Abstract
Purpose
Lung adenocarcinoma (AdC) and lung squamous cell carcinoma (SCC) are the most common non-small cell lung cancer (NSCLC) subtypes. This study was designed to determine whether reduced expression of transforming growth factor β type II receptor (TGFβRII) promotes lung AdC and SCC carcinogenesis.
Experimental Design
We examined TGFβRII expression at the protein and mRNA levels in human NSCLC samples and assessed the relationship between TGFβRII expression and clinico-pathologic parameters. To determine if sporadic TGFβRII deletion in airway epithelial cells induces NSCLC formation, we targeted TGFβRII deletion alone and in combination with oncogenic KrasG12D to murine airways using a keratin 5 (K5) promoter and inducible Cre recombinase.
Results
Reduced TGFβRII expression in human NSCLC is associated with male gender, smoking, SCC histology, reduced differentiation, increased tumor stage, increased nodal metastasis, and reduced survival. Homozygous or heterozygous TGFβRII deletion in mouse airway epithelia increases the size and number of KrasG12D-initiated AdC and SCC. TGFβRII deletion increases proliferation, local inflammation, and TGFβ ligand elaboration; TGFβRII knockdown in airway epithelial cells increases migration and invasion.
Conclusions
Reduced TGFβRII expression in human NSCLC is associated with more aggressive tumor behavior and inflammation that is at least partially mediated by increased TGFβ1 expression. TGFβRII deletion in mouse airway epithelial cells promotes AdC and SCC formation, indicating that TGFβRII loss plays a causal role in lung carcinogenesis. That TGFβRII demonstrates haploid insufficiency, suggests that a 50% TGFβRII protein reduction would negatively impact lung cancer prognosis.
Keywords: lung adenocarcinoma, lung squamous cell carcinoma, TGFβ, tumor progression, mouse model
Introduction
Although over 175,000 cases of non-small cell lung cancer (NSCLC) are diagnosed annually in the United States, the five-year survival rate remains less than 20% (1). Lung adenocarcinoma (AdC) and lung squamous cell carcinoma (SCC) are the most common histologic subtypes and although most (~85%) lung cancer is smoking related, lung SCC is more strongly associated with smoking exposure (2). It is hypothesized that lung AdC arises from the distal airway epithelial progenitor (3), while lung SCC arises from the keratin-positive basal cell population that contains the upper airway epithelial progenitor (4). Transforming growth factor β (TGFβ) is a multi-functional cytokine that promotes epithelial differentiation and inhibits cell growth (5) and defective TGFβ signaling is often associated with more aggressive tumor behavior (6). TGFβ binds a heterodimer of TGFβ type I and type II receptors (TGFβRI and TGFβRII) that signal by phosphorylating Smad family transcription factors (5).
In previous small studies with poorly defined patient populations, reduced TGFβRII expression was reported in 40–80% of NSCLC at the protein and/or mRNA level (7–9). Although TGFβRII mutations are uncommon (10), both microsatellite instability and promoter methylation have been associated with reduced TGFβRII expression (8, 11). From these studies, it is unknown whether reduced TGFβRII expression in human NSCLC is associated with specific histologic subtypes, more aggressive tumor behavior, or reduced patient survival. In lung cancer cell lines, TGFβRII restoration reduces proliferation, anchorage-independent growth, and xenograft growth (7) while TGFβRII knockdown increases transwell migration (12). In mouse models, TGFβRII deletion promotes the development of oncogene-initiated malignancies in the pancreas, colon, breast, and oral epithelium (13–16) and a recent study shows that TGFβRII loss increases the invasiveness of Kras-initiated lung adenocarcinomas (17). However, because most lung cancer mouse models produce predominantly, if not exclusively, adenomas and adenocarcinomas (18), it is unknown whether TGFβ signaling disruption affects development of other NSCLC subtypes.
In this study we evaluated TGFβRII expression at the protein and mRNA level in a large number of human NSCLC samples and show that reduced TGFβRII expression is associated with more aggressive tumor behavior, including reduced tumor differentiation, higher tumor stage, increased nodal metastases, and reduced survival. To generate a model that can produce lung SCCs and to determine if TGFβRII loss initiates or promotes lung carcinogenesis, we targeted TGFβRII deletion to the conducting airway epithelium with the keratin-5 Cre*PR transgene (K5Cre*PR) (19). The K5 promoter targets the multi-potent basal cell progenitor of the upper airway (20). Cre*PR is a humanized Cre recombinase-progesterone receptor (Cre*PR) fusion protein that can be activated by RU486 but has low ligand-independent activity (21). Tracheal RU486 activates Cre*PR in sporadic airway epithelial cells (21), hence oncogenic alterations driven by this construct allow for the clonal expansion of cells with somatic mutations, analogous to what occurs in human cancer. Using this system, we show that TGFβRII deletion markedly promotes the development of Kras-initiated lung tumors, indicating that TGFβRII loss plays a causal role during lung carcinogenesis. Our studies in human samples show that reduced TGFβRII expression is negative prognostic marker in lung cancer.
Methods
NSCLC samples
After IRB approval, 38 NSCLC samples were obtained from the Oregon Health & Sciences University (OHSU) Department of Pathology Tumor Bank. Demographic and pathologic data were recorded when banked; histology and grade were confirmed by a second pathologist (S. White). RNA was extracted from13 corresponding frozen samples. We also purchased a tissue microarray (T8235724, lot A606098, Biochain, Hayward, CA) comprised of 64 NSCLC sections with accompanying demographic and pathologic data. With IRB approval, 85 NSCLC samples were obtained from the Colorado Lung SPORE Tissue Bank Core. Pathologic diagnosis of these samples was confirmed by a second pathologist (W. Franklin). Additional clinical and pathological data was extracted from pathology reports and medical records; survival data was obtained by searching the social security death index (SSDI) database. Of these samples, 77 had paraffin blocks available for immunostaining and 37 had matched tumor and non-malignant lung RNA available for analysis.
Immunostaining and qRT-PCR of human NSCLC samples
Immunostaining was performed as previously described (14) with anti-TGFβRII antibody (1:200 Santa Cruz C16) and staining intensity was graded as 3+ (intensity greater than normal airway) 2+ (equal to normal airway), 1+ (reduced compared to normal airway) or 0 (absent) by two independent observers (SPM and SMH). Due to the low frequency of samples with increased (3+) or absent (0) staining, samples were grouped as having preserved (2+ or 3+) or reduced (0 or 1+) staining for analysis. Immunostaining for lymphocytes and macrophages was performed with anti-CD3 antibody (Abcam #5690 at 1:250) and anti-MAC387 (Abcam #22506 at 1:200). Staining was quantified by counting the number of positive cells on four random 10x fields in at least 24 independent human NSCLC samples. RNA was harvested by homogenization in RNAzol (InVitrogen, Carlsbad, CA) then purification with RNeasy columns (Qiagen, Valencia, CA). RT-qPCR was performed with the Brilliant II kit (Stratagene, Santa Clara, CA) using a GAPDH internal control and Taqman probe Hs00559657 (Applied Biosystems, Foster City, CA) on an ABI 7900 thermal cycler. Data were analyzed by the ΔΔCt method and expressed as fold reduction compared to paired non-malignant lung. When paired non-malignant lung was unavailable expression in tumor samples was compared to grouped non-malignant samples run on the same plate. Differences in immunostaining frequency between groups were analyzed by a chi-squared or Fisher’s exact test depending on the expected frequency values. Differences in mRNA expression between groups were analyzed by non-parametric methods (Mann-Whitney or Kruskal-Wallis). Survival curves were obtained using the Kaplan-Meier method and comparisons across groups were made with logrank and stratified logrank tests. Analyses were performed with Prism5 (Graph Pad, La Jolla, CA) and SAS version 9.2 (SAS Institute, Inc., Cary, North Carolina).
