparison of these homology maps suggests that when a new inhibitor is developed and resources are limited, it may ultimately be more informative to test for off-target activity against kinases which are closely related by active site as opposed to the entire kinase domain. Certainly, testing a small molecule against the largest fraction of the human kinome as possible is more desirable when resources permit, because off-target activity can be exceptionally unpredictable, with inhibitors demonstrating potency for kinases that are very poorly related to the intended target. If a limited subset of kinases must be selected, profiling inhibitors against a panel of active site relatives may be more representative of overall selectivity. It is worthwhile to note that this simplification may have caveats, as a few kinases which are completely identical in their active site residues by our analysis still demonstrate differential preference for small molecules inhibitors. For example, RSK1, RSK2 and RSK4 contain identical active site pseudosequences, yet 21, 22, 27 and 29 exhibited at least 30% more inhibition for one or two of these kinases over the others.
Conclusions
Herein, we have reported the inhibition profiles of 27 AGC kinases with a library of 80 commercially available protein kinase inhibitors, with the aim of contributing to publicly available knowledge of compound selectivities. The small molecule profiles against the AGC family may aid in the design of new inhibitors that target this family and at the same time allow for understanding the biological effects of these compounds arising from off-target activities described herein. As small molecule intervention continues to play an important part in resolving the physiological role of protein kinases in signal transduction and disease, the level of confidence applied to cell-based assays studying the modulation of kinase signaling and the predictability of kinase related off-target toxicity caused by therapeutics is of concern. The most expedient method of improving confidence in experimental conclusions is through more thorough inhibitor screens for small molecule promiscuity. Off-target kinases expressed ubiquitously and at a high level are the most likely candidates to interfere with experimental results reliant upon selectively inhibiting a particular protein kinase with a small molecule inhibitor. Cell-based signaling studies can overcome this issue by using cell lines that don’t express these enzymes or by confirming results with two or more structurally distinct small molecules.66 Poorly selective molecules may still prove useful for implicating a target kinase in certain cellular processes,15 but in the absence of truly comprehensive inhibitor profiling, only limited conclusions can be drawn regarding a more fundamental role for a specific kinase in a given signal transduction pathway. Relatively little is known about several of the kinases tested here, like STK32B and PRKX, and a suitable probe would prove efficacious toward the identification of physiological substrates and revealing the mechanism of action. Furthermore, selective inhibitors do not yet exist for the vast majority of AGC kinases.5 Though the last several years have seen the publication of the most comprehensive kinase inhibitor screens to date, a great number of commercially available reagents have yet to be comprehensively screened for the frequency and potency of off-target interactions.
We have shown that the screening of a group of closely related kinases is useful for identifying patterns of inhibition and in confirming the structural determinants of ligand binding. While the staurosporine-like series of compounds tested were promiscuous, many of the molecules demonstrated limited off-target associations, with about half of the library compounds demonstrating minimal activity toward any of the AGC kinases tested here. As trends in therapeutic inhibitor design continue to move toward a multi-targeted approach,11,67 the ability to avoid off-target interactions will be improved by screens that identify molecular starting points that do not inhibit a large number of kinases. One general trend that can be identified within these results is that like inhibitors frequently exhibit activity toward like kinases. Nearly identical small molecules differing only by minor functional groups would be anticipated to have subtly modulated effects on the inhibition of a given target. This is reinforced by the correlation between high sequence identity and the frequency with which related kinases are inhibited by the same set of small molecules. As an example, 16 different compounds inhibited at least one of the RSKs >25%, with more than half of those molecules hitting at least five of the six kinases in that family. An analysis of the results of small molecule screens supported by kinase-ligand co-crystal structures can also provide explanations for why seemingly similar inhibitors show differential activity for certain targets, as in the case of ML-9 (14), and how selectivity can be dictated by specific active site residues, as in the case of PP1 (16). Our confidence in predicting inhibitor selectivity and promiscuity will surely increase with future work aimed towards an extensive profiling of this and other compound classes against larger kinase libraries made possible with the development of simple, inexpensive and high-throughput screens.
Experimental Section
Construction of the Fusion Proteins and mRNA Synthesis
Kinase and Fos constructs attached to luciferase fragments were prepared as previously reported.22 Briefly, DNA fragments encoding their respective proteins were generated by PCR and cloned into either pETDuet-1 or pRSFDuet-1 vectors (Novagen). Each fusion construct was linked to its respective luciferase fragment via a 13 residue (GGS)n linker. PCR fragments were prepared with appropriate primers from template sequences, and cloning results were verified by dideoxyoligonucleotide sequencing. A full list of the luciferase constructs and kinase NCBI reference sequence numbers can be found in the Supporting Information, Table S1. A PCR product of each fusion construct was generated with primers containing a T7 RNA polymerase promoter, a mammalian Kozak sequence, and a 3′ hairpin loop68 as a template for in vitro mRNA synthesis. RiboMax Large Scale RNA Production System-T7 kits (Promega) were used to prepare mRNA from PCR fragments.