NSCLC mouse model
All animal studies were IACUC approved. The TGFβRII conditional allele, lox-stop-lox-KrasG12D conditional allele, and K5Cre*PR transgene have been previously described (16, 19, 22, 23). Mice were treated with tracheal RU486 (500μg in 25μl 10% acetone/90% sesame oil) between 4–6 weeks of age as previously described (21). Mice were monitored weekly and euthanized at one year of age or if they lost >15% of their body weight. When euthanized, a blood sample was collected by cardiac puncture, tumors were enumerated and measured, and a bronchioalvelolar lavage (BAL) sample was collected by instillation of two 1mL aliquots of sterile phosphate buffered saline (PBS). The left lung was inflated with 10% formalin then paraffin embedded. Blood was allowed to clot then serum isolated by centrifugation (5900g, 15′, 25°C) and stored at −80°C. After 300μl of BAL was cytospun (500rpm, 5′, 25°C) onto a slide, remaining BAL cells were separated by centrifugation (5900g, 15′, 25°C) and BAL supernatant stored at −80°C. BAL cells in four 40x objectives were scored by morphologic criteria and differences between genotypes compared by unpaired t tests.
Analysis of mouse NSCLC
Lung tumors were separated from grossly normal lung and DNA extracted with the DNeasy kit (Qiagen); PCR for the recombinant TGFβRII allele was performed as previously described (16). RNA extraction, RT-qPCR, and TGFβRII IHC were performed as described above. Total TGFβ1 in BAL and serum was quantified by ELISA after acid activation per the manufacturer’s instructions (R&D Systems, Minneapolis, MN) and differences between groups were compared by one-way ANOVA or unpaired t test. Lesions were classified as atypical adenomatous hyperplasia (AAH), adenoma, adenocarcinoma, or squamous carcinoma on H&E sections by two investigators (SPM and SMH) using previously described criteria (24) with independent confirmation by a pathologist (DM) on a subset (~10%) of lesions. Tumors were immunostained with antibodies against keratin 5 (K5, 1:1000, Abcam #53121) and thyroid transcription factor (TTF, 1:100, Abcam #40880) to further confirm histologic subtype. Images were acquired on a Nikon Eclipse 80i and analyzed with Nikon Elements Software. Lesions were quantified per lung section and per mm2 lung and differences compared by one-way ANOVA or unpaired t test. Immunostaining for proliferating cellular antigen (PCNA, 1:100, Santa Cruz SC-56) and leukocytes (1:500 anti-CD3, Abcam #5690) was performed as described above and quantified by counting the number of positive cells per tumor area.
Migration and invasion assays in Beas2b cells with TGFβRII knockdown
Beas2B human bronchial epithelial cells (ATCC, Manassas, VA) were cultured in BEGM basal media supplemented with BEGM SingleQuot growth factors (Lonza, Basel, Switzerland) under standard conditions. Experiments were performed between the 50th and 65th passages. A stable TGFβRII knockdown line was produced by transfecting the pcPUR+U6 plasmid containing the shRNA against TGFβRII (25) with the pcPUR+U6 cassette as a control (iGene Therapeutics, Tokyo, Japan). Cells were transfected at 60% confluence with 16μg/mL plasmid DNA and 40μL/mL Lipofectamine 2000 (Invitrogen); 48h post transfection, puromycin (1.0 μg/mL; InvivoGen, San Diego, CA) selection was initiated. TGFβRII knockdown was confirmed by qPCR and Western blotting against TGFβRII (1:200; SC400 Santa Cruz Biotechnology) with GAPDH (1:40000; Abcam #8245) control. Scratch assays were performed by seeding 4 × 105 cells into six well plates, allowing cells to grow to confluence then wounding with a P200 pipette tip. Wounds were photographed at 0h and 24h and wound closure quantified as a percentage of the original wound area (26). Transwell assays were performed using 200μL Blindwell Boyden chambers (50mm2 area) and PVP-free polycarbonate membranes with 12μm pores (Neuro Probe, Gaithersburg, MD) (27). Migration membranes were coated with 0.01% porcine gelatin; invasion membranes were coated with 5% growth factor reduced Matrigel (BD Biosciences, Minneapolis, MN). Media conditioned for 24h with 3T3 cells was used as a chemo-attractant in the lower chamber and 3.5 × 104 cells were seeded into the upper chamber. After 24h, non-migrating cells were removed, membranes were fixed, stained with Diff-Quik (Siemens Healthcare Diagnostics Inc, Newark, DE), and photographed at 100X. Cells from 5 fields of view per insert were counted, experiments were repeated five times, and differences between groups were compared by unpaired t-tests.
Results
Reduced TGFβRII expression is more common in males, smokers, and lung SCC
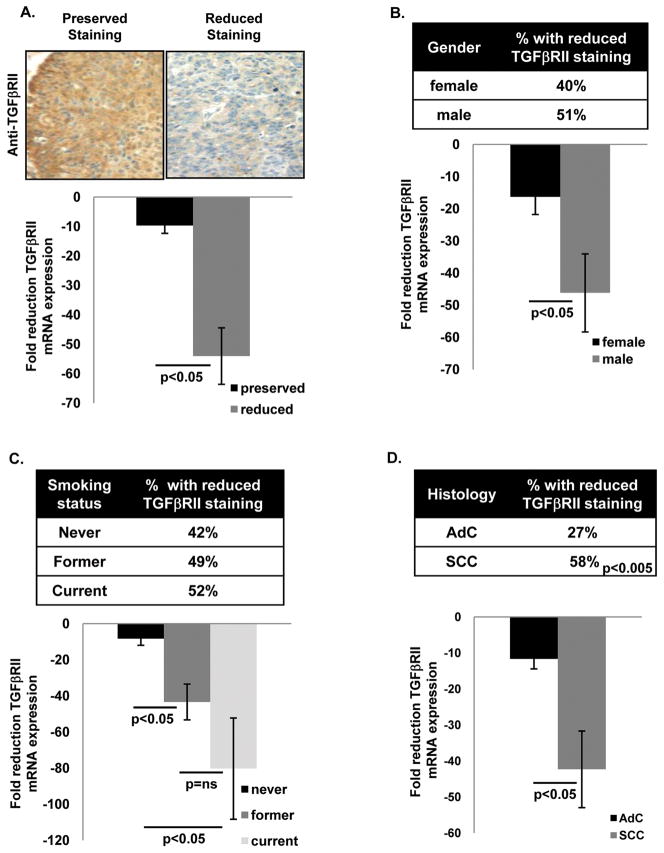
Reduced TGFβRII expression has been previously reported in a small study with limited demographic data (7). We analyzed TGFβRII expression in 187 NSCLC samples (full demographic data shown in Supplemental Table 1) using a combination of immunostaining and qRT-PCR. Immunostaining was performed on 172/187 samples and each sample was scored as having either preserved (intensity equal to or greater than normal airway) or reduced (intensity less than 50% of normal airway in at least 50% of cells) immunostaining. mRNA expression was analyzed in 48/187 samples and was compared to TGFβRII expression in either paired non-malignant lung (37/48 samples) or grouped non-malignant lung (11/48 samples). In Figs. 1–2 immunostaining data are expressed as the fraction (%) of NSCLC samples with reduced staining and mRNA expression data are expressed as fold reduction of TGFβRII expression compared to non-malignant lung. We found that 45% of NSCLC samples had reduced TGFβRII immunostaining and that samples with reduced TGFβRII immunostaining also had reduced TGFβRII mRNA expression (Fig. 1A). Although reduced TGFβRII immunostaining was somewhat more common in males and smokers this did not reach statistical significance (Fig. 1B–1C top panels); in contrast, both males and smokers had significantly reduced TGFβRII mRNA expression (Fig. 1B–1C bottom panels) and the degree of reduced TGFβRII expression was weakly correlated with smoking exposure (Supplemental Fig. 1). Interestingly, reduced TGFβRII immunostaining and mRNA expression were markedly more common in SCCs compared to AdCs (Fig. 1D).