Synthesis of the Jun-staurosporine conjugate
The peptide-ligand conjugate (>95% purity by HPLC) used here has been previously reported in the literature and used as such.22,24,25
Small Molecule Inhibitor Profiling
Plate-based small molecule screens were performed as previously reported.22 mRNA for each of the Cfluc-kinase fusions was co-translated with mRNA for Fos-Nfluc in rabbit reticulocyte lysate (Luceome Biotechnologies) at a sufficient volume to take measurements of each control and assay point in duplicate. Bulk translations (~4.5 mL of lysate) were divided into 400 μL aliquots and incubated at 30 °C for 90 min. After incubation, aliquots were stored at −80 °C overnight before being thawed on ice, collected, and assayed. Several 24 μL aliquots from the recollected bulk solution were set aside and treated with 1 μL of Buffer A (6.76 mM Tris HCl, 3.38 mM Mg(OAc)2, pH 7.45) per aliquot to serve as a negative control. The remaining lysate was treated with 3.125 μM 2 in Buffer A, to a final concentration of 125 nM. Treated lysate was then aliquoted into appropriate wells of a 96-well Lumitrac 200 plate (Greiner Bio-one) containing either 1 μL of DMSO for controls or 1 μL of an inhibitor diluted to 250 μM in DMSO. All of the inhibitors tested were taken from the Tocris Kinase Inhibitor Toolbox (Tocris Biosciences) with the exception of PKC-412 (LC Laboratories), Sunitinib (LC Laboratories), Flavopiridol (Alexis Biochemicals), and Roscovitine (LC Laboratories). The final concentrations of 2 and inhibitor prior to the addition of a luciferase reagent were 120 nM and 10 μM, respectively. Plates were covered and allowed to incubate 1 h at room temperature prior. Luminescence measurements were taken immediately upon addition of 80 μL of a luciferin assay reagent to each well using a Centro XS LB 960 plate reader (Berthold Technologies) and a 1 s integration time.
Percent Inhibition Calculations
Percent inhibition values for each inhibitor were calculated by first normalizing to the relevant controls. The luminescence measured for each negative control (no 2) was subtracted from the raw positive control (with 2) and inhibitor values. Measurements for each inhibitor were normalized to the positive control and subtracted from 1 to generate percent inhibition values. A control of dimerized Fos-Nfluc and Cfluc-Jun was used to identify small molecule activity against reassembled luciferase, and the measured percent inhibition values of each inhibitor for Fos/Jun were subtracted from the corresponding inhibition values for each kinase, with percent inhibition values <0 adjusted to 0% inhibition. Some molecular scaffolds, such as quinolines, are known to act as potent inhibitors of kinases69 as well as luciferase,70 and the observance of activity toward luciferase in library screens has been estimated to be at least 3% of compounds.70,71 Eight of the initial 80 compounds tested were excluded from the final analysis because they affected luciferase activity in the Fos/Jun control, and their structures can be found in the Supporting Information, Figure S1. The full table of percent inhibition values is located in the Supporting Information, Table S2. The results for PKA and AKT1 are reproduced from a previously published report.22
Kinase Sequence Identity and Homology Mapping
The kinase domain sequences used in alignments were taken from the corresponding Swiss-Prot annotations found at the UniProt website (http://www.uniprot.org). Pairwise percent identity scores were generated using a ClustalW2 alignment tool hosted by the European Bioinformatics Institute. Residues within 6 Å of an ATP analog were identified using the aligned structures of PKA (PDB ID: 3O7L), AKT2 (PDB ID: 1O6K), and AURKA (PDB ID: 2DWB) in PyMOL (http://www.pymol.org). The 34 amino acids retrieved by this search were used to define a pseudosequence for these three kinases. This pseudosequence was extrapolated to the other 24 kinases by identifying homologous residues in an alignment of all of the kinase domains. Active site pseudosequences were aligned to obtain percent identity scores as previously mentioned. Full tables of the identity scores acquired for both the kinase domain and the active site pseudosequence alignments can be found in the Supporting Information (Tables S3 and S4). The homology maps were created by importing the tables of identity scores into Cytoscape (http://www.cytoscape.org) and filtering out the lowest ~90% of identity scores.
Supplementary Material
Acknowledgments
The authors thank the NIH (R21CA141974 and R44GM087807) for support. Additional thanks goes to Dr. Reena Zutshi, Dr. Matthew Cordes, Dr. Christian Roessler and members of the Ghosh laboratory.