Figure 1. Reduced TGF β RII immunostaining and mRNA expression in human NSCLC is more common in males, smokers, and SCC histology.
(A) Example of reduced TGFβRII immunostaining in human NSCLC. Staining intensity (compared to normal airway) was characterized as preserved (equal to or greater than normal airway) or reduced. Reduced TGFβRII immunostaining was observed in 45% of NSCLC and correlated with reduced expression by qPCR (bottom panel A). (B and C) Reduced TGFβRII immunostaining and mRNA expression are more common in males and current smokers although only reduced mRNA expression reached significance in these comparisons. Immunostaining data are expressed as the fraction (%) of samples with reduced TGFβRII immunostaining. (D) Reduced TGFβRII immunostaining and mRNA expression are both more common in SCC than AdC.
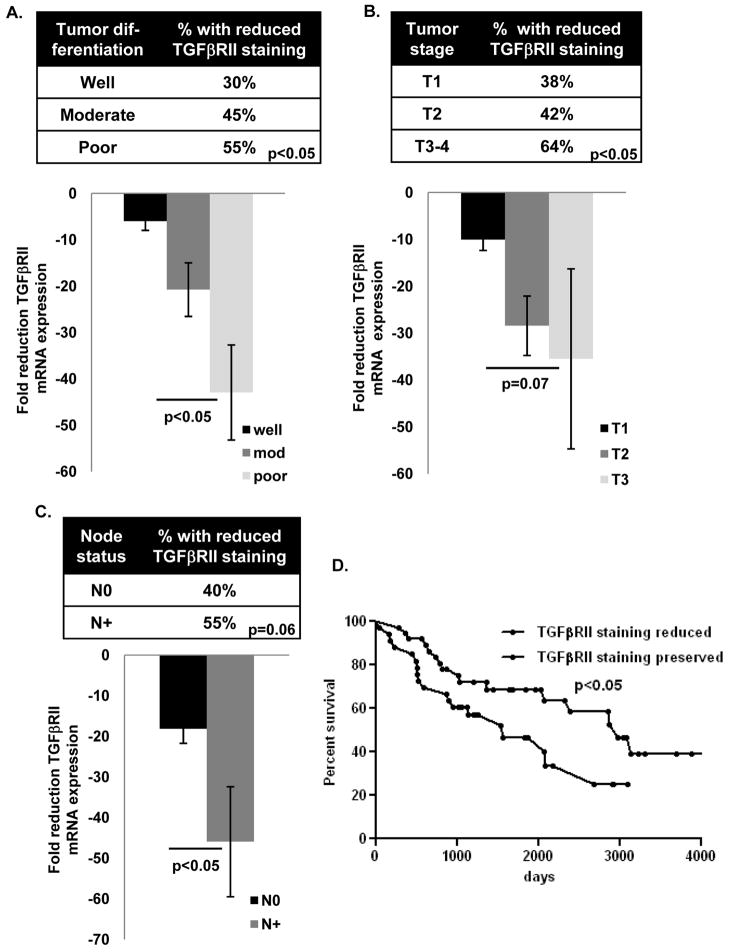
Figure 2. Reduced TGF βRII expression in human NSCLC is associated with more aggressive tumor behavior.
(A–D) Reduced TGFβRII immunostaining and mRNA expression are more common with in NSCLC with reduced differentiation, higher T stage, and nodal involvement. Data were analyzed by chi-squared or Fisher’s exact test (for immunostaining data) and non-parametric t test or ANOVA (for expression data). (D) Reduced TGFβRII immunostaining is associated with reduced NSCLC survival by Kaplan-Meier test and remained significant when adjusted for stage in a Cox proportional hazards survival model (not shown).
Reduced TGFβRII expression is associated with more aggressive NSCLC behavior
Although reduced TGFβRII immunostaining or expression has been associated with increased invasion in human lung AdC (12, 17) and more aggressive tumor behavior in other human malignancies (28–30) the relationship between reduced TGFβRII expression and clinical tumor behavior in human NSCLC has not been defined. We found that reduced TGFβRII immunostaining and mRNA expression were more common in more poorly differentiated tumors (Fig. 2A), tumors with higher tumor (T) stage (Fig. 2B), and tumors with nodal metastases (Fig. 2C). Interestingly there was no association between the degree of reduced TGFβRII mRNA expression and tumor size as a continuous variable (not shown), suggesting that reduced TGFβRII expression with higher tumor stage may be driven more by the invasive parameters that dictate clinical tumor stage (e.g., invasion) as opposed to size. Since tumor stage and nodal status dictate overall clinical stage, reduced TGFβRII immunostaining was also more common with increasing clinical stage (not shown) and reduced TGFβRII immunostaining was associated with reduced overall survival in NSCLC (Fig. 2D). After adjusting for stage in a Cox proportional hazards survival model, preserved TGFβRII immunostaining remained associated with improved survival with a hazard ratio of 0.49 (95% CI: 0.24–0.98, p=0.04).