Abbreviations
- AKT
RAC serine/threonine-protein kinase
- AURK
aurora kinase
- c-MET
hepatocyte growth factor receptor
- CDK
cyclin-dependent kinase
- Cfluc
C-terminal fragment of split-firefly luciferase
- CHK1
checkpoint kinase 1
- DMPK
myotonic dystrophy protein kinase
- IKK
inhibitor of nuclear factor kappa-B kinase
- MNK1
MAP-kinase interacting serine/threonine-protein kinase 1
- MSK
nuclear mitogen- and stress-activated protein kinase
- Nfluc
N-terminal fragment of split-firefly luciferase
- PDPK1
3-phosphoinositide-dependent protein kinase 1
- PKC
protein kinase C
- PKG
cGMP-dependent protein kinase
- PKA
cAMP-dependent protein kinase catalytic subunit alpha
- PRKX
protein kinase X
- ROCK1
rho-associated protein kinase 1
- RSK
ribosomal protein S6 kinase
- SGK
serum/glucocorticoid-regulated kinase
- Src
proto-oncogene tyrosine kinase Src
- STK32B
serine/threonine-protein kinase 32B
- TGFβR1
transforming growth factor-beta receptor 1
Footnotes
Supporting Information Available: A complete list of kinases tested, the structures of luciferase effectors, and full data tables can be found in Tables S1-S4 and Figure S1. This material is available free of charge via the Internet at http://pubs.acs.org.
References
- 1.Manning G, Whyte D, Martinez R, Hunter T, Sudarsanam S. The protein kinase complement of the human genome. Science. 2002;298:1912–1934. doi: 10.1126/science.1075762. [DOI] [PubMed] [Google Scholar]
- 2.Cohen P. Protein kinases - the major drug targets of the twenty-first century? Nat Rev Drug Discov. 2002;1:309–315. doi: 10.1038/nrd773. [DOI] [PubMed] [Google Scholar]
- 3.Bain J, Plater L, Elliott M, Shpiro N, Hastie C, McLauchlan H, Klevernic I, Arthur J, Alessi D, Cohen P. The selectivity of protein kinase inhibitors: a further update. Biochem J. 2007;408:297–315. doi: 10.1042/BJ20070797. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Hanks SK, Hunter T. Protein kinases 6. The eukaryotic protein-kinase superfamily: kinase (catalytic) domain-structure and classification. FASEB J. 1995;9:576–596. [PubMed] [Google Scholar]
- 5.Pearce LR, Komander D, Alessi DR. The nuts and bolts of AGC protein kinases. Nat Rev Mol Cell Bio. 2010;11:9–22. doi: 10.1038/nrm2822. [DOI] [PubMed] [Google Scholar]
- 6.Datta SR, Dudek H, Tao X, Masters S, Fu HA, Gotoh Y, Greenberg ME. Akt phosphorylation of BAD couples survival signals to the cell-intrinsic death machinery. Cell. 1997;91:231–241. doi: 10.1016/s0092-8674(00)80405-5. [DOI] [PubMed] [Google Scholar]
- 7.Brunet A, Park J, Tran H, Hu LS, Hemmings BA, Greenberg ME. Protein kinase SGK mediates survival signals by phosphorylating the forkhead transcription factor FKHRL1 (FOXO3a) Mol Cell Biol. 2001;21:952–965. doi: 10.1128/MCB.21.3.952-965.2001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Carpten JD, Faber AL, Horn C, Donoho GP, Briggs SL, Robbins CM, Hostetter G, Boguslawski S, Moses TY, Savage S, Uhlik M, Lin AM, Du J, Qian YW, Zeckner DJ, Tucker-Kellogg G, Touchman J, Patel K, Mousses S, Bittner M, Schevitz R, Lai MHT, Blanchard KL, Thomas JE. A transforming mutation in the pleckstrin homology domain of AKT1 in cancer. Nature. 2007;448:439–U431. doi: 10.1038/nature05933. [DOI] [PubMed] [Google Scholar]
- 9.Maurer M, Su T, Saal LH, Koujak S, Hopkins BD, Barkley CR, Wu JP, Nandula S, Dutta B, Xie YL, Chin YR, Kim DI, Ferris JS, Gruvberger-Saal SK, Laakso M, Wang XM, Memeo L, Rojtman A, Matos T, Yu JS, Cordon-Cardo C, Isola J, Terry MB, Toker A, Mills GB, Zhao JJ, Murty VVVS, Hibshoosh H, Parsons R. 3-phosphoinositide-dependent kinase 1 potentiates upstream lesions on the phosphatidylinositol 3-kinase pathway in breast carcinoma. Cancer Res. 2009;69:6299–6306. doi: 10.1158/0008-5472.CAN-09-0820. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Liu PX, Cheng HL, Roberts TM, Zhao JJ. Targeting the phosphoinositide 3-kinase pathway in cancer. Nat Rev Drug Discov. 2009;8:627–644. doi: 10.1038/nrd2926. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Knight ZA, Lin H, Shokat KM. Targeting the cancer kinome through polypharmacology. Nat Rev Cancer. 2010;10:130–137. doi: 10.1038/nrc2787. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Force T, Kolaja KL. Cardiotoxicity of kinase inhibitors: the prediction and translation of preclinical models to clinical outcomes. Nat Rev Drug Discov. 2011;10:111–126. doi: 10.1038/nrd3252. [DOI] [PubMed] [Google Scholar]
- 13.Kerkela R, Woulfe KC, Durand JB, Vagnozzi R, Kramer D, Chu TF, Beahm C, Chen MH, Force T. Sunitinib-Induced Cardiotoxicity Is Mediated by Off-Target Inhibition of AMP-Activated Protein Kinase. Clin Transl Sci. 2009;2:15–25. doi: 10.1111/j.1752-8062.2008.00090.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Giamas G, Man YL, Hirner H, Bischof J, Kramer K, Khan K, Ahmed SSL, Stebbing J, Knippschild U. Kinases as targets in the treatment of solid tumors. Cell Signal. 2010;22:984–1002. doi: 10.1016/j.cellsig.2010.01.011. [DOI] [PubMed] [Google Scholar]
- 15.Davies S, Reddy H, Caivano M, Cohen P. Specificity and mechanism of action of some commonly used protein kinase inhibitors. Biochem J. 2000;351:95–105. doi: 10.1042/0264-6021:3510095. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Bain J, McLauchlan H, Elliott M, Cohen P. The specificities of protein kinase inhibitors: an update. Biochem J. 2003;371:199–204. doi: 10.1042/BJ20021535. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Fabian MA, Biggs WH, 3rd, Treiber DK, Atteridge CE, Azimioara MD, Benedetti MG, Carter TA, Ciceri P, Edeen PT, Floyd M, Ford JM, Galvin M, Gerlach JL, Grotzfeld RM, Herrgard S, Insko DE, Insko MA, Lai AG, Lelias JM, Mehta SA, Milanov ZV, Velasco AM, Wodicka LM, Patel HK, Zarrinkar PP, Lockhart DJ. A small molecule-kinase interaction map for clinical kinase inhibitors. Nat Biotechnol. 2005;23:329–336. doi: 10.1038/nbt1068. [DOI] [PubMed] [Google Scholar]
- 18.Karaman M, Herrgard S, Treiber D, Gallant P, Atteridge C, Campbell B, Chan K, Ciceri P, Davis M, Edeen P, Faraoni R, Floyd M, Hunt J, Lockhart D, Milanov Z, Morrison M, Pallares G, Patel H, Pritchard S, Wodicka L, Zarrinkar P. A quantitative analysis of kinase inhibitor selectivity. Nat Biotechnol. 2008;26:127–132. doi: 10.1038/nbt1358. [DOI] [PubMed] [Google Scholar]
- 19.Bamborough P, Drewry D, Harper G, Smith GK, Schneider K. Assessment of chemical coverage of kinome space and its implications for kinase drug discovery. J Med Chem. 2008;51:7898–7914. doi: 10.1021/jm8011036. [DOI] [PubMed] [Google Scholar]
- 20.Bamborough P, Brown MJ, Christopher JA, Chung CW, Mellor GW. Selectivity of kinase inhibitor fragments. J Med Chem. 2011;54:5131–5143. doi: 10.1021/jm200349b. [DOI] [PubMed] [Google Scholar]
- 21.Metz JT, Johnson EF, Soni NB, Merta PJ, Kifle L, Hajduk PJ. Navigating the kinome. Nat Chem Biol. 2011;7:200–202. doi: 10.1038/nchembio.530. [DOI] [PubMed] [Google Scholar]
- 22.Jester BW, Cox KJ, Gaj A, Shomin CD, Porter JR, Ghosh I. A coiled-coil enabled split-luciferase three-hybrid system: applied toward profiling inhibitors of protein kinases. J Am Chem Soc. 2010;132:11727–11735. doi: 10.1021/ja104491h. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Kannan N, Haste N, Taylor SS, Neuwald AF. The hallmark of AGC kinase functional divergence is its C-terminal tail, a cis-acting regulatory module. Proc Natl Acad Sci USA. 2007;104:1272–1277. doi: 10.1073/pnas.0610251104. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Meyer S, Shomin C, Gaj T, Ghosh I. Tethering small molecules to a phage display library: discovery of a selective bivalent inhibitor of protein kinase A. J Am Chem Soc. 2007;129:13812–13813. doi: 10.1021/ja076197d. [DOI] [PubMed] [Google Scholar]