TGFβRII deletion promotes growth and multiplicity of Kras-induced NSCLC
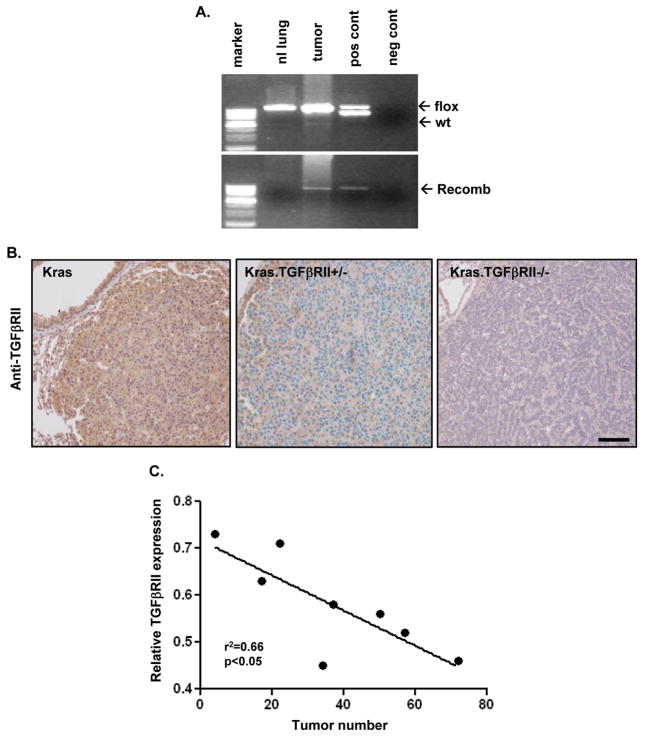
TGFβRII deletion promotes development and malignant conversion of Kras-initiated tumors in both oral epithelium (14) and pancreas (13). To address the role of TGFβRII loss in lung cancer development in vivo, we developed a mouse model combining TGFβRII deletion with oncogenic KrasG12D activation. We used the K5Cre*PR transgene that contains an RU486-inducible Cre recombinase (19) and tracheal RU486 to direct TGFβRII deletion (16) and oncogenic KrasG12D (22) activation to the airways (21). As expected, the recombinant TGFβRII allele could be detected in tumors but not in adjacent grossly normal lung (Fig. 3A) and TGFβRII immunostaining was reduced in tumors with TGFβRII deletion (hereafter referred to as Kras.TGFβRII +/− and Kras.TGFβRII −/−) compared to Kras tumors (Fig. 3B). In addition, TGFβRII mRNA expression in whole lung homogenate negatively correlated with increasing tumor burden (Fig. 3C)
Figure 3. TGFβRII deletion in Kras induced mouse tumors.
(A) The recombinant TGFβRII allele can be detected in lung tumors but not in grossly normal adjacent lung. Genotyping is shown in the upper gel; floxed and wild type alleles are indicated. (B) Reduced TGFβRII immunostaining in Kras.TGFβRII+/− tumors and Kras.TGFβRII−/− tumors compared to Kras tumors; scale bar is 100μm. (C) Reduced TGFβRII expression (in whole lung homogenate) is inversely correlated with increasing tumor burden in Kras.TGFβRII−/− animals.
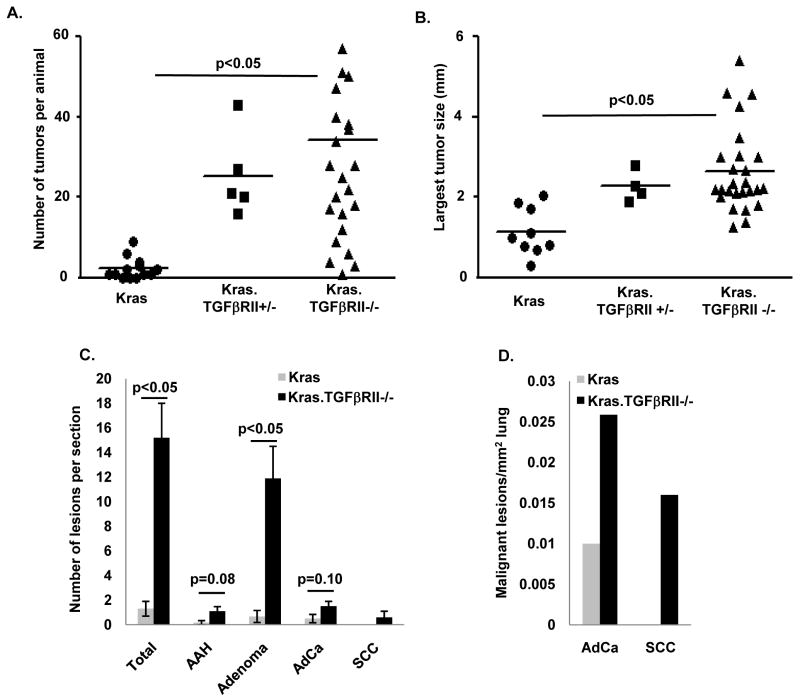
We only observed one lung tumor in 30 K5Cre*PR.TGFβRII ff animals (hereafter referred to as TGFβRII−/−), however approximately 65% of animals harboring both the K5Cre*PR transgene and a lox-stop-lox KrasG12D allele (K5Cre*PR.LSL-Kras; hereafter referred to as Kras) developed lung tumors after tracheal RU486 (Supplemental Table 2). Kras animals typically developed 2–3 tumors per animal; deletion of one or both TGFβRII alleles increased tumor multiplicity by approximately 10-fold (Fig. 4A) and also increased overall tumor size (Supplemental Fig. 2). Because of the increased number of small tumors with TGFβRII deletion, we analyzed the largest single tumor in each animal to assess the effect of TGFβRII deletion on established tumor growth (Fig. 4B); this analysis showed that TGFβRII deletion increased the growth of established Kras-initiated lung tumors. Although there was a low level of tumor formation in vehicle treated Kras.TGFβRII−/− animals, there was a clear, dose-dependent increase in tumor formation with increasing tracheal RU486 dose (Supplemental Table 2). These data demonstrate that the K5Cre*PR transgene can be used for lung targeting and that TGFβRII deletion greatly increases both the number and size of Kras-initiated lung tumors.
Figure 4. TGFβRII deletion in mouse airway epithelia increases formation of both adenocarcinomas and squamous cell carcinomas.
(A) Deletion of one or both TGFβRII alleles increases the number of Kras-initiated lung tumors. Each data point represents the number of tumors observed in an individual animal; the mean overlies individual tumor data points. Kras animals had 3.2 ± 1.0 tumors per animal while Kras.TGFβRII+/− animals had 25.4 ± 4.7 tumors per animal and Kras.TGFβRII−/− animals had 34.4 ± 5.9 tumors per animal (both significant compared to Kras). (B) Deletion of one or both TGFβRII alleles increases the size of established Kras-initiated lung tumors. Each data point represents the single largest tumor from an individual animal; Kras tumors were 1.15 ± 0.20 mm while Kras.TGFβRII+/− tumors were 2.28 ± 0.19 mm and Kras.TGFβRII−/− tumors were 2.64 ± 0.20 mm (both significant compared to Kras). A separate analysis that includes all tumors is shown in Supplemental Fig. 2. (C–D) TGFβRII deletion increased the number of benign and malignant lung lesions per tumor section and the number of malignant lesions per mm2 lung. Because SCCs were not observed in Kras animals, p values could not be calculated for these lesions. Additional data regarding the age, number of animals, tracheal RU486 dosage, and tumor penetrance is presented in Supplemental Table 2.
Kras.TGFβRII−/− mice develop both adenocarcinomas and squamous cell carcinomas
In our mouse model that employs a keratin 5 promoter and tracheal RU486 to target our genetic manipulations (19, 21), we observed the formation of both adenocarcinomas and squamous cell carcinomas. Adenocarinomas displayed typical glandular morphology and stained negative for keratin 5 (K5) and positive for thyroid transcription factor (TTF) while squamous cell carcinomas displayed keratinization and intercellular bridges and were K5 positive and TTF negative (Supplemental Fig. 3). To analyze the effect of TGFβRII deletion on the formation of specific tumor types, we classified individual lesions according to consensus recommendations (24) and found that TGFβRII deletion increased the number of both benign (atypical adenomatous hyperplasias (AAH) and adenomas) and malignant (adenocarinoma and squamous cell carcinoma) lesions (Fig. 4C–D). In sum, TGFβRII deletion increases KrasG12D-dependent tumor initiation and allows for the formation of a spectrum of malignant lesions that includes the two most common human NSCLC subtypes.
TGFβRII deletion increases proliferation and inflammation in vivo
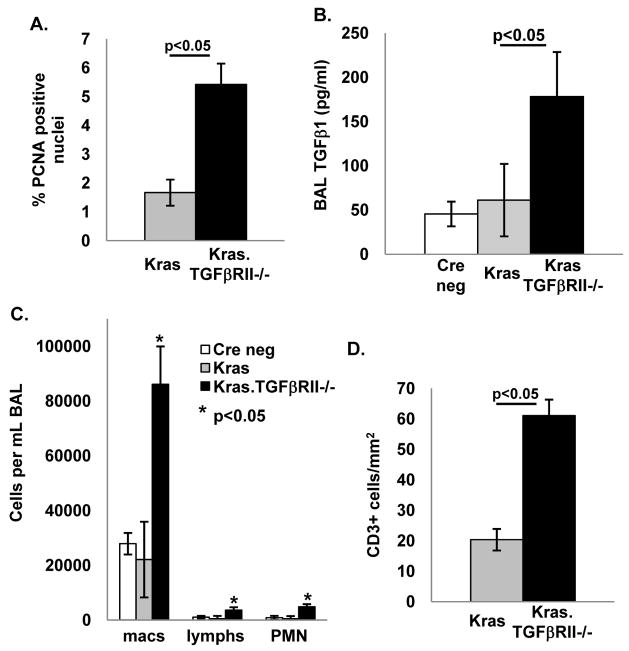
Because TGFβ signaling inhibits epithelial proliferation (5), we compared proliferation in Kras and Kras.TGFβRII−/− tumors and found that Kras.TGFβRII−/− tumors had a 5-fold increase in staining for the proliferation marker proliferating cell nuclear antigen (PCNA; Fig. 5A, immunostaining example shown in Supplemental Fig. 4A). Loss of functional TGFβ signaling can cause a compensatory increase in TGFβ1 ligand production (14, 31) which can then promote tumor development through increased angiogenesis and inflammation in the tumor microenvironment (14, 32, 33). We found that TGFβRII deletion increased TGFβ1 ligand in the bronchioalveolar lavage (BAL) fluid (Fig. 5B) and that increasing tumor burden correlated with higher BAL TGFβ1 (Supplemental Fig. 4B). TGFβRII deletion was also associated with increased serum TGFβ1 ligand levels (not shown) similar to the increased serum TGFβ1 levels reported in humans with NSCLC (34). Similarly, TGFβRII knockdown in human Beas2B cells increased TGFβ1 ligand production (Supplemental Fig. 5C). We did not find increased angiogenesis in TGFβRII−/− lung tumors (not shown), but observed that TGFβRII deletion increased the number of macrophages, lymphocytes, and neutrophils in the BAL (Fig. 5C) and that the BAL macrophage count correlated with increasing tumor burden (Supplemental Fig. 4C). In addition, compared to Kras tumors, Kras.TGFβRII−/− tumors had increased infiltration of CD3+ lymphocytes (Fig. 5D, immunostaining example shown in Supplemental Fig. 4D). To determine whether a similar relationship between reduced TGFβRII expression and increased inflammation exists in human lung cancer, we immunostained human NSCLC samples for both lymphocytes and macrophages and found that human lung cancers with reduced TGFβRII expression had increased numbers of infiltrating CD3+ lymphocytes but not macrophages (Supplemental Fig. 6). In sum, these data show that TGFβRII deletion in a murine lung cancer model increases both proliferation and local inflammation and human lung cancer with reduced TGFβRII expression also have increased inflammation.
Figure 5. TGFβRII deletion in mouse airway epithelia increases proliferation, TGFβ1 ligand elaboration, and inflammation in vivo.
(A) Kras.TGFβRII−/− tumors have increased proliferation (PCNA staining) compared to Kras tumors (examples of immunostaining shown in Supplemental Fig. 4A). (B) TGFβRII deletion increases total TGFβ1 in the BAL. (C) TGFβRII deletion increases macrophages, lymphocytes, and neutrophils in the BAL. (D) TGFβRII deletion increases recruitment of CD3+ lymphocytes to lung tumors (examples of immunostaining shown in Supplemental Fig. 4D).
TGFβRII knockdown in bronchial epithelial cells increases migration and invasion
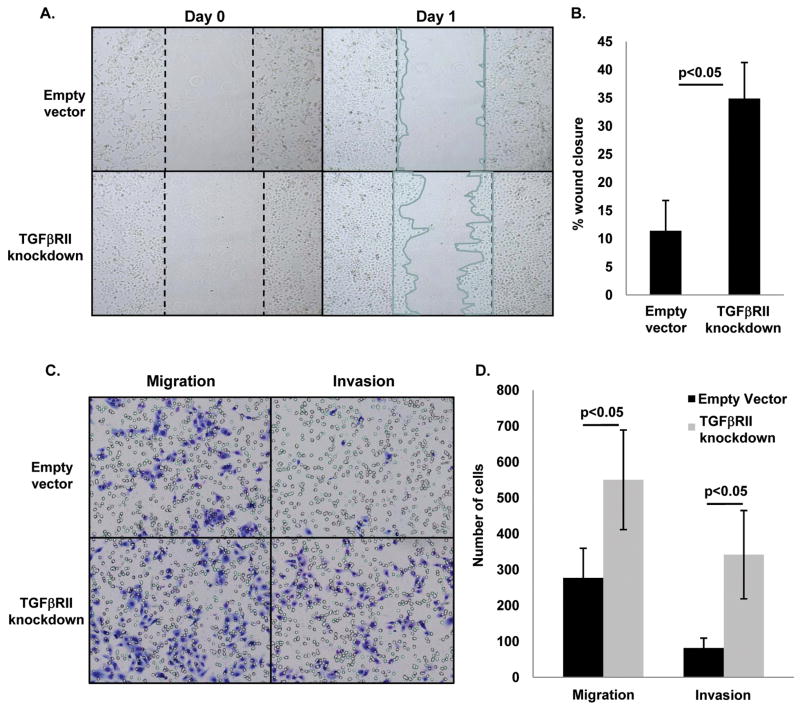
That reduced TGFβRII expression was associated with both increased aggressiveness of human NSCLC and progression of murine lung tumors, prompted us to assess whether TGFβRII knockdown promotes cell migration and invasion in vitro. We stably knocked down TGFβRII in a human bronchial epithelial cell line (Beas2B) using a small-hairpin RNA approach (25) and achieved reduced TGFβRII expression at both the RNA and protein level (Supplemental Fig. 5A). Interestingly, TGFβRII knockdown did not affect proliferation of Beas2B cells (Supplemental Fig. 5D), potentially because these cells are SV40 T antigen immortalized and hence not sensitive to TGFβ-mediated growth inhibition. In a scratch assay, TGFβRII knockdown increased in vitro wound closure by almost 3-fold (Fig 6A–B). In a Matrigel transwell invasion assay, TGFβRII knockdown increased migration by approximately 2-fold and invasion by approximately 3-fold (Fig. 6C–D). These data suggest that increased lung tumor aggressiveness in tumors with reduced TGFβRII expression is at least partially mediated through increased invasion and migration of tumor cells.