- 25.Shomin CD, Meyer SC, Ghosh I. Staurosporine tethered peptide ligands that target cAMP-dependent protein kinase (PKA): Optimization and selectivity profiling. Bioorgan Med Chem. 2009;17:6196–6202. doi: 10.1016/j.bmc.2009.07.056. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Wagner J, von Matt P, Sedrani R, Albert R, Cooke N, Ehrhardt C, Geiser M, Rummel G, Stark W, Strauss A, Cowan-Jacob SW, Beerli C, Weckbecker G, Evenou JP, Zenke G, Cottens S. Discovery of 3-(1H-indol-3-yl)-4-[2-(4-methylpiperazin-1-yl)quinazolin-4-yl]pyrrole-2,5-dione (AEB071), a potent and selective inhibitor of protein kinase C isotypes. J Med Chem. 2009;52:6193–6196. doi: 10.1021/jm901108b. [DOI] [PubMed] [Google Scholar]
- 27.Eathiraj S, Palma R, Volckova E, Hirschi M, France DS, Ashwell MA, Chan TCK. Discovery of a novel mode of protein kinase inhibition characterized by the mechanism of inhibition of human mesenchymal-epithelial transition factor (c-Met) protein autophosphorylation by ARQ 197. J Biol Chem. 2011;286:20666–20676. doi: 10.1074/jbc.M110.213801. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Jackson JR, Gilmartin A, Imburgia C, Winkler JD, Marshall LA, Roshak A. An indolocarbazole inhibitor of human checkpoint kinase (Chk1) abrogates cell cycle arrest caused by DNA damage. Cancer Res. 2000;60:566–572. [PubMed] [Google Scholar]
- 29.Davis PD, Hill CH, Keech E, Lawton G, Nixon JS, Sedgwick AD, Wadsworth J, Westmacott D, Wilkinson SE. Potent selective inhibitors of protein kinase C. FEBS Lett. 1989;259:61–63. doi: 10.1016/0014-5793(89)81494-2. [DOI] [PubMed] [Google Scholar]
- 30.Toullec D, Pianetti P, Coste H, Bellevergue P, Grandperret T, Ajakane M, Baudet V, Boissin P, Boursier E, Loriolle F, Duhamel L, Charon D, Kirilovsky J. The Bisindolylmaleimide Gf-109203x Is a Potent and Selective Inhibitor of Protein-Kinase-C. J Biol Chem. 1991;266:15771–15781. [PubMed] [Google Scholar]
- 31.Sanchez-Martinez C, Shih C, Faul MM, Zhu GX, Paal M, Somoza C, Li TC, Kumrich CA, Winneroski LL, Xun Z, Brooks HB, Patel BKR, Schultz RM, DeHahn TB, Spencer CD, Watkins SA, Considine E, Dempsey JA, Ogg CA, Campbell RM, Anderson BA, Wagner J. Aryl[a]pyrrolo[3,4-c]carbazoles as selective cyclin D1-CDK4 inhibitors. Bioorg Med Chem Lett. 2003;13:3835–3839. doi: 10.1016/s0960-894x(03)00791-1. [DOI] [PubMed] [Google Scholar]
- 32.Palmer BD, Thompson AM, Booth RJ, Dobrusin EM, Kraker AJ, Lee HH, Lunney EA, Mitchell LH, Ortwine DF, Smaill JB, Swan LM, Denny WA. 4-phenylpyrrolo[3,4-c]carbazole-1,3(2H,6H)-dione inhibitors of the checkpoint kinase Wee1. Structure-activity relationships for chromophore modification and phenyl ring substitution. J Med Chem. 2006;49:4896–4911. doi: 10.1021/jm0512591. [DOI] [PubMed] [Google Scholar]
- 33.Kouroedov A, Eto M, Joch H, Volpe M, Luscher TF, Cosentino F. Selective inhibition of protein kinase C beta(2) prevents acute effects of high glucose on vascular cell adhesion molecule-1 expression in human endothelial cells. Circulation. 2004;110:91–96. doi: 10.1161/01.CIR.0000133384.38551.A8. [DOI] [PubMed] [Google Scholar]
- 34.Niggli V. Rho-kinase in human neutrophils: a role in signalling for myosin light chain phosphorylation and cell migration. FEBS Lett. 1999;445:69–72. doi: 10.1016/s0014-5793(99)00098-8. [DOI] [PubMed] [Google Scholar]