Figure 6. TGFβRII knockdown in Beas2B human airway epithelial cells increases migration and invasion in vitro.
(A–B) Stable TGFβRII knockdown increases migration in an in vitro would closure assay. (C–D) TGFβRII knockdown increases both migration and invasion in a Matrigel transwell invasion assay.
Discussion
Reduced TGFβRII expression in human NSCLC is associated with more aggressive tumor behavior
In this study we found that reduced TGFβRII expression is common in NSCLC and occurs more frequently in males, smokers, and tumors with SCC histology; this grouping of associations is not surprising given that lung SCCs are more common in males and are more strongly associated with tobacco exposure (35), however, this is the first description clearly linking reduced TGFβRII expression with smoking exposure. Consistent with previous reports that TGFβRII expression in NSCLC can be reduced through promoter methylation or microsatellite instability (8, 11), we found both reduced TGFβRII at both the mRNA and protein levels, suggesting that reduced TGFβRII expression occurs pretranslationally. We also show that reduced TGFβRII expression is associated with more aggressive NSCLC behavior including reduced differentiation, higher T stage, nodal metastases, and reduced patient survival. This is consistent with TGFβRII functioning as a NSCLC tumor suppressor, although other molecules clearly also contribute to malignant progression. These associations suggest a causal role of TGFβRII reduction in NSCLC progression, which is further supported by our animal study in which TGFβRII deletion increases NSCLC development in vivo.
Targeting TGFβRII deletion to keratin 5-positive airway cells promotes formation of multiple NSCLC subtypes
We targeted TGFβRII deletion to the conducting airway epithelium using a K5-driven, RU486-inducible Cre recombinase and tracheal RU486 (19, 21). Because keratin expression in the lung is limited to basal cells which are thought to contain the SCC progenitor cell (4, 36), we expected these animals to develop exclusively SCCs. To our surprise, while these animals did develop some SCCs, they predominantly developed adenomas and adenocarcinomas. In the murine airway, K5/K14 positive basal cells function as stem cells or facultative progenitors capable of giving rise to multiple cell types (20, 37); our data suggest that K5 positive cells contain both AdC and SCC progenitors.
TGFβRII deletion alone in the airway epithelium results in a very low incidence of lung tumors; this is consistent with the observation that targeting TGFβRII deletion to a large fraction of lung epithelial cells by AdCre also fails to initiate tumor formation (17). In contrast, TGFβRII deletion markedly increases Kras-initiated lung tumor number and size, suggesting that intact TGFβ signaling inhibits lung tumor growth and that TGFβ signaling disruption increases both tumor initiation and progression. Increased proliferation and tumor size is likely driven by loss of TGFβ-mediated growth inhibition while increased tumor multiplicity may be a result of impaired immune surveillance from increased TGFβ. These data are consistent with TGFβRII function in other oncogene-initiated cancer models (13–16) as well as a recent report showing that invasive lung adenocarcinoma is modeled by Kras activation and TGFβRII deletion targeted by adenoviral-delivered Cre recombinase (AdCre) (17). Our data demonstrate that TGFβRII deletion in mice increases tumor number, tumor size, and the number of malignant lesions; these observations are consistent with our observation that reduced TGFβRII expression in human NSCLC is associated with more aggressive tumor behavior. Our study shows that in addition to abrogation of TGFβ-induced growth arrest, TGFβRII loss also promotes tumor cell migration and invasion. Although Kras.TGFβRII−/− tumors had little evidence of epithelial to mesenchymal transition (EMT), we have previously shown that TGFβRII loss causes an EMT-independent migratory and invasive phenotype in keratinocytes (38).
Interestingly, deleting one TGFβRII allele had an effect similar to deleting both TGFβRII alleles, indicating that TGFβRII exhibits haploid insufficiency, and that a 50% reduction (i.e., roughly the amount used to score human NSCLCs) is sufficient to promote lung cancer growth. This is similar to what was observed in both a head and neck cancer model (14) and a pancreatic cancer model (13), where deletion of a single TGFβRII allele caused intermediate phenotypes in terms of tumor penetrance or survival, respectively. In another study, TGFβ1 haploid insufficiency also increased progression of Kras-initiated lung tumors and shortened survival (39), suggesting that reduced TGFβ signaling in tumor epithelial cells promotes lung tumor progression.
TGFβRII deletion in airway epithelia increases inflammation that could promote malignant progression
In our mouse model, TGFβRII deletion increased TGFβ1 ligand elaboration, BAL inflammation, and tumor-associated macrophages and lymphocytes. In human lung cancer samples, reduced TGFβRII expression was associated with increased lymphocyte infiltration but not with increased macrophage infiltration. Because macrophages are recruited to areas of tissue damage, it is possible that macrophage infiltration in human lung samples may predominantly be driven by environmental factors (e.g., smoking) as opposed to reduced TGFβRII expression. In contrast, increased TGFβ1 ligand produced by TGFβRII negative tumors may recruit other inflammatory cells, e.g., CD3 positive T lymphocytes, to the local environment where these cells could promote lung tumor progression. Supporting this notion, increased lymphocyte infiltration (of CD4 cells, CD8 cells, and B cells) was also reported with AdCre-mediated Kras.TGFβRII−/− lung tumorigenesis (17). In contrast to AdCre mediated targeting that likely affects multiple cell types, targeting in our model was restricted to sporadic epithelial cells, thus inflammation was likely a result of secreted pro-inflammatory cytokines/chemokines, such as TGFβ1, by targeted epithelial cells. TGFβ1 can recruit myeloid cells (16) and induce development of Treg and Th17 lymphocytes (41), all of which can facilitate tumor growth. However, the paracrine effects of inflammatory cytokines in our model were insufficient to elicit the intense fibroblastic stromal response seen in AdCre-initiated KrasG12D.TGFβRII−/− tumors (17), presumably because AdCre targeting results in recombination in much higher proportion of epithelial cells than does K5Cre*PR targeting. Finally, although TGFβRII deletion and TGFβ1 overexpression have been associated with increased angiogenesis in other systems (14, 33), we did not observe increased angiogenesis in Kras.TGFβRII−/− lung tumors, potentially because the lung has a higher oxygen tension that limits the need for tumor neovascularization, particularly of small tumors.
In summary, our study demonstrates that reduced TGFβRII expression in human NSCLC is a negative prognostic marker associated with more aggressive tumor behavior and worse clinical outcome. In addition, TGFβRII loss plays a causal role in promoting the development of multiple NSCLC subtypes. Our study should instigate investigation into mechanisms underlying the effects of reduced TGFβRII expression on both tumor epithelium and tumor stroma. This could potentially facilitate selection of patients for therapies targeting events downstream of TGFβRII loss.
Supplementary Material
Translational Relevance.
Although lung cancer is a common malignancy, the 5 year survival remains low, illustrating a need for an improved understanding of basic molecular mechanisms that can subsequently be translated into novel therapeutic strategies. In this study, we found that reduced TGFβRII expression in human NSCLC samples is associated with more aggressive tumor behavior and reduced patient survival. Our mouse model shows that TGFβRII deletion in sporadic airway epithelial cells promotes formation of both adenocarcinomas and squamous carcinomas, supporting a causal role of TGFβRII loss in lung carcinogenesis and suggesting that reduced TGFβRII expression is a negative prognostic marker in lung cancer. Our mouse model will be a useful resource for testing novel therapeutic approaches directed toward NSCLC with reduced TGFβRII expression.
Acknowledgments
This work was supported by the NIH/NCI (F32 CA119467 and K08 CA131483 to S.P.M.) and NIH/NIDCR (R01 DE015953 to X.J.W.). S.P.M. was supported by T32 CA106195, the Oregon Medical Research Foundation, and the National Lung Cancer Partnership. The OHSU BioLibrary is supported by P30 CA 069533 and the Colorado Lung SPORE Tissue Bank is supported by P30 CA046934 and P50 CA058187. We would particularly like to thank Cara Poage, Carolyn Gendron, and Christopher Corless at the OHSU BioLibrary and Jerry Haney, Christina Nall, and Marina Lewis at the Colorado Lung SPORE for their assistance with acquisition of patient samples. We also thank Sandra White for confirming tumor grade in OHSU samples.
This work was supported by the NIH/NCI (F32 CA119467 and K08 CA131483 to S.P.M.) and NIH/NIDCR (R01 DE015953 to X.J.W.). S.P.M. was also supported by T32 CA106195, the Oregon Medical Research Foundation, and the National Lung Cancer Partnership.
Footnotes
The authors declare no conflict of interest.
References
- 1.Jemal A, Siegel R, Ward E, Hao Y, Xu J, Thun MJ. Cancer statistics, 2009. CA Cancer J Clin. 2009;59:225–49. doi: 10.3322/caac.20006. [DOI] [PubMed] [Google Scholar]
- 2.Kenfield SA, Wei EK, Stampfer MJ, Rosner BA, Colditz GA. Comparison of aspects of smoking among the four histological types of lung cancer. Tob Control. 2008;17:198–204. doi: 10.1136/tc.2007.022582. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Kim CF, Jackson EL, Woolfenden AE, Lawrence S, Babar I, Vogel S, et al. Identification of bronchioalveolar stem cells in normal lung and lung cancer. Cell. 2005;121:823–35. doi: 10.1016/j.cell.2005.03.032. [DOI] [PubMed] [Google Scholar]
- 4.Ooi AT, Mah V, Nickerson DW, Gilbert JL, Ha VL, Hegab AE, et al. Presence of a putative tumor-initiating progenitor cell population predicts poor prognosis in smokers with non-small cell lung cancer. Cancer Res. 70:6639–48. doi: 10.1158/0008-5472.CAN-10-0455. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Bierie B, Moses HL. TGF-beta and cancer. Cytokine Growth Factor Rev. 2006;17:29–40. doi: 10.1016/j.cytogfr.2005.09.006. [DOI] [PubMed] [Google Scholar]
- 6.Levy L, Hill CS. Alterations in components of the TGF-beta superfamily signaling pathways in human cancer. Cytokine Growth Factor Rev. 2006;17:41–58. doi: 10.1016/j.cytogfr.2005.09.009. [DOI] [PubMed] [Google Scholar]
- 7.Anumanthan G, Halder SK, Osada H, Takahashi T, Massion PP, Carbone DP, et al. Restoration of TGF-beta signalling reduces tumorigenicity in human lung cancer cells. Br J Cancer. 2005;93:1157–67. doi: 10.1038/sj.bjc.6602831. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Zhang HT, Chen XF, Wang MH, Wang JC, Qi QY, Zhang RM, et al. Defective expression of transforming growth factor beta receptor type II is associated with CpG methylated promoter in primary non-small cell lung cancer. Clin Cancer Res. 2004;10:2359–67. doi: 10.1158/1078-0432.ccr-0959-3. [DOI] [PubMed] [Google Scholar]
- 9.Xu JB, Bao Y, Liu X, Liu Y, Huang S, Wang JC. Defective expression of transforming growth factor beta type II receptor (TGFBR2) in the large cell variant of non-small cell lung carcinoma. Lung Cancer. 2007;58:36–43. doi: 10.1016/j.lungcan.2007.04.019. [DOI] [PubMed] [Google Scholar]
- 10.Tani M, Takenoshita S, Kohno T, Hagiwara K, Nagamachi Y, Harris CC, et al. Infrequent mutations of the transforming growth factor beta-type II receptor gene at chromosome 3p22 in human lung cancers with chromosome 3p deletions. Carcinogenesis. 1997;18:1119–21. doi: 10.1093/carcin/18.5.1119. [DOI] [PubMed] [Google Scholar]
- 11.Kim WS, Park C, Hong SK, Park BK, Kim HS, Park K. Microsatellite instability(MSI) in non-small cell lung cancer(NSCLC) is highly associated with transforming growth factor-beta type II receptor(TGF-beta RII) frameshift mutation. Anticancer Res. 2000;20:1499–502. [PubMed] [Google Scholar]
- 12.Borczuk AC, Kim HK, Yegen HA, Friedman RA, Powell CA. Lung adenocarcinoma global profiling identifies type II transforming growth factor-beta receptor as a repressor of invasiveness. Am J Respir Crit Care Med. 2005;172:729–37. doi: 10.1164/rccm.200504-615OC. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Ijichi H, Chytil A, Gorska AE, Aakre ME, Fujitani Y, Wright CV, et al. Aggressive pancreatic ductal adenocarcinoma in mice caused by pancreas-specific blockade of transforming growth factor-beta signaling in cooperation with active Kras expression. Genes Dev. 2006;20:3147–60. doi: 10.1101/gad.1475506. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Lu SL, Herrington H, Reh D, Weber S, Bornstein S, Wang D, et al. Loss of transforming growth factor-beta type II receptor promotes metastatic head-and-neck squamous cell carcinoma. Genes Dev. 2006;20:1331–42. doi: 10.1101/gad.1413306. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Munoz NM, Upton M, Rojas A, Washington MK, Lin L, Chytil A, et al. Transforming growth factor beta receptor type II inactivation induces the malignant transformation of intestinal neoplasms initiated by Apc mutation. Cancer Res. 2006;66:9837–44. doi: 10.1158/0008-5472.CAN-06-0890. [DOI] [PubMed] [Google Scholar]
- 16.Forrester E, Chytil A, Bierie B, Aakre M, Gorsaka AE, Sharif-Afshar AR, et al. Effect of conditional knockout of the type II TGF-beta receptor gene in mammary epithelia on mammary gland development and polyomavirus middle T antigen induced tumor formation and metastasis. Cancer Res. 2005;65:2296–302. doi: 10.1158/0008-5472.CAN-04-3272. [DOI] [PubMed] [Google Scholar]
- 17.Borczuk AC, Sole M, Lu P, Chen J, Wilgus ML, Friedman RA, et al. Progression of human bronchioloalveolar carcinoma to invasive adenocarcinoma is modeled in a transgenic mouse model of K-ras-induced lung cancer by loss of the TGF-{beta} type II receptor. Cancer Res. doi: 10.1158/0008-5472.CAN-11-1590. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Meuwissen R, Berns A. Mouse models for human lung cancer. Genes Dev. 2005;19:643–64. doi: 10.1101/gad.1284505. [DOI] [PubMed] [Google Scholar]
- 19.Caulin C, Nguyen T, Longley MA, Zhou Z, Wang XJ, Roop DR. Inducible activation of oncogenic K-ras results in tumor formation in the oral cavity. Cancer Res. 2004;64:5054–8. doi: 10.1158/0008-5472.CAN-04-1488. [DOI] [PubMed] [Google Scholar]