- 35.Shimokawa H, Seto M, Katsumata N, Amano M, Kozai T, Yamawaki T, Kuwata K, Kandabashi T, Egashira K, Ikegaki I, Asano T, Kaibuchi K, Takeshita A. Rho-kinase-mediated pathway induces enhanced myosin light chain phosphorylations in a swine model of coronary artery spasm. Cardiovasc Res. 1999;43:1029–1039. doi: 10.1016/s0008-6363(99)00144-3. [DOI] [PubMed] [Google Scholar]
- 36.Uehata M, Ishizaki T, Satoh H, Ono T, Kawahara T, Morishita T, Tamakawa H, Yamagami K, Inui J, Maekawa M, Narumiya S. Calcium sensitization of smooth muscle mediated by a Rho-associated protein kinase in hypertension. Nature. 1997;389:990–994. doi: 10.1038/40187. [DOI] [PubMed] [Google Scholar]
- 37.Breitenlechner C, Gassel M, Hidaka H, Kinzel V, Huber R, Engh RA, Bossemeyer D. Protein kinase a in complex with rho-kinase inhibitors Y-27632, fasudil, and H-1152P: Structural basis of selectivity. Structure. 2003;11:1595–1607. doi: 10.1016/j.str.2003.11.002. [DOI] [PubMed] [Google Scholar]
- 38.Jacobs M, Hayakawa K, Swenson L, Bellon S, Fleming M, Taslimi P, Doran J. The structure of dimeric ROCK I reveals the mechanism for ligand selectivity. J Biol Chem. 2006;281:260–268. doi: 10.1074/jbc.M508847200. [DOI] [PubMed] [Google Scholar]
- 39.Hanke JH, Gardner JP, Dow RL, Changelian PS, Brissette WH, Weringer EJ, Pollok K, Connelly PA. Discovery of a novel, potent, and Src family-selective tyrosine kinase inhibitor - Study of Lck- and FynT-dependent T cell activation. J Biol Chem. 1996;271:695–701. doi: 10.1074/jbc.271.2.695. [DOI] [PubMed] [Google Scholar]
- 40.Liu Y, Bishop A, Witucki L, Kraybill B, Shimizu E, Tsien J, Ubersax J, Blethrow J, Morgan DO, Shokat KM. Structural basis for selective inhibition of Src family kinases by PP1. Chem Biol. 1999;6:671–678. doi: 10.1016/s1074-5521(99)80118-5. [DOI] [PubMed] [Google Scholar]
- 41.Schindler T, Sicheri F, Pico A, Gazit A, Levitzki A, Kuriyan J. Crystal structure of Hck in complex with a Src family-selective tyrosine kinase inhibitor. Mol Cell. 1999;3:639–648. doi: 10.1016/s1097-2765(00)80357-3. [DOI] [PubMed] [Google Scholar]
- 42.Jaeschke A, Karasarides M, Ventura JJ, Ehrhardt A, Zhang C, Flavell RA, Shokat KM, Davis RJ. JNK2 is a positive regulator of the cJun transcription factor. Mol Cell. 2006;23:899–911. doi: 10.1016/j.molcel.2006.07.028. [DOI] [PubMed] [Google Scholar]
- 43.Knauf U, Tschopp C, Gram H. Negative regulation of protein translation by mitogen-activated protein kinase-interacting kinases 1 and 2. Mol Cell Biol. 2001;21:5500–5511. doi: 10.1128/MCB.21.16.5500-5511.2001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Gaestel M, Mengel A, Bothe U, Asadullah K. Protein kinases as small molecule inhibitor targets in inflammation. Curr Med Chem. 2007;14:2214–2234. doi: 10.2174/092986707781696636. [DOI] [PubMed] [Google Scholar]
- 45.Waelchli R, Bollbuck B, Bruns C, Buhl T, Eder J, Felfel R, Hersperger R, Janser P, Revesz L, Zerwes HG, Schlapbach A. Design and preparation of 2-benzamido-pyrimidines as inhibitors of IKK. Bioorg Med Chem Lett. 2006;16:108–112. doi: 10.1016/j.bmcl.2005.09.035. [DOI] [PubMed] [Google Scholar]
- 46.Chang YT, Gray NS, Rosania GR, Sutherlin DP, Kwon S, Norman TC, Sarohia R, Leost M, Meijer L, Schultz PG. Synthesis and application of functionally diverse 2,6,9-trisubstituted purine libraries as CDK inhibitors. Chem Biol. 1999;6:361–375. doi: 10.1016/S1074-5521(99)80048-9. [DOI] [PubMed] [Google Scholar]
- 47.Gray NS, Wodicka L, Thunnissen AMWH, Norman TC, Kwon SJ, Espinoza FH, Morgan DO, Barnes G, LeClerc S, Meijer L, Kim SH, Lockhart DJ, Schultz PG. Exploiting chemical libraries, structure, and genomics in the search for kinase inhibitors. Science. 1998;281:533–538. doi: 10.1126/science.281.5376.533. [DOI] [PubMed] [Google Scholar]