- 20.Rock JR, Onaitis MW, Rawlins EL, Lu Y, Clark CP, Xue Y, et al. Basal cells as stem cells of the mouse trachea and human airway epithelium. Proc Natl Acad Sci U S A. 2009;106:12771–5. doi: 10.1073/pnas.0906850106. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Malkoski SP, Cleaver TG, Lu SL, Lighthall JG, Wang XJ. Keratin promoter based gene manipulation in the murine conducting airway. Int J Biol Sci. 6:68–79. doi: 10.7150/ijbs.6.68. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Johnson L, Mercer K, Greenbaum D, Bronson RT, Crowley D, Tuveson DA, et al. Somatic activation of the K-ras oncogene causes early onset lung cancer in mice. Nature. 2001;410:1111–6. doi: 10.1038/35074129. [DOI] [PubMed] [Google Scholar]
- 23.Wunderlich FT, Wildner H, Rajewsky K, Edenhofer F. New variants of inducible Cre recombinase: a novel mutant of Cre-PR fusion protein exhibits enhanced sensitivity and an expanded range of inducibility. Nucleic Acids Res. 2001;29:E47. doi: 10.1093/nar/29.10.e47. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Nikitin AY, Alcaraz A, Anver MR, Bronsn RT, Cardiff RD, Dixon D, et al. Classification of proliferative pulmonary lesions of the mouse: recommendations of the mouse models of human cancers consortium. Cancer Res. 2004;64:2307–16. doi: 10.1158/0008-5472.can-03-3376. [DOI] [PubMed] [Google Scholar]
- 25.Jazag A, Kanai F, Ijichi H, Tateishi K, Ikenoue T, Tanaka Y, et al. Single small-interfering RNA expression vector for silencing multiple transforming growth factor-beta pathway components. Nucleic Acids Res. 2005;33:e131. doi: 10.1093/nar/gni130. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Owens P, Engelking E, Han G, Haeger SM, Wang XJ. Epidermal Smad4 deletion results in aberrant wound healing. Am J Pathol. 176:122–33. doi: 10.2353/ajpath.2010.090081. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Albini A, Benelli R. The chemoinvasion assay: a method to assess tumor and endothelial cell invasion and its modulation. Nat Protoc. 2007;2:504–11. doi: 10.1038/nprot.2006.466. [DOI] [PubMed] [Google Scholar]
- 28.Mincione G, Di Marcantonio MC, Artese L, Vianale G, Piccirelli A, Piccrilli M, et al. Loss of expression of TGF-beta1, TbetaRI, and TbetaRII correlates with differentiation in human oral squamous cell carcinomas. Int J Oncol. 2008;32:323–31. [PubMed] [Google Scholar]
- 29.Paiva CE, Drigo SA, Rosa FE, Moraes Neto FA, Caldeira JR, Soares FA, et al. Absence of transforming growth factor-beta type II receptor is associated with poorer prognosis in HER2-negative breast tumours. Ann Oncol. 21:734–40. doi: 10.1093/annonc/mdp518. [DOI] [PubMed] [Google Scholar]
- 30.Mamiya T, Yamazaki K, Masugi Y, Mori T, Effendi K, Du W, et al. Reduced transforming growth factor-beta receptor II expression in hepatocellular carcinoma correlates with intrahepatic metastasis. Lab Invest. 90:1339–45. doi: 10.1038/labinvest.2010.105. [DOI] [PubMed] [Google Scholar]
- 31.Bornstein S, White R, Malkoski S, Oka M, Han G, Cleaver T, et al. Smad4 loss in mice causes spontaneous head and neck cancer with increased genomic instability and inflammation. J Clin Invest. 2009;119:3408–19. doi: 10.1172/JCI38854. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Blobe GC, Schiemann WP, Lodish HF. Role of transforming growth factor beta in human disease. N Engl J Med. 2000;342:1350–8. doi: 10.1056/NEJM200005043421807. [DOI] [PubMed] [Google Scholar]
- 33.Lu SL, Reh D, Li AG, Woods J, Corless CL, Kulesz-Martin M, et al. Overexpression of transforming growth factor beta1 in head and neck epithelia results in inflammation, angiogenesis, and epithelial hyperproliferation. Cancer Res. 2004;64:4405–10. doi: 10.1158/0008-5472.CAN-04-1032. [DOI] [PubMed] [Google Scholar]
- 34.Kumar S, Guleria R, Mohan A, Singh V, Ali A, Bharti AC, et al. Utility of plasma tumour necrosis factor-alpha and transforming growth factor-beta1 as predictors of survival and treatment outcome in advanced non-small cell lung carcinoma. Biomarkers. 15:446–53. doi: 10.3109/1354750X.2010.485699. [DOI] [PubMed] [Google Scholar]
- 35.Khuder SA. Effect of cigarette smoking on major histological types of lung cancer: a meta-analysis. Lung Cancer. 2001;31:139–48. doi: 10.1016/s0169-5002(00)00181-1. [DOI] [PubMed] [Google Scholar]
- 36.Hegab AE, Ha VL, Gilbert JL, Zhang KX, Malkoski SP, Chon AT, et al. Novel stem/progenitor cell population from murine tracheal submucosal gland ducts with multipotent regenerative potential. Stem Cells. 29:1283–93. doi: 10.1002/stem.680. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Hong KU, Reynolds SD, Watkins S, Fuchs E, Stripp BR. Basal cells are a multipotent progenitor capable of renewing the bronchial epithelium. Am J Pathol. 2004;164:577–88. doi: 10.1016/S0002-9440(10)63147-1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Han G, Lu SL, Li AG, He W, Corless CL, Kulesz-Martin M, et al. Distinct mechanisms of TGF-beta1-mediated epithelial-to-mesenchymal transition and metastasis during skin carcinogenesis. J Clin Invest. 2005;115:1714–23. doi: 10.1172/JCI24399. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Pandey J, Umphress SM, Kang Y, Angdisen J, Naumova A, Mercer KL, et al. Modulation of tumor induction and progression of oncogenic K-ras-positive tumors in the presence of TGF-b1 haploinsufficiency. Carcinogenesis. 2007;28:2589–96. doi: 10.1093/carcin/bgm136. [DOI] [PubMed] [Google Scholar]
- 40.Bierie B, Moses HL. Transforming growth factor beta (TGF-beta) and inflammation in cancer. Cytokine Growth Factor Rev. 21:49–59. doi: 10.1016/j.cytogfr.2009.11.008. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Ruffell B, DeNardo DG, Affara NI, Coussens LM. Lymphocytes in cancer development: polarization towards pro-tumor immunity. Cytokine Growth Factor Rev. 21:3–10. doi: 10.1016/j.cytogfr.2009.11.002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Detterbeck FC, Boffa DJ, Tanoue LT. The new lung cancer staging system. Chest. 2009;136:260–71. doi: 10.1378/chest.08-0978. [DOI] [PubMed] [Google Scholar]
- 43.Mountain CF. The international system for staging lung cancer. Semin Surg Oncol. 2000;18:106–15. doi: 10.1002/(sici)1098-2388(200003)18:2<106::aid-ssu4>3.0.co;2-p. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.