- 48.Vesely J, Havlicek L, Strnad M, Blow JJ, Donelladeana A, Pinna L, Letham DS, Kato J, Detivaud L, Leclerc S, Meijer L. Inhibition of Cyclin-Dependent Kinases by Purine Analogs. Eur J Biochem. 1994;224:771–786. doi: 10.1111/j.1432-1033.1994.00771.x. [DOI] [PubMed] [Google Scholar]
- 49.Havlicek L, Hanus J, Vesely J, Leclerc S, Meijer L, Shaw G, Strnad M. Cytokinin-derived cyclin-dependent kinase inhibitors: Synthesis and cdc2 inhibitory activity of olomoucine and related compounds. J Med Chem. 1997;40:408–412. doi: 10.1021/jm960666x. [DOI] [PubMed] [Google Scholar]
- 50.Kubo K, Shimizu T, Ohyama S, Murooka H, Iwai A, Nakamura K, Hasegawa K, Kobayashi Y, Takahashi N, Takahashi K, Kato S, Izawa T, Isoe T. Novel potent orally active selective VEGFR-2 tyrosine kinase inhibitors: Synthesis, structure-activity relationships, and antitumor activities of N-phenyl-N ′-{4-[4-quinolyloxy)phenyl}ureas. J Med Chem. 2005;48:1359–1366. doi: 10.1021/jm030427r. [DOI] [PubMed] [Google Scholar]
- 51.Ditchfield C, Johnson VL, Tighe A, Ellston R, Haworth C, Johnson T, Mortlock A, Keen N, Taylor SS. Aurora B couples chromosome alignment with anaphase by targeting BubR1, Mad2, and Cenp-E to kinetochores. J Cell Biol. 2003;161:267–280. doi: 10.1083/jcb.200208091. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Weller M, Uhl M, Aulwurm S, Wischhusen J, Weiler M, Ma JY, Almirez R, Mangadu R, Liu YW, Platten M, Herrlinger U, Murphy A, Wong DH, Wick W, Higgins LS. SD-208, a novel transforming growth factor beta receptor I kinase inhibitor, inhibits growth and invasiveness and enhances immunogenicity of murine and human glioma cells in vitro and in vivo. Cancer Res. 2004;64:7954–7961. doi: 10.1158/0008-5472.CAN-04-1013. [DOI] [PubMed] [Google Scholar]
- 53.Wick W, Grimmel C, Wild-Bode C, Platten M, Arpin M, Weller M. Ezrin-dependent promotion of glioma cell clonogenicity, motility, and invasion mediated by BCL-2 and transforming growth factor-beta 2. J Neurosci. 2001;21:3360–3368. doi: 10.1523/JNEUROSCI.21-10-03360.2001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Podolin PL, Callahan JF, Bolognese BJ, Li YH, Carlson K, Davis TG, Mellor GW, Evans C, Roshak AK. Attenuation of murine collagen-induced arthritis by a novel, potent, selective small molecule inhibitor of I kappa B kinase 2, TPCA-1 (2-[(aminocarbonyl)amino]-5-(4-fluorophenyl)-3-thiophenecarboxamide), occurs via reduction of proinflammatory cytokines and antigen-induced T cell proliferation. J Pharmacol Exp Ther. 2005;312:373–381. doi: 10.1124/jpet.104.074484. [DOI] [PubMed] [Google Scholar]
- 55.Lansing TJ, McConnell RT, Duckett DR, Spehar GM, Knick VB, Hassler DF, Noro N, Furuta M, Emmitte KA, Gilmer TM, Mook RA, Cheung M. In vitro biological activity of a novel small-molecule inhibitor of polo-like kinase 1. Mol Cancer Ther. 2007;6:450–459. doi: 10.1158/1535-7163.MCT-06-0543. [DOI] [PubMed] [Google Scholar]
- 56.Christensen JG, Schreck R, Burrows J, Kuruganti P, Chan E, Le P, Chen J, Wang XY, Ruslim L, Blake R, Lipson KE, Ramphal J, Do S, Cui JRJ, Cherrington JM, Mendel DB. A selective small molecule inhibitor of c-Met kinase inhibits c-Met-dependent phenotypes in vitro and exhibits cytoreductive antitumor activity in vivo. Cancer Res. 2003;63:7345–7355. [PubMed] [Google Scholar]
- 57.Losiewicz MD, Carlson BA, Kaur G, Sausville EA, Worland PJ. Potent inhibition of cdc2 kinase-activity by the flavonoid L86-8275. Biochem Bioph Res Co. 1994;201:589–595. doi: 10.1006/bbrc.1994.1742. [DOI] [PubMed] [Google Scholar]
- 58.Lackey K, Cory M, Davis R, Frye SV, Harris PA, Hunter RN, Jung DK, McDonald OB, McNutt RW, Peel MR, Rutkowske RD, Veal JM, Wood ER. The discovery of potent cRaf1 kinase inhibitors. Bioorg Med Chem Lett. 2000;10:223–226. doi: 10.1016/s0960-894x(99)00668-x. [DOI] [PubMed] [Google Scholar]
- 59.Sawyer JS, Anderson BD, Beight DW, Campbell RM, Jones ML, Herron DK, Lampe JW, McCowan JR, McMillen WT, Mort N, Parsons S, Smith ECR, Vieth M, Wier LC, Yan L, Zhang FM, Yingling JM. Synthesis and activity of new aryl- and heteroaryl- substituted pyrazole inhibitors of the transforming growth factor-b type I receptor kinase domain. J Med Chem. 2003;46:3953–3956. doi: 10.1021/jm0205705. [DOI] [PubMed] [Google Scholar]
- 60.Ciardiello F, Caputo R, Bianco R, Damiano V, Pomatico G, De Placido S, Bianco AR, Tortora G. Antitumor effect and potentiation of cytotoxic drugs activity in human cancer cells by ZD-1839 (Iressa), an epidermal growth factor receptor-selective tyrosine kinase inhibitor. Clin Cancer Res. 2000;6:2053–2063. [PubMed] [Google Scholar]
- 61.Kawakami T, Kawakami Y, Hartman SE, Kinoshita E, Suzuki H, Kitaura J, Yao L, Inagaki N, Franco A, Hata D, Maeda-Yamamoto M, Fukamachi H, Nagai H. Terreic acid, a quinone epoxide inhibitor of Bruton’s tyrosine kinase. P Natl Acad Sci USA. 1999;96:2227–2232. doi: 10.1073/pnas.96.5.2227. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 62.Underwood DC, Osborn RR, Kotzer CJ, Adams JL, Lee JC, Webb EF, Carpenter DC, Bochnowicz S, Thomas HC, Hay DWP, Griswold DE. SB 239063, a potent p38 MAP kinase inhibitor, reduces inflammatory cytokine production, airways eosinophil infiltration, and persistence. J Pharmacol Exp Ther. 2000;293:281–288. [PubMed] [Google Scholar]
- 63.Cuenda A, Rouse J, Doza YN, Meier R, Cohen P, Gallagher TF, Young PR, Lee JC. SB-203580 is a specific inhibitor of a MAP kinase homolog which is stimulated by cellular stresses and interleukin-1. FEBS Lett. 1995;364:229–233. doi: 10.1016/0014-5793(95)00357-f. [DOI] [PubMed] [Google Scholar]
- 64.Niederhauser O, Mangold M, Schubenel R, Kusznir EA, Schmidt D, Hertel C. NGF ligand alters NGF signaling via p75(NTR) and TrkA. J Neurosci Res. 2000;61:263–272. doi: 10.1002/1097-4547(20000801)61:3<263::AID-JNR4>3.0.CO;2-M. [DOI] [PubMed] [Google Scholar]
- 65.Gschwendt M, Muller HJ, Kielbassa K, Zang R, Kittstein W, Rincke G, Marks F. Rottlerin, a novel protein-kinase inhibitor. Biochem Bioph Res Co. 1994;199:93–98. doi: 10.1006/bbrc.1994.1199. [DOI] [PubMed] [Google Scholar]
- 66.Hardy LW, Peet NP. The multiple orthogonal tools approach to define molecular causation in the validation of druggable targets. Drug Discov Today. 2004;9:117–126. doi: 10.1016/S1359-6446(03)02969-6. [DOI] [PubMed] [Google Scholar]
- 67.Hajduk PJ, Metz JT. Rational approaches to targeted polypharmacology: creating and navigating protein-ligand interaction networks. Curr Opin Chem Biol. 2010;14:498–504. doi: 10.1016/j.cbpa.2010.06.166. [DOI] [PubMed] [Google Scholar]
- 68.Porter JR, Stains CI, Jester BW, Ghosh I. A general and rapid cell-free approach for the interrogation of protein-protein, protein-DNA, and protein-RNA interactions and their antagonists utilizing split-protein reporters. J Am Chem Soc. 2008;130:6488–6497. doi: 10.1021/ja7114579. [DOI] [PubMed] [Google Scholar]
- 69.Boschelli DH. 4-anilino-3-quinolinecarbonitriles: an emerging class of kinase inhibitors. Curr Top Med Chem. 2002;2:1051–1063. doi: 10.2174/1568026023393354. [DOI] [PubMed] [Google Scholar]
- 70.Auld DS, Southall NT, Jadhav A, Johnson RL, Diller DJ, Simeonov A, Austin CP, Inglese J. Characterization of chemical libraries for luciferase inhibitory activity. J Med Chem. 2008;51:2372–2386. doi: 10.1021/jm701302v. [DOI] [PubMed] [Google Scholar]
- 71.Thorne N, Auld DS, Inglese J. Apparent activity in high-throughput screening: origins of compound-dependent assay interference. Curr Opin Chem Biol. 2010;14:315–324. doi: 10.1016/j.cbpa.2010.03.020. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